Abstract
Chemical agents that cleave HIV genome can be potentially used for anti-HIV therapy. In this report, the cleavage of the upper stem-loop region of HIV-1 TAR RNA was studied in a variety of buffers containing organic catalysts. trans-(±)-Cyclohexane-1,2-diamine was found to cleave the RNA with the highest activity (31%, 37°C, 18 h). Cleavage of the RNA in trans-(±)-cyclohexane-1,2-diamine buffer was also studied when the RNA was hybridized with complementary DNAs. A pyrene-modified C3 spacer was incorporated to the DNA strand to facilitate the formation of a RNA bulge loop in the RNA/DNA duplex. In contrast, unmodified DNAs cannot efficiently generate RNA bulge loops, regardless of the DNA sequences. The results showed that the pyrene-stablized RNA bulge loops were efficiently and site-specifically cleaved by trans-(±)-cyclohexane-1,2-diamine.
1. Background
RNA cleaving agents are promising next-generation therapeutic tools. Ribozyme and RNA interference have been found to cleave RNA in high efficiencies [1-4]. Artificial ribonuclease is another appealing candidate due to the flexibility to introduce chemical modifications [5-8]. These modifications can play crucial roles in enhancing cleavage activities or increasing the stability of artificial ribonucleases in vivo. The catalytic group of an artificial nuclease is often a metal ion or a charged organic functional group. Compared to metal complexes, organic molecules have better pharmacological profiles. Therefore, it is of great interest to develop a highly efficient and sequence-specific non-metalloribonuclease. Many efforts have been exerted to attach a catalytic moiety to antisense deoxyribonucleotide to achieve specific sequence targeting [9-17]. The catalytic moiety often contains cationic nitrogen atoms such as those of oligoamine, imidazole, and guanidine. The effectiveness of such a strategy is highly dependent on several factors. The first factor is the DNA sequence which determines whether the DNA forms a stable duplex with the target RNA, especially when the RNA is highly structured, and whether the duplex formation affects the RNA phosphodiester group on its susceptibility to cleavage. The second factor is the inherent activity of the catalytic group. The third factor is the chemical linkage that positions the catalytic moieties around the cleavage site. Systematic investigation of the above three factors simultaneously is difficult. Fortunately, the first two factors can be studied independently. Indeed, studies have been conducted on DNA/RNA duplex cleavage in the presence of high concentrations of metal ions and imidazole [18,19]. Once the ideal DNA sequence and catalytic functionality are identified, the chemical linkage can be selected from a library that contains linkers of randomized length and orientation.
Here we report a detailed study of the cleavage of the hairpin region of HIV-1 trans-activation response element (TAR) in various buffers of organic catalysts in the absence and presence of various complementary DNAs. Among the examined conditions, a two-base bulge loop stabilized by a pyrene intercalator is the most cleavage-prone DNA/RNA duplex structure. trans-(±)-Cyclohexane-1,2-diamine is the most active catalytic moiety. These results will be important for future design of sequence-specific artificial ribonuclease for HIV-1 TAR.
2. Results and Discussion
2.1 Cleavage of RNA in the presence of different organic catalysts
The RNA sequence (RNA 1) used in this study contains a hairpin and a four-base stem region of HIV TAR, an important target for the future HIV-1 therapy (Figure 1a) [20]. The 3'-end of 1 was labeled with 6-carboxyfluorescein (FAM). Cleavage of the RNA was studied in 0.5 M buffers containing different catalysts at pH 8.0. Ethylenediamine, propane-1,3-diamine, trans-(±)-cyclohexane-1,2-diamine, glycine, and imidazole were used as the catalysts. Ethylenediamine differs from propane-1,3-diamine by the distance between the two amino groups, and from trans-(±)-cyclohexane-1,2-diamine by the geometric constraints added to the two amino groups. Under physiological conditions, only one of the two amino groups is expected to be protonated. The protonated amino group will function as the activator of phosphodiester bond, while the unprotonated amino group is expected to serve as the basic catalyst to activate the 2'-OH. Unlike the diamine catalysts, glycine contains a carboxylate, which may also function as the basic catalytic group to cleave RNA. Although not a bifunctional catalyst, imidazole has been widely used in the design of artificial ribonucleases and thus was also included in this study.
Figure 1.
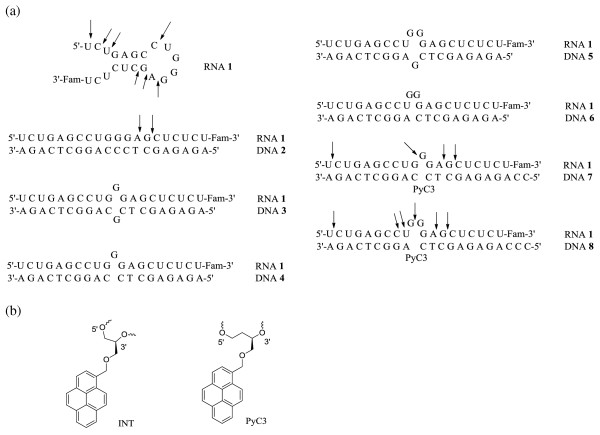
Oligonucleotide sequences and structure. (a) Oligonucleotide sequences and major cleavage sites. Arrow indicates the major cleavage sites when the RNA or RNA/DNA duplex was treated with trans-(±)-cyclohexane-1,2-diamine. The cleavage of 1/3, 1/4, 1/5, and 1/6 were not tested due to incomplete hybridization. (b) Chemical modifications of C3-spacer.
RNA 1 was incubated in the above buffers at 37°C for 18 h. The cleavage was analyzed by visualizing the fluorescent RNA bands after gel electrophoresis (Figure 2). Ethylenediamine and trans-(±)-cyclohexane-1,2-diamine showed the most prominent cleavage effects. The higher activity observed for trans-(±)-cyclohexane-1,2-diamine (31%) compared to ethylenediamine (24%) was perhaps a result of two preorganized amino groups constrained by the cyclohexane ring. In contrast, propane-1,3-diamine only generated 5% of cleavage. A previous study on the hydrolysis of a short single-stranded RNA suggested that propane-1,3-diamine was more active than ethylenediamine [21]. However, for RNA 1, ethylenediamine and trans-(±)-cyclohexane-1,2-diamine, both containing an ethylene linkage between the two amino groups, were much more active. The discrepancy between the two studies may result from the difference between the highly structured geometry of RNA 1 and the random coil of the short single-stranded RNA used in the previous study.
Figure 2.
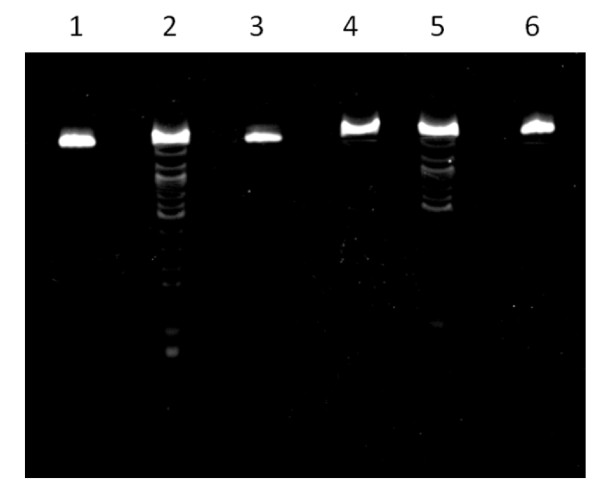
Cleavage of RNA 1 in buffers containing 20 mM NaCl, 1 mM Tris-EDTA (pH 8.0), and different organic catalysts (0.5 M, pH 8.0). Lane 1, no catalyst. Lane 2, trans-(±)-cyclohexane-1,2-diamine. Lane 3, glycine, Lane 4, propane-1,3-diamine. Lane 5, ethylenediamine. Lane 6, imidazole. Overall cleavage: Lane 1, 0%. Lane 2, 31%. Lane 3, 0%. Lane 4, 5%. Lane 5, 24%. Lane 6, < 1%.
Glycine has been used to cleave RNA when conjugated with an RNA intercalator [22]. However, it was the least active catalyst shown in this study. No cleaved band can be observed when the RNA was incubated in glycine buffer (Figure 2). This result demonstrates the importance of the basic group of the bifunctional catalyst, although the likelihood of glycine being more difficult to access the cleavage site than 1,2-diamines due to the charge differences cannot be completely excluded. The less basic carboxylate group of glycine may have led to lower efficiency in activating the 2'-OH for in-line attack compared to the more basic amino groups of 1,2-diamines. It is rather difficult to design a moderately basic but unprotonated group under physiological conditions. The 1,2-diamino compounds are ideal small molecular candidates to fill the role of providing basic groups to activate the 2'-OH of RNA nucleotides.
Surprisingly, imidazole, which has been widely used in the design of artificial ribonucleases, showed very low cleaving activity (< 1%). Many artificial ribonucleases as well as the natural RNase A contain a pair of imidazoles to function cooperatively [10,11]. The activity of such a bis-imidazole varies and is highly dependent on the locations and orientations of the two imidazole rings. In contrast, 1,2-diamino compounds are smaller and structurally less complicated, and more suitable for developing a small library of RNA-cleaving agents.
The superior RNA cleavage activity of trans-(±)-cyclohexane-1,2-diamine was also observed in our studies of other RNA sequences (J. Rana, H. Huang, Cleavage of DNA/RNA duplexes by trans-cyclohexane-1,2-diamine, unpublished). We tested the low concentration limit in using this catalyst. Significant cleavage of 1 (5%) can still be observed in 0.05 M trans-(±)-cyclohexane-1,2-diamine, but not in further lower catalyst concentrations.
2.2 Design and synthesis of oligonucleotides containing pyrene-modified C3 spacer
After establishing that trans-(±)-cyclohexane-1,2-diamine is the best small molecular catalyst for the cleavage of 1, we sought to study the influence of the complementary DNA on the cleavage. It has been reported that RNA bulge loop region of an RNA/DNA duplex is susceptible to hydrolysis [10,15,18,19]. We first tested five DNA sequences (DNA 2, 3, 4, 5 and 6) for their impact on RNA cleavage rate. The antisense DNA strands were mixed with RNA 1 using standard hybridization conditions prior to the treatment of trans-(±)-cyclohexane-1,2-diamine. However, 1/3, 1/4, 1/5, and 1/6 were only partially hybridized, as indicated by non-denaturing gel electrophoresis. The percentages of unhybridized RNA at room temperature are 24%, 22%, 57%, and 67% for 1/3, 1/4, 1/5, and 1/6, respectively, when DNA is present in 1.2 fold excess. The perfectly matching antisense DNA 2 formed a stable duplex with 1. Because unhybridized RNA will cause ambiguity in analyzing cleavage products, we looked for structurally modified DNA that can bind RNA more strongly. Although various oligonucleotide backbone modifications have been invented for more efficient binding to complementary RNA [23,24], these modifications require substitution of multiple nucleotides. We intended to restrict the modification to only one nucleotide and keep the rest with native DNA structure. This chemical modification will also serve as the tagging site for future conjugation with RNA cleaving agents. We designed a pyrene-modified C-3 spacer (PyC3) to approach this goal (Figure 1b). The PyC3 spacer is one carbon longer than the previously reported pyrene-glycol spacer (INT™), which stabilizes DNA duplex but destabilizes RNA duplex when introduced as a bulge [25]. We anticipated that INT can somewhat stabilize DNA/RNA duplex when introduced opposite an RNA bulge. However, glycol nucleic acids are known to pair with DNA or RNA weakly [26], indicating the importance of keeping three carbon atoms between the phosphate groups. Accordingly, the INT structure was modified to PyC3 by slightly increasing the length of the spacer.
The phosphoramidite of PyC3 was synthesized from (R)-butane-1,2,4-triol (Scheme 1). The 1- and 4- hydroxyl groups were protected with p-methoxybenzylidene (PMB). The remaining free 2- hydroxyl group was reacted with 1-bromomethylpyrene to form 9. The phosphoramidite was synthesized using a standard procedure and incorporated to DNA 7 and 8. DNA 7 and 8 contain extra nucleotides in the 5' end in order to distinguish the possible fluorescent DNA band from the RNA bands on the gel. Hybridization between DNA 7/8 and RNA was examined by non-denaturing gel electrophoresis. DNA 7 was completely hybridized with RNA 1. The hybridization between 1 and 8 was also nearly complete, with only 2% of RNA remained unhybridized.
Scheme 1.
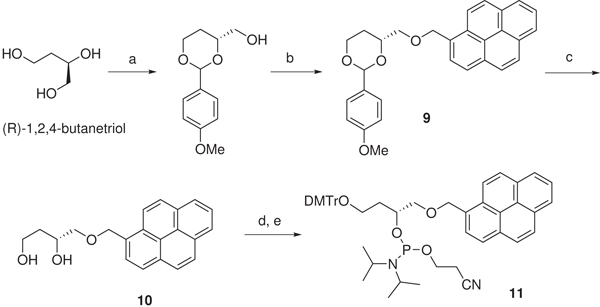
Synthesis of the phosphoramidite of the PyC3 spacer. Regents and conditions: (a) p-anisaldehyde, TsOH, CH2Cl2, 12 h, 83%. (b) NaH, 1-bromomethylpyrene, THF/toluene, 48 h, 77%. (c) TsOH, MeOH/CH2Cl2, 30 h, 30%. (d) Dimethoxytrityl chloride, triethylamine, DMAP, CH2Cl2, 12 h, 65%. (e) N, N'-diisopropyl 2-cyanoethyl phosphoramidic chloride, N, N'-diisopropylethylamine, CH2Cl2, 2 h, 100%.
Melting studies on RNA 1 and duplexes 1/2, 1/7, and 1/8 (the 3'-Fam is not attached to the RNA) indicates that the melting temperature of the looped RNA is significantly higher than those of the duplexes (Table 1). The perfectly matching duplex (1/2) is the most thermodynamically stable duplex. The duplex containing a single unpaired RNA nucleotide (1/7) is less stable than 1/2, but more stable than the duplex containing double unpaired RNA nucleotides (1/8). The ΔG298 values of 1/7 and 1/8 suggest that both duplexes are stable at room temperature, consistent with the non-denaturing gel electrophoresis results.
Table 1.
Thermodynamic parameters of RNA/DNA duplexes 1/2, 1/7, and 1/8.a, b
| Duplex | ΔH (kcal/mol) | ΔS (cal/mol·K) | Tm (°C)c | ΔG298 (kcal/mol) |
|---|---|---|---|---|
| 1/2 | -214.7 | -607 | 68.8 ± 0.3 | -33.8 |
| 1/7 | -126.0 | -365 | 55.7 ± 0.4 | -17.2 |
| 1/8 | -83.5 | -238 | 48.5 ± 0.3 | -12.6 |
a. RNA is not labeled with FAM.
b. Buffer: 10 mM PIPES (pH 7.5), 100 mM NaCl, 1 mM Tris·EDTA.
c. Duplex concentration: 2 μM.
2.3 Cleavage of RNA/DNA duplexes
Duplexes 1/2, 1/7, and 1/8 were incubated in trans-(±)-cyclohexane-1,2-diamine at 37°C for 18 h (Figure 3 and Table 2). Gel electrophoresis of the reaction mixtures showed that the pyrene-containing DNA strands were not visible under the conditions of visualization. The percent cleavage of the 1/2 duplex was significantly lower than 1 alone. This result confirmed that RNA nucleotides in duplex regions are more chemically stable than unpaired RNA nucleotides. Duplex 1/7 showed lower cleavage rate than 1, but higher rate than duplex 1/2. The preferred cleavage sites of the RNA were significantly affected by the presence of DNA 7. The phosphodiester bond between G10 and G11 became sensitive to hydrolysis, which was not observed for 1 alone. Duplex 1/8 showed overall cleavage rate comparable to 1. The higher cleavage rate of 1/8 in the region between U9 and G11 compared to 1 indicated that duplex 1/8 was still stably formed at elevated temperature (37°C), despite the fact that about 2% RNA was not completely hybridized at room temperature. The higher rate of 1/8 compared to 1/7 suggests that a bulge formed by a single unpaired RNA base is not as prone to hydrolysis as that formed by multiples unpaired RNA bases. For this particular RNA, the maximum number of unpaired RNA bases in a DNA/RNA duplex is two. Formation of more than two unpaired RNA base will greatly reduce the thermodynamic stability of the duplex. In other words, shorter complementary DNAs will not be able to unwrap the duplex stem of the hairpin RNA. Overall, the two-base bulge loop structure is the best form to maintain high cleavage activity without affecting the duplex stability and site selectivity. We also tested the cleavage of duplex 1/8 in buffers containing various organic catalysts. trans-(±)-Cyclohexane-1,2-diamine remained the most active catalysts, consist with the studies of the hairpin RNA.
Figure 3.
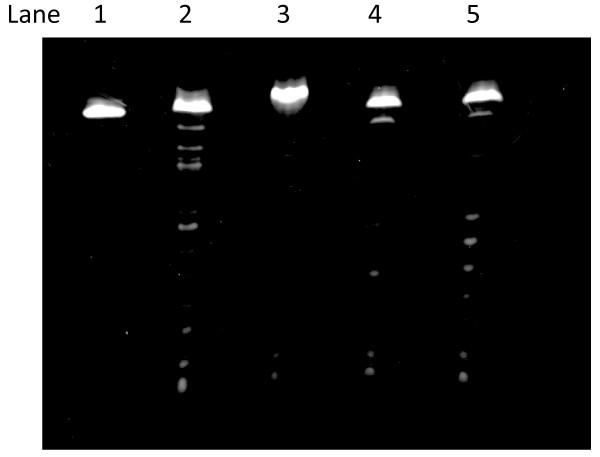
Cleavage of DNA/RNA duplexes in trans-(±)-cyclohexane-1,2-diamine (0.5 M), 20 mM NaCl, and 1 mM Tris-EDTA (37°C, pH 8.0, 18 h). Lane 1, RNA 1 in the absence of the catalyst. Lane 2, RNA 1 in the presence of the catalyst. Lane 3, duplex 1/2, Lane 4, duplex 1/7, Lane 5, duplex 1/8.
Table 2.
Product distribution of the RNA cleavage reaction in trans-(±)-cyclohexane-1,2-diaminea
| RNA 1 (%) | Duplex 1/2 (%) | Duplex 1/7 (%) | Duplex 1/8 (%) | |
|---|---|---|---|---|
| Full-length | 70.3 ± 5.1 | 88.6 ± 2.3 | 75.6 ± 6.5 | 68.9 ± 1.7 |
| n-1 | 4.0 ± 0.5 | < 1 | 6.9 ± 1.0 | 4.4 ± 2.1 |
| n-2 | 3.8 ± 0.4 | < 1 | < 1 | < 1 |
| n-3 | 4.5 ± 0.5 | < 1 | < 1 | < 1 |
| n-8 | < 1 | < 1 | < 1 | 3.6 ± 0.8 |
| n-9 | < 1 | < 1 | < 1 | 4.7 ± 0.2 |
| n-10 | < 1 | < 1 | 3.4 ± 1.2 | 4.0 ± 0.1 |
| n-11 | 3.6 ± 0.3 | < 1 | < 1 | < 1 |
| n-12 | 3.1 ± 1.0 | < 1 | < 1 | < 1 |
| n-13 | 2.9 ± 1.1 | 4.2 ± 0.9 | 2.6 ± 0.3 | 4.6 ± 2.3 |
| n-14 | 3.6 ± 1.3 | 3.6 ± 0.9 | 3.3 ± 1.1 | 4.8 ± 1.1 |
a. Buffer: 0.5 M trans-(±)-cyclohexane-1,2-diamine, 20 mM NaCl, and 1 mM Tris-EDTA (37°C, pH 8.0, 18 h).
Use of intercalators to enforce RNA bulge formation has been documented before. Kuzuya et al used DNA containing acridine to generate a single unpaired RNA nucleotide and studied its cleavage in the presence of zinc ions [27]. Our results suggest that when hybridization is efficient, more than one unpaired RNA base will be more useful in cleaving RNA nucleotides.
3. Conclusion
We have identified trans-(±)-cyclohexane-1,2-diamine as the most active organic catalyst in cleaving the upper stem-loop region of HIV-1 TAR among a group of organic molecules. The simple cyclohexane ring may geometrically constrain the two amino groups in a preorganized state for catalysis. We also designed a pyrene-modified C3 spacer to enhance the binding of a complementary DNA to the highly structured RNA. The RNA/DNA duplex showed the highest cleavage rate when two unpaired RNA nucleotides were formed. Although the overall cleavage rate for this RNA/DNA duplex was comparable to the native hairpin RNA, the cleavage sites were altered. This study demonstrated the feasibility of using trans-(±)-cyclohexane-1,2-diamine and PyC3 antisense DNA as the basic structural moieties to design artificial ribonucleases to site-specifically target HIV TAR.
4. Experimental
4.1 General
All reagents were purchased from Sigma-Aldrich or Fisher Scientific unless specified. NMR were recorded at Bruke 300 MHz facility. Mass spectra were obtained at the mass spectrometry facility of Rutgers University (Newark). RNA and structurally unmodified DNA were purchased from Integrated DNA Technology. Pyrene-containing oligonucleotides were synthesized on ABI 394 DNA synthesizer. Water used for electrophoresis was purified using Millipore DirectQ system with a BioPak Filter. Water used for other purpose was purchased from Fisher as RNAase free water. PAGE gel was prepared from 19:1 acrylamide/bisacrylamide.
4.2 RNA cleavage assay
The RNA/DNA (1/2, 1/7, 1/8) duplexes were hybridized in 100 mM NaCl and 1 mM tris-EDTA (pH 7.5). The RNA concentration was 50 μM. The DNA concentration was 60 μM. The hybridization was examined by 20% nondenaturaing PAGE electrophoresis. The gel was visualized using AlphaImager® EP.
A solution of 10 μM single stranded RNA (1) or DNA/RNA duplex (1/3, 1/7, 1/8) was incubated in 0.5 M cleaving agents at 37°C for 18 h. The buffer contained 20 mM NaCl and 1 mM Tris-EDTA (pH 8.0) and one of the following reagents: ethylenediamine, propane-1,3-diamine, trans-(±)-cyclohexane-1,2-diamine, glycine, and imidazole. Formamide loading buffer was added. The products were resolved on a 0.4 mm 20% denaturing PAGE gel. The gel was visualized using AlphaImager® EP at excitation wavelength 360 nm. The intensities of the bands were quantified using Alphaview 3.0.
4.3. Compound 9
The PMB acetal of (R)-1,2,4-butanetriol was synthesized from (R)-1,2,4-butanetriol and p-anisaldehyde using an established procedure [28]. To a solution of the PMB acetal of (R)-1,2,4-butanetriol (157 mg, 0.7 mmol) in 3 mL of THF, 28 mg of NaH (60%, 0.7 mmol) was added. The suspension was stirred at room temperature for 15 min. A solution of 1-bromomethylpyrene (200 mg, 0.97 mmol, prepared from 1-pyrenemethanol and phosphorus tribromide) in toluene (2 mL) was added. The reaction mixture was stirred at room temperature overnight before quenched with saturated sodium bicarbonate solution. A mixture of ethyl acetate and hexanes (1:1, 200 mL) was added for extraction. The organic layer was washed with saturated sodium chloride solution and dried over magnesium sulfate. The crude material was purified by column chromatography (hexanes/ethyl acetate 3:1) to yield 9 (216 mg, 77%). 1H NMR (CDCl3) δ 7.95 - 8.41 (m, 10H), 7.37 (d, 2H, J = 10 Hz), 6.83 (d, 2H, J = 10 Hz), 5.48 (s, 1H), 5.27 (d, 1H, J = 6 Hz), 4.34 (dd, 1H, J = 6 Hz, J = 3 Hz), 4.12 (m, 1H), 3.93 (m, 1H), 3.78 (s, 3H), 3.72 (dd, 1H, dd, 1H, J = 6 Hz, J = 3 Hz), 3.66 (dd, 1H, J = 6 Hz, J = 3 Hz), 1.89 (m, 1H), 1.54 (m, 1H). 13C NMR (CDCl3) δ 159.6, 131.1, 131.0, 130.9, 130.6, 129.3, 127.5, 127.2(5), 127.2(2), 127.1(9), 127.0, 125.7, 124.9(9), 124.9(8), 124.7, 124.5, 124.2, 123.4, 113.3, 100.9, 76.2, 72.8, 72.0, 66.5, 55.1, 27.8. ESI-MS (M-H) m/z Calcl for C29H25O4, 437.18 found 437.19.
4.4 Compound 10
A solution of 9 (210 mg, 0.5 mmol), p-toluenesulfonic acid monohydrate (10 mg) in a mixture of methanol and methylene chloride (1:1, 30 ml) was stirred at room for 30 h. The solvent was evaporated and the residue was purified by column chromatography (5% methanol in methylene chloride) to yield 10 (50 mg, 30%). 1H NMR (CD3OD) δ 8.27 (1H, d, J = 10 Hz), 7.81-8.02 (m, 8 H), 5.07 (s, 2H), 3.92 (m, 1H), 3.66 (m, 2H), 3.52 (m, 2H), 1.64 (m, 2H). 13C NMR (CD3OD) δ 131.0, 130.9, 130.4, 128.9, 126.9, 126.7, 126.5, 125.4, 124.6, 124.2, 124.1, 123.8, 122.8, 74.2, 71.0, 67.3, 58.2, 35.7. ESI-MS (M+Na) m/z Calcl for C21H20O3Na, 343.13, found 343.13.
4.5 Dimethoxytritylation of 10
A solution of 10 (50 mg, 0.16 mmol), dimethoxytrityl chloride (70 mg, 0.21 mmol), 4-dimethylaminopyridine (2 mg), triethylamine (46 μL, 0.32 mmol) in methylene chloride (2 mL) was stirred at room temperature overnight. The reaction was quenched with 100 μL of methanol. The solvent was evaporated. The crude material was purified by column chromatography (hexanes/ethyl acetate 2:1) to yield tritylated 10 (65 mg, 65%). 1H NMR (CDCl3) δ 8.39 (d, 1H, J = 10 Hz), 8.03-8.28 (m, 8H), 7.42 (d, 2H, J = 8 Hz), 7.24 (m, 7H), 6.73 (d, 4H, J = 8 Hz), 5.37 (s, 2H), 4.08 (m, 1H), 3.75 (s, 6H), 3.59 (m, 1H), 3.50 (m, 1H), 3.19-3.23 (m, 2H), 1.77 (m, 2H). 13C NMR (CDCl3) δ 158.1, 144.7, 135.9, 135.8, 131.1, 131.0, 130.9, 130.6, 129.7, 129.2, 127.8, 127.6, 127.2(3), 127.1(8), 126.9, 126.5, 125.7, 125.0(4), 125.0(3), 124.7, 124.5, 124.2, 123.1, 112.8, 86.1, 74.0, 71.7, 69.2, 60.8, 54.9, 33.1, 14.0. ESI-MS (M+Na) m/z Calcl for C42H38O5Na, 645.26, found 645.25.
4.6 Compound 11
To a solution of tritylated 10 (55 mg, 0.088 mmol), N, N'-diisopropylethyldiamine (72 μL, 0.41 mmol) in methylene chloride (2 mL), N, N'-diisopropyl 2-cyanoethyl phosphoramidic chloride (55 μL, 0.24 mmol) was added at 0°C. The solution was stirred at room temperature for 2 h. The reaction was diluted with ethyl acetate (100 mL) and washed with saturated sodium bicarbonate and brine. The organic layer was dried over magnesium sulfate. The crude material was purified by column chromatography (hexanes/ethyl acetate 5:1) to yield 11 (80 mg, 100%). 1H NMR (CDCl3) δ 8.37 (d, 1H), 8.21(d, 2H), 7.95-8.13 (m, 6H), 4.48 (m, 2H), 7.22 (m, 7H), 6.88 (m, 4H), 5.24 (m, 2H), 4.20 (m, 1H) 3.73 (s, 6H), 3.42-3.85 (m, 6H), 3.12 (m, 2H), 2.30-2.85 (m, 2H), 1.98 (m, 2H), 0.82-1.39 (m, 12H). 13P NMR (CDCl3) δ 149.6, 149.2. ESI-MS (M+Li) m/z Calcl for C51H55N2O6PLi, 829.40, found 829.40.
4.7 DNA synthesis and purification
Oligonucleotides were synthesized on an ABI 394 DNA synthesizer. Phosphoramidites of Bz-A, dmf-G, Ac-C, and T were purchased from Glen Research. The coupling time of pyrene-modified C3 spacer was extended to 15 min. Oligonucleotides were deprotected in concentrated ammonia at 55°C for 12 h. The crude oligonucleotides were purified by 20% denaturing PAGE. The gel band that contains the desired oligonucleotide was cut, crushed, and soaked in a solution of 100 mM NaCl and 1 mM EDTA overnight. The extract was centrifuged and the supernatant was desalted using a C-18 Sep-Pak cartridge (Waters). The concentrations of DNA were quantified using UV absorbance at 260 nm on a Shimadzu UV-1700 spectrometer. ESI-MS spectra of the fluorescent base-modified oligonucleotides were obtained in Rutgers University (Newark) mass facility. DNA 7 (M+K, monoisotope, positive ion mode), m/z calculated, 6843.1605, found, 6843.1959. DNA 8 (M+H, monoisotope, positive ion mode), m/z calculated, 6805.2046, found, 6805.2645.
4.8 Tm measurement
UV melting curves were recorded on a Shimadzu UV-1700 spectrometer equipped with TMSPC-8 UV melting system. A solution of the DNA/RNA duplex (0.5-3 μM) containing 100 mM NaCl and 10 mM PIPES (pH 7.5) in a total volume of 100 μL was heated from 20 to 85°C at 0.5°C/min. UV absorbance at 260 nm was taken every 1°C. The melting temperatures were calculated from the first derivative of the curve using the LabSolution software. Experiments were performed in triplicate. ΔH and ΔS were derived from Van't Hoff plots using the equation 1/Tm = (R/ΔH)ln[C]total + (ΔS-Rln4)/ΔH. [C]total is the total concentration of the RNA and the DNA strands.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SP and Jyoti Roy carried out the RNA cleavage assays. Jagruti Rana carried out the melting studies. HH synthesized the modified oligonucleotides. All authors read and approved the final manuscript.
Contributor Information
Sejal Patel, Email: sejalpa@gmail.com.
Jagruti Rana, Email: jdr6@njit.com.
Jyoti Roy, Email: jyotiroy@gmail.com.
Haidong Huang, Email: haidong.huang@njit.edu.
Acknowledgements
We thank New Jersey Institute of Technology for financial support.
References
- Foster AE, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Lewin AS, Hauswirth WW. Ribozyme gene therapy: applications for molecular medicine. Trends Mol Med. 2001;7:221–228. doi: 10.1016/S1471-4914(01)01965-7. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;441:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;475:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JR, Iranzo O. Synthetic metallonucleases for RNA cleavage. Curr Opin Chem Biol. 2004;8:192–200. doi: 10.1016/j.cbpa.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Niittymäki T, Lönnberg H. Artificial ribonucleases. Org Biomol Chem. 2006;4:15–25. doi: 10.1039/b509022a. [DOI] [PubMed] [Google Scholar]
- Kuzuya A, Komiyama M. Site-selective artificial ribonucleases and their applications. Curr Org Chem. 2007;11:1450–1459. doi: 10.2174/138527207782418717. [DOI] [Google Scholar]
- Trawick BN, Daniher AT, Bashkin JK. Inorganic mimics of ribonucleases and ribozymes: From random cleavage to sequence-specific chemistry to catalytic antisense-drugs. Chem Rev. 1998;98:939–960. doi: 10.1021/cr960422k. [DOI] [PubMed] [Google Scholar]
- Mironova NL, Pyshnyi DV, Shtadler DV, Fedorova AA, Vlassov VV, Zenkova MA. RNase T1 mimicking artificial ribonuclease. Nucleic Acids Res. 2007;35:2356–2367. doi: 10.1093/nar/gkm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh AD, Miller PS. Hydrolysis of bulged nucleotides in hybrids formed by RNA and imidazole-derivatized oligo-2'-O-methylribonucleotides. Nucleos Nucleot Nucl. 2011;30:235–255. doi: 10.1080/15257770.2011.569810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova NG, Fabani MM, Zenkova MA, Bichenkova EV, Polushin NN, Sil'nikov VV, Douglas KT, Vlassov VV. Sequence-specific artificial ribonucleases. I. Bis-imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy. Nucleic Acids Res. 2004;32:3887–3897. doi: 10.1093/nar/gkh702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Endo M, Komiyama M. Lanthanide complex-oligo-DNA hybrid for sequence-selective hydrolysis of RNA. J Chem Soc, Chem Commun. 1994. pp. 2019–2020.
- Endo M, Azuma Y, Saga Y, Kuzuya A, Kawai G, Komiyama M. Molecular design for a pinpoint RNA scission. Interposition of oligoamines between two DNA oligomers. J Org Chem. 1997;62:846–852. doi: 10.1021/jo9611780. [DOI] [Google Scholar]
- Kalek M, Benediktson P, Vester B, Wengel J. Identification of efficient and sequence specific biomolecular artificial ribonucleases by a combinatorial approach. Chem Commun. 2008. pp. 762–764. [DOI] [PubMed]
- Murtola M, Wenska M, Strömberg R. PNAzymes that are artificial RNA restriction enzymes. J Am Chem Soc. 2010;132:8984–8990. doi: 10.1021/ja1008739. [DOI] [PubMed] [Google Scholar]
- Verheijen JC, Deiman B, Yeheskiely E, van der Marel GA, van Boom JH. Efficient hydrolysis of RNA by a PNA-diethylenetriamine adduct. Angew Chem Int Ed. 2000;39:369–372. doi: 10.1002/(SICI)1521-3773(20000117)39:2<369::AID-ANIE369>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Gnaccarini C, Peter S, Scheffer U, Vonhoff S, Klussmann S, Göbel MW. Site-specific cleavage of RNA by a metal-free artificial nuclease attached to antisense oligonucleotides. J Am Chem Soc. 2006;128:8063–8067. doi: 10.1021/ja061036f. [DOI] [PubMed] [Google Scholar]
- Kuznetsova IL, Zenkova MA, Gross HJ, Vlassov VV. Enhanced RNA cleavage within bulge-loops by an artificial ribonuclease. Nucleic Acids Res. 2005;33:1201–1212. doi: 10.1093/nar/gki264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagórowska I, Kuusela S, Lönnberg H. Metal ion-dependent hydrolysis of RNA phosphodiester bonds within hairpin loops. A comparative kinetic study on chimeric ribo/24-O-methylribo oligonucleotides. Nucleic Acids Res. 1998;26:3392–3396. doi: 10.1093/nar/26.14.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J, Keen NJ, Churcher MJ, Aboul-ela F, Varani G, Hamy F, Felder ER, Heizmann G, Klimkait T. The HIV tat-TAR interaction, a novel target for drug discovery. Trends in Drug Research II. 1998;29:121–132. [Google Scholar]
- Bibillo A, Figlerowicz M, Kierzek R. The non-enzymatic hydrolysis of oligonucleotides VI. The role of biogenic polyamines. Nucleic Acids Res. 1999;27:3931–3937. doi: 10.1093/nar/27.19.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Hirata K, Ihara T, Sueda S, Takagi M, Komiyama M. RNA hydrolysis by the cooperation of carboxylate ion and ammonium ion. J Am Chem Soc. 1996;118:5478–5479. doi: 10.1021/ja960009u. [DOI] [Google Scholar]
- Takahashi T, Hamasaki K, Kumagai I, Ueno A, Mihara H. Design of a nucleobase-conjugated peptide that recognizes HIV-1 RRE IIBRNA with high affinity and specificity. Chem Commun. 2000. pp. 349–350.
- Summerton J. Morpholino antisense oligomers: the case for an RNA H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Christensen UB, Pedersen EB. Intercalating nucleic acids containing insertions of 1-O-(1-pyrenylmethyl)glycerol: Stabilization of dsDNA and discrimination of DNA over RNA. Nucleic Acids Res. 2002;30:4918–4925. doi: 10.1093/nar/gkf624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Chen J, Szostak JW. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc Natl Acad Sci USA. 2007;104:14598–14603. doi: 10.1073/pnas.0704211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuya A, Mizoguchi R, Sasayama T, Zhou JM, Komiyama M. Selective activation of two sites in RNA by acridine-bearing oligonucleotides for clipping of designated RNA fragments. J Am Chem Soc. 2004;124:1430–1436. doi: 10.1021/ja0389568. [DOI] [PubMed] [Google Scholar]
- Goldring WPD, Pattenden G, Rimmington SL. Synthesis of a tetraoxy-bis-nortaxadiene, en route to taxol, using a cascade radical cyclisation sequence. Tetrahedron. 2009;65:6670–6681. doi: 10.1016/j.tet.2009.04.021. [DOI] [Google Scholar]