Abstract
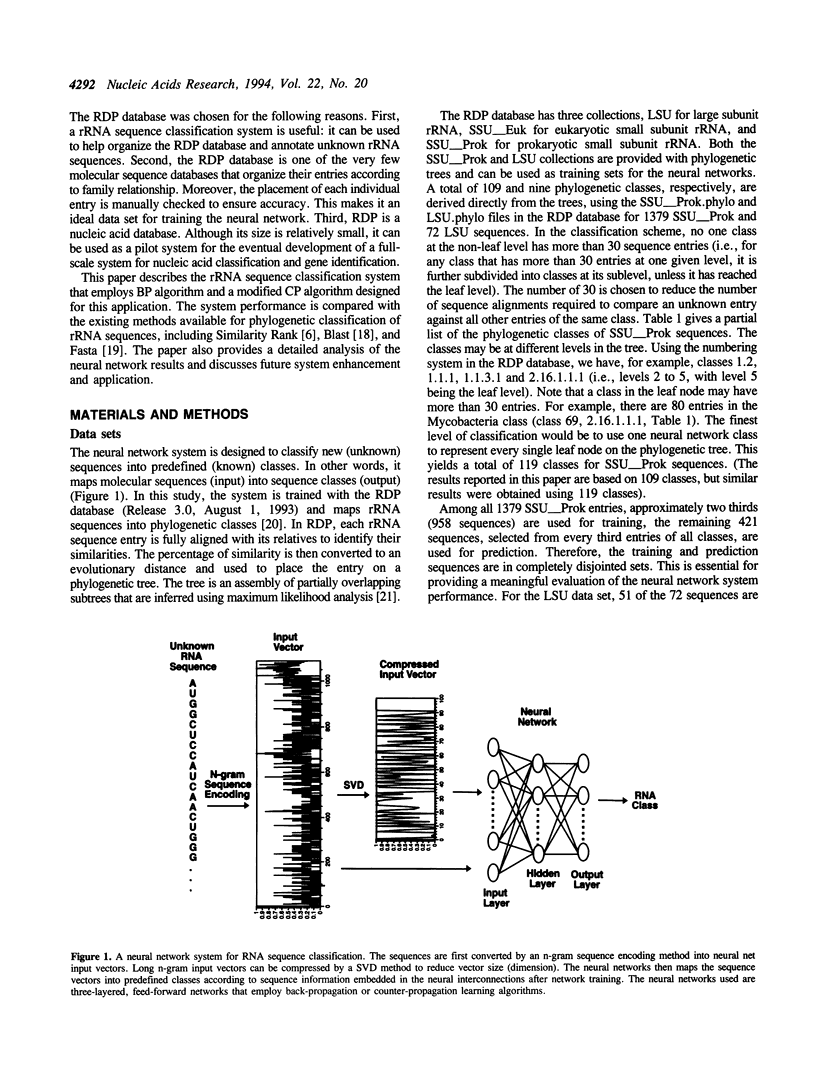
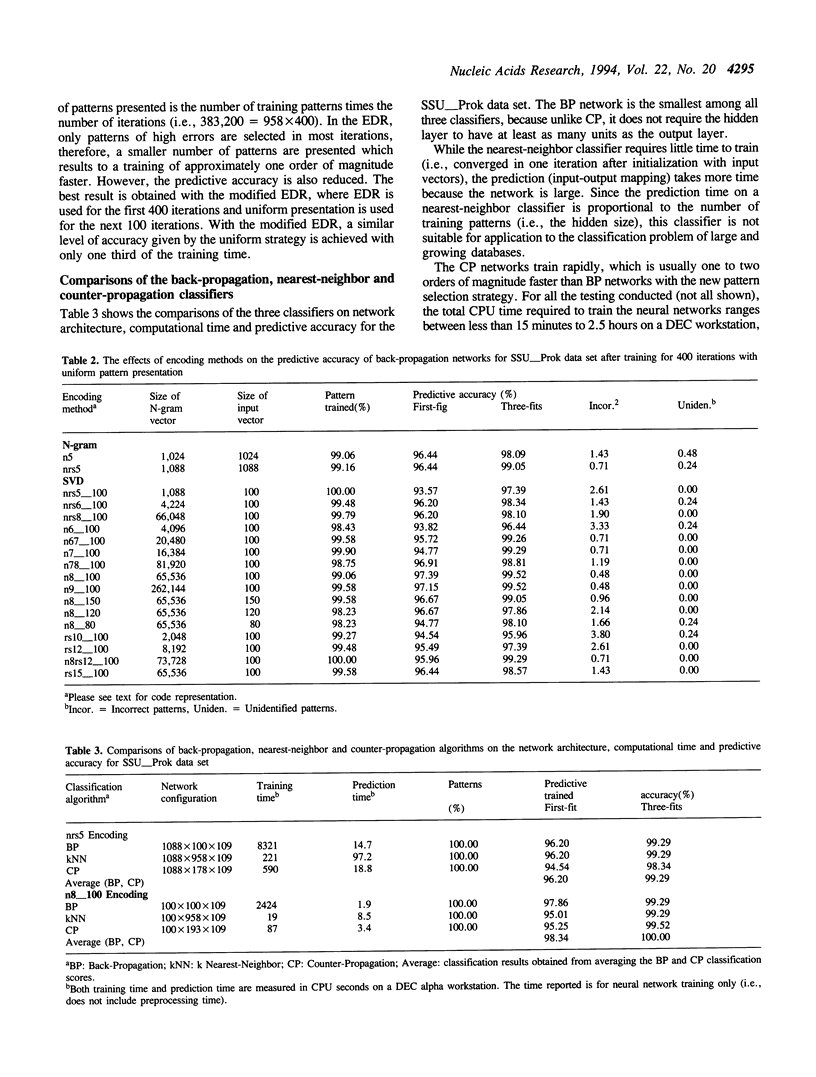
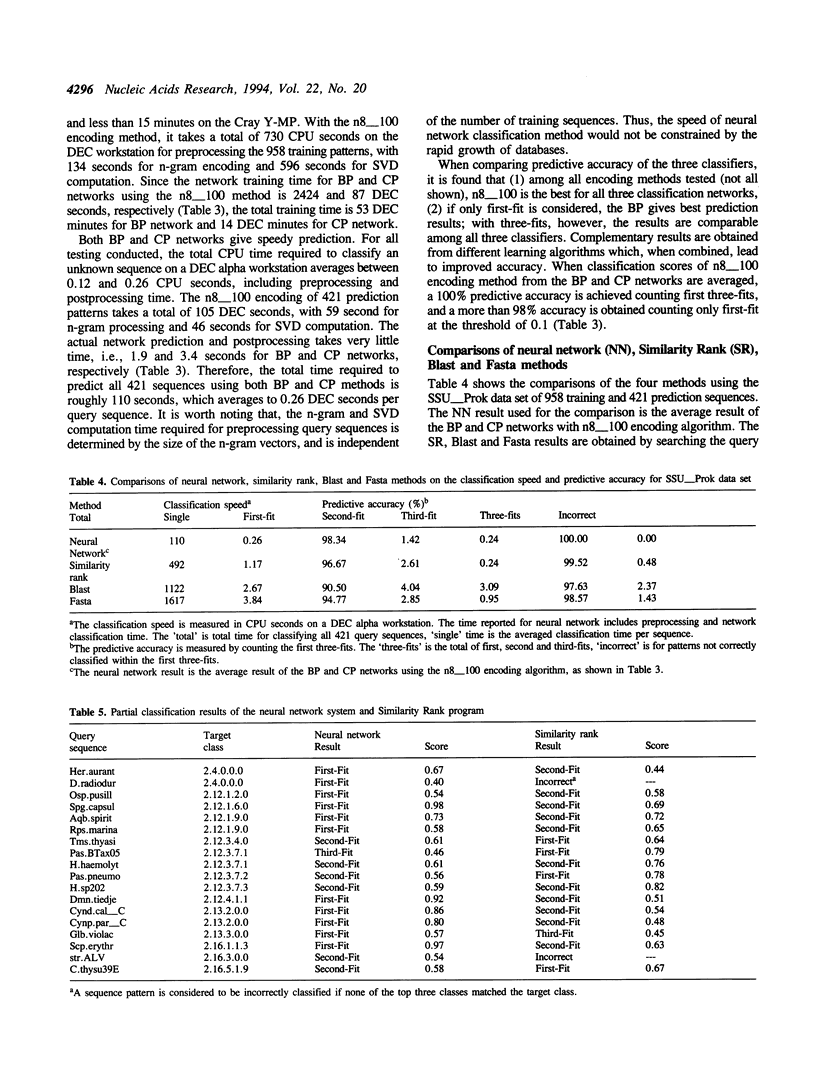
A neural network system has been developed for rapid and accurate classification of ribosomal RNA sequences according to phylogenetic relationship. The molecular sequences are encoded into neural input vectors using an n-gram hashing method. A SVD (singular value decomposition) method is used to compress and reduce the size of long and sparse n-gram input vectors. The neural networks used are three-layered, feed-forward networks that employ supervised learning paradigms, including the back-propagation algorithm and a modified counter-propagation algorithm. A pedagogical pattern selection strategy is used to reduce the training time. After trained with ribosomal RNA sequences of the RDP (Ribosomal Database Project) database, the system can classify query sequences into more than one hundred phylogenetic classes with a 100% accuracy at a rate of less than 0.3 CPU second per sequence on a workstation. When compared to other sequence similarity search methods, including Similarity Rank, Blast and Fasta, the neural network method has a higher classification accuracy at a speed of about an order of magnitude faster. The software tool will be made available to the biology community, and the system may be extended into a gene identification system for classifying indiscriminately sequenced DNA fragments.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker W. C., George D. G., Mewes H. W., Pfeiffer F., Tsugita A. The PIR-International databases. Nucleic Acids Res. 1993 Jul 1;21(13):3089–3092. doi: 10.1093/nar/21.13.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber R., Lapedes A., Sirotkin K. Determination of eukaryotic protein coding regions using neural networks and information theory. J Mol Biol. 1992 Jul 20;226(2):471–479. doi: 10.1016/0022-2836(92)90961-i. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Ferrán E. A., Pflugfelder B., Ferrara P. Self-organized neural maps of human protein sequences. Protein Sci. 1994 Mar;3(3):507–521. doi: 10.1002/pro.5560030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. D., Sternberg M. J. Prediction of structural and functional features of protein and nucleic acid sequences by artificial neural networks. Biochemistry. 1992 Aug 18;31(32):7211–7218. doi: 10.1021/bi00147a001. [DOI] [PubMed] [Google Scholar]
- Kneller D. G., Cohen F. E., Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990 Jul 5;214(1):171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters: neural networks develop distinct descriptions in learning to search for promoters of different spacing classes. Nucleic Acids Res. 1992 Jul 11;20(13):3471–3477. doi: 10.1093/nar/20.13.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian N., Sejnowski T. J. Predicting the secondary structure of globular proteins using neural network models. J Mol Biol. 1988 Aug 20;202(4):865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- Uberbacher E. C., Mural R. J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Whitson G., McLarty J., Ermongkonchai A., Chang T. C. Protein classification artificial neural system. Protein Sci. 1992 May;1(5):667–677. doi: 10.1002/pro.5560010512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. A new family of powerful multivariate statistical sequence analysis techniques. J Mol Biol. 1991 Aug 20;220(4):877–887. doi: 10.1016/0022-2836(91)90360-i. [DOI] [PubMed] [Google Scholar]


