Summary
Genes underlying repeated adaptive evolution in natural populations are still largely unknown. Stickleback fish (Gasterosteus aculeatus) have undergone a recent dramatic evolutionary radiation, generating numerous examples of marine–freshwater species-pairs, and a small number of benthic–limnetic species-pairs found within single lakes [1]. We have developed a new genome-wide SNP genotyping array to study patterns of genetic variation in sticklebacks over a wide geographic range, and to scan the genome for regions that contribute to repeated evolution of marine–freshwater or benthic–limnetic species-pairs. Surveying 34 global populations with 1159 informative markers revealed substantial genetic variation, with predominant patterns reflecting demographic history and geographic structure. After correcting for geographic structure and filtering for neutral markers, we detect large repeated shifts in allele frequency at some loci, identifying both known and novel loci likely contributing to marine–freshwater and benthic–limnetic divergence. Several novel loci fall close to genes implicated in epithelial barrier or immune functions, which have likely changed as sticklebacks adapt to contrasting environments. Specific alleles differentiating sympatric benthic–limnetic species-pairs are shared in nearby solitary populations, suggesting an allopatric origin for adaptive variants, and selection pressures unrelated to sympatry in the initial formation of these classic vertebrate species-pairs.
Many traits in plants and animals evolve repeatedly in response to similar environmental conditions, suggesting they are adaptive and shaped by natural selection [2]. Marine threespine sticklebacks colonized and adapted to a large number of new freshwater environments at the end of the last ice age, providing an outstanding opportunity for identifying the genetic and genomic basis of repeated evolutionary change in natural populations. A variety of molecular tools have recently been developed for sticklebacks, including microsatellite based linkage maps, physical maps, restriction-site associated DNA polymorphisms, and a whole genome sequence assembly [3–5]. To facilitate the further genetic and genomic characterization of stickleback diversity, we designed a genotyping array with 3,072 well-spaced single nucleotide polymorphisms identified from expressed sequence tags and genomic sequences from different populations (Supplementary Methods). We used this platform to study patterns of variation found in 196 individuals from a world-wide collection of 34 different stickleback populations, bracketing both the large geographic range and extreme morphological diversity of the species complex ([1, 6, 7] Figure 1A; Table S1). A total of 1,159 SNPs gave good genotyping signals and were variable among the populations surveyed (Figure S1A&B, Table S2). Based on the overall level of polymorphism observed within and among global populations (Figure 1A&B, Figure S1B), the arrays should provide a useful platform both for mapping phenotypic traits in laboratory crosses ([8, 9]; Supplementary Methods), and for studying relationships between populations using a genome wide set of markers.
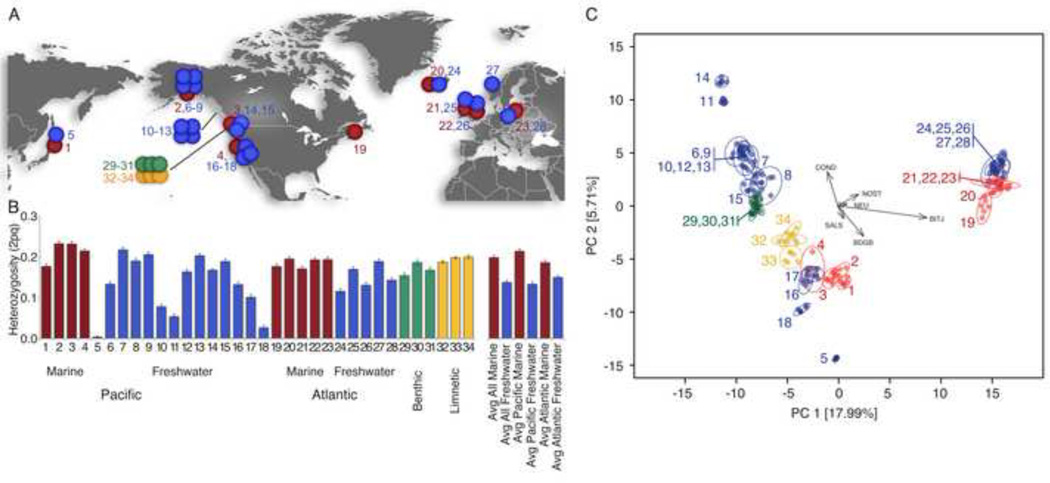
Figure 1. Levels of heterozygosity and patterns of genetic variation among global stickleback populations.
(A) Marine (red) and freshwater (blue) sticklebacks were sampled across the northern hemisphere. Benthic (green) and limnetic (yellow) species-pairs are restricted to a few lakes in British Columbia. See Table S1 for population information. (B) Mean heterozygosity levels (2pq +/− SEM) within freshwater populations are significantly lower than marine populations. (C) Principal component analysis of global populations reveals the first major axis of variation separating Pacific (left) from Atlantic populations (right cluster), and a second axis separating Northern Pacific freshwater populations from Southern Pacific freshwater and Pacific marine populations. SNP ascertainment vectors are shown as grey arrows (see Experimental Procedures). See also Figure S1, and Tables S1 & S2.
Patterns of variation within and among global stickleback populations
Our survey of 34 populations shows that mean heterozygosity is significantly higher in marine than in freshwater populations (0.200 vs. 0.139, p=0.017), with some freshwater populations showing very low diversity (Figure 1B). Lower heterozygosity in freshwater may reflect smaller effective population sizes and varying demographic histories involving bottlenecks during the recent colonization of freshwater habitats (see also [10–12]), but was not significantly associated with lake size, elevation, or distance from the sea in the current study (p>0.05 all tests).
High phenotypic diversity among sticklebacks is mirrored by a relatively high degree of genetic differentiation among populations (mean pairwise FST=0.193; range 0.031–0.383, Figure S1C). Principal Component Analysis (PCA) revealed a geographic split between Atlantic and Pacific basins as a major axis of variation in genome-wide SNPs (18 percent variation explained, PVE, Figure 1C). This is consistent with a deep split between Atlantic and Pacific lineages observed with a smaller set of nuclear markers [13]. PC2 describes variation among Pacific populations and divides northern freshwater from marine and southern freshwater populations (Figure 1C).
Both neutral and selective processes can lead to phenotypic and genomic differentiation. To identify signatures of adaptive genomic differences, we looked for loci consistently differentiated in stickleback species-pairs that have evolved repeatedly in multiple locations, including marine–freshwater pairs found in many different marine and lake/stream environments; and benthic–limnetic pairs found in a small number of isolated lakes in British Columbia.
Genome scans in marine–freshwater species-pairs
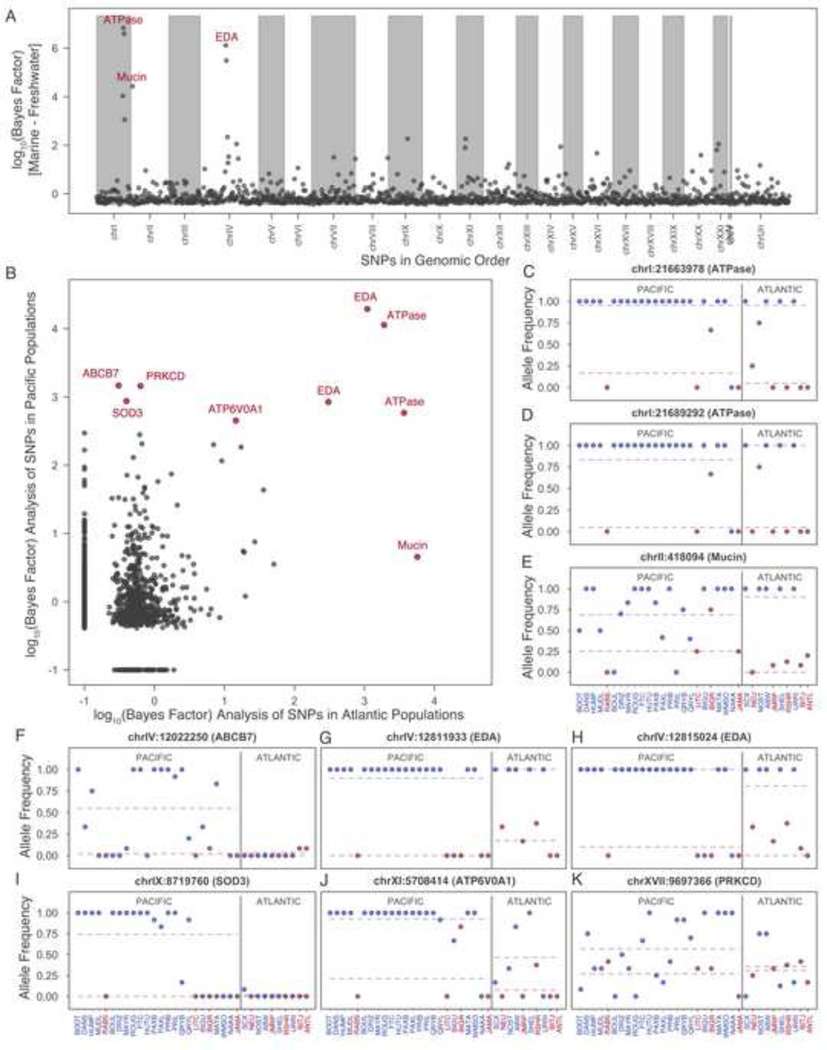
Marine and freshwater sticklebacks represent genetically, morphologically, and behaviorally distinct ecotypes that have evolved repeatedly throughout the Northern hemisphere [7, 14]. To identify potential adaptive loci that underlie repeated ecological differentiation, we searched for SNPs that consistently distinguish marine and freshwater ecotypes, using the method of Coop et al. [15] to correct for overall structure between populations (Figure 2, Table S3). Five of the top six scoring SNPs fell near or within genomic regions previously associated with marine and freshwater ecotypes [4, 13, 16, 17], including two SNPs (chrIV:12,811,933 and chrIV:12,815,024, candidate class) near the ectodysplasin (EDA) region underlying variation in armor plate phenotypes, and three SNPs (chrI:21,663,978, chrI:21,689,292, and 21,487034 candidate class) near the Na/K ATPase (ATP1a1) gene for salinity tolerance [18] that shows strong differentiation along a marine–freshwater salinity gradient in Scotland [16]. Although these genes have already been linked to morphological or physiological differences between particular marine and freshwater populations, allele frequencies have not previously been characterized in multiple individuals from a global set of populations. The repeated differentiation of these regions in many marine and freshwater sticklebacks provides strong additional support for the adaptive significance of these loci even after correcting for potential non-independence of populations–a confounding factor not explicitly adjusted for in previous studies [4, 13].
Figure 2. Bayesian scan for genomic regions consistently differentiated between marine and freshwater environments.
(A) Bayes factor scores for all markers in the genome, with the top scoring regions (ATPase, EDA, and Mucin gene cluster regions on chromosomes I, II, and IV, respectively) indicated in red. (All SNPs with log10 Bayes factors >=1.5 are listed in Table S3) (B) Bayes factors scores found when Pacific and Atlantic Ocean sticklebacks are analyzed as separate regional groups. SNPs with Bayes factors >2.5 in either population are labeled with corresponding gene region in red. (C–K) Allele frequencies in each of the 34 global populations for top ranking SNPs highlighted in panels A and B (freshwater populations: blue points; marine populations: red points; mean freshwater and marine allele frequency: blue and red dashed lines, respectively; Pacific and Atlantic populations grouped on the left and right side of the vertical solid grey line).
Our analysis also identified a new marker associated with marine–freshwater differentiation on chromosome 2. This outlier SNP (chrII:418,094, candidate class) is located near multiple, potentially duplicated, genes belonging to the mucin gene family, Table S3, [19, 20]). Mucus secretions in fish play important roles in osmoregulation, locomotion, and protection against pathogens [21]. Genetic differences in epithelial barrier functions seem likely in marine and freshwater fish, and we propose that variation in the mucin cluster contributes to repeated transformation of marine sticklebacks to freshwater forms living in low ionic-strength environments.
To determine if we detect similar or different adaptive loci in Pacific and Atlantic sticklebacks, we also performed analyses separately in fish from each ocean basin. SNPs in the EDA and ATPase region showed high Bayes factor scores (BF >3.0) in sticklebacks from both basins (Figure 2B). In Atlantic sticklebacks the mucin gene region SNP scored even higher than EDA and ATPase loci, but was only weakly differentiated in Pacific sticklebacks. In contrast, four SNPs located on chromosomes IV, IX, XI and XVII had BF >2.5 in Pacific, but not Atlantic populations. These SNPs are located near genes that influence iron metabolism in humans and fish (chrIV:12,022,250, general class, Figure 2F, ABCB7 gene [22, 23]), alter blood pressure and protect the heart, vasculature, and lungs from oxidative stress (chrIX:8,719,760, candidate class, Figure 2I, SOD3 gene [24–26]), control ion gradients involved in sperm motility, resorption of bone, and digestion of microbes by phagocytes (chrXI:5,708,414, candidate class, Figure 2J, ATP6V0A1 gene [27, 28]), and influence immune functions, pathogen clearing, and expression levels of a cell surface mucin gene required for epithelial barrier function and protection against infections (chrXVII:9,697,366, general class, Figure 2K, PRKCD gene [29–33]). Given the low density of the genotyping array, the actual targets of selection might be other linked genes (see Table S3). However we find it interesting that SNPs linked to mucin functions were recovered in both basins, highlighting the likely importance of epithelial barrier changes during recurrent evolution of marine and freshwater sticklebacks.
Recurrent genetic differentiation suggests that marine–freshwater stickleback adaptation proceeds, in part, by large shifts in allele frequency at some loci shared across multiple populations (Figure 2). Many additional adaptive variants may exist that are specific to individual populations, and these would not be detected by the current method. Given the average spacing between markers in our study (~one marker per 400kb), we may have missed other strongly selected loci in the genome, even if they are repeatedly used in different populations. Previous studies show that linkage disequilibrium extends approximately 20–40 kb around major genes controlling armor and pelvic traits in natural populations [11, 13]. Selective sweeps that are tightly linked to the genotyped SNPs, and sweep events that are young, strong, or fall within regions of suppressed recombination are therefore the most likely to be detected in genome scans for adaptive loci using this SNP genotyping platform. Our results do not preclude the possibility that marine–freshwater adaptation might also involve other loci with smaller shifts in allele frequency (we note that a number of other markers show high or moderately high Bayes factors; Figure 2B, Table S3). Nevertheless, these findings highlight how repeated evolution can be used to filter genetic drift from the signatures of adaptive loci, and show the potential of this approach for future studies using even higher densities of markers or whole genome sequencing.
Genome scans in benthic–limnetic species-pairs
A second example of species-pair in sticklebacks is much less common than marine–freshwater pairs, but provides one of the best-studied examples of ecological speciation in vertebrates. In a few lakes in British Columbia, sticklebacks occur as distinct open-water (limnetic) and bottom-dwelling (benthic) forms [14, 34–36]. These sympatric forms are distinguishable by heritable differences in size, shape, feeding morphology, body armor, mate preference, and behavior, which confer fitness advantages when tested in the corresponding open water and near shore environments [37, 38]. Although fish with hybrid morphology can be found at low frequencies in the species-pair lakes, hybrids appear less fit than parental forms in both environments [37, 39]. Patterns of variation in mitochondrial and a small number of autosomal microsatellite markers suggest that these species-pairs have evolved multiple times in different lakes, rather than originating once and spreading to multiple lakes (although the data are also inconsistent with benthic-limnetic monophyly within each lake [12, 40, 41]). We reinvestigated this classic example of recurrent ecological speciation on a genome-wide scale, scanning for loci that remain differentiated between benthic and limnetic forms, despite ongoing homogenization of neutral regions of the genome due to hybridization in sympatry [41].
We used FST outlier tests [42, 43] to identify loci with higher or lower levels of differentiation between limnetic and benthic pairs than expected under a neutral model. Separate tests were performed on each of three lakes and identified 46 SNPs as outliers linked to putatively selected regions (FDR 0.05, Table S4). These comprise 2.5, 4.3, and 1.9% of polymorphic SNPs surveyed in Paxton, Priest and Little Quarry (hereafter, “Quarry”) lakes respectively, indicating that multiple loci (but a small proportion overall) are involved in benthic–limnetic species-pair divergence via large shifts in allele frequency. Fifteen SNPs were outliers in two or more lakes (12 general class, 3 candidate class), four of which were outliers in all three lakes (3 general class, 1 candidate class) (Figure 3 and Table S4). For comparison, only 1.4 and 0.01 SNPs would be expected to be parallel selected by chance in two and three lakes respectively. As parallel outliers across lakes, these SNPs tag regions that are common and therefore likely important to the repeated evolution of benthic–limnetic species-pairs (see Table S5 for a summary of nearby genes).
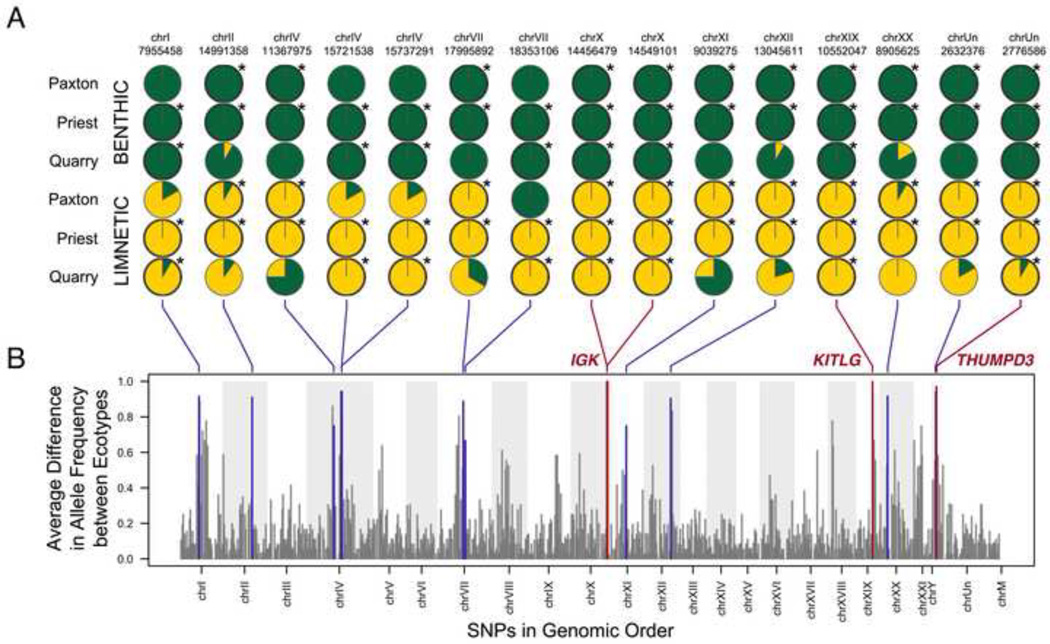
Figure 3. FST outlier analyses in benthic–limnetic populations.
(A) Allele frequency of FST outlier SNPs are shown in pie charts for three species-pair lakes. Asterisks and dark outlines around pies indicate lakes in which SNP was found to be an FST outlier. Four SNPs appeared to be under selection in all three lakes (red lines, corresponding gene regions indicated) and a further 11 SNPs in two of three lakes (blue lines). (B) Genome graph of allele frequency differences between pooled ecotypes indicating chromosomal position of FST outliers. See Table S4 and Figure S2 for more details.
One outlier SNP (chrXIX:10,552,047, candidate class) is linked to KITLG, a gene previously shown to control lighter gill and skin pigmentation in Paxton Benthic fish [44]. We find that KITLG is strongly differentiated in all three species-pair lakes, suggesting that genetic variation at this locus controls an adaptive trait repeatedly selected during formation of all benthic–limnetic species-pairs (perhaps color matching to different light and dark water backgrounds typical of near shore and open water environments [45]).
Two other outlier SNPs (chrX:14,456,479 and chrX:14,549,101; general class) showing benthic-limnetic differences in all three lakes fall within a region that contains genes with roles in immune function and vision and flanked by large repeated clusters of >24 variable and constant domains of immunoglobulin light chain genes (IGK; Figure S2). Benthic and limnetic ecotypes are known to differ in parasite loads [46]. Differential use of microhabitats and food sources likely leads to differential exposure to a variety of pathogens, and strong selection on host defenses. Our results suggest that differences in immunity play a significant role in the evolution of benthic–limnetic species-pairs, and identify a specific genomic region likely contributing to immunological differences between species.
Evolution of benthic–limnetic species-pairs
Factors influencing the origin and restricted distribution of benthic–limnetic species-pairs in threespine sticklebacks are not well understood. Previous studies have hypothesized that the limited distribution of benthic–limnetic species-pairs could be due to rare ‘double-invasion’ events fluctuations in sea level in British Columbia [12, 47]. The genome-wide data showed two features consistent with separate invasion events: lower average heterozygosity within the postulated older (benthic) members of the species-pairs (Figure 1B; mean benthic and limnetic heterozygosity: 0.196 vs. 0.170; p=0.030), and closer clustering of the postulated younger (limnetic) members of the species-pairs with marine fish in the global PCA (Figure 1C). However, these differences could also arise from differences in effective population size of benthic and limnetic ecotypes, and recent geological data [48] suggest that the magnitude of local sea-level changes may not have been sufficient to mediate double invasions.
To further investigate species-pair relationships we performed a PCA restricted to benthic and limnetic populations (Figure 4A). The first PC (15.15% PVE) clearly separated benthic and limnetic fish by different lakes of origin, consistent with the independent evolution of pairs in each of the three lakes. However, PC2, accounting for nearly as much variation (13.41%), separated all three benthic from limnetic types, suggesting similar genetic variants underlying benthic and limnetic differences in all three lakes.
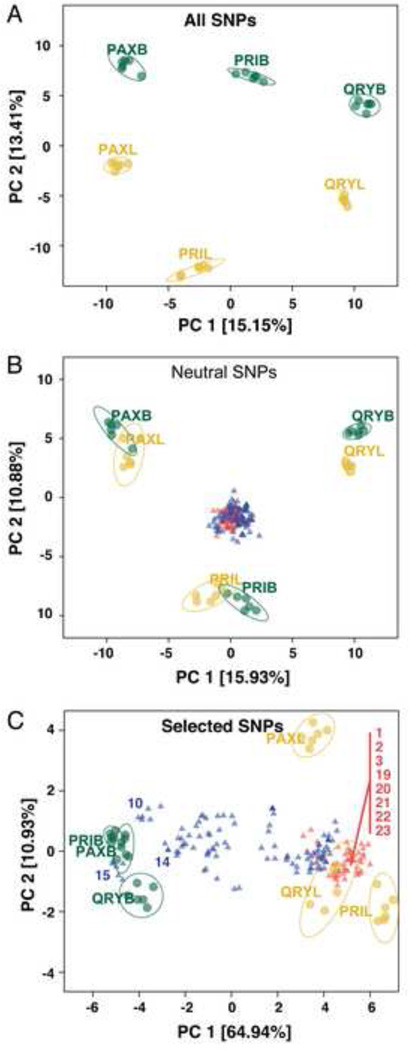
Figure 4.
Principal component analyses of benthic (green) and limnetic (yellow) individuals using (A) all 925 polymorphic SNPs, (B) 879 putatively neutral SNPs or (C) 46 putatively selected SNPs. Ellipses represent 95% onfidence intervals for each ecotype within a lake. Global populations projected onto benthic–limnetic variation space in B and C are shown as red (marine) and blue (freshwater) triangles, with the benthic-like or limnetic-like populations labeled with a number code. See Table S1 for population information, Table S4 for information on SNPs linked to adaptive loci in each of the three lakes, and Figure S3 for the global distribution of alleles at SNPs linked to putatively adaptive loci.
When adaptive divergence occurs in the face of gene flow, the evolutionary history inferred from patterns of neutral variation can be very different from the evolutionary history inferred from variation at adaptive loci [13, 49]. We therefore divided SNPs into a set of putatively selected (46 adaptive loci identified in any of the three different lake genome-scans described above) and neutral markers (879 polymorphic SNPs among benthic and limnetic individuals from the three lakes). PCA of putatively neutral SNPs revealed clustering by lake rather than ecotype (Figure 4B). In contrast, PC1 of ‘selected’ SNPs separates benthic and limnetic fish into ecotypic clusters (65% PVE, Figure 4C) highlighting the importance of a shared origin to adaptive genetic variation in these species-pairs. Interestingly, when global populations were projected onto adaptive benthic–limnetic PC space, we observed a number of freshwater and marine populations that clearly overlap with the divergent benthic (e.g., Pop15/FTC) and limnetic (e.g., Pop3/LITC) clusters respectively (Figure 4C, see Table S1 for population details). In contrast, none of the global populations showed affinity with the clusters of genetic variation observed in PCA of neutral markers (Figure 4B).
Clustering of benthic and limnetic fish by ecotype suggests that some of the adaptive genetic variation in each lake arises from shared adaptive variants, rather than novel mutations that have occurred separately in each lake. The allopatric single-ecotype populations that cluster with benthic and limnetic fish identify new examples of solitary populations that also carry similar allele assemblages. Interestingly, these solitary populations are located in geographic proximity to the lakes with species-pairs. Such populations could represent allopatric source populations from which adaptive benthic and limnetic alleles originally arose and spread, or single-species populations in which ecological selection has also occurred for benthic-like or limnetic-like phenotypes, resulting in similar fixation of shared genetic variants that contribute to particular traits. Regardless, our results indicate that selection pressures unrelated to sympatry can also generate a substantial part of characteristic benthic or limnetic allele assemblies, and suggest a more important role of allopatric adaptive divergence and reuse of standing genetic variation than previously recognized for the repeated evolution of the classic benthic–limnetic species pairs.
Summary
Studies in many organisms have demonstrated the benefits of being able to type multiple individuals at many markers in a fast and affordable way. The development of an informative genome-wide SNP array for sticklebacks will facilitate additional genetic mapping studies in this emerging model organism, and further studies of natural population diversity. Our genome scans for adaptive loci using the genome-wide array show that repeated formation of species-pairs occurs in part by recurrent strong shifts in allele frequency at particular loci. These data confirm that strong differentiation has repeatedly occurred at some previously known loci (EDA, ATP1a1, KITLG). In addition, we identify many new regions that may contribute to repeated differentiation of either marine–freshwater ecotypes (e.g., markers linked to ABCB7, ATP6V0A1, mucin gene cluster) or benthic–limnetic ecotypes (e.g., markers near the immunoglobulin light chain gene cluster (IGK) that are differentiated in all three species-pair lakes), which can now be further investigated to better understand the basis of selection and ecological adaptation. Finally our studies illustrate how genotyping of a large number of loci in many different populations can identify novel relationships among the gene assemblages that have evolved in different geographic locations. Many of the outlier loci identified in benthic–limnetic genome scans are not only shared in two or more lakes, but also found in similar assemblages in nearby allopatric solitary populations. The markers identified in these studies provides a new foundation for studying the molecular basis of specific benthic–limnetic phenotypes, as well as the larger historic and geographic factors that have led to this classic example of recurrent ecological speciation in vertebrates.
EXPERIMENTAL PROCEDURES
SNPs were ascertained primarily from large EST collections of marine and freshwater populations. Potential SNPs were genotyped on 196 individuals using an Illumina GoldenGate genotyping array platform. Analyses of allele frequency, heterozygosity and FST [50] were performed using Perl scripts. Principal components analysis was performed in R (v2.11.1). Genome scans for SNPs with allelic correlation to marine-freshwater environments were performed using BayEnv [15] which allowed for correction for the potential non-independence among populations. Genome scans for FST outliers among benthic-limnetic species-pairs was performed using BayeScan 2.01 [42, 43]. See Supplementary Experimental Procedures for more details
Supplementary Material
Acknowledgments
We thank Jenny Gow, Rick Taylor, Mike Bell, Jeff McKinnon, Seiichi Mori, and Bjarni Jónsson for samples. This work was supported by grant P50 HG002568 from the US NIH (JS, JG, DA, RM and DMK), an Affymetrix Bio-X Graduate Fellowship to YFC and a NSERC operating grant to TER (NRC2354). DS is a Canada Research Chair supported by grants from NSERC and CFI. DMK is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
SNPs reported herein have been deposited in NCBI dbSNP database with accession numbers specified in Table S2.
SUPPLEMENTARY INFORMATION
Detailed experimental procedures, analysis methodologies, five supplementary tables and three supplementary figures are available with this paper online at http://xxx.
References
- 1.Bell MA, Foster SA. In: The Evolutionary Biology of the Threespine Stickleback. Bell MA, Foster SA, editors. New York: Oxford University Press; 1994. [Google Scholar]
- 2.Wake DB, Wake MH, Specht CD. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science. 2011;331:1032–1035. doi: 10.1126/science.1188545. [DOI] [PubMed] [Google Scholar]
- 3.Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-spine Stickleback. Boca Raton, FL: CRC Press; 2007. pp. 41–81. [Google Scholar]
- 4.Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000862. e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broad Institute. Stickleback Draft Genome Assembly: gasAcu1.0. 2007 Available at: http://www.broadinstitute.org/models/stickleback.
- 6.Reimchen TE, Stinson EM, Nelson JS. Multivariate differentiation of parapatric and allopatric populations of threespine stickleback in the Sangan River watershed, Queen Charlotte Islands. Can. J. Zool. 1985;63:2944–2951. [Google Scholar]
- 7.McKinnon JS, Rundle HD. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 2002;17:480–488. [Google Scholar]
- 8.Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenwood A, Jones F, Chan Y, Brady S, Absher D, Grimwood J, Schmutz J, Myers R, Kingsley D, Peichel C. The genetic basis of divergent pigment patterns in juvenile threespine sticklebacks. Heredity. 2011;1:1–12. doi: 10.1038/hdy.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mäkinen H, Cano J, Merila J. Genetic relationships among marine and freshwater populations of the European three-spined stickleback (Gasterosteus aculeatus) revealed by microsatellites. Mol. Ecol. 2006;15:1519–1534. doi: 10.1111/j.1365-294X.2006.02871.x. [DOI] [PubMed] [Google Scholar]
- 11.Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor EB, McPhail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proc. R. Soc. Lond. B, Biol. Sci. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 14.McPhail JD. Speciation and the evolution of reproductive isolation in the sticklebacks (Gasterosteus) of south-western British Columbia. In: Bell MA, Foster SA, editors. The Evolutionary Biology of the Threespine Stickleback. New York: Oxford University Press; 1994. pp. 399–437. [Google Scholar]
- 15.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones F, Brown C, Pemberton J, Braithwaite V. Reproductive isolation in a threespine stickleback hybrid zone. J. Evol. Biol. 2006;19:1531–1544. doi: 10.1111/j.1420-9101.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- 17.McCairns R, Bernatchez L. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution. 2010;64:1029–1047. doi: 10.1111/j.1558-5646.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 18.Herrera VL, Xie HX, Lopez LV, Schork NJ, Ruiz-Opazo N. The alpha1 Na,K-ATPase gene is a susceptibility hypertension gene in the Dahl salt-sensitive HSD rat. J Clin Invest. 1998;102:1102–1111. doi: 10.1172/JCI3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devine PL, McKenzie IFC. Mucins: structure, function, and associations with malignancy. Bioessays. 1992;14:619–625. doi: 10.1002/bies.950140909. [DOI] [PubMed] [Google Scholar]
- 20.Rose C. Mucins: structure, and role in pulmonary diseases. Am. J. Physiol. Lung Cell. Mol. Physiol. 1992;263:L413–L429. doi: 10.1152/ajplung.1992.263.4.L413. [DOI] [PubMed] [Google Scholar]
- 21.Shephard KL. Functions for fish mucus. Reviews in fish biology and fisheries. 1994;4:401–429. [Google Scholar]
- 22.Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum. Mol. Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- 23.Miyake A, Higashijima S, Kobayashi D, Narita T, Jindo T, Setiamarga DHE, Ohisa S, Orihara N, Hibiya K, Konno S, et al. Mutation in the abcb7 gene causes abnormal iron and fatty acid metabolism in developing medaka fish. Dev. Growth Differ. 2008;50:703–716. doi: 10.1111/j.1440-169X.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- 24.Lu z, Xu X, Hu X, Zhu G, Zhang P, Deel EDV, Joel P, Fassett JT, Oury TD, Bache RJ, et al. Extracellular superoxide dismutase deficiency exacerbates pressure overload-induced left ventricular hypertrophy and dysfunction. Erasmus. 2008;51:19–25. doi: 10.1161/HYPERTENSIONAHA.107.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gongora MC, Lob HE, Landmesser U, Guzik TJ, Martin WD, Ozumi K, Wall SM, Wilson DS, Murthy N, Gravanis M, et al. Loss of extracellular superoxide dismutase leads to acute lung damage in the presence of ambient air: a potential mechanism underlying adult respiratory distress syndrome. Am. J. Pathol. 2008;173:915–926. doi: 10.2353/ajpath.2008.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Guo H, Xu X, Lu Z, Fassett J, Hu X, Xu Y, Tang Q, Hu D, Somani A, et al. Exacerbated pulmonary arterial hypertension and right ventricular hypertrophy in animals with loss of function of extracellular superoxide dismutase. Hypertension. 2011;58:303–309. doi: 10.1161/HYPERTENSIONAHA.110.166819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor-Soler N, Pietrement C, Breton S. Role of acid/base transporters in the male reproductive tract and potential consequences of their malfunction. Physiology. 2005;20:417–428. doi: 10.1152/physiol.00036.2005. [DOI] [PubMed] [Google Scholar]
- 28.Peri F, Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, et al. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 30.Ren J, Li Y, Kufe D. Protein kinase C delta regulates function of the DF3/MUC1 carcinoma antigen in beta-catenin signaling. J. Biol. Chem. 2002;277:17616–17622. doi: 10.1074/jbc.M200436200. [DOI] [PubMed] [Google Scholar]
- 31.Park J-A, Crews AL, Lampe WR, Fang S, Park J, Adler KB. Protein kinase C delta regulates airway mucin secretion via phosphorylation of MARCKS protein. Am. J. Pathol. 2007;171:1822–1830. doi: 10.2353/ajpath.2007.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwegmann A, Guler R, Cutler AJ, Arendse B, Horsnell WGC, Flemming A, Kottmann AH, Ryan G, Hide W, Leitges M, et al. Protein kinase C delta is essential for optimal macrophage-mediated phagosomal containment of Listeria monocytogenes. Proc. Natl. Acad. SciU.SA. 2007;104:16251–16256. doi: 10.1073/pnas.0703496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117:2313–2324. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): evidence for a species-pair in Paxton Lake, Texada Island, British Columbia. Can. J. Zool. 1992;70:361–369. [Google Scholar]
- 35.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): Morphological and genetic evidence for a species pair in Enos Lake, British Columbia. Can. J. Zool. 1984;62:1402–1408. [Google Scholar]
- 36.Gow JL, Rogers SM, Jackson M, Schluter D. Ecological predictions lead to the discovery of a benthic–limnetic sympatric species pair of threespine stickleback in Little Quarry Lake, British Columbia. Can. J. Zool. 2008;86:564–571. [Google Scholar]
- 37.Schluter D. Adaptive radiation in sticklebacks: trade-offs in feeding performance and growth. Ecology. 1995;76:82–90. [Google Scholar]
- 38.Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution. 1999;53:866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- 39.Gow JL, Peichel CL, Taylor EB. Ecological selection against hybrids in natural populations of sympatric threespine sticklebacks. J. Evol. Biol. 2007;20:2173–2180. doi: 10.1111/j.1420-9101.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- 40.Taylor EB, McPhail J. Evolutionary history of an adaptive radiation in species pairs of threespine sticklebacks (Gasterosteus): insights from mitochondrial DNA. Biol. J. Linn. Soc. Lond. 1999;66:271–291. [Google Scholar]
- 41.Gow JL, Peichel CL, Taylor EB. Contrasting hybridization rates between sympatric three-spined sticklebacks highlight the fragility of reproductive barriers between evolutionarily young species. Mol. Ecol. 2006;15:739–752. doi: 10.1111/j.1365-294X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- 42.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer MC, Foll M, Excoffier L, Heckel G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis) Mol. Ecol. 2011;20:1450–1462. doi: 10.1111/j.1365-294X.2011.05015.x. [DOI] [PubMed] [Google Scholar]
- 44.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clarke JM, Schluter D. Colour plasticity and background matching in a threespine stickleback species pair. Biol. J. Linn. Soc. Lond. 2011;102:902–914. [Google Scholar]
- 46.MacColl ADC. Parasite burdens differ between sympatric three-spined stickleback species. Ecography. 2009;32:153–160. [Google Scholar]
- 47.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): origin of the species pairs. Can. J. Zool. 1993;71:515–523. [Google Scholar]
- 48.Hutchinson I, James T, Clague J, Barrie JV, Conway K. Reconstruction of late Quaternary sea-level change in southwestern British Columbia from sediments in isolation basins. Boreas. 2004;33:183–194. [Google Scholar]
- 49.Via S. Natural selection in action during speciation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;589:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.