Abstract
Cardiovascular disease encompasses a range of conditions extending from myocardial infarction to congenital heart disease most of which are heritable. Enormous effort has been invested in understanding the genes and specific DNA sequence variants responsible for this heritability. Here, we review the lessons learned for monogenic and common, complex forms of cardiovascular disease. We also discuss key challenges that remain for gene discovery and for moving from genomic localization to mechanistic insights with an emphasis on the impact of next generation sequencing and the use of pluripotent human cells to understand the mechanism by which genetic variation contributes to disease.
Introduction
Cardiovascular disease (CVD) is a leading health problem, affecting over 80,000,000 individuals in the United States alone. CVD encompasses a broad range of disorders including diseases of the vasculature, the myocardium, the heart’s electrical circuit, and congenital heart disease (Roger et al., 2012). For nearly all of these disorders, inherited DNA sequence variants play a role in conferring risk for disease. For example, in the general population, a history of premature atherosclerotic CVD in a parent confers ~3.0-fold increase in CVD risk to offspring (Lloyd-Jones et al., 2004). The precise magnitude of the role of inheritance, however, varies by disease and by other factors such as age of disease onset and subtype of disease.
Over the past century, a key goal of biomedical research has been to correlate genotype with phenotype, i.e., to identify the specific genes and DNA sequence variants responsible for trait variation in humans. What is the principal reason to pursue this goal? Naturally occurring genetic variation has the unique potential to reveal causal biologic mechanisms in humans. This is particularly important as some diseases like myocardial infarction (MI) are poorly modeled in non-human species.
In this review, we consider the approaches used to discover genes for human CVD, the lessons learned from the study of Mendelian and of common, complex forms of CVD, and take a look forward at research driven by next generation techniques, including sequencing and modeling human genetic disease in cells.
Approaches to discover genes for cardiovascular disease
To discover genes for CVD and its risk factors in humans, two major approaches - linkage analysis and genetic association - have been utilized. The choice of approach has depended on the pattern of segregation, whether consistent with the ratios described by Mendel or more complex. Some forms of CVD exhibit a simple pattern of inheritance suggestive of a single causal gene that confers a large effect on phenotype. For many of these Mendelian forms of CVD, direct DNA sequencing and/or linkage analysis has successfully yielded the causal gene and mutation. In 1985, Lehrman and colleagues directly sequenced the low-density lipoprotein receptor (LDLR) gene in a patient with homozygous familial hypercholesterolemia and uncovered a 5 kilobase deletion that eliminated several exons, representing the first demonstration of a mutation for Mendelian CVD (Lehrman et al., 1985). In 1989, linkage analysis was used to localize the chromosomal position of a causal gene for hypertrophic cardiomyopathy and in the subsequent year, mutations in the beta cardiac myosin heavy chain were discovered as causal for the phenotype (Geisterfer-Lowrance et al., 1990; Jarcho et al., 1989). Other prominent examples in the CVD field include long QT syndrome, severe hypercholesterolemia, Mendelian forms of hypertension, Marfan’s syndrome, and several forms of congenital heart disease including septal defects and valve defects (Abifadel et al., 2003; Basson et al., 1997; Berge et al., 2000; Curran et al., 1995; Dietz et al., 1991; Garcia et al., 2001; Garg et al., 2003; Garg et al., 2005; Lifton et al., 2001; Schott et al., 1998; Soria et al., 1989; Tartaglia et al., 2001).
However, most CVD traits such MI or concentrations of plasma LDL cholesterol show complex inheritance, suggestive of an interplay between multiple genes and non-genetic factors. Mapping gene loci associated with complex traits requires substantial levels of information and analysis, but since 2007 approaches to accomplish this goal have matured, and genetic mapping for complex traits in humans has become a reality. The intellectual foundations that enabled a systematic genome-wide screen of common variants (termed genome wide association study or GWAS) and results from this approach were recently reviewed (Altshuler et al., 2008; O’Donnell and Nabel, 2011). The tools and methods included catalogs of polymorphisms, techniques to genotype these DNA sequence variants, and the analytical framework to distinguish true association signal from false positives. The initial focus has been on utilizing common DNA sequence variants (variants with allele frequency > 1:20) as a discovery tool, largely because it was practical to do so; recent advances in DNA sequencing and genotyping technology will allow interrogation of less frequent variants and will be considered below. The National Human Genome Research Institute hosts a catalog of published GWASs and as of January 14, 2012, the catalog includes 1143 publications and 5585 single nucleotide polymorphisms (SNPs) with association evidence at P < 10−5 (Hindorff et al., 2009).
What have we learned from these gene discovery efforts? Below, we discuss lessons that have emerged from genotype to phenotype correlation studies for Mendelian diseases followed by lessons from studies of common, complex diseases.
Lessons learned from the study of Mendelian forms of CVD
Rare variants lead to broadly relevant insights
Since Mendelian diseases are rare in the population, there was initial skepticism about whether the genes and mechanisms that cause these diseases would inform our understanding of common forms of CVD. Linkage studies involve identifying and recruiting families with unique, often severe phenotypes, isolating a chromosomal segment that tracks with disease status in the family, and then pinpointing the causal gene and mutation in the linked segment. For a range of conditions, the genes identified by these linkage studies have transformed our understanding of CVD. Selected examples of Mendelian diseases, the responsible genes, and the gleaned biological and clinical insights are detailed in Table 1. Of particular significance is monogenic severe hypercholesterolemia where the six responsible genes have led to fundamental new biological concepts and supported the development of new therapies.
Table 1. Selected examples of Mendelian diseases and insights derived from the study of causal genes.
| Mendelian condition | Causal genes | Key biological and clinical insight(s) | Reference(s) |
|---|---|---|---|
| Severe hypercholesterolemia |
LDLR, APOB, ABCG5, ABCG8, ARH, PCSK9 |
|
(Abifadel et al., 2003; Brown and Goldstein, 1986; Garcia et al., 2001; Lehrman et al.,1985) |
| Familial hypobetalipoproteinemia |
APOB, PCSK9, ANGPTL3 |
|
(Cohen et al., 2006; Musunuru et al., 2010a; Soria et al., 1989) |
| Mendelian forms of low and high blood pressure |
SLC12A3, SLC12A1, KCNJ1, CLCNKB, NR3C2, SCNN1A, SCNN1B, SCNN1G CYP11B2, CYP11B1, HSD11B2, NR3C2, SCNN1B, SCNN1G, WNK1, WNK4, KLHL3, CUL3 |
|
(Boyden et al., 2012; Chang et al., 1996; Geller et al., 2000; Geller et al., 1998; Hansson et al., 1995; Lifton et al., 1992a; Lifton et al., 1992b; Lifton et al., 2001; Mune et al., 1995; Shimkets et al., 1994; Simon et al., 1997; Simon et al., 1996a; Simon et al., 1996b; Simon et al., 1996c; Wilson et al., 2001) |
| Hypertrophic cardiomyopathy |
MYH7, TNNT2, TPM1, TNNI3, MYL2, MYBPC3, ACTC, MYL3 |
|
(Bonne et al., 1995; Carrier et al., 1993; Geisterfer-Lowrance et al., 1990; Kimura et al., 1997; Olson et al., 2000; Poetter et al., 1996; Seidman and Seidman, 2001; Thierfelder et al., 1994; Watkins et al., 1995) |
| Marfan’s syndrome | FBN1 |
|
(Dietz et al., 1991; Lindsay and Dietz, 2011) |
| Atrial or ventricular septal defects |
NKX2-5, GATA-4, TBX5 |
|
(Basson et al., 1997; Garg et al., 2003; Schott et al., 1998) |
| Bicuspid aortic valve, Calcific aortic valve disease |
NOTCH1 |
|
(Garg et al., 2005) |
Genotype-phenotype correlation can be complex in monogenic disorders
Although there are cases where single gene mutations lead to straightforward genotype-phenotype associations, other, more complex relationships exist as well. This complexity can arise from three distinct genetic phenomena: pleiotropy, penetrance, and expressivity. Sometimes, mutations in a single gene can influence multiple phenotypic traits (i.e., pleiotropy). In 1995, Wang, Keating and colleagues identified the alpha subunit of the type V voltage-gated sodium channel (SCN5A) as the cause of inherited long QT syndrome type 3 (Wang et al., 1995). Since then, mutations in the same gene have been demonstrated to cause Brugada syndrome (right precordial ST-segment elevation and increased risk for ventricular arrhythmias), cardiac conduction system disease, and dilated cardiomyopathy (Chen et al., 1998; McNair et al., 2004; Schott et al., 1999). This range of disease phenotypes may reflect the underlying functionality of the channel.
Among carriers of a Mendelian mutation in a given family, some may exhibit the condition and others may not. Penetrance refers to the proportion of individuals with a given genotype who exhibit the phenotype associated with the genotype. In many Mendelian cardiovascular conditions inherited in an autosomal dominant manner, there is evidence for incomplete penetrance. For example, Hobbs and colleagues reported that in a pedigree with familial hypercholesterolemia due to a point mutation in LDLR, only 12 of 18 heterozygotes had high LDL cholesterol (>95th percentile) whereas some of the remaining 6 had LDL cholesterol as low as 28th percentile for the population (Hobbs et al., 1989). The lack of a high cholesterol phenotype given the same genotype may be due to modifier genes or environmental influences.
Individuals with the same Mendelian genotype can also show different degrees of the same phenotype. Expressivity is the degree to which trait expression differs among individuals. Marfan’s syndrome is a multi-system Mendelian disorder that can include a range of signs and symptoms involving the skeletal system (pectus excavatum, increased arm span to height ratio, craniofacial alterations), ocular system (eye lens dislocation, flat cornea), and cardiovascular system (aortic aneurysm, dissection of the ascending aorta, mitral valve prolapse), among others (Canadas et al., 2010). Dietz and colleagues identified mutations in the FBN1 gene encoding the extracellular matrix protein fibrillin 1 as responsible for Marfan’s syndrome (Dietz et al., 1991). When a specific mutation in the fibrillin 1 (FBN1) gene causes Marfan’s syndrome in a family, carriers of the same mutation can display variable clinical manifestations (Faivre et al., 2007).
Pleiotropy, penetrance, expressivity, and non-genetic factors conspire to ensure that even in a single gene disorder, genotype does not “equal” a specific phenotype. This complexity has several consequences. First, gene discovery is more difficult as genotype may not segregate perfectly with phenotype, thereby reducing the power of linkage. Second, there is intense interest in identifying modifiers - genetic or environmental - that may modulate the relationship between genotype and phenotype. Finally, because of this complexity, in many Mendelian diseases, it has been difficult to develop genotype-specific prognostic or treatment recommendations.
The long road from genotype to mechanism to treatment
For Marfan’s syndrome, the path from the discovery of FBN1 as the causal gene to a breakthrough in the molecular understanding of the disease has spanned more than two decades. Historically, Marfan’s syndrome had been viewed as a structural disease due to a defect in elastic fibers (Lindsay and Dietz, 2011). The identification of mutations in an extracellular matrix protein seemed to confirm this view. However, more recent studies suggest that microfibrils normally bind the large latent complex of the cytokine transforming growth factor β (TGF-β) and that failure of this event to occur results in increased TGF-β activation and signaling. Now, investigators are exploring the hypothesis that blocking TGF-β signaling will ameliorate the growth of aortic aneurysms in Marfan’s syndrome. For further examples of therapeutic approaches derived from the study of Mendelian disorders, we refer the reader to a recent review on this topic (Dietz, 2010).
Lessons learned from the study of common, complex forms of heart disease
Gene variants across the spectrum of allele frequency contribute to most complex, common diseases and quantitative traits
Variants associated with common, complex traits range in frequency from common (>1:20 frequency), to low-frequency (1:1000 to 1:20), to very rare (< 1:1000). In other words, genetic heterogeneity from variants across the frequency spectrum may be the rule. Consider the example of plasma triglycerides, a phenotype that marks triglyceride-rich lipoproteins including very-low density lipoprotein particles, chylomicrons, and remnant products of their metabolism. Roughly 50% of the inter-individual variability in plasma triglycerides is estimated to be on the basis of DNA sequence variants. Johansen, Hegele, and colleagues studied individuals from the extremes of the plasma triglyceride distribution (438 individuals with high triglycerides (mean triglycerides = 14.2 mmol/l) and 327 individuals with low triglycerides (mean triglycerides = 1.2 mmol/l)) (Johansen et al., 2010) using both GWAS and resequencing of selected genes. In the GWAS, common variants at seven loci were associated with plasma triglycerides, and in the re-sequencing study, there was an excess of rare, non-synonymous variants across four genes in individuals with high triglycerides when compared with those with low triglycerides. A comprehensive logistic regression model including clinical variables and both common and rare genetic variants explained 42% of total variation in hypertriglyceridemia diagnosis: clinical variables explained 20%, common genetic variants in seven loci explained 21%, and rare genetic variants in four loci explained 1%. The genetic architecture for triglycerides in the population appears to be that of a mosaic comprised of large-effect variants rare in frequency, small-effect variants common in frequency, and environmental influences.
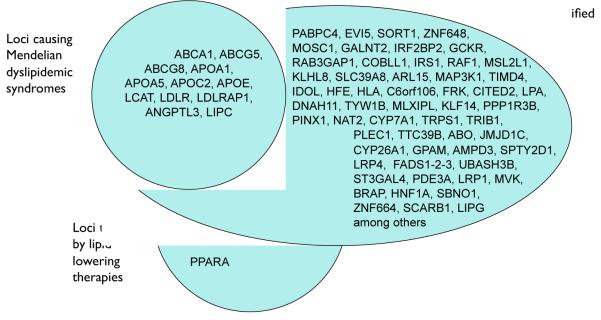
More generally, the concept of a mosaic model is supported by the fact that for many cardiovascular traits and diseases, there is strong overlap between the genes mapped using GWAS and those identified earlier through Mendelian families. Nineteen genes have been identified as monogenic causes of extremely low or high levels of LDL cholesterol, high-density lipoprotein (HDL) cholesterol, and triglycerides; loci harboring 16 of these genes were also mapped using GWAS (Figure 1) (Teslovich et al., 2010). Rare mutations in FBN1 cause the thoracic aortic aneurysms and dissections seen in Marfan’s syndrome, whereas common SNPs in the introns of FBN1 are the top association result in a GWAS for spontaneous, non-syndromic thoracic aortic aneurysm and dissection (Lemaire et al., 2011). Rare mutations in SCN5A, KCNQ1, KCNH2, KCNE1, and KCNJ2 cause monogenic long QT syndrome, whereas common SNPs in these five genes are associated with QT interval measured on electrocardiograms in the population (Newton-Cheh et al., 2009).
Figure 1. Overlap of genetic loci causing Mendelian dyslipidemic syndromes, those targeted by lipid lowering therapies, and those identified by GWAS.
Each GWAS locus is named according to a plausible biologic candidate in the locus or the gene nearest to the lead single nucleotide polymorphism (SNP). Note that for most GWAS loci, the causal gene is not yet proven. Of 19 genes previously implicated in Mendelian lipid disorders, 16 of the genes underlying these Mendelian disorders lie within 100 kilobases of one of the lead SNPs mapped by GWAS, including nine that lie within 10 kilobases of the nearest lead SNP (Teslovich et al., 2010).
New biological insights from GWAS genes
Plasma lipids, platelets, and sickle cell disease represent three fields where there has been progress toward new biology based on GWAS. GWASs for plasma LDL cholesterol, HDL cholesterol and triglycerides have evaluated >100,000 participants and mapped 95 distinct loci associated with at least one of these traits at a stringent statistical threshold (P < 5 × 10−8) (Kathiresan et al., 2008; Kathiresan et al., 2009b; Pollin et al., 2008; Teslovich et al., 2010; Willer et al., 2008). Approximately one-third of the loci harbored genes previously appreciated to play a role in lipoprotein metabolism including five targets of lipid-modifying therapies - HMGCR (statins), NPC1L1 (ezetimibe), APOB (mipomersen), CETP (anacetrapib, dalcetrapib, and evacetrapib), and PCSK9 (therapies in development by several pharmaceutical companies) (Figure 1).
Of note, the proportion of overall phenotypic variance explained by a genetic variant may have little correlation with the ultimate therapeutic or biological value of the gene mapped by the variant. Phenotypic variance explained by a variant is a function of two key parameters: allele frequency and effect size. For Mendelian diseases, the causal variants typically confer large effects but explain a small proportion of trait variance due to their rare frequencies. Variants from GWAS are common but explain a small proportion of trait variance due to modest effects. Nevertheless, variants that explain a small proportion of phenotypic variance may provide substantial biological or therapeutic insights. This has been highlighted for Mendelian genes in Table 1. Two examples for common SNPs include variants in the introns of HMGCR and NPC1L1 (Teslovich et al., 2010). These SNPs confer a small effect on plasma LDL cholesterol at 3 mg/dl and 2 mg/dl, respectively; however, targeting of these genes with statins or ezetimibe, respectively, has a much more dramatic effect on LDL cholesterol. And to date, there have been no rare, large-effect Mendelian mutations described in HMGCR, presumably because such mutations are highly deleterious and not tolerated. Thus, some disease genes may only be discoverable through common, small-effect variants.
Approximately two-thirds of the 95 loci discovered for plasma lipid traits harbored genes not previously appreciated to play a role in the biology of lipoproteins. Several genes at the “novel” loci have now been manipulated in the mouse and this manipulation led to plasma lipid changes analogous to that suggested by the human genetics. An example is the 8q24 locus containing the tribbles homolog 1 (TRIB1) gene (Burkhardt et al., 2010). DNA sequence variants downstream of the TRIB1 gene were initially associated with plasma lipids, with the minor allele having association with lower plasma triglycerides, lower plasma LDL cholesterol, and higher plasma HDL cholesterol. Given this pattern of plasma lipid changes, minor allele carriers would be expected to have lower risk for coronary heart disease and several groups have confirmed this expectation (Varbo et al., 2011). Targeted deletion of Trib1 in mice led to elevated levels of plasma triglycerides and cholesterol, whereas liver-specific overexpression of Trib1 reduced levels of plasma triglycerides and cholesterol. SORT1, GALNT2, PPP1R3B and TTC39B are four other genes implicated in lipoprotein regulation by GWAS and confirmed in a similar manner using mouse models (Musunuru et al., 2010b; Teslovich et al., 2010).
Analogous to plasma lipids, well-powered GWAS and experimental followup in cells and model organisms have identified new regulators of thrombopoiesis and/or erythropoiesis. A GWAS study for blood platelet count and volume mapped 58 loci (Gieger et al., 2011). For genes at 11 mapped loci, gene silencing in either the zebrafish or Drosophila led to alterations in thrombopoiesis and/or erythropoiesis. The precise mechanisms by which these genes alter blood cell formation remain to be defined through additional studies; however, we now have a plethora of new genes for experimental follow-up, all with a foundation of relevance to human phenotypes.
The identification of BCL11A as a transcriptional regulator of fetal hemoglobin synthesis and as a potential therapeutic target for sickle cell disease represents more biology learned from GWAS. Sickle cell disease results from the substitution of a valine for glutamic acid in the beta-globin chain of adult hemoglobin. The mutated hemoglobin (HbS) undergoes conformational change and polymerization upon deoxygenation, leading to hemolysis, red blood cell deformation, and pathology due to microvascular occlusion. Hereditary persistence of fetal hemoglobin (HbF) decreases the severity of sickle cell disease and the level of HbF in adults is inherited as a quantitative trait. As a result, induction of HbF in adults has been a long-standing goal of therapies for sickle cell disease. In 2007, GWAS for the HbF phenotype revealed a single strong locus, with sequence variants in the intron of a transcription factor, B-cell CLL/lymphoma 11A (BCL11A) (Menzel et al., 2007). A series of elegant studies led by Orkin and colleagues have now established that BCL11A represses the transcription of HbF expression in erythroid cells (Sankaran et al., 2008) (Sankaran et al., 2009; Xu et al., 2010) and demonstrated that inactivation of BCL11A in a mouse model of sickle cell disease leads to pancellular induction of HbF and corrected the hematologic and pathologic defects of the disease (Xu et al., 2011).
Despite these successes, for the vast majority of mapped loci, it has been a challenge to move from genomic localization to biologic mechanism. Why? First, as discussed earlier in the context of discovery of Mendelian disease genes, inferring new biology from human genetics takes time and only about five years have elapsed since the initial GWAS publications. Second, gene mapping and experimental follow-up require unique skill sets and expertise. In the examples highlighted above, collaborations between researchers focused on human genetics and experimentalists has been crucial to making progress, yet skepticism regarding the validity and value of discoveries made from GWAS may have dampened the enthusiasm for such collaborations (McClellan and King, 2010).
Third, genetic mapping by association gives us gene regions and not necessarily specific causal variants or causal genes. For each SNP and locus mapped by GWAS, we can generally conclude that within ~100,000 bases of the locus, there exists a causal gene. Ideally, at each discovered locus, we need to understand: 1) the causal variant; 2) the causal gene; 3) the mechanism by which the variant affects the gene; and 4) the mechanism by which the gene affects the phenotype. At the 1p13 locus for LDL cholesterol and MI, finemapping, re-sequencing and manipulation of positional candidate genes in cell culture and model organisms have addressed these key questions (Musunuru et al., 2010b). However, at most loci, the answers remain a mystery.
Several features of genetic mapping using common variants have contributed to the difficulties. Local correlation (linkage disequilibrium) and weak effect size of common variants have made it difficult to identify causal variants. Mapped SNP variants have weak or modest effect (odds ratios of 1.05 to 1.40 for a dichotomous trait and <1% of variance explained for a continuous traits) and these weak effects mean that in order to statistically distinguish between two correlated variants, the sample sizes need to be inordinately large. Nearly all of the mapped SNP variants are non-coding and our ability to interpret the non-coding portion of the genome remains limited. Mechanisms by which a non-coding variant may affect a nearby gene include, affecting a transcription factor or microRNA binding site and regulating the local chromatin state, among other possibilities.
In only a few instances has there been direction demonstration that a specific non-coding, GWAS SNP affects the relevant gene through one of these mechanisms (Musunuru et al., 2010b). The example of chromosome 9p21 and risk for MI exemplifies the challenge. In 2007, several independent GWASs identified SNPs on chromosome 9p21 as associated with MI or coronary artery disease, with ~50% of the population carrying a risk allele and each copy of the risk allele conferring ~29% increase in risk for MI/coronary artery disease (Helgadottir et al., 2007; McPherson et al., 2007; Samani et al., 2007). The most strongly associated SNPs were non-coding and more than 100 kilobases downstream of the nearest protein-coding genes CDKN2B (cyclin-dependent kinase inhibitor 2B encoding the protein p15INK4b) and CDKN2A (cyclin-dependent kinase inhibitor 2A encoding the proteins p16INK4a and p14ARF). Re-sequencing and fine-mapping studies in the gene region have identified a set of SNPs with indistinguishable statistical evidence, but not one causal variant (Shea et al., 2011). By what mechanism does the risk allele alter the relevant gene at the locus and what is the gene responsible for atherosclerosis susceptibility? Answers to these two questions have been particularly elusive. Several mechanisms by which the non-coding variant might impact phenotype have been explored including altering a non-coding RNA in the region (antisense noncoding RNA in the INK4 locus (ANRIL) or a synonym is CDKN2B antisense RNA (CDKN2BAS)), serving as an enhancer of regional gene expression, and disrupting the binding of a transcription factor STAT1 (Harismendy et al., 2011; Holdt and Teupser, 2012). No definitive answers have emerged. Finally, a non-coding gene region homologous to the human associated interval on 9p21 was deleted in mice and this targeted deletion led to near absence of CDKN2A and CDKN2B expression in several tissues, increased rate of tumors, and increased proliferation of aortic smooth muscle cells (Visel et al., 2010). However, these mice did not develop increased atherosclerosis on a diet rich in fat and cholesterol. For the 9p21 locus, the path from genomic localization to functional insights has not been a straightforward one, likely due to our limited ability at present to interpret the non-coding portions of the human genome. However, as recently reviewed (Raychaudhuri, 2011), integration of GWAS findings with expression quantitative trait loci (eQTL; eQTLs are genetic variants that correlate with the transcript level of a gene) and genome-wide maps of chromatin state dynamics may improve our ability to interpret GWAS findings and understand how non-coding variation regulates genes (Ernst et al., 2011; Schadt et al., 2008).
Gene variants may distinguish causal from non-causal biomarkers
Hypotheses concerning etiologic agents for complex diseases have often initially come from observational epidemiology. In 1961, in a paper entitled “Factors of Risk in the Development of Coronary Heart Disease”, Dr. William Kannel and colleagues at the Framingham Heart Study established an association of plasma total cholesterol with future risk for coronary heart disease (Kannel et al., 1961). Since then, hundreds of soluble biomarkers have similarly been associated with risk for coronary artery disease. How many of these biomarkers directly cause coronary artery disease and how many simply reflect other causal processes and why is this question important? Both causal and non-causal biomarkers may be helpful in terms of predicting risk for future disease. However, only a causal biomarker may be appropriate as a target of therapy. The ultimate proof of causality in humans is a randomized controlled trial testing whether a treatment that alters the biomarker will affect disease risk. However, as clinical trials are expensive and time-consuming, it would be helpful to have evidence in humans prior to engaging in a clinical trial.
In a technique termed Mendelian randomization, DNA variants are used to address the question of whether an epidemiological association between a risk factor and disease reflects a causal influence of the former on the latter (Davey Smith and Ebrahim, 2003; Gray and Wheatley, 1991; Katan, 1986). In principle, if a DNA variant is known to directly affect an intermediate phenotype (e.g., a variant in the promoter of a gene encoding a biomarker, affecting its expression) and the intermediate phenotype truly contributes to the disease, then the DNA variant should be associated with the disease to the extent predicted by (a) the size of the effect of the variant on the phenotype and (b) the size of the effect of the phenotype on the disease (Musunuru and Kathiresan, 2010). If in an adequately powered sample, the predicted association between the variant and disease were not observed, it would argue against a purely causal role for the intermediate phenotype in the pathogenesis of the disease. The study design is akin to a prospective randomized clinical trial in that the randomization for each individual occurs at the moment of conception---genotypes of DNA variants are randomly “assigned” to gametes during meiosis, a process that should be impervious to the typical confounders observed in observational epidemiological studies. For example, a parent’s disease status or socioeconomic status should not affect which of the parent’s two alleles at a given SNP is passed to a child, with each allele having an equal (50%) chance of being transmitted via the gamete to the zygote. Thus, Mendelian randomization should be unaffected by confounding or reverse causation. Mendelian randomization has potential shortcomings, including (a) the technique is only as reliable as the robustness of the estimates of the effect sizes of the variant on the phenotype and of the phenotype on disease, and (b) it assumes that the DNA variant does not influence the disease by means other than the intermediate phenotype being studied (pleiotropy), which may not be true. Nevertheless, Mendelian randomization has the potential to be as informative as a traditional randomized clinical trial.
Several Mendelian randomization studies have confirmed a causal relationship between LDL cholesterol and coronary artery disease. Nonsense variants in the PCSK9 gene that significantly reduce plasma LDL cholesterol concentrations were observed to be associated with reduced incidence of coronary artery disease in an African American cohort (Cohen et al., 2006). Similarly, in European Americans a common missense variant in PCSK9 associated with lower LDL cholesterol levels was also found to be associated with lower risk of MI. These observations suggested that lower LDL cholesterol is sufficient to provide protection against coronary artery disease. Similar to LDL cholesterol, several recent genetic studies have confirmed prior observations that plasma lipoprotein(a) causally relates to coronary artery disease (Clarke et al., 2009; Kamstrup et al., 2009; Wensley et al., 2011).
Unlike the results with plasma LDL cholesterol concentrations and plasma lipoprotein(a), three recent, large Mendelian randomization studies of C-reactive protein gene (CRP) variants that affect plasma CRP concentrations, performed in thousands of individuals, did not show an association between these variants and ischemic vascular disease or coronary artery disease (Elliott et al., 2009; Wensley et al., 2011; Zacho et al., 2008). It is unlikely that these studies were confounding or affected by pleiotropy, since the tested variants were within the CRP gene itself rather than being in other loci that might secondarily affect CRP levels. Although these studies cannot definitively rule out a causal role of CRP in cardiovascular disease, they strongly suggest that high CRP levels are indirectly rather than directly related to coronary artery disease.
A parallel line of genetic evidence also casts doubt on the notion that inflammatory biomarkers such as CRP are critical mediators of MI and coronary artery disease. Of 33 loci most highly associated with MI and coronary artery disease (Table 2), nine are related to plasma LDL cholesterol or lipoprotein(a), arguing for a strong causal relationship between LDL [or a modified LDL particle such as lipoprotein(a)] and disease (Schunkert et al., 2011). None of the other 24 loci are clearly related to inflammation. Overall, with the recent explosion in our ability to measure both soluble biomarkers (including metabolites and proteins) and genetic variation, Mendelian randomization will likely be increasingly utilized approach to distinguish causal biomarkers from non-causal ones.
Table 2. Genetic loci mapped by GWAS for myocardial infarction or coronary artery disease.
| Unique locus |
Chr | SNP | Risk allele frequency |
Odds ratio (95% CI) per risk allele |
Gene(s) of interest within or near associated interval |
Association of SNP or proxy with other phenotypes |
Reference(s) |
|---|---|---|---|---|---|---|---|
| Also associated with established risk factor - plasma LDL cholesterol | |||||||
|
| |||||||
| 1 | 1p13 | rs599839 | 78% | 1.11 (1.08-1.15) | CELSR2, PSRC1, SORT1 | eQTL for SORT1, CELSR2, and PSRC1 transcript levels in liver |
(Samani et al., 2007; Schunkert et al., 2011) |
| 2 | 19p13 | rs6511720 | 89% | 1.18 (1.11-1.25) | LDLR | (Consortium, 2011; Kathiresan et al., 2009a) | |
| 3 | 1p32 | rs11206510 | 82% | 1.08 (1.05-1.11) | PCSK9 | (Kathiresan et al., 2009a) | |
| 4 | 6q26- 6q27 |
rs3798220 rs10455872 |
2% 7% |
1.51 (1.33-1.70) 1.68 (1.43-1.98) |
LPA | lipoprotein (a) | (Clarke et al., 2009) |
| 5 | 9q34 | rs579459 | 21% | 1.10 (1.07-1.13) | ABO | venous thromboembolism, ACE enzyme activity, plasma Eselectin level, plasma vWF level, among others |
(Reilly et al., 2011; Schunkert et al., 2011) |
| 6 | 11q23 | rs964184 | 13% | 1.13 (1.10-1.16) | ZNF259, APOA5, APOA1 | triglycerides, HDL cholesterol | (Schunkert et al., 2011) |
| 7 | 8q24 | rs17321515 | 53% | 1.06 (1.03-1.10) | TRIB1 | triglycerides, HDL cholesterol | (Consortium, 2011; Varbo et al., 2011) |
| 8 | 2p21 | rs4299376 | 32% | 1.07 (1.04-1.11) | ABCG5, ABCG8 | serum phytosterols | (Consortium, 2011; Teupser et al., 2010) |
| 9 | 19q13 | rs2075650 | 14% | 1.14 (1.09-1.19) | APOE | Alzhemier’s dementia | (Consortium, 2011) |
|
| |||||||
| Also associated with established risk factor - blood pressure | |||||||
|
| |||||||
| 10 | 12q24 | rs3184504 | 44% | 1.07 (1.04-1.10) | SH2B3 | LDL cholesterol, type 1 diabetes, platelet count, plasma eosinophil count, celiac disease, among others |
(Gudbjartsson et al., 2009; Schunkert et al., 2011) |
| 11 | 10q24 | rs12413409 | 89% | 1.12 (1.08-1.16) | CYP17A1, CNNM2, NT5C2 | intracranial aneurysm | (Schunkert et al., 2011; Yasuno et al., 2010) |
|
| |||||||
| Unknown mechanism by which variant confers risk for myocardial infarction or coronary artery disease | |||||||
|
| |||||||
| 12 | 9p21 | rs4977574 | 46% | 1.29 (1.23-1.36) | CDKN2A, CDKN2B, ANRIL | coronary artery calcification, intracranial aneurysm, abdominal aortic aneurysm, among others |
(2007; Helgadottir et al., 2007; McPherson et al., 2007) |
| 13 | 21q22 | rs9982601 | 15% | 1.18 (1.12-1.24 | SLC5A3, MRPS6, KCNE2 | eQTL for MRPS6 transcript level in blood |
(Kathiresan et al., 2009a) |
| 14 | 1q41 | rs17465637 | 74% | 1.14 (1.09-1.20) | MIA3 | (Samani et al., 2007) | |
| 15 | 10q11 | rs1746048 | 87% | 1.09 (1.07-1.13) | CXCL12 | plasma CXCL12 level | (Samani et al., 2007) |
| 16 | 6p24 | rs12526453 | 67% | 1.10 (1.06-1.13) | PHACTR1 | coronary artery calcification | (Kathiresan et al., 2009a) |
| 17 | 2q33 | rs6725887 | 15% | 1.14 (1.09-1.19) | WDR12, NBEAL1 | eQTL for NBEAL1 transcript level in aortic media |
(Kathiresan et al., 2009a) |
| 18 | 3q22 | rs9818870 | 18% | 1.12 (1.07-1.16) | MRAS | eQTL for MRAS transcript level in aortic media |
(Erdmann et al., 2009) |
| 19 | 1p32 | rs17114036 | 91% | 1.17 (1.13-1.22) | PPAP2B | (Schunkert et al., 2011) | |
| 20 | 6p21 | rs17609940 | 75% | 1.07 (1.05-1.10) | ANKS1A | (Schunkert et al., 2011) | |
| 21 | 6q23 | rs12190287 | 62% | 1.08 (1.06-1.10) | TCF21 | eQTL for TCF21 transcript level in liver, fat |
(Schunkert et al., 2011) |
| 22 | 7q32 | rs11556924 | 62% | 1.09 (1.07-1.12) | ZC3HC1 | (Schunkert et al., 2011) | |
| 23 | 13q34 | rs4773144 | 44% | 1.07 (1.05-1.09) | COL4A1, COL4A2 | (Schunkert et al., 2011) | |
| 24 | 14q32 | rs2895811 | 43% | 1.07 (1.05-1.10) | HHIPL1 | (Schunkert et al., 2011) | |
| 25 | 15q25 | rs3825807 | 57% | 1.08 (1.06-1.10) | ADAMTS7 | (Reilly et al., 2011; Schunkert et al., 2011) | |
| 26 | 17p13 | rs216172 | 37% | 1.07 (1.05-1.09) | SMG6 | (Schunkert et al., 2011) | |
| 27 | 17p11 | rs12936587 | 56% | 1.07 (1.05-1.09) | RASD1, PEMT, RAI1 | eQTL for RASD1 and PEMT transcript levels in monocytes |
(Schunkert et al., 2011) |
| 28 | 17q21 | rs46522 | 53% | 1.06 (1.04-1.08) | UBE2Z | eQTL for UBE2Z transcript level in blood |
(Schunkert et al., 2011) |
| 29 | 7q22 | rs10953541 | 80% | 1.08 (1.05-1.11) | BCAP29, DUS4L | (2011) | |
| 30 | 10p11 | rs2505083 | 38% | 1.07 (1.04-1.09) | KIAA1462 | (2011) | |
| 31 | 11q22 | rs974819 | 32% | 1.07 (1.04-1.09) | PDGFD | eQTL for PDGFD transcript level in aortic media |
(2011) |
| 32 | 10q23 | rs1412444 | 42% | 1.09 (1.07-1.12) | LIPA | eQTL for LIPA transcript level in monocytes |
(2011; Wild et al., 2011) |
| 33 | 6p24 | rs6903956 | 7% | 1.65 (1.44-1.90) | c6orf105 | (Wang et al., 2011) | |
Next Generation of Approaches
Massively parallel sequencing as a discovery tool
As described above, considerable progress has been made in correlating genotype to phenotype for both Mendelian and common, complex diseases over the past several decades. Over the next decade, we are in a unique position to “finish the job.” For Mendelian diseases, the goal is nothing short of attempting to solve all Mendelian diseases not yet mapped. For common, complex diseases, the goal is to extend the range of genetic variation evaluated from common (>1:20 frequency) to low-frequency (1:1000 - 1:20 frequency) and very rare (<1:1000 frequency).
Dramatic advances in sequencing and genotyping make such ambitious goals feasible. Next-generation sequencing platforms have markedly decreased the cost of DNA sequencing when compared with Sanger sequencing. Hybridization approaches have enabled selection of the portion of the genome that is protein-coding (roughly 1% of the 3.2 billion bases), the so-called “exome.” Sequencing just the exome (rather than the entire genome) is well justified in the search for genetic causes of rare inherited disorders (Ng et al., 2010). This is because the majority of alleles responsible for Mendelian disorders disrupt protein-coding sequence and a large fraction of rare missense mutations in the human genome are predicted to be deleterious (Kryukov et al., 2007; Stenson et al., 2009).
Next-generation sequencing and Mendelian disease
Over the past three years, whole exome and whole genome sequencing has been successfully utilized to identify new genes for several Mendelian forms of CVD. Most studies have sequenced the exomes of one or a few individuals affected with the disorder. Variants seen in these individuals are typically compared with those from reference individuals (unaffected individuals who are family members or unrelated). Variants that are shared by affected individuals and not present in the unaffected population are considered causal candidates. With this strategy, investigators identified ANGPTL3 mutations as a cause of familial combined hypolipidemia, BAG mutations for dilated cardiomyopathy, NT5E mutations for arterial calcification, KCNJ5 mutations for hereditary hypertension due to aldosterone-producing adenomas, and KLHL3 or CUL3 mutations for hypertension and hyperkalemia (Boyden et al., 2012; Choi et al., 2011; Musunuru et al., 2010a; Norton et al., 2011; St Hilaire et al., 2011).
The discovery of KLHL3 as a cause of pseudohypoaldosteronism type II, a Mendelian form of hypertension, is an elegant example of the value of exome sequencing (Boyden et al., 2012). The gene was identified by directly sequencing the exomes of 11 unrelated index cases with pseudohypoaldosteronism type II and contrasting with exome sequences from 699 population-based, unrelated controls without hypertension. Of 22 case chromosomes, 5 harbored a rare, protein-altering mutation in KLHL3 whereas only 2 of 1398 control chromosomes did so, leading to a low probability that this observation was due to chance (P = 1 × 10−8). Such successes have raised expectations for the exome sequencing approach.
However, the true yield of exome sequencing in solving Mendelian disorders is difficult to know at present, as negative results have not been routinely reported (Bamshad et al., 2011). It can be difficult to arrive at a single causal mutation after exome sequencing due to the following reasons. First, the causal variant may not be protein-coding. Second, the causal variant may be protein coding but the relevant gene not successfully captured and sequenced. Approximately 5-10% of all exons may be poorly sequenced due to genomic features such as high GC content. Third, if the causal mutation is not fully penetrant, it will be present in individuals who are both phenotypically affected and unaffected and thus, may be “filtered” out. Fourth, the segregation of a phenotype in a family may be due to non-genetic factors rather than a single gene of large effect. This problem is particularly true for CVD conditions that are common in the population like MI and atrial fibrillation. If one observes a multi-generation family where multiple individuals are affected with MI, is this pattern due to a new Mendelian gene or poor lifestyle habits shared by the family? Fifth, in contrast to single nucleotide substitutions, methods for calling small insertion-deletions and copy number changes from short-read sequence data are in need of improvement. If the causal mutation is a one or two base-pair insertion or deletion, it might be overlooked. Finally, exome sequencing and filtering is more efficient at reducing the number of candidate causal mutations for recessive rather than dominant disorders. After exome sequencing and filtering for a dominantly inherited condition, the number of mutations compatible with causing disease can be large.
Next-generation sequencing and common, complex disease
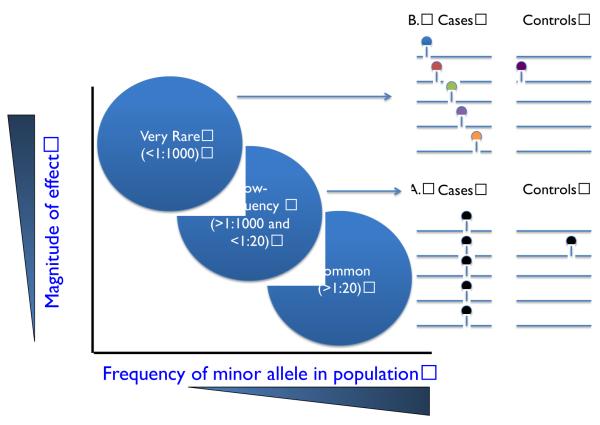
What about exome or whole genome sequencing to identify rare variants that confer a large effect on common, complex traits and diseases? The optimal study design for complex traits will depend on the frequency of the genetic variant(s) that are the source of the association signal. Though the term “rare” is used in the literature to refer to variants less than 1:20 frequency, we find it useful to distinguish between two types of “rare” variants, those that range in frequency from 1:1000 to 1:20 and denoted as “low-frequency” and those that are <1:1000 frequency and denoted as “very rare” (Figure 2). This categorical distinction is important because the approach to discover and replicate the two signals can differ.
Figure 2.
Association signals arising from rare variants. A. and B. Each line represents DNA sequence from a different individual and each circle represents a DNA sequence variant. Each color denotes a DNA sequence variant allele at a different nucleotide site. A. “Low-frequency” variants can be defined as variants in frequency between 1:1000 and 1:20. Each variant can be assayed typically by genotyping and tested for association individually using single marker tests of association. B. “Very Rare” variants can be defined as variants in frequency less than 1:1000 frequency and often the variant may be seen only in a single person (i.e., a singleton). Variants in this frequency range need to be grouped together and tested in aggregate. Association signals from a burden of such very rare variants are typically best validated using re-sequencing of the same genomic interval in additional individuals. Figure adapted from (Manolio et al., 2009)
For low-frequency variants, it is now possible to catalogue nearly all coding variants segregating in a population at a frequency of 1:1000 or greater, directly genotype these variants, and test for association with disease (Figure 2A). For example, data from 12,000 human exome sequences has been mined and all coding sites where the alternate allele is seen at least three times (twice for nonsense and splice) and in two separate studies have been catalogued. Investigators have designed a custom genotyping array - the ‘exome array’ - to directly genotype these ~300,000 variants at low cost. This genotyping array will allow for comprehensive testing of low-frequency coding variants for association with cardiovascular traits and diseases.
In contrast, a signal emerging from a burden of very rare variants can best be discovered by sequencing and can often only be replicated by sequencing (Figure 2B). The promise and challenges of this approach are evident in the candidate gene sequencing studies completed over the past few years. In seminal work, Cohen, Hobbs, and colleagues tested the hypothesis that rare, coding variation in candidate genes contributes to plasma HDL cholesterol variation in the population (Cohen et al., 2004). Three genes that cause Mendelian forms of low HDL cholesterol (ABCA1, APOA1, and LCAT) were sequenced in 128 individuals ascertained from the lowest 5th percentile (mean, 31 mg/dl) and 128 individuals from the highest 95th percentile of the HDL cholesterol distribution (mean, 91 mg/dl) in a population-based cohort study. The investigators aggregated rare variants that met three criteria: 1) present within a gene; 2) changed amino acid; and 3) exclusively present in either the low or high group. Twenty such rare alleles were found in the low group and two such rare alleles were found in the high group (Fisher’s exact P=0.00006). This observation was replicated in an independent study and, additionally, cells from mutation carriers had reduced cholesterol effux rates, a key function of ABCA1. This study established that rare non-synonymous mutations in ABCA1 with large phenotypic effects contribute to low HDL cholesterol in the population.
The challenge now is to utilize sequencing to enable de novo discovery of new genes that contribute to complex traits. It is now practical to extend the experiment described above from the sequencing of 3 genes to ~20,000 genes. Exomes can be rapidly sequenced and single nucleotide substitutions can be accurately called; however, will such sequencing for complex traits readily yield new discoveries? The key barriers at present are statistical. DNA sequence variants that are very rare (e.g., a mutation seen in a single person or a singleton) cannot be tested individually for association with phenotype. Instead, they need to be aggregated with similar rare variants to be tested collectively for association with phenotype, due to their rarity (Figure 2B).
This requirement immediately brings forward several questions. Do we only aggregate variants within a single gene or should it be extended to collections of genes in a pathway? Do we only group variants of a certain annotation - non-synonymous, nonsense, synonymous, splice-site, etc.? Should we impose a frequency threshold to define ‘rare’ (i.e., only aggregate variants that are less than 1% frequency) and if so, what should this frequency threshold be? The answers to many of these questions are in development. We refer the reader to several recent publications that address these issues (Li and Leal, 2008; Madsen and Browning, 2009; Neale et al., 2011; Price et al., 2010).
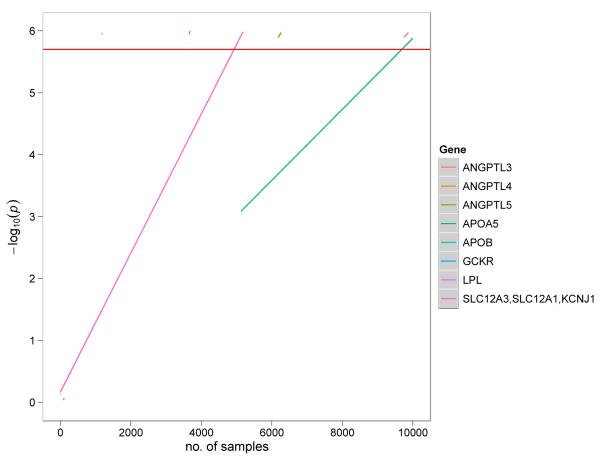
Another key question is statistical power. What sample size will be required to identify a new disease gene based on a burden of rare coding mutations? An early computer simulation study conducted by Kryukov, Sunyaev, and colleagues suggested that 10,000 individuals (5000 drawn from the lowest 5th percentile and 5000 above the 95th percentile) will be needed to discover another gene with a mutation burden similar to ABCA1 (Kryukov et al., 2009). The principal reason for this large sample size is the stringent statistical threshold that needs to be imposed when testing 20,000 genes. A first approximation of the appropriate statistical threshold would be Bonferonni correction for all the genes sequenced (P value of 2.5 × 10−6). Our extrapolation of effect sizes and frequencies from published studies shows (Figure 3) that thousands of individuals are required to reach acceptable statistical power.
Figure 3. Estimate of sample sizes required for gene discovery in exome sequencing studies of complex traits.
Extrapolations were performed using gene re-sequencing counts from published candidate gene studies. Fisher exact test was performed for gene burden testing on mutation carrier status within each study. The red horizontal line indicates the genome-wide P-value threshold (P = 2.5 × 10−6) for gene burden of rare variants with minor allele frequencies < 1%. The extrapolations show that thousands of samples are needed to exceed statistical significance at a genome-wide level. Rare variant counts for ANGPTL3, ANGPTL4 and ANGPTL5 for plasma triglycerides were obtained from Romeo and colleagues (Romeo et al., 2009); APOA5, APOB, GCKR and LPL for hypertriglyceridemia from Johansen and colleagues (Johansen et al., 2010); SLC12A3/SLC12A1/KCNJ1 from Ji and colleagues (Ji et al., 2008).
Statistical power would improve dramatically if we were able to distinguish rare mutations that affected protein function from those that did not. The bulk of discovered rare mutations are missense. Of all missense, it is estimated that ~1/5 are as deleterious as nonsense mutations, ~1/2 are mildly deleterious, and ~1/4 do not affect protein function (Kryukov et al., 2007). If one restricted mutation counts to only functional mutations, noise would be removed. At present, functional consequence of variants can be predicted based on comparative sequence analysis and protein structure analysis. Several software tools are available and the accuracy of these methods remains a subject of debate (Sunyaev et al., 2001).
An alternative method is experimental evaluation of whether a given mutation is functional. Such experimental evaluation is possible for genes with known function and where that function can be readily evaluated with moderate or high throughput approaches. However, such evaluation of mutations assumes that the parameter evaluated is an appropriate surrogate for the mechanism by which the gene impacts the human phenotype. For complex phenotypes such as MI that arise after decades of pathology, it is often difficult to know if this is the case. Nevertheless, distinguishing functional versus neutral missense mutations will be a key to improving statistical power.
Modeling human genetic disease in reprogrammed cells
As an increasing number of genetic variants become associated with human disease, it will be essential to develop effective and accurate model systems to understand the mechanism of disease. This step is a prerequisite to translate genetic findings into new targets for therapy. Animal models have traditionally been used to mimic human mutations through gene knock-in approaches or study of null mutations (Bruneau et al., 2001; Lindsay et al., 2001). While this approach has been of value and led to many discoveries, it is more common that mouse models fail to effectively recapitulate the human phenotype. This situation is even true for Mendelian traits, where heterozygous deletion of human disease genes is often well-tolerated in mice, while homozygous deletions can have catastrophic consequences, failing to model the human condition. For many human disease genes, this has been a bottleneck, limiting the value of gene-hunting approaches described above.
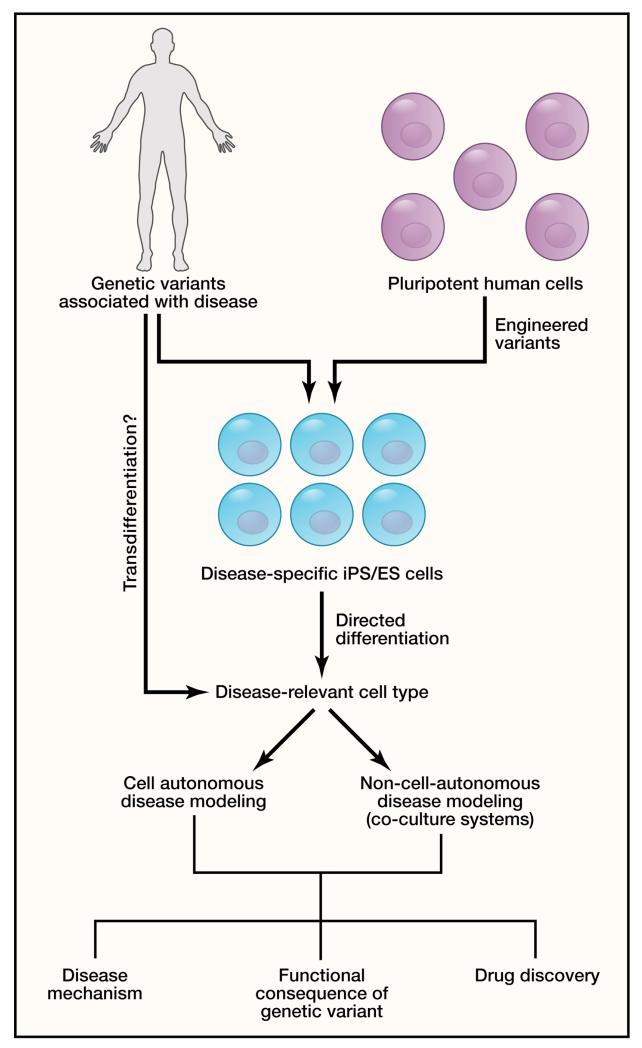
The recent revolution in cellular reprogramming technology provides a potential solution to the need for disease modeling (Figure 4). The striking observation by Yamanaka and colleagues that human adult somatic cells could be reprogrammed into a pluripotent state by expressing four transcription factors not only revolutionized the field of stem cell biology, but also has major consequences for the study of human genetics (Takahashi et al., 2007). Investigators can now reprogram skin or blood cells from patients with defined genetic variants/mutations into induced pluripotent stem (iPS) cells, and then differentiate the iPS cells into the specific cell type affected by disease. This approach provides an unprecedented opportunity to investigate the consequences of human genetic variation on cellular phenotypes that may contribute to disease. There have been several reports of partial modeling of CVD using human iPS cells in the last two years, although success to date has been limited to diseases with discrete cell-autonomous cellular defects. We will consider a few examples below and highlight the advantages and limitations of this approach.
Figure 4. Modeling human genetic disease in reprogrammed cells.
Reprogrammed cells derived from skin fibroblasts or blood cells of individuals with genetic variants associated with disease can be used for disease modeling, drug discovery, and to determine the impact of sequence variants on cellular biology. Reprogramming to induced pluripotent stem (iPS) cells and subsequent directed differentiation to relevant cell types can yield large numbers of human disease-related cell types for investigation. Specific genetic variants can also be engineered into human pluripotent stem cells using zinc-finger nucleases or TALENs to generate iPS or embryonic stem (ES) cells harboring disease-associated variants. Future approaches involving transdifferentiation may allow for a more direct approach to generate relevant cell types for study of CVD. In some cases diseases will be cell autonomous, but in others may require co-culture of two or more cell types to recapitulate the disease process.
Autosomal dominant long QT syndrome (LQTS) that predisposes to cardiac arrhythmias and sudden death involves single gene mutations of sodium or potassium channels that provide a discrete and quantitative phenotype in iPS-derived cardiomyocytes. In LQTS, repolarization of cardiomyocytes is delayed resulting in a prolonged action potential duration. Several groups have reported successful generation of iPS-derived cardiomyocytes from patients with LQTS, and an elongation of the action potential duration has been reported for a subset of genetically-induced LQTS (Itzhaki et al., 2011; Moretti et al., 2010; Yazawa et al., 2011). LQTS can also be drug-induced in selected individuals, and pluripotent stem cell-derived cardiomyocytes appear to be an effective model system to detect prolongation of action potentials due to cardiotoxic drugs (Braam et al., 2010). It is exciting to consider that identification of common genetic variants that predispose to drug-induced LQTS might be modeled in iPS-derived cardiomyocytes, thereby allowing a system in which to screen for this toxic side effect prior to in vivo use of new drugs.
Despite these successes, certain types of LQTS have been more difficult to model due to the incomplete maturation of iPS-derived cardiomyocytes and appropriate expression of ion channels in a consistent manner. It is possible that recent direct reprogramming strategies involving transdifferentiation, which do not involve a progenitor intermediate, will allow modeling of diseases where a mature cell type is required (Ieda et al., 2010). The inability to expand mature, transdifferentiated cells is currently a rate-limiting factor for this approach, but future improvements in efficiency of reprogramming may overcome this hurdle.
Another example of iPS-based modeling recently came from the study of Noonan and LEOPARD syndromes, autosomal dominant conditions that involve pulmonary valve stenosis and hypertrophic cardiomyopathy in a subset of patients (Gelb and Tartaglia, 2011). The genetic cause of Noonan and LEOPARD syndromes involves activating mutations in members of the Ras pathway. Correspondingly, iPS-derived cardiomyocytes from patients with cardiomyopathy associated with LEOPARD syndrome exhibit higher levels of Ras signaling that appear to contribute to excessive growth of cardiac cells (Carvajal-Vergara et al., 2010). While such findings successfully model the pathway known to be affected, the ultimate goal will be to identify new therapeutics that can disrupt abnormal pathways and restore normal physiology. This has remained elusive thus far, in part because most iPS-based disease models lead to broad variability in phenotype, whereas high-throughput screening assays typically require a consistent phenotype with a narrow range of variability. As protocols for generating iPS cells improve and directed differentiation methods become more consistent, “disease in a dish” models promise to alter the landscape of drug discovery (Figure 4).
Besides modeling the Mendelian syndromes described above, another potential use of iPS technology will be to annotate the function of SNPs identified by GWAS or rare mutations discovered by sequencing. iPS cells are currently being generated from large populations with defined genetic variants and CVD in the hope that certain aspects of the phenotype could be modeled in iPS-derived cells. It may prove difficult to identify consistent phenotypic differences associated with genetic variants of small effect; however, it is possible that the genetic background present in affected individuals will be sufficient to reveal a cellular defect.
The contribution of individual sequence variants in the context of complex genetic background issues can be interrogated through the use of recently developed DNA-correction approaches in human pluripotent stem cells. The use of zinc-finger nucleases and transcription activator-like effector nuclease (TALEN) technology to specifically introduce or remove a sequence variant in stem cells will reveal the contribution of individual variants to cellular phenotypes (Yusa et al., 2011). Addition of environmental stresses to the cellular system may be necessary to provoke phenotypes and will be an important tool in disease modeling in iPS cells. Such studies are currently underway and we look forward to the challenges and opportunities offered by this new approach to modeling human genetic variants.
Summary
The tools to decode human genetic variants on a large population scale have finally arrived and should begin to reveal the genetic contribution to disease at an exponential pace. Rigorous analysis of the functional consequences of sequence variants associated with disease will be critical, and new approaches leveraging stem cell technologies may facilitate study of human variants in relevant human cell types. Ultimately, the convergence of human genetics with functional biology will reveal new therapeutic targets for cardiovascular disease and a new generation of drug discovery efforts.
Acknowledgements
The authors wish to thank Drs. Ron Do, Nathan O. Stitziel, Gina M. Peloso, and Kiran Musunuru for assistance with analyses and critical review of the manuscript. S.K. was supported by grants from NHLBI/NIH, Foundation Leducq, and the Howard Goodman Fellowship from Massachusetts General Hospital; D.S. was supported grants from the NHLBI/NIH, the California Institute for Regenerative Medicine, the Younger Family Foundation, the Roddenberry Foundation and the Whittier Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Burkhardt R, Toh SA, Lagor WR, Birkeland A, Levin M, Li X, Robblee M, Fedorov VD, Yamamoto M, Satoh T, et al. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest. 2010;120:4410–4414. doi: 10.1172/JCI44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadas V, Vilacosta I, Bruna I, Fuster V. Marfan syndrome. Part 1: pathophysiology and diagnosis. Nat Rev Cardiol. 2010;7:256–265. doi: 10.1038/nrcardio.2010.30. [DOI] [PubMed] [Google Scholar]
- Carrier L, Hengstenberg C, Beckmann JS, Guicheney P, Dufour C, Bercovici J, Dausse E, Berebbi-Bertrand I, Wisnewsky C, Pulvenis D, et al. Mapping of a novel gene for familial hypertrophic cardiomyopathy to chromosome 11. Nat Genet. 1993;4:311–313. doi: 10.1038/ng0793-311. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Grunder S, Hanukoglu A, Rosler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, et al. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Yue P, Bjorklund P, Zhao B, Nelson-Williams C, Ji W, Cho Y, Patel A, Men CJ, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Boerwinkle E, Mosley TH, Jr., Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Consortium IKC. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Dietz HC. New therapeutic approaches to mendelian disorders. N Engl J Med. 2010;363:852–863. doi: 10.1056/NEJMra0907180. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre L, Collod-Beroud G, Loeys BL, Child A, Binquet C, Gautier E, Callewaert B, Arbustini E, Mayer K, Arslan-Kirchner M, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Tartaglia M. RAS signaling pathway mutations and hypertrophic cardiomyopathy: getting into and out of the thick of it. J Clin Invest. 2011;121:844–847. doi: 10.1172/JCI46399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, Lifton RP. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science. 2000;289:119–123. doi: 10.1126/science.289.5476.119. [DOI] [PubMed] [Google Scholar]
- Geller DS, Rodriguez-Soriano J, Vallo Boado A, Schifter S, Bayer M, Chang SS, Lifton RP. Mutations in the mineralocorticoid receptor gene cause autosomal dominant pseudohypoaldosteronism type I. Nat Genet. 1998;19:279–281. doi: 10.1038/966. [DOI] [PubMed] [Google Scholar]
- Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, Serbanovic-Canic J, Elling U, Goodall AH, Labrune Y, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R, Wheatley K. How to avoid bias when comparing bone marrow transplantation with chemotherapy. Bone Marrow Transplant. 1991;7 [PubMed] [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs HH, Leitersdorf E, Leffert CC, Cryer DR, Brown MS, Goldstein JL. Evidence for a dominant gene that suppresses hypercholesterolemia in a family with defective low density lipoprotein receptors. J Clin invest. 1989;84:656–664. doi: 10.1172/JCI114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- IBC 50K CAD Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, Levi T, Donis-Keller H, Seidman JG, Seidman CE. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 2009;301:2331–2339. doi: 10.1001/jama.2009.801. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J., 3rd Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009a;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009b;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SA, McDonald ML, Guo DC, Russell L, Miller CC, 3rd, Johnson RJ, Bekheirnia MR, Franco LM, Nguyen M, Pyeritz RE, et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat Genet. 2011;43:996–1000. doi: 10.1038/ng.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel JM. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992a;355:262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- Lifton RP, Dluhy RG, Powers M, Rich GM, Gutkin M, Fallo F, Gill JR, Jr., Feld L, Ganguly A, Laidlaw JC, et al. Hereditary hypertension caused by chimaeric gene duplications and ectopic expression of aldosterone synthase. Nat Genet. 1992b;2:66–74. doi: 10.1038/ng0992-66. [DOI] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Lindsay ME, Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. doi: 10.1038/nature10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones DM, Nam BH, D’Agostino RB, Sr., Levy D, Murabito JM, Wang TJ, Wilson PW, O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Mune T, Rogerson FM, Nikkila H, Agarwal AK, White PC. Human hypertension caused by mutations in the kidney isozyme of 11 beta-hydroxysteroid dehydrogenase. Nat Genet. 1995;10:394–399. doi: 10.1038/ng0895-394. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Kathiresan S. Genetics of coronary artery disease. Annu Rev Genomics Hum Genet. 2010;11:91–108. doi: 10.1146/annurev-genom-082509-141637. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010a;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010b;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Zuchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88:273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011;365:2098–2109. doi: 10.1056/NEJMra1105239. [DOI] [PubMed] [Google Scholar]
- Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:1687–1694. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers JR, Fananapazir L, Epstein ND. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Executive Summary: Heart Disease and Stroke Statistics--2012 Update: A Report From the American Heart Association. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Shea J, Agarwala V, Philippakis AA, Maguire J, Banks E, Depristo M, Thomson B, Guiducci C, Onofrio RC, Kathiresan S, et al. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat Genet. 2011;43:801–805. doi: 10.1038/ng.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Jr., Ulick S, Milora RV, Findling JW, et al. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- Simon DB, Bindra RS, Mansfield TA, Nelson-Williams C, Mendonca E, Stone R, Schurman S, Nayir A, Alpay H, Bakkaloglu A, et al. Mutations in the chloride channel gene, CLCNKB, cause Bartter’s syndrome type III. Nat Genet. 1997;17:171–178. doi: 10.1038/ng1097-171. [DOI] [PubMed] [Google Scholar]
- Simon DB, Karet FE, Hamdan JM, DiPietro A, Sanjad SA, Lifton RP. Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet. 1996a;13:183–188. doi: 10.1038/ng0696-183. [DOI] [PubMed] [Google Scholar]
- Simon DB, Karet FE, Rodriguez-Soriano J, Hamdan JH, DiPietro A, Trachtman H, Sanjad SA, Lifton RP. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996b;14:152–156. doi: 10.1038/ng1096-152. [DOI] [PubMed] [Google Scholar]
- Simon DB, Nelson-Williams C, Bia MJ, Ellison D, Karet FE, Molina AM, Vaara I, Iwata F, Cushner HM, Koolen M, et al. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet. 1996c;12:24–30. doi: 10.1038/ng0196-24. [DOI] [PubMed] [Google Scholar]
- Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci U S A. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Hilaire C, Ziegler SG, Markello TC, Brusco A, Groden C, Gill F, Carlson-Donohoe H, Lederman RJ, Chen MY, Yang D, et al. NT5E mutations and arterial calcifications. N Engl J Med. 2011;364:432–442. doi: 10.1056/NEJMoa0912923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teupser D, Baber R, Ceglarek U, Scholz M, Illig T, Gieger C, Holdt LM, Leichtle A, Greiser KH, Huster D, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3:331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Varbo A, Benn M, Tybjaerg-Hansen A, Grande P, Nordestgaard BG. TRIB1 and GCKR polymorphisms, lipid levels, and risk of ischemic heart disease in the general population. Arterioscler Thromb Vasc Biol. 2011;31:451–457. doi: 10.1161/ATVBAHA.110.216333. [DOI] [PubMed] [Google Scholar]
- Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, Shah T, Engert JC, Clarke R, Davey-Smith G, Nordestgaard BG, et al. Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild PS, Zeller T, Schillert A, Szymczak S, Sinning CR, Deiseroth A, Schnabel RB, Lubos E, Keller T, Eleftheriadis MS, et al. A Genome-wide Association Study Identifies LIPA as a Susceptibility Gene for Coronary Artery Disease. Circ Cardiovasc Genet. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, Simon M, Krex D, Arlier Z, Nayak N, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacho J, Tybjaerg-Hansen A, Jensen JS, Grande P, Sillesen H, Nordestgaard BG. Genetically elevated C-reactive protein and ischemic vascular disease. N Engl J Med. 2008;359:1897–1908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]