Abstract
The Eph receptors are a large family of receptor tyrosine kinases. Their kinase activity and downstream signaling ability are stimulated by the binding of cell surface-associated ligands, the ephrins. The ensuing signals are bidirectional because the ephrins can also transduce signals (known as reverse signals) following their interaction with Eph receptors. The ephrin-binding pocket in the extracellular N-terminal domain of the Eph receptors and the ATP-binding pocket in the intracellular kinase domain represent potential binding sites for peptides and small molecules. Indeed, a number of peptides and chemical compounds that target Eph receptors and inhibit ephrin binding or kinase activity have been identified. These molecules show promise as probes to study Eph receptor/ephrin biology, as lead compounds for drug development, and as targeting agents to deliver drugs or imaging agents to tumors. Current challenges are to find (1) small molecules that inhibit Eph receptor-ephrin interactions with high binding affinity and good lead-like properties and (2) selective kinase inhibitors that preferentially target the Eph receptor family or subsets of Eph receptors. Strategies that could also be explored include targeting additional Eph receptor interfaces and the ephrin ligands.
Keywords: ephrin, tyrosine kinase, cancer, angiogenesis, nerve regeneration, protein-protein interface, kinase inhibitor
1. Introduction
EphA and EphB receptors together with ephrin-A and ephrin-B ligands play a wide variety of physiological and pathological roles in many tissues ([1,2] and articles in this issue). Therefore, molecules that modulate their activities can serve as probes to study the complex biology of the Eph/ephrin system and potentially as therapeutic agents to treat a variety of diseases. They could be useful, for example, for inhibiting cancer progression and pathological forms of angiogenesis [2–4] or promoting nerve regeneration in the injured nervous system [1,5]. The roles of the Eph/ephrin system in stem cell proliferation and differentiation could also be exploited in regenerative medicine [6]. Furthermore, interfering with the EphA/ephrin-A system in the nervous system may also be useful to treat diseases where excessive extracellular levels of the neurotransmitter glutamate cause hyperexcitability or toxicity [7], while enhancing EphB2 function may help treat Alzheimer’s disease [8]. Other applications of Eph/ephrin-targeting molecules could involve modulation of bone remodeling and glucose homeostasis [1], and reduction of heart damage following myocardial infaction [9]. Finally, EphA2 and ephrin-B ligands could be targets for anti-viral therapies [1,10]. Targeted delivery of imaging agents or chemotherapeutic drugs and toxins to tumors expressing high levels of certain Eph/ephrin family members also offers medical promise for diagnosis or therapy.
Several types of molecules that target Eph receptors and ephrins have been identified. Recombinant monomeric Eph receptor extracellular domains and chimeric proteins containing the extracellular domain of an Eph receptor or ephrin fused to the Fc portion of an antibody have also been used to functionally modulate the Eph/ephrin system [2]. However, these molecules lack selectivity because of the promiscuity in Eph receptor-ephrin interactions [11]. More selective targeting agents include antibodies that recognize Eph receptor or ephrin extracellular epitopes and peptides that bind to Eph receptors. Small molecules that bind to the Eph receptor extracellular or kinase domain have also been identified. Whereas antibodies can bind with high affinity to a variety of sites, binding of peptides and small molecules typically requires the presence of suitable cavities, such as the ephrin-binding pocket or the ATP-binding pocket of the Eph receptors (Fig. 1). Such cavities are not evident in the ephrins, which therefore may be less suitable for targeting with this type of molecule.
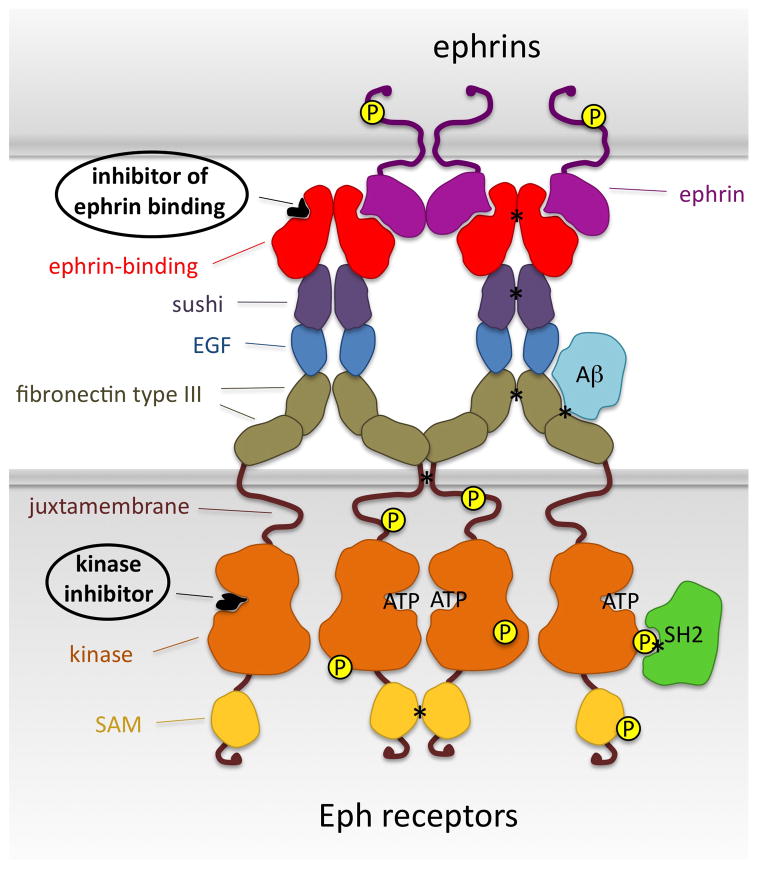
Fig. 1.
Current and potential strategies to target Eph receptors with peptides and small molecules. Molecules that target the ephrin-binding pocket inhibit ephrin binding, while molecules that target the ATP-binding site inhibit kinase activity. Other interfaces that could be targeted are those between the ephrin-binding domains, sushi domains, transmembrane segments and sterile alpha motifs (SAM) of two neighboring Eph receptor molecules (asterisks). Furthermore, inhibiting the binding of amyloid-β (A-β) or cytoplasmic signaling proteins, such as those containing SH2 domains, could more selectively affect only some Eph receptor activities.
This review focuses on peptides and small molecules that bind to Eph receptors and inhibit either ephrin ligand binding or kinase activity. Other types of Eph/ephrin-targeting molecules have been reviewed elsewhere [2,3,12].
2. Peptides that bind to Eph receptors and inhibit ephrin binding
A stretch of 15 consecutive amino acids within the ephrin sequence (known as the G-H loop) mediates high-affinity binding to Eph receptors by interacting with a deep pocket in their N-terminal ephrin-binding domain (Fig. 1). Although the isolated ephrin G-H loop binds poorly to Eph receptors [13], phage display screens have identified a number of 12 amino acid-long peptides that bind to the Eph receptor extracellular domain with low micromolar affinity [13–15]. The most potent peptides block ephrin binding, suggesting that they target the ephrin-binding pocket of the Eph receptors. This has been confirmed for two peptides that were crystallized in complex with EphB2 or EphB4. Although each ephrin binds to most or all Eph receptors of the same A or B class [11], some of the peptides show remarkable selectivity and bind only to one Eph receptor.
2.1. EphA2
Two related peptides identified by phage display selectively bind to EphA2: YSA (YSAYPDSVPMMS) and SWL (SWLAYPGAVSYR; identical amino acids in the two peptides are underlined) [13]. Amino acid replacements and truncations in the YSA peptide revealed that five of the six N-terminal amino acids are essential for high affinity binding to EphA2, while the last three amino acids are the least critical [16]. Interestingly, a shorter version of YSA comprising only the first 5 amino acids is still able to bind selectively to EphA2, albeit very weakly.
YSA and SWL both inhibit ephrin binding with an IC50 value of ~1 μM in ELISA assays, and computer modeling suggests that YSA binds in the ephrin-binding pocket of EphA2 [13,16]. In cells, both YSA and SWL induce EphA2 tyrosine phosphorylation (activation), leading to receptor endocytosis and downstream signaling. Similar to the ephrin-A1 ligand, the two peptides can also cause inhibition of two major oncogenic signaling cascades, the PI3 kinase-Akt and Ras-MAP kinase pathways, and cell retraction [13,16]. It is surprising that the two monomeric peptides act as agonists, rather than antagonists, given that Eph receptor activation is thought to involve clustering [11].
EphA2 is highly expressed in many types of cancers and in tumor blood vessels, while it is not expressed in quiescent vasculature and is present at low levels in most adult tissues [3,12,17]. Hence, the YSA and SWL peptides could be useful for targeted delivery of anticancer drugs. YSA–functionalized nanogels have indeed been used to deliver siRNA that downregulates the epidermal growth factor (EGF) receptor into EphA2-expressing ovarian cancer cells, causing their sensitization to chemotherapy [18,19]. Moreover, adenoviruses engineered to express YSA in their viral capsid have improved ability to transduce EphA2-expressing pancreatic cancer cell lines and resection specimens [20]. The YSA peptide conjugated to magnetic nanoparticles has also been used to capture ovarian cancer cells expressing EphA2 from the mouse peritoneal cavity and human ascites fluids [21,22]. A similar approach might be useful to capture circulating tumor cells for diagnostic purposes.
2.2. EphA4
Three peptides identified by phage display preferentially bind to EphA4: KYL (KYLPYWPVLSSL), APY (APYCVYRGSWSC) and VTM (VTMEAINLAFPG) [15]. KYL and APY have related sequences (identical amino acids are underlined), although the conformation of APY is likely constrained by a disulfide bond between its two cysteines. All three peptides compete with each other for binding to EphA4 and inhibit ephrin binding, suggesting that they target the ephrin-binding pocket. However, their interaction with EphA4 remains to be structurally characterized.
KYL is the most potent of the three peptides. It inhibits ephrin-A binding to EphA4 with an IC50 value of ~1 μM in ELISA assays and blocks ephrin-A-induced EphA4 tyrosine phosphorylation in cells and hippocampal slices at ~15 μM. KYL has been used in organotypic cultures to investigate the role of EphA4 in neural crest cell migration and the topographic arborization of hippocampal mossy fibers [15,23]. Furthermore, in cortical neurons and explants from nasal retina, KYL prevents ephrin-induced growth cone collapse and axon retraction [24,25]. These effects, together with evidence that KYL promotes axon sprouting and functional recovery when infused in the injured rat spinal cord [24], suggest that inhibition of EphA4 may enhance axon regrowth after injury. KYL was also shown to increase adhesion of T lymphocytes to endothelial cells [26], implying that targeting EphA4 could also be useful for modulating immune responses.
2.3 EphB2
A number of EphB2-targeting peptides have been identified by phage display [14]. Among them, SNEW (SNEWIQPRLPQH) is the most potent, with an IC50 value for inhibition of ephrin-B2 binding of ~15 μM in ELISA assays and a Kd of 6 μM in isothermal titration calorimetry assays [27]. SNEW completely inhibits ephrin binding to EphB2 in ELISA assays and ephrin-induced EphB2 tyrosine phosphorylation in cells at ~100 μM. The crystal structure of SNEW in complex with EphB2 revealed that the peptide binds in the ephrin-binding pocket of EphB2 [27]. All the amino acids in SNEW (except L9 and H12) contribute to the interaction with the receptor. Furthermore, structure-guided in silico combinatorial mutagenesis identified a Q6 to L amino acid change that results in a 2-fold increased EphB2 binding affinity.
SNEW inhibits the binding of phage clones displaying most of the other EphB2-binding peptides identified, suggesting that these peptides also target the ephrin-binding pocket [14]. While SNEW selectively binds to EphB2, many of the other phage-displayed peptides identified by panning on EphB2 – and EphB1 – bind to both receptors, underlying the close similarity in their ephrin-binding pockets [14]. Peptides inhibiting EphB2-ephrin interaction may be useful to inhibit pathological forms of angiogenesis and the progression of cancers driven by EphB2 activation [1,2].
2.4. EphB4
Many peptides that selectively bind to EphB4 have been identified by phage display [14]. TNYL (TNYLFSPNGPIA) was the most potent among several synthetic peptides examined, with an IC50 value of 50–150 μM for inhibition of EphB4-ephrin-B2 interaction in ELISA assays. However, a modified version that contains at the C terminus the RAW motif found in other EphB4-binding peptides (TNYL-RAW) has dramatically improved potency, with a 10,000 fold decrease in IC50 and a low nanomolar binding affinity [14,28,29]. Consistent with this, the crystal structure of TNYL-RAW in complex with EphB4 revealed that the peptide occupies the ephrin-binding pocket and forms many interactions that stabilize binding [28]. The conformation of TNYL-RAW is governed by turns induced by P7 and the G9P10 motif, which is conserved in many of the other EphB4-binding peptides [14], as well as by the pseudohelix formed by the RAW motif. Surprisingly, the FSPN sequence of TNYL-RAW binds in an opposite N- to C-terminal orientation compared to the same sequence in the ephrin-B2 G-H loop.
T1 and N2 in TNYL-RAW are dispensable for the interaction with EphB4 [28] and can therefore be modified for the attachment of drugs or imaging agents. Indeed, TNYL-RAW has been recently used to image EphB4-positive cancer xenografts in mice. TNYL-RAW was labeled with 64Cu for positron emission tomography (PET) and attached to polymeric micellar nanoparticles containing a fluorescent dye and 111I for dual imaging by near-infrared fluorescence and single photon emission computed tomography (SPECT) [29,30].
Blocking EphB4-ephrin-B2 binding would be expected to inhibit the pro-angiogenic effects of not only EphB4 signaling but also ephrin-B2 reverse signaling [2,31–33]. Indeed, TNYL-RAW lacking the first two amino acids (in combination with the EphB2-inhibitory peptide SNEW) was shown to disrupt the assembly of endothelial cells and pericytes into vascular structures, concomitant with inhibition of ephrin-B2 reverse signaling [34]. However, high TNYL-RAW concentrations (10–100 μM) are needed to inhibit EphB4 phosphorylation in cells [14,34], likely because the presence of R13 makes the peptide particularly susceptible to protease digestion.
2.5 Other Eph receptors
Peptides that bind to the EphA5, EphA7 and EphB1 receptors have also been identified by phage display [14,15]. Of these, the EWLS peptide selectively binds to EphB1 and inhibits ephrin-B2 binding in ELISA assays with an IC50 value of ~10 μM. The EphA5- and EphA7-binding peptides remain to be characterized as isolated peptides. Moreover, a 18-amino acid peptide derived from azurin, a bacterial protein of the cupredoxin family (which is structurally related to the ephrin family), was reported to bind to EphB2, EphA6 and other Eph receptors and to inhibit EphB2 activation by ephrin-B2 in cultured cells [35]. Finally, a 13-amino acid cyclic peptide was computationally designed based on the structure of the ephrin-B2 G-H loop in complex with EphB4 [36]. If further studies demonstrate that this peptide indeed binds to EphB4 with good affinity, this would support the feasibility of structure-guided computational approaches to design novel Eph receptor-targeting peptides. However, the selectivity of ephrin-based peptides may be low, given the promiscuity of Eph receptor-ephrin interactions.
3. Small molecules that bind to Eph receptors and inhibit ephrin binding
The identification of small molecules capable of disrupting protein-protein interfaces is a challenging endeavour [37–39]. Difficulties include the often large size of the protein interacting surfaces, which may lack deep indentations where small molecules could bind with high affinity, and the poor suitability of traditional small molecule libraries used in high-throughput screening. The ephrin-binding pocket of Eph receptors, however, seems to present favorable features for high-affinity binding of small molecules [37]. Consistent with this, a few small molecules that inhibit Eph receptor-ephrin interaction have been recently reported (Fig. 1 and Table 1), although some appear to function through non-classical mechanisms.
Table 1.
Small molecule inhibitors of Eph receptors
| Compounds | Eph receptors tested | Inhibition (biochemical assays) | Inhibition (cell-based assays) | Kinases1 targeted/ tested | Method of identification | Ref |
|---|---|---|---|---|---|---|
|
Inhibitors of Eph receptor-ephrin interaction
| ||||||
| salicylic acid-dimethylpyrrole derivatives | A2 A42 | ~10 μM* | ~100–300 μM* | 0/1 | HTS and SAR analysis | [25] |
| A3 A5 A6 B6 | 30–50% at 50 μM*** | nd | ||||
| A7 B1 B2 B3 B43 | <10% at 50 μM*** | nd | ||||
|
| ||||||
| disalicylic acid-furanyl derivative | A2 A4 | <5 μM* | 50–100 μM* | nd | screen of 46 salicylates | [41] |
| A5 A6 A7 B4 B6 | 15–25 μM* | nd | ||||
| A3 B1 B3 | >100 μM* | nd | ||||
|
| ||||||
| lithocholic acid | A1 A2 A3 A4 A5 A6 A8 | 25–50 μM* | <100 μM* (A2) | 0/4 | screen of ~200 cmpd4 collection | [42] |
| A7 B1 B2 B3 B4 B6 | 50–100 μM* | ~200 μM* (B4) | ||||
|
| ||||||
|
Inhibitors of kinase activity
| ||||||
| catechol derivatives | A2 | 0.5–2 μM* | nd | 85/19 | HTS | [61] |
|
| ||||||
| trifluoromethylphenyl derivatives (ALW-II-49-7 = cmpd 9) | A2 A4 A8 B2 B4 | 100–200 nM** | 40–1,000 nM* (B2) | 116/345 | screen of combinatorial library of type 2 inhibitors | [48] |
| A3 A5 B3 | ~500 nM** | |||||
| A6 A7 B1 | >8μM** | |||||
|
| ||||||
| imidazo[1,2-a]pyridines | B3 | 80 nM* | ~85% at 10 μM*** (B3) | 347/276 | HTS, optimization | [49] |
| pyrazolo[1,5-a]pyridines (LDN-211904 = cmpd 32) | A1–A5 A8 B1–B4 | >90% at 5 μM*** | ||||
| A6, A7 | <10% at 5 μM*** | |||||
|
| ||||||
| 2,4-bis-anilinopyrimidines (cmpd 28) | B2 B4 | 50–90 nM* | 190 nM* (B4) | 18/15 | screen of kinase inhibitors, structure guided optimization | [50] |
|
| ||||||
| 2,4-bis-anilinopyrimidines (cmpd 20) | B4 | 1.3 nM* | 9 nM* | 129/68 | structure-guided optimization | [52] |
|
| ||||||
| 2,4-bis-anilinopyrimidines (cmpd 12) | B4 | 4 nM* | 12 nM* | 39/70 | structure-guided optimization | [51] |
|
| ||||||
| thieno[3,2-c]pyridine derivatives (cmpds 9c–9f) | B4 | 20–60 nM* | 20–30 nM* | nd | pharmacophore model | [53] |
|
| ||||||
| xanthine derivatives (cmpd 66) | A1 A2 A4 A5 A8 B1 B2 B4 | 1–5 nM* | nd | 2110/130 | fragment based docking, optimization | [55] |
| A3 B3 | 15–40 nM* | |||||
| A7 | 1 μM* | |||||
|
| ||||||
| imidazo[1,2-a]pyrazine diaryl ureas | A2 B4 | 30–60 nM* (cmpd 2) | 50 nM* (B4; cmpds 45,59) | 38/26 (cmpd 2) | scaffold-based strategy, optimization | [54] |
|
| ||||||
| phenylthiopene derivatives (cmpd 1) | A2 B4 | <10 μM** | nd | 1111/83 | high-throughput docking | [56] |
|
| ||||||
| pyrazolo[3,4-d]pyrimidine NVP-BHG712 | B4 | nd | 25 nM* | 08/40 | molecular modeling, optimization | [32] |
| A2 B2 B3 | <100 nM* | |||||
| A3 | <500 nM* | |||||
|
| ||||||
| imidazo[1,2-a]pyrazines (ONC-101, ONC-102) | A1 A6 B2 B3 B4 | 0.3–0.9 μM** | 100% at 10 μM*** (B4) | 98/40 | screen for kinase inhibitors using a yeast growth assay | [33] |
| A2 A3 A4 A5 A8 B1 | 1–6 μM** | |||||
| A7 | 29 μM** | |||||
|
| ||||||
| furo[3,2-c]pyridine (cmpd 15c) | B4 | 0.4 nM* | nd | 18/2 | VEGFR24 inhibitor | [53] |
|
| ||||||
| imidazopyridines (LY2457546) | A5 A8 B1 | 10–30 nM* | 10 nM* (B4) | 88/10 | inhibitor of angiogenic RTKs4 | [57] |
| A2 B2 B4 | 40–50 nM* | |||||
| A1 A3 A4 A7 | 100–600 nM* | |||||
|
| ||||||
| thiazolylamino-pyrimidine (dasatinib) | A3 A5 A8 B1 B2 B4 | 0.01–0.5 nM** | ~100 nM* (A2) | 168/277 | Src/Bcr-Abl inhibitor | [45,62] |
| A2 A4 | 0.8–1.2 nM** | |||||
| A1 B3 | 4–7 nM** | |||||
| A6 A7 | >10 μM** | |||||
|
| ||||||
| phenylamino-pyrimidine (nilotinib) | B1 B2 | 95% at 10 μM*** | 4–15 nM* | 58/27 | Bcr-Abl inhibitor | [58] |
| B4 | 27% at 10 μM*** | 37 nM* | ||||
|
| ||||||
| quinazoline (EXEL-7647) | B4 | 1.4 nM* | nd | 38/69 | EGFR/VEGFR4 inhib | [63] |
|
| ||||||
| FMK4 | A2 | 0.76 μM* | nd | 412/71 | Rsk4 inhibitor | [59] |
|
| ||||||
| pyrido[2-3]pyrimidine (PD173955) | A2 B2 B4 | 18–34 nM* | nd | 48,13/all | Src/FGFR4 inhibitor | [64] |
|
| ||||||
| B1 | 120 nM* | |||||
IC50;
Kd;
% inhibition.
This column does not include Eph receptors.
The Eph receptor used for screening is indicated in bold.
Eph receptors that are least inhibited are indicated in gray.
Abbreviations: cmpd, compound; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; FMK, fluoromethylketone-modified version of the low potency tyrosine kinase inhibitor PP1; Rsk, ribosomal S6 kinase; RTK, receptor tyrosine kinase; VEGFR, vascular endothelial growth factor receptor.
Kinases inhibited ≥ 50% by 10 μM of at least 2 of the compounds.
Kinases with a competition score < 10 fold that for the most potently inhibited Eph receptor tested.
Kinases inhibited ≥ 90% by 5 μM compound.
Kinases inhibited with IC50 or Kd value < 10 fold that for the most potently inhibited Eph receptor tested.
Kinases inhibited ≥ 30% by 10 μM compound.
Kinases inhibited ≥ 90% by 1 μM compound.
Kinases inhibited ≥ 30% by 10 μM compound (compared to ~60% inhibition for EphA2).
Inhibited similarly to EphA2.
Identified as positive in a cell-based screen of a cDNA library. A black line separates kinase inhibitors identified in screens for Eph receptor inhibitors from some examples of inhibitors identified by screening other kinases.
3.1 Salicylates
A high-throughput screen measuring inhibition of EphA4 receptor binding to the KYL peptide followed by structure-activity relationship (SAR) analysis identified two isomeric salicylic acid-dimethylpyrrole derivatives that preferentially inhibit ephrin binding to the EphA2 and EphA4 receptors with Ki values of ~10 μM in ELISA assays [25]. Nuclear magnetic resonance (NMR) characterization was consistent with the binding of the two salicylates in the ephrin-binding pocket of EphA4 and cell-based assays suggested that the compounds inhibit ephrin-dependent EphA2 and EphA4 activation and biological responses, although with low potency [25,40]. However, recent studies have revealed that some modification of the original compounds, which may involve polymerization or oxidative processes, is required for their inhibitory activity [41].
A small molecule containing two salicylic acid-furanyl groups was also found to target the ephrin-binding pocket of EphA4 and inihibit ephrin binding to EphA2, EphA4 and several other Eph receptors through a mechanism that may involve irreversible binding [41]. The ability of the three salicylates to inhibit the activation of a subset of Eph receptors and their effects in cells, including endothelial capillary-like tube formation, suggest that these compounds could be used as chemical tools to study Eph receptor functions. In addition, salicylic acid derivatives with better pharmacological properties may be developed as Eph receptor-targeting agents.
3.2 Lithocholic acid
The recent screen of a collection of ~200 chemicals, including some natural compounds, identified the bile acid lithocholic acid as an inhibitor of EphA2-ephrin-A1 interaction [42]. Lithocholic acid behaves as a reversible competitive inhibitor of EphA2, with a Ki value of ~50 μM, while other related bile acids were found to be inactive. Lithocholic acid inhibits ephrin binding to both EphA and EphB receptors in biochemical assays as well as ephrin-induced EphA2 and EphB4 tyrosine phosphorylation in cell-based assays, while it does not target several other receptor tyrosine kinase families. It will be interesting to investigate whether lithocholic acid plays a role in intestinal homeostasis through interference with Eph receptor-ephrin interactions, given its millimolar fecal concentration.
3.2 Other compounds
A recent screen of 133 plant extracts showed that extracts that are rich in polyphenols inhibit EphA2-ephrin-A1 interaction in ELISA assays and ephrin-A1-induced EphA2 phosphorylation in cell-based assays with potencies in the μg/ml range [43]. This is consistent with previous studies reporting that in endothelial cells low micromolar concentrations of epigallocatechin gallate, a green tea polyphenol with antitumorigenic and antiangiogenic properties, inhibit ephrin-A1-dependent EphA2 tyrosine phosphorylation, cell migration and capillary-like tube formation [44]. However, given the many reported activities of epigallocatechin gallate, more work is needed to establish whether this compound acts only by inhibiting ephrin binding or also by inhibiting kinase activity and/or through other mechanisms.
4. Kinase inhibitors
Many inhibitors that bind with nanomolar affinity to the ATP-binding pocket of Eph receptors (Fig. 1) have been identified, either in screens focused on Eph receptors or when profiling the selectivity of compounds known to inhibit other kinases (Table 1). In fact, given the high conservation of the ATP-binding pocket, kinase inhibitors generally target more than a few of the 518 kinases in the human kinome [45–47].
A variety of approaches have been used to search for novel kinase inhibitors that target Eph receptors. Several compounds that show relatively good selectivity for the Eph family were identified by screening a combinatorial library of inhibitors designed to bind to the inactive kinase conformation (type 2 inhibitors) in a cell-based assay measuring phosphorylation of an EphB2 chimeric receptor [48]. A series of imidazo[1,2-a]pyrimidines and pyrazolo[1,5-a]pyridines that preferentially target tyrosine kinases over serine/threonine kinases were identified in a high-throughput screen measuring inhibition of EphB3 kinase activity [49].
The EphB4 receptor, however, has been the focus of most efforts because it is regarded as a promising target for inhibition of tumor angiogenesis [2,4,32]. Several classes of 2,4-bis-anilinopyrimidine derivatives were identified by screening libraries of kinase inhibitors followed by crystal structure-guided optimization [50–52]. Some of these compounds are active in biochemical and cell-based assays at low nanomolar concentrations and have a good selectivity profile, although they also inhibit Src family kinases and some other kinases. EphB4 is also a target for various classes of inhibitors designed based on a pharmacophore model for ATP competitive inhibitors or around a known scaffold [53,54], and for inhibitors identified by high-throughput in silico docking to a model of the EphB4 kinase domain [55,56].
A particularly promising inhibitor is NVP-BHG712, which was identified by computer design using a model of the EphB4 kinase domain followed by optimization based on inhibition of EphB4 phosphorylation in cells [32]. This compound shows substantial selectivity for the Eph family, with some preference for EphB4, and has good pharmacokinetic properties. It inhibits EphB4 phosphorylation in tissues after oral administration and vascular endothelial growth factor (VEGF)-driven angiogenesis in vivo. Because NVP-BHG712 has little effect on VEGF receptor kinase activity, its effects suggest that Eph receptors play an important role in VEGF-induced angiogenesis. This work demonstrates the usefulness of pharmacological tools for elucidating Eph biological activities. Eph kinase inhibitors could also help discriminate between effects mediated by Eph receptor or ephrin signaling [32,33].
Several kinase inhibitors show some selectivity for certain Eph receptors (Table 1). In particular, EphA6 and EphA7 are often poorly inhibited by compounds that target other Eph receptors. This is likely due to their different “gatekeeper”, a residue that controls access of inhibitors to a deep hydrophobic pocket adjacent to the ATP binding site [47]. The gatekeeper residue is a threonine in most Eph receptors, but it is a valine in EphA6 and a bulkier isoleucine in EphA7. Interestingly Src, Abl and platelet-derived growth factor (PDGF) and EGF receptors also have a threonine gatekeeper residue. This is consistent with the selectivity profile observed for many of the kinase inhibitors targeting Eph receptors. For example, dasatinib and nilotinib were first identified as Src and Abl inhibitors but also potently target Eph receptors [57,58].
Despite the propensity of kinase inhibitors to be promiscuous, several strategies may be used to improve selectivity. Crystal structures of inhibitors in complex with the kinase domain of Eph receptors provide information on how to maximize interactions with specific residues both within and near the ATP-binding pocket [46–48,50]. A computational analysis of key residues in the ATP binding site of all human kinases suggests that it may be possible to design selective inhibitors for EphB4, besides EphA6 and EphA7 [46]. Certain cysteines in the ATP-binding pocket may also be exploited by irreversible inhibitors that contain mildly reactive groups capable of forming a covalent bond [47]. For example, a cysteine near the P loop of most Eph receptors is found in only a small subset of other kinases. This cysteine may enable inhibition of Eph receptors by FMK, an irreversible Rsk inhibitor developed by modifying the tyrosine kinase inhibitor PP1 with a fluoromethylketone reactive group [47,59]. Moreover, a cysteine in the hinge region of EphB3 is found in only two other kinases [47]. Allosteric inhibitors should also be more selective.
High selectivity may not always be desirable, however, and inhibitors that target multiple kinases involved in tumor angiogenesis, cancer progression or nerve regeneration may have increased effectiveness and better ability to circumvent resistance mechanisms [47,57]. For example, a series of imidazo[1,2-a]pyrazine diaryl ureas and a furo[3,2-c]pyridine derivatives target EphB4, EphA2, VEGF receptor 2 and the Tie2 receptor [53,54]. These angiogenic kinases, and the PDGF receptor, are also targeted by LY2457546. This orally bioavailable multikinase inhibitor has anti-angiogenic activity at nanomolar concentrations in cell culture assays and inhibits tumor growth in preclinical mouse xenograft models [57,58].
5. Conclusions and perspectives
A number of peptides and small molecules that bind to Eph receptors and inhibit ephrin binding or kinase activity have been successfully identified in the last decade. In future studies, it will be important to increase the binding affinity and resistance to proteases of existing peptides as well as screen for peptides that target other Eph receptors. Additional small molecule inhibitors of the Eph receptor-ephrin interaction will likely also emerge from the optimization of existing compounds as well as from new high-throughput, computational, biophysical and NMR-based screens. In silico screens and rational design of targeting peptides and small molecules may also represent viable options, given the rapidly increasing crystallographic structural information becoming available for Eph receptors in complex with diverse ligands.
Other interfaces that could be targeted to inhibit Eph receptor function include receptor-receptor interfaces in the extracellular, transmembrane and cytoplasmic regions (Fig. 1) in order to inhibit the clustering that promotes cross-phosphorylation between receptor molecules [11,60]. Alternatively, inhibition of Eph receptor association with specific cytoplasmic or extracellular binding partners would allow selective silencing of downstream signaling pathways or other effects. However, these interfaces have less favorable features for the binding of small molecules.
The development of chemical libraries more suitable for the identification of inhibitors of protein-protein interactions would be particularly useful for facilitating the discovery of new modulators of the Eph/ephrin system [38,39]. It would also be interesting to investigate whether non-classical mechanisms of inhibition, such as those employed by mildly reactive small molecules that can form covalent bonds in the targeted binding pocket, represent a viable strategy to increase potency and selectivity in the inhibition of Eph receptors [41,47].
Deciding how best to target Eph receptors for therapeutic applications represents another challenge, due to the complex and incompletely understood biology of the Eph/ephrin system [1,2]. For example, Eph receptors and ephrins can mediate bidirectional signals and engage in crosstalk with other signaling systems, and have diverse effects in different cellular contexts. Indeed, not only inhibiting but also enhancing Eph receptor activation may be useful in some cases. The ensuing increased Eph kinase activity could stimulate the tumor suppressor effects of Eph receptors in certain cancers or enhance the beneficial effects of Eph receptor signaling in the diseased nervous system [1,2,8]. Eph receptor activation might be enhanced, for example, by molecules disrupting the inhibitory association of the juxtamembrane domain with the kinase domain. Despite the challenges, progress has been made in targeting Eph receptors with peptides and small molecules, and further exciting advances are expected in the years to come.
Highlights.
The Eph receptor tyrosine kinases are an emerging family of drug targets.
Small molecules and peptides have been identified that target these receptors.
Some of these molecules bind to the ligand-binding pocket and inhibit ligand binding.
Others target the ATP-binding site and inhibit kinase activity.
These molecules show promise for research, diagnostic and therapeutic applications.
Acknowledgments
The authors thank N. Cosford for reading the manuscript. Work in the authors’ laboratory is supported by grants from the NIH, DOD and the University of California TRDRP.
Abbreviations
- EGF
epidermal growth factor
- NMR
nuclear magnetic resonance
- PDGF
platelet-derived growth factor
- PET
positron emission tomography
- SAR
structure-activity relationship
- SPECT
single photon emission computed tomography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brantley-Sieders DM, Chen J. Eph receptor tyrosine kinases in angiogenesis: from development to disease. Angiogenesis. 2004;7:17–28. doi: 10.1023/B:AGEN.0000037340.33788.87. [DOI] [PubMed] [Google Scholar]
- 4.Kuijper S, Turner CJ, Adams RH. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med. 2007;17:145–51. doi: 10.1016/j.tcm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Du J, Fu C, Sretavan DW. Eph/ephrin signaling as a potential therapeutic target after central nervous system injury. Current pharmaceutical design. 2007;13:2507–18. doi: 10.2174/138161207781368594. [DOI] [PubMed] [Google Scholar]
- 6.Genander M, Frisen J. Ephrins and Eph receptors in stem cells and cancer. Curr Opin Cell Biol. 2010;22:611–6. doi: 10.1016/j.ceb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Carmona MA, Murai KK, Wang L, Roberts AJ, Pasquale EB. Glial ephrin-A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proc Natl Acad Sci U S A. 2009;106:12524–9. doi: 10.1073/pnas.0903328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cisse M, Halabisky B, Harris J, Devidze N, Dubal DB, Sun B, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dries JL, Kent SD, Virag JA. Intramyocardial administration of chimeric ephrinA1-Fc promotes tissue salvage following myocardial infarction in mice. J Physiol. 2011;589:1725–40. doi: 10.1113/jphysiol.2010.202366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–95. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–75. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 12.Tandon M, Vemula SV, Mittal SK. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert opinion on therapeutic targets. 2011;15:31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem. 2002;277:46974–9. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- 14.Koolpe M, Burgess R, Dail M, Pasquale EB. EphB receptor-binding peptides identified by phage display enable design of an antagonist with ephrin-like affinity. J Biol Chem. 2005;280:17301–11. doi: 10.1074/jbc.M500363200. [DOI] [PubMed] [Google Scholar]
- 15.Murai KK, Nguyen LN, Koolpe M, McLennan R, Krull CE, Pasquale EB. Targeting the EphA4 receptor in the nervous system with biologically active peptides. Mol Cell Neurosci. 2003;24:1000–11. doi: 10.1016/j.mcn.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Mitra S, Duggineni S, Koolpe M, Zhu X, Huang Z, Pasquale EB. Structure-activity relationship analysis of peptides targeting the EphA2 receptor. Biochemistry. 2010;49:6687–95. doi: 10.1021/bi1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landen CN, Kinch MS, Sood AK. EphA2 as a target for ovarian cancer therapy. Expert opinion on therapeutic targets. 2005;9:1179–87. doi: 10.1517/14728222.9.6.1179. [DOI] [PubMed] [Google Scholar]
- 18.Blackburn WH, Dickerson EB, Smith MH, McDonald JF, Lyon LA. Peptide-Functionalized Nanogels for Targeted siRNA Delivery. Bioconjugate chemistry. 2009;20:960–8. doi: 10.1021/bc800547c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickerson EB, Blackburn WH, Smith MH, Kapa LB, Lyon LA, McDonald JF. Chemosensitization of cancer cells by siRNA using targeted nanogel delivery. BMC Cancer. 2010;10:10. doi: 10.1186/1471-2407-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Geer MA, Brevoord D, Kuhlmann KF, Bakker CT, Mizuguchi H, Wesseling JG, et al. A fiber modified adenovirus vector that targets to the EphrinA2 receptor reveals enhanced gene transfer to ex vivo pancreatic cancer. Int J Oncol. 2010;36:233–44. [PubMed] [Google Scholar]
- 21.Scarberry KE, Dickerson EB, McDonald JF, Zhang ZJ. Magnetic nanoparticle-peptide conjugates for in vitro and in vivo targeting and extraction of cancer cells. J Am Chem Soc. 2008;130:10258–62. doi: 10.1021/ja801969b. [DOI] [PubMed] [Google Scholar]
- 22.Scarberry KE, Dickerson EB, Zhang ZJ, Benigno BB, McDonald JF. Selective removal of ovarian cancer cells from human ascites fluid using magnetic nanoparticles. Nanomedicine. 2010;6:399–408. doi: 10.1016/j.nano.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Galimberti I, Bednarek E, Donato F, Caroni P. EphA4 signaling in juveniles establishes topographic specificity of structural plasticity in the hippocampus. Neuron. 2010;65:627–42. doi: 10.1016/j.neuron.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Fabes J, Anderson P, Brennan C, Bolsover S. Regeneration-enhancing effects of EphA4 blocking peptide following corticospinal tract injury in adult rat spinal cord. Eur J Neurosci. 2007;26:2496–505. doi: 10.1111/j.1460-9568.2007.05859.x. [DOI] [PubMed] [Google Scholar]
- 25.Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, et al. Small Molecules Can Selectively Inhibit Ephrin Binding to the EphA4 and EphA2 Receptors. J Biol Chem. 2008;283:29461–72. doi: 10.1074/jbc.M804103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharfe N, Nikolic M, Cimpeon L, Van De Kratts A, Freywald A, Roifman CM. EphA and ephrin-A proteins regulate integrin-mediated T lymphocyte interactions. Mol Immunol. 2008;45:1208–20. doi: 10.1016/j.molimm.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Chrencik JE, Brooun A, Recht MI, Nicola G, Davis LK, Abagyan R, et al. Three-dimensional Structure of the EphB2 Receptor in Complex with an Antagonistic Peptide Reveals a Novel Mode of Inhibition. J Biol Chem. 2007;282:36505–13. doi: 10.1074/jbc.M706340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chrencik JE, Brooun A, Recht MI, Kraus ML, Koolpe M, Kolatkar AR, et al. Structure and thermodynamic characterization of the EphB4/Ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure. 2006;14:321–30. doi: 10.1016/j.str.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Xiong C, Huang M, Zhang R, Song S, Lu W, Flores L, 2nd, et al. In vivo small-animal PET/CT of EphB4 receptors using 64Cu-labeled peptide. J Nucl Med. 2011;52:241–8. doi: 10.2967/jnumed.110.081943. [DOI] [PubMed] [Google Scholar]
- 30.Zhang R, Xiong C, Huang M, Zhou M, Huang Q, Wen X, et al. Peptide-conjugated polymeric micellar nanoparticles for Dual SPECT and optical imaging of EphB4 receptors in prostate cancer xenografts. Biomaterials. 2011;32:5872–9. doi: 10.1016/j.biomaterials.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noren NK, Pasquale EB. Paradoxes of the EphB4 receptor in cancer. Cancer Res. 2007;67:3994–7. doi: 10.1158/0008-5472.CAN-07-0525. [DOI] [PubMed] [Google Scholar]
- 32.Martiny-Baron G, Holzer P, Billy E, Schnell C, Brueggen J, Ferretti M, et al. The small molecule specific EphB4 kinase inhibitor NVP-BHG712 inhibits VEGF driven angiogenesis. Angiogenesis. 2010;13:259–67. doi: 10.1007/s10456-010-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–6. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 34.Salvucci O, Maric D, Economopoulou M, Sakakibara S, Merlin S, Follenzi A, et al. EphrinB reverse signaling contributes to endothelial and mural cell assembly into vascular structures. Blood. 2009;114:1707–16. doi: 10.1182/blood-2008-12-192294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhari A, Mahfouz M, Fialho AM, Yamada T, Granja AT, Zhu Y, et al. Cupredoxin-cancer interrelationship: azurin binding with EphB2, interference in EphB2 tyrosine phosphorylation, and inhibition of cancer growth. Biochemistry. 2007;46:1799–810. doi: 10.1021/bi061661x. [DOI] [PubMed] [Google Scholar]
- 36.London N, Raveh B, Movshovitz-Attias D, Schueler-Furman O. Can self-inhibitory peptides be derived from the interfaces of globular protein-protein interactions? Proteins. 2010;78:3140–9. doi: 10.1002/prot.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fry DC, Vassilev LT. Targeting protein-protein interactions for cancer therapy. J Mol Med. 2005;83:955–63. doi: 10.1007/s00109-005-0705-x. [DOI] [PubMed] [Google Scholar]
- 38.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–8. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 39.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 40.Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal Structure and NMR Binding Reveal That Two Small Molecule Antagonists Target the High Affinity Ephrin-binding Channel of the EphA4 Receptor. J Biol Chem. 2008;283:29473–84. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noberini R, De SK, Zhan Z, Wu B, Raveendra-Panickar D, Chen V, et al. A disalicylic acid-furanyl derivative inhibits ephrin binding to a subset of Eph receptors. Chemical Biology & Drug Design. doi: 10.1111/j.1747-0285.2011.01199.x. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giorgio C, Hassan Mohamed I, Flammini L, Barocelli E, Incerti M, Lodola A, et al. Lithocholic Acid Is an Eph-ephrin Ligand Interfering with Eph-kinase Activation. PLoS One. 2011;6:e18128. doi: 10.1371/journal.pone.0018128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohamed IH, Giorgio C, Bruni R, Flammini L, Barocelli E, Rossi D, et al. Polyphenol-rich botanicals used as food supplements interfere with the EphA2-ephrin-A1 system. doi: 10.1016/j.phrs.2011.06.008. in press. [DOI] [PubMed] [Google Scholar]
- 44.Tang FY, Chiang EP, Shih CJ. Green tea catechin inhibits ephrin-A1-mediated cell migration and angiogenesis of human umbilical vein endothelial cells. J Nutr Biochem. 2007;18:391–9. doi: 10.1016/j.jnutbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 46.Huang D, Zhou T, Lafleur K, Nevado C, Caflisch A. Kinase selectivity potential for inhibitors targeting the ATP binding site: a network analysis. Bioinformatics. 2010;26:198–204. doi: 10.1093/bioinformatics/btp650. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 48.Choi Y, Syeda F, Walker JR, Finerty PJ, Jr, Cuerrier D, Wojciechowski A, et al. Discovery and structural analysis of Eph receptor tyrosine kinase inhibitors. Bioorganic & medicinal chemistry letters. 2009;19:4467–70. doi: 10.1016/j.bmcl.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiao L, Choi S, Case A, Gainer TG, Seyb K, Glicksman MA, et al. Structure-activity relationship study of EphB3 receptor tyrosine kinase inhibitors. Bioorganic & medicinal chemistry letters. 2009;19:6122–6. doi: 10.1016/j.bmcl.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bardelle C, Cross D, Davenport S, Kettle JG, Ko EJ, Leach AG, et al. Inhibitors of the tyrosine kinase EphB4. Part 1: Structure-based design and optimization of a series of 2,4-bis-anilinopyrimidines. Bioorganic & medicinal chemistry letters. 2008;18:2776–80. doi: 10.1016/j.bmcl.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Barlaam B, Ducray R, Brempt CL, Ple P, Bardelle C, Brooks N, et al. Inhibitors of the tyrosine kinase EphB4. Part 4: Discovery and optimization of a benzylic alcohol series. Bioorganic & medicinal chemistry letters. 2011;21:2207–11. doi: 10.1016/j.bmcl.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Bardelle C, Barlaam B, Brooks N, Coleman T, Cross D, Ducray R, et al. Inhibitors of the tyrosine kinase EphB4. Part 3: identification of non-benzodioxole-based kinase inhibitors. Bioorganic & medicinal chemistry letters. 2010;20:6242–5. doi: 10.1016/j.bmcl.2010.08.100. [DOI] [PubMed] [Google Scholar]
- 53.Miyazaki Y, Nakano M, Sato H, Truesdale AT, Stuart JD, Nartey EN, et al. Design and effective synthesis of novel templates, 3,7-diphenyl-4-amino-thieno and furo-[3,2-c]pyridines as protein kinase inhibitors and in vitro evaluation targeting angiogenetic kinases. Bioorganic & medicinal chemistry letters. 2007;17:250–4. doi: 10.1016/j.bmcl.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell SA, Danca MD, Blomgren PA, Darrow JW, Currie KS, Kropf JE, et al. Imidazo[1,2-a]pyrazine diaryl ureas: inhibitors of the receptor tyrosine kinase EphB4. Bioorganic & medicinal chemistry letters. 2009;19:6991–5. doi: 10.1016/j.bmcl.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 55.Lafleur K, Huang D, Zhou T, Caflisch A, Nevado C. Structure-based optimization of potent and selective inhibitors of the tyrosine kinase erythropoietin producing human hepatocellular carcinoma receptor B4 (EphB4) Journal of medicinal chemistry. 2009;52:6433–46. doi: 10.1021/jm9009444. [DOI] [PubMed] [Google Scholar]
- 56.Zhou T, Caflisch A. High-throughput virtual screening using quantum mechanical probes: discovery of selective kinase inhibitors. ChemMedChem. 2010;5:1007–14. doi: 10.1002/cmdc.201000085. [DOI] [PubMed] [Google Scholar]
- 57.Burkholder TP, Clayton JR, Rempala ME, Henry JR, Knobeloch JM, Mendel D, et al. Discovery of LY2457546: a multi-targeted anti-angiogenic kinase inhibitor with a novel spectrum of activity and exquisite potency in the acute myelogenous leukemia-Flt-3-internal tandem duplication mutant human tumor xenograft model. Invest New Drugs. 2011 doi: 10.1007/s10637-011-9640-6. [DOI] [PubMed] [Google Scholar]
- 58.Melnick JS, Janes J, Kim S, Chang JY, Sipes DG, Gunderson D, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A. 2006;103:3153–8. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kajanajmudeen A. Receptor tyrosine kinase transmembrane domain interactions: potential target for “interceptor” therapy. Sci Signal. 2010;3:jc6. doi: 10.1126/scisignal.3138jc6. [DOI] [PubMed] [Google Scholar]
- 61.Stroylov VS, Rakitina TV, Novikov FN, Stroganov OV, Chilov GG, Lipkin AV. Novel fragment-like inhibitors of EphA2 obtained by experimental screening and modelling. Mendeleev Commun. 2010;20:263–5. [Google Scholar]
- 62.Chang Q, Jorgensen C, Pawson T, Hedley DW. Effects of dasatinib on EphA2 receptor tyrosine kinase activity and downstream signalling in pancreatic cancer. Br J Cancer. 2008;99:1074–82. doi: 10.1038/sj.bjc.6604676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gendreau SB, Ventura R, Keast P, Laird AD, Yakes FM, Zhang W, et al. Inhibition of the T790M gatekeeper mutant of the epidermal growth factor receptor by EXEL-7647. Clin Cancer Res. 2007;13:3713–23. doi: 10.1158/1078-0432.CCR-06-2590. [DOI] [PubMed] [Google Scholar]
- 64.Caligiuri M, Molz L, Liu Q, Kaplan F, Xu JP, Majeti JZ, et al. MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem Biol. 2006;13:711–22. doi: 10.1016/j.chembiol.2006.05.008. [DOI] [PubMed] [Google Scholar]