Abstract
The conformational properties of an active-site loop segment, defined by residues Ser161-Glu162-Asn163-Ser164, have been shown to be important for modulating the intrinsic reactivity of Mn(II) in the active site of Bacillus subtilis oxalate decarboxylase. We now detail the functional and structural consequences of removing a conserved Arg/Thr hydrogen bonding interaction by site-specific mutagenesis. Hence, substitution of Thr-165 by a valine residue gives an OxDC variant (T165V) that exhibits impaired catalytic activity. Heavy-atom isotope effect measurements, in combination with the X-ray crystal structure of the T165V OxDC variant, demonstrate that the conserved Arg/Thr hydrogen bond is important for correctly locating the side chain of Glu-162, which mediates a proton-coupled electron transfer (PCET) step prior to decarboxylation in the catalytically competent form of OxDC. In addition, we show that the T165V OxDC variant exhibits a lower level of oxalate consumption per dioxygen molecule, consistent with the predictions of recent spin-trapping experiments (Imaram et al., 2011, Free Rad. Biol. Med. 50, 1009–1015). This finding implies that dioxygen might participate as a reversible electron sink in two putative PCET steps and is not merely used to generate a protein-based radical or oxidized metal center.
Keywords: Oxalate Decarboxylase, Heavy Atom Isotope Effects, Proton-Coupled Electron Transfer, Enzyme Catalysis
Recent work has identified an active-site loop segment Ser161-Glu162-Asn163 (SENS)1,2 that plays a critical role in the control of catalytic function in Bacillus subtilis oxalate decarboxylase (OxDC) (1), an enzyme catalyzing the Mn(II)-dependent conversion of oxalate into CO2 and formate (2–5). The X-ray crystal structure of wild-type (WT) OxDC shows the existence of an intriguing hydrogen bond network when the SENS loop segment adopts a “closed” conformation, which includes an interaction between the side chains of Arg-92 and Thr-165 (Fig. 1) (4). Both of these residues are stringently conserved and Arg-92 has been shown to be functionally important during turnover (4, 6). Thus, replacing Arg-92 by a lysine residue gives an OxDC R92K variant with altered substrate binding kinetics and a lower rate of decarboxylation, which is probably associated with altered substrate polarization in the Michaelis complex of the R92K OxDC variant (6). We have investigated the consequences of removing the Arg-92/Thr-165 hydrogen bond by expressing and purifying two OxDC variants in which Thr-165 is replaced by either valine or serine (T165V and T165S, respectively). Spin-trapping experiments using these OxDC variants show that the N-terminal Mn(II) site of the T165V OxDC variant remains solvent accessible during catalytic turnover (7). Herein, we report a detailed structural and kinetic characterization of the T165V OxDC variant that demonstrates that the Arg-92/Thr-165 hydrogen bond is important for correctly locating an active site glutamate side chain, which mediates a proton-coupled electron transfer (PCET) step prior to decarboxylation, in the catalytically competent form of OxDC.
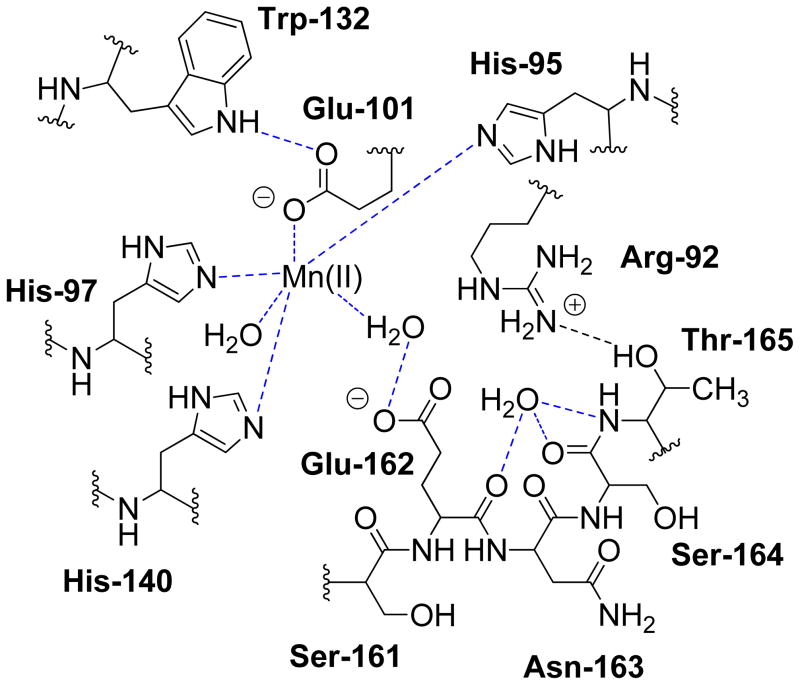
Fig. 1.
Schematic representation of hydrogen bonding interactions (dotted lines) between residues in the active site loop Ser161-Glu162-Asn163 and other conserved residues in the N-terminal Mn(II) binding site when the loop adopts a “closed” conformation (4). [Used with permission.]
MATERIALS AND METHODS
General information
Unless otherwise stated, all chemicals were purchased from Sigma (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) and were of the highest available purity. Nickel-nitrilotriacetic acid agarose (Ni-NTA) was obtained from Qiagen (Germantown, MD). Protein concentrations were determined using the Bradford Assay (Pierce, Rockford, IL) (8). Spectra were recorded on an Agilent 8453 spectrophotometer. All DNA primers were obtained from Integrated DNA Technologies, Inc. (Coralville, IA), and DNA sequencing was performed by the core facility in the Interdisciplinary Center for Biotechnology Research at the University of Florida. ICP-MS determinations of metal content were performed at the University of Wisconsin Soil and Plant Analysis Laboratory (Verona, WI). Oxygen consumption experiments were performed using the NeoFox® sensor system from Ocean Optics (Dunedin, FL).
Expression and purification of C-terminally tagged WT OxDC and OxDC variants
A plasmid containing the gene encoding C-terminally tagged OxDC was generously provided by Dr. Stephen Bornemann (John Innes Center, Norwich, UK). The recombinant, His6-tagged WT enzyme was expressed and purified following published procedures (4, 9) except that expression was induced in the presence of 5 mM MnCl2 after heat shocking the bacteria for 18 min at 42 °C. Site-directed mutagenesis to obtain the genes encoding the T165S and T165V OxDC variants was performed by overlap extension (10) using standard primers (Supporting information). After cell lysis and centrifugation, the desired enzymes were purified by metal affinity chromatography on an Ni-NTA column, and stored in 50 mM Tris buffer, pH 8.5, containing 0.5 M NaCl. Samples used for EPR analysis were concentrated to 6–15 mg/mL using an Amicon Centriprep filter unit from Millipore (Billerica, MA).
Steady-state kinetic assays
The catalytic properties of the His6-tagged WT OxDC and the OxDC mutants were determined by measuring formate production using an end-point assay, as described elsewhere (11). Measurements were made at specific substrate and enzyme concentrations in triplicate, and the data were analyzed to obtain the values of V and V/K using standard computer-based methods (12).
Isotope effect nomenclature
The nomenclature used herein is due to Northrop (13), where 13(V/K) represents the ratio of V/K for the 12C-containing species relative to the 13C-containing species, and 18(V/K) is the same ratio for 16O and 18O.
Isotope effect measurements
Heavy atom isotope effects were determined using procedures employing isotope-ratio mass spectrometry (IRMS) (6, 11) with minor modifications. Briefly, the partial enzymatic reactions were performed by incubation of the T165V mutant of OxDC with oxalate (25 mM) and o-phenylenediamine (0.2 mM) at 22°C in either 0.1 M 1,4-bis-(2-hydroxyethyl)piperazine buffer, pH 4.2, or piperazine buffer, pH 5.7 (790 μL total volume). All reaction solutions were sparged with nitrogen, with the exception of oxalate solutions, which were sparged with dioxygen prior to use. Reactions in which oxalate was fully converted to formate and CO2 were performed in a similar manner to those described above except that the oxalate concentration was only 12.5 mM. Control experiments established that when a partial reaction was performed in the presence of H218O, formate and unreacted oxalate incorporated less than 0.2% of 18O, as observed for untagged samples of recombinant WT OxDC (11).
Crystallization of the C-terminally tagged T165V OxDC mutant
Purified, His6-tagged T165V OxDC was concentrated to 6 mg/mL in 1 mM HEPES buffer, pH 7.5, for crystallization. Crystallization conditions were determined using the Crystal Screen HT from Hampton Research (Aliso Viejo, CA) via the sitting drop vapor diffusion method, in which drops were formed by mixing 1.2 μL of protein solution with 1.2 μL of precipitant solution and equilibrated against the well solution at 290K. The condition comprised of 10% w/v polyethylene glycol 6000 and 2 M NaCl gave crystals in ~ 1 week. Crystals were used in diffraction data collection directly from the crystallization screen without further optimization.
Data collection, structure determination and crystallographic refinement
Diffraction data were collected at 100 K on crystals without further cryo-protection on a Bruker AXS Proteum-R instrument with a MICROSTAR rotating anode X-ray source and PLATINUM135 CCD area detector, located in the Department of Chemistry at Boston University, and processed using the Proteum2 software suite. The His6-tagged T165V OxDC mutant crystallized in space group R32 with unit cell dimensions a = b = 154.76 Å and c = 121.50 Å. Data from a crystal with dimensions 0.2 × 0.1 × 0.02 mm, were collected to 2.31 Å resolution. Data collection, refinement, and final model statistics are reported in Table 1. Phases from the structure of native OxDC (PDB accession code 1UW8) (4) were used to solve the T165V structure by molecular replacement. The programs MOLREP (14) and PHASER in the CCP4 program suite (15) were used to solve the rotation and translation functions, yielding a solution with one enzyme protomer in the asymmetric unit. Alternating rounds of manual rebuilding were performed using the molecular graphics program COOT (16) followed by minimization and simulated annealing in PHENIX (17). During the refinement procedure, water molecules and metal ions were added only after Rfree dropped below 30%. The final protein model was analyzed using composite-omit electron-density maps calculated with CNS (18) and stereochemistry was monitored using PROCHECK (19).
Table 1.
Crystallographic data collection and refinement statistics.a
| Data Collection | |
|---|---|
| Resolution (highest resolution shell) (Å) | ∞-2.31 (2.40–2.31) |
| X-ray Source | CuKα |
| Wavelength (Å) | 1.5418 |
| Space Group | R32 |
| Cell dimension (Å) | a = b = 154.76, c = 121.50 |
| Reflections observed (unique) | 141418 (46701) |
| Completeness (%) | 99.8 (100) |
| Rmerge (%)b | 18.6 (34.3) |
| I/σ (I) | 5.4 (2.3) |
| Redundancy | 5.7 (4.1) |
| Refinement | |
| Protein residues/water atoms per asu | 377/392 |
| Other ligands per asu | 8 |
| Reflections (work/free) | 41624/2114 |
| Rwork/Rfree (%) | 18.9/25.3 |
| Resolution (Å) | 23.61–2.31 |
| Average B-factor (Å2) | 7.1 |
| Protein (Å2) | 7.0 |
| Mn(II) (Å2) | 10.9 |
| Carbonate(Å2) | 3.1 |
| Water (Å2) | 15.8 |
| RMSDc Bond lengths (Å) | 0.006 |
| RMSD Bond angles (°) | 1.009 |
Data for the highest resolution shell is in parenthesis.
Rmerge = Σhkl Σi| Ihkl, i-<Ihkl> I | / ΣhklΣi | Ihkl, i |, where <Ihkl> is the mean intensity of the multiple Ihkl, i observations for symmetry-related reflections.
RMSD: root mean square deviation.
The final model of the T165V OxDC variant, including residues 6-382 in the protomer, 392 waters, one carbonate and two Mn(II) ions, was refined to 2.31 Å resolution with a Rwork of 18.9% and Rfree of 25.3%. Analysis of the Ramachandran plot showed that 94.4% of residues fall in the most favored regions with 4.7% in the additionally allowed and generously allowed regions and 0.9% of residues falling in the disallowed regions. All residues are well-defined with an average B-factor for the protein atoms of 7.0 Å2. The low average B-factor is consistent with that calculated from the Wilson plot. Figures were produced using PyMOL ((DeLano Scientific Software LLC, Palo Alto, CA) or Molscript (20), and were rendered in POVRAY (21).
Spin trapping measurements for the T165V OxDC variant
Samples for spin trapping were prepared by mixing 100 mM α-phenyl-t-butylnitrone (20 μL), 1M aq. potassium oxalate (10 μL) and the OxDC T165V variant (40 μL of a 8 mg/mL solution) dissolved in 1M NaOAc buffer containing 300 mM NaCl, pH 4.1 (final volume 100 μL). The sample was quickly pulled up into a quartz capillary (1 mm ID), which was sealed with Cha-Seal clay (Fisher Scientific, Pittsburgh PA) and transferred into the EPR resonator. Spectra were taken on this sample (25 °C) at a series of time points, and the evolution of the signal determined by simulation of the low-field line and double-integration of the simulated line, as reported previously (7). EPR spectra were recorded using a Bruker Elexsys E580 EPR spectrometer with a high-Q cavity (Bruker ER 4123SHQE). Instrumental parameters were: 100 kHz modulation frequency, 1 G modulation amplitude, 20 mW microwave power, 9.87104 GHz microwave frequency, 81.92 ms time constant, and 163.84 ms conversion time/point. The magnetic field was measured using a calibrated Hall probe.
Measurements of dioxygen consumption by wild type OxDC and the T165V OxDC variant
All dioxygen consumption experiments were performed using the NeoFox sensor system from Ocean Optics (Dunedin, FL) (22). In these experiments, WT OxDC or the T165V OxDC variant (at a final concentration of 1.0–1.4 μM) was dissolved in 50 mM acetate buffer, pH 4.2, containing 0.2% Triton X-100. After equilibration in the sensor system (5–7 min), catalysis was initiated by the addition of potassium oxalate, to give a final concentration of 2–200 mM (total volume 200 μL), and the rate of oxygen consumption was monitored for either 20 min or until the oxalate substrate had been completely consumed.
RESULTS
Steady-state kinetics of the T165S and T165V OxDC variants
C-terminally, His6-tagged T165V and T165S OxDC variants were prepared by overlap extension (10) and purified using metal-affinity chromatography (4, 9). Steady-state kinetic measurements, using an end-point assay for formate production (11, 23), showed that the T165S OxDC variant exhibited similar catalytic activity to wild-type (WT) OxDC when adjusted for the amount of Mn(II) incorporated into the recombinant protein (Table 2) (5). In contrast, removal of the hydrogen–bonding interaction gave an enzyme with impaired catalytic activity. Thus, kcat/KM for the T165V OxDC variant was decreased ~15-fold relative to that of WT OxDC after normalizing for Mn(II) content. Heavy-atom IE and structural studies were therefore undertaken to establish how the loss of the putative active-site hydrogen bonding interaction might have affected the chemical steps prior to irreversible loss of CO2.
Table 2.
Steady-state kinetic parameters for the decarboxylase activity of C-terminally His6-tagged, WT OxDC, and the T165S and T165V OxDC variants.a
| Enzyme | Km (mM) | kcat (s−1) | Mn/monomerb | kcat/Km/Mn (M−1s−1) |
|---|---|---|---|---|
| WT OxDC | 18 ± 2 | 30 ± 1 | 1.4 | 1190 |
| T165S | 9 ± 1 | 20 ± 1 | 0.7 | 3170 |
| T165V | 30 ± 9 | 3.8 ± 0.4 | 1.8 | 70 |
Reaction mixtures consisted of enzyme (4 μg) and potassium oxalate (2.5 – 100 mM) dissolved in 50 mM NaOAc, pH 4.2, containing 0.5 mM o-phenylenediamine and 0.2% Triton-X100 (100 μL total volume). Turnover was initiated by the addition of enzyme. After incubation at 25 °C, the reaction was quenched by the addition of 1.1 M aq. NaOH (10 μL), and the formate quantified using the NADH absorption at 340 nm in an assay mixture containing formate dehydrogenase (23).
Heavy-atom isotope effect measurements
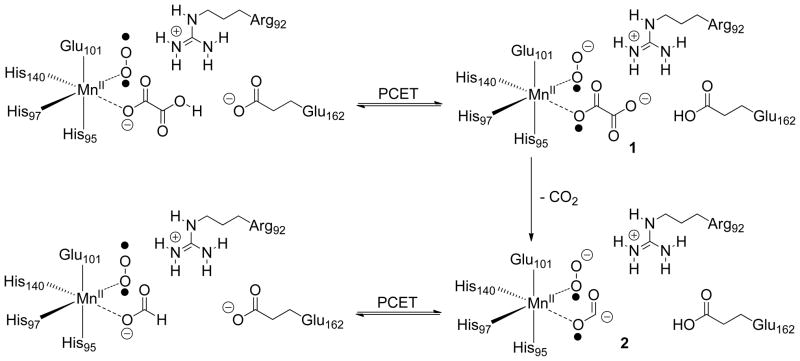
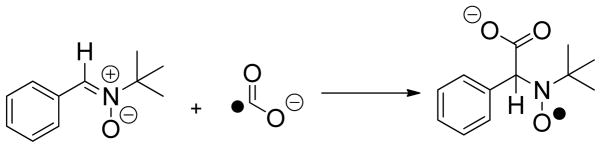
Although many details of the catalytic mechanism remain to be elucidated, heavy-atom isotope effect (IE) measurements support a multi-step process in which Mn(II)-bound, mono-protonated oxalate is activated for decarboxylation by an initial proton-coupled electron transfer (PCET) step (11). The resulting oxalate radical anion 1 then undergoes an irreversible decarboxylation, which takes place via heterolytic cleavage of the C-C bond, to give a new radical anion intermediate 2 that is converted to formate in a second PCET step (Scheme 1). A series of 13C and 18O IE measurements using the T165V OxDC mutant, at both pH 4.2 and 5.7, were performed in order to evaluate the extent to which each (or both) of these two catalytic steps had been affected by removal of the Arg-92/Thr-165 hydrogen bond (Table 3). We note that the primary 13C and secondary 18O isotope effects for the T165V OxDC-catalyzed decarboxylation reaction were measured using the internal competition method (24), and hence these isotope effects represent the effects of isotopic substitution on V/Koxalate. Hence, these values are associated only with steps up to and including loss of CO2, the first irreversible step in the mechanism (Scheme 1).
Scheme 1.
Proposed mechanism for oxalate decarboxylase showing the putative involvement of dioxygen in proton-coupled electron transfer (PCET) (11). Although spin-trapping experiments support radical anion 2 as an intermediate,7 the metal oxidation state(s) and location of the dioxygen binding site during catalytic turnover remain to be experimentally defined.
Table 3.
13C and 18O isotope effects on the decarboxylase reaction catalyzed by C-terminally His6-tagged, WT OxDC and the T165V OxDC mutant.a,b
| 13(V/K), % | 18(V/K),c % | |||
|---|---|---|---|---|
| Enzyme (pH) | CO2 | HCO2− | CO2 | HCO2− |
| WT OxDC (4.2) | 1.005 ± 0.001 | 1.015 ± 0.001 | 0.998 ± 0.002 | 1.011 ± 0.002 |
| T165V (4.2) | 0.998 ± 0.001 | 1.008 ± 0.001 | 0.991 ± 0.001 | 1.004 ± 0.001 |
| WT OxDC (5.7) | 1.008 ± 0.001 | 1.019 ± 0.001 | 0.993 ± 0.002 | 1.010 ± 0.001 |
| T165V (5.7) | 0.997 ± 0.001 | 1.009 ± 0.001 | 0.984 ± 0.001 | 1.006 ± 0.001 |
Partial enzymatic reactions were performed by incubation of the T165V OxDC variant with oxalate (25 mM) and o-phenylenediamine (0.2 mM) at 22°C in either 0.1 M 1,4-bis-(2-hydroxyethyl)piperazine buffer (pH 4.2) or piperazine buffer (pH 5.7) (790 μL total volume).
Kinetic isotope effect values for WT OxDC at both pH 4.2 and 5.7 have been published previously (11) and are included here for ease of comparison.
Value is given per two oxygen atoms.
At pH 5.7, the observed 13(V/K) IE on the formation of CO2 (0.3%) for the T165V OxDC variant is markedly different from that seen in studies using other site-specific mutants (R92K, R270K, E162Q, and E333D) for which this IE is more normal (1.1% – 1.5%) (6) in comparison to WT OxDC (0.8%) (11). Indeed, all of the observed 13(V/K) and 18(V/K) IEs for both cleavage products (CO2 and HCO2−) are decreased for the T165V OxDC variant when compared with the cognate values for WT enzyme. Given that kcat/KM values are smaller and the observed 13(V/K) and 18(V/K) IEs are larger at pH 5.7, external commitments to catalysis that are pH-dependent should be less at pH 5.7 that those measured at the pH optimum (4.2) for the enzyme (11).
In our earlier analysis for WT OxDC, the IEs on both the PCET and the C-C cleavage steps contributed to the observed IEs in product formate, and the 13(V/K) IE in product CO2 was attributed mostly to the decarboxylation step. In the reaction catalyzed by the T165V OxDC variant, 13(V/K) on the formation of CO2, already well masked from an intrinsic value of 3–5% (for rate-limiting C-C bond cleavage reactions) (25) by a commitment when compared with the PCET step in WT OxDC, becomes even smaller and is barely rate limiting. For the T165V-catalyzed reaction, the 13(V/K) IE in product formate (0.9%) is due largely to the PCET step. 18(V/K) values for CO2 production are more inverse for the T165V OxDC variant than for WT enzyme. In general, an inverse 18(V/K) is expected for decarboxylases, reflecting the change in bond order (1.5 to 2.0) as the carboxylate moiety is converted to CO2. WT OXDC and a number of other site-specific mutants exhibit 18(V/K) IEs of −0.2% to −0.7%, similar to decarboxylases for which C-C bond cleavage is rate limiting.
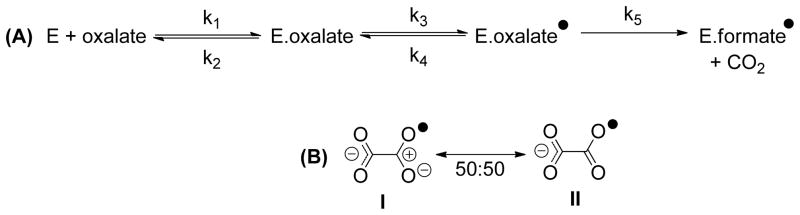
The observed IEs were interpreted using a minimal, two step kinetic mechanism for OxDC-catalyzed decarboxylation (PCET and decarboxylation) leading to the following expression (Eqn. 1) (6, 11):
| (Eqn. 1) |
where x = 13 or 18 for a 13C or 18O IE, respectively, and isotopic substitution is assumed to affect k3, k4, and k5 (Scheme 2A) (11). This quantitative analysis (Supporting information) was accomplished using similar procedures to those already detailed in our previous work on WT OxDC (11). As with other carboxylic acids (26), proton removal from mono-protonated oxalate likely proceeds with a negligible 13C IE, allowing the values of 13k3 and 13Keq3 to be set to unity.3 From the observed IEs at pH 5.7 where the external commitments (k3/k2) are small, Eqn. 1 can be solved by taking an average value of 1.04 of the 13C IE (13k5) for decarboxylation (25, 27). This gives a value of k5/k4 = 12.3 for the T165V OxDC mutant at pH 5.7, which is four-fold greater than that determined previously for WT enzyme (11). Thus, the decarboxylation step was proceeding either with a larger rate constant in the T165V OxDC variant (increased k5) or the “back-reaction” of 1 to regenerate the Michaelis complex was slowed (decreased k4) relative to that of WT enzyme.
Scheme 2.
Minimal kinetic mechanism for the OxDC-catalyzed reaction up to, and including, the first irreversible step.
To calculate the forward 18O isotope effect (18k3) on the PCET step, the 18O isotope effect at the carbon center that becomes CO2 was interpreted using Eqn 1. First, a correction of the observed 18(V/K) by 0.98 was applied for 18O enrichment due to the required protonation of oxalate to the monoanion.4 Making the same estimates for the 18O IE effect on the equilibrium constant (18Keq3) for reversible formation of the intermediate prior to decarboxylation and 18k5 as employed in a previous analysis of data obtained for WT OxDC (1.02 and 0.983, respectively) (11) yielded an expression for determining the value of 18k3 (Eqn. 2).
| (Eqn. 2) |
Solving this equation gives a value of 1.0042 for 18k3, which is substantially smaller than that determined in the reaction catalyzed by WT OxDC (1.0159) (11).
Separate analyses of the 13C and 18O IEs for the T165V-catalyzed reaction in the substrate fragment that becomes formate gave fractionation factor values of 13Keq3 =1.0125 and 18Keq3 =1.0107, in turn correspond to C-O bond orders in the transition state for decarboxylation of 1.27 and 1.25, respectively (Fig. S1, Supporting information) (11). The decreased C-O bond order is due to the oxidative transfer of a single electron from oxalate to a protein-bound acceptor, such as the metal or bound dioxygen. The average value for the C-O bond order of 1.26 implies that the structure of the oxalate radical is a 50:50 hybrid of the two resonance forms I and II (Scheme 2B). This differs from the 70:30 hybrid of I and II that was determined in similar heavy-atom IE experiments on WT OxDC (11) and argues against the idea that the rate constant for decarboxylation rate k5 might be greater for the T165V OxDC variant because the amount of positive charge is lower on the central carbon that is converted to formate. The normal 18O IE on the formation of formate is therefore due to a decreased C-O bond order in the transition state for decarboxylation.
The smaller heavy-atom IEs that are observed at pH 4.2 in the T165V-catalyzed reaction suggest that k3/k2 may no longer be negligible. We therefore used Eqn. 1 to interpret the 18O isotope effect on the substrate fragment that becomes CO2 by substituting the kinetic constants and IEs obtained above to obtain a calculated value of k3/k2 = 3.2 (Supporting information). This large value, however, appears questionable for reasons that are discussed below.
X-ray crystal structure of the T165V OxDC variant
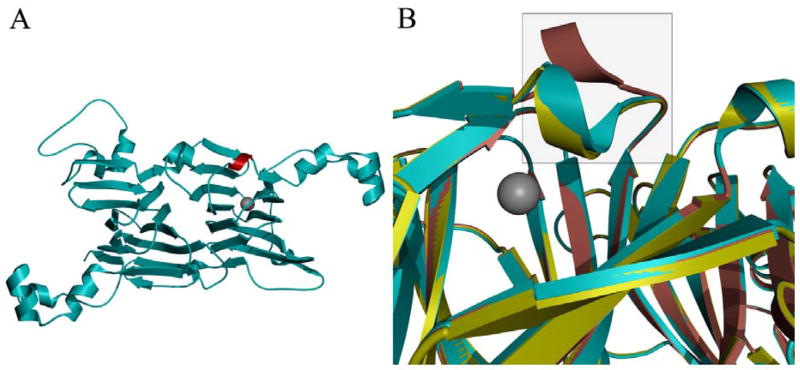
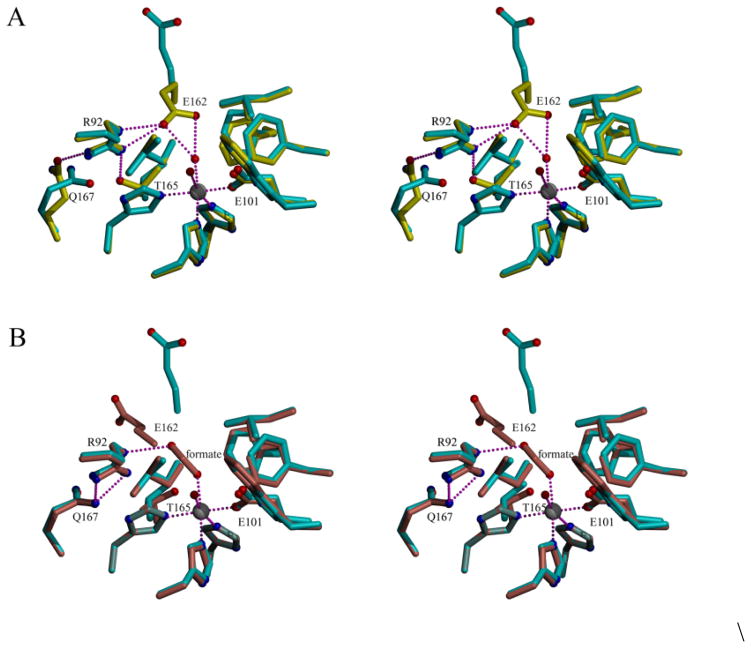
Direct evidence that the conformational preferences of the active-site loop had indeed been modified by removal of the Thr-165/Arg-92 hydrogen bond was obtained by determining the X-ray crystal structure of the T165V OxDC variant at a resolution of 2.31 Å. A sparse-matrix screen search gave conditions (10% w/v polyethylene glycol 6000, 2 M NaCl) that yielded crystals for T165V, which were very similar to those reported previously for WT enzyme (8% polyethylene glycol 8000, 100 mM Tris, pH 8.5). Moreover, both WT and T165V crystals exhibited identical unit cell parameters (Table 1), consistent with the finding that the overall hexameric and protomeric structure of the T165V OxDC variant is similar to that observed for WT enzyme when the SENS loop in the latter adopts either its “open” (rmsd 0.327 Å) (27) or “closed” (rmsd 0.227 Å) (4) forms (Fig. 2). These differences are comparable to that seen when the two structures of WT OxDC are superimposed (rmsd 0.227 Å). In the latter case, rearrangement of the SENS loop segment in the WT enzyme makes only a small local perturbation and there is little change in the relative positions of the two cupin domains. The B-factors for the Mn ions in the X-ray crystal structure were both low (10 Å2; Table 1) suggesting that the metal occupancy of 1.0 used in refinement was correct for both sites. This is also true for both of the WT OxDC structures determined in other laboratories (4, 27). Since the T165V OxDC variant was found to contain 1.8 Mn/monomer (Table 2) it appears that the fully occupied enzyme selectively crystallizes from solution. Perhaps surprisingly, there was no significant displacement of Arg-92 in the active site of the T165V OxDC variant (Fig. 3) even though the original Thr-165/Arg-92 hydrogen bond was absent. Indeed, the Val-165 side chain was superimposable on that of Thr-165 in WT OxDC with the SENS loop in its “closed” conformation (Fig. 3A). In a similar manner, the hydrogen bond network involving Arg-92, Gln-167 and the Tyr-200 backbone carbonyl in the T165V OxDC variant is uninterrupted, and the introduction of the valine side chain causes no significant displacement of the metal cofactor and its surrounding network of hydrogen bonds when compared with WT enzyme (Fig. 4C). However, disruption of the electronic environment by replacement of –OH with –CH3 does cause a displacement of the Glu-162 side-chain away from the active site. The re-positioning of Glu-162 is not, however, the result of the entire SENS loop in the T165V OxDC variant adopting the “open” conformation that is observed in the OxDC/formate complex (28) and the side chain of Glu-162 does not occupy the same location as seen in this structure (Fig. 3B). Indeed, the conformation of the SENS segment in T165V is nearly identical to that seen in the “closed” form of WT OxDC, with a single water molecule replacing the Glu-162 side chain carboxylate. In addition, the accessibility of the active site of the T165V variant in the conformation visualized in the X-ray crystal structure is similar to that of the “closed” form of WT OxDC, with no direct channel between bulk solvent and the active site. This finding has implications for recent spin trapping experiments (Scheme 3), which have provided the first direct evidence for involvement of the formate radical anion 2 in the catalytic mechanism of OxDC (7). In this earlier work, it was proposed that removal of the Arg-92/Thr-165 bond causes the SENS loop in the T165V OxDC variant to adopt a conformation that increases solvent accessibility of the active site, thereby permitting escape of the formate radical anion into solution. However, this suggestion needs to be modified in light of the X-ray crystal structure. Our working model is that the lifetime of radical anion 2 is extended by the inability of the Glu-162 side chain to adopt an active conformation thereby facilitating loss of this species into solution.
Fig. 2.
Ribbon representation of the protomer structure for the T165V OxDC variant (cyan) (A) with the SENS loop highlighted in red and the active site indicated by the location of the Mn(II) cofactor (grey sphere) and (B) overlaid with that of WT OxDC with the SENS loop (enclosed in square) in either the “open” (pink) or “closed” (yellow) conformation.
Fig. 3.
Overlay of the active sites of the T165V OxDC variant (cyan) with that of (A) WT OxDC in the “closed” (yellow) and (B) “open” (pink) conformation bound to formate (ball and stick). In both images, waters are represented by red spheres and Mn(II) is shown as a grey sphere.
Fig. 4.
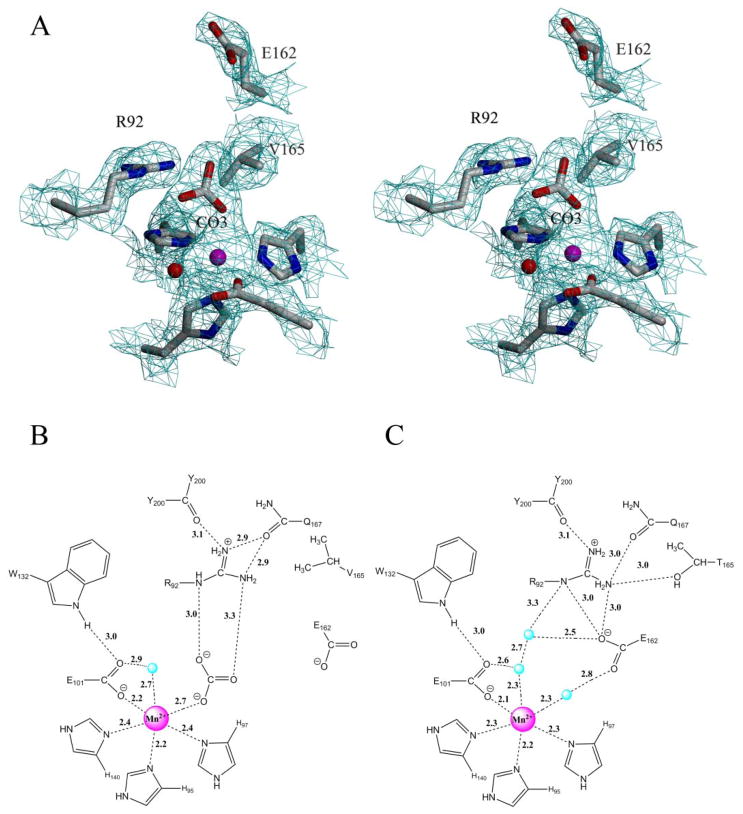
(A) Stereo view of the N-terminal active site of the T165V OxDC variant showing a simulated annealing omit map calculated with coefficients Fo-Fc and contoured at 1 σ (cyan cages). Mn(II) is shown as a magenta sphere and the water molecule as a red sphere. Schematic representations of the N-terminal active site of (B) the T165V OxDC variant with bound CO32− and (C) WT OxDC with the SENS loop in the “closed” conformation (1UW8) (4). In both images, Mn(II) is shown as a magenta sphere, water molecules are shown as blue spheres, and distances are given in Å.
Scheme 3.
The reaction of α-phenyl-t-butylnitrone (PBN) with formate radical anion in spin-trapping experiments.
In a further notable observation, electron density was observed within coordinate bond distance of the Mn(II) ion in the N-terminal domain of the T165V OxDC variant, which could not be refined as water because of the close proximity of the three lobes (Fig. 4A). As a result, and given that the pH of the crystallization medium was 7.5, this density was modeled as a carbonate anion located in the position occupied by formate in a previous X-ray structure of WT OxDC (Fig. 4B) (27). Support for this assignment has been provided by preliminary data from membrane-inlet mass spectrometric measurements (29, 30), which suggest that OxDC is inhibited by bicarbonate at pH 5.2 (Moral et al., unpublished results).
Spin trapping studies and dioxygen consumption by the T165V OxDC variant
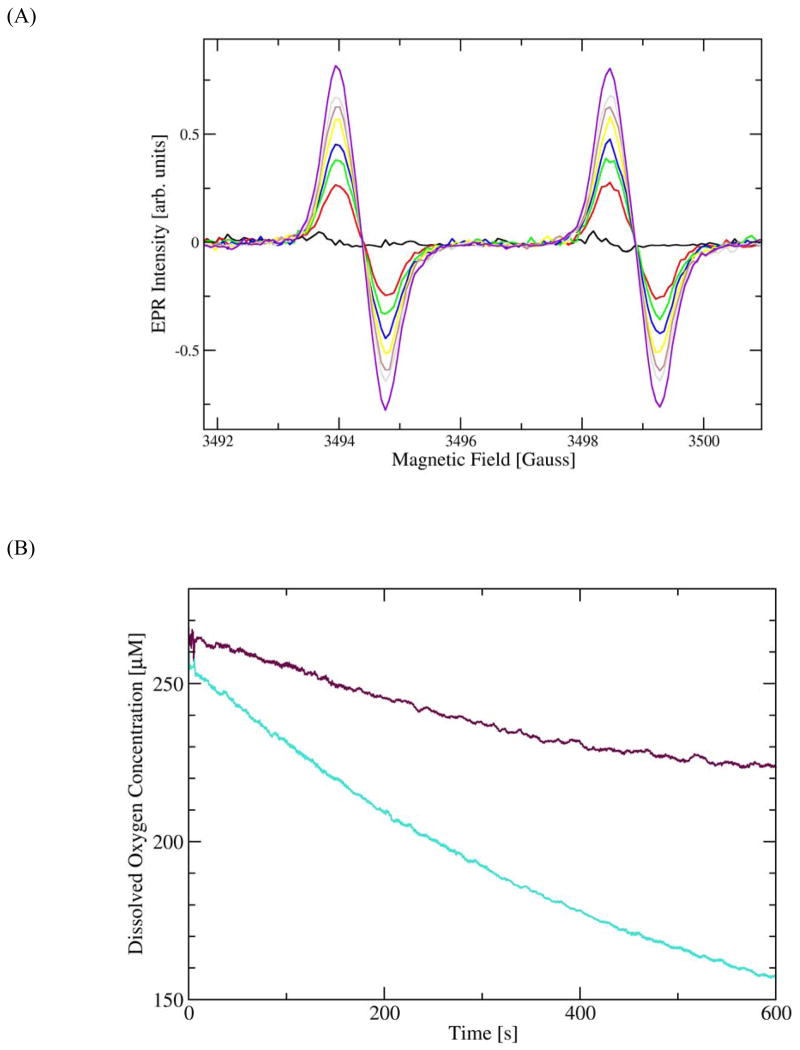
By monitoring the time-dependence of the build-up of the stable radical formed in the spin-trapping reaction (Fig. 5A), we were able to estimate that loss of 2 from the T165V OxDC variant takes place with a rate constant of 1.1 × 10−3 s−1 at pH 4.1 (Supporting information). If dioxygen is participating as a reversible electron sink during catalysis (Scheme 1), however, then loss of the formate radical anion 2 must be accompanied by the release of superoxide anion from the active site if the enzyme is to resume the catalytic cycle. We therefore compared the rate of dioxygen consumption by the WT OxDC and the T165V OxDC variant given that the WT enzyme does not release 2 to any significant extent during catalytic turnover (7). In these experiments, changes in dissolved dioxygen concentration were determined by monitoring the fluorescence lifetime of an immobilized ruthenium complex (475 nm) (22).5 Under our standard conditions (pH 4.1 in NaOAc buffer) WT OxDC and the T165V variant were found to consume dioxygen with rates of 238 nM.s−1 and 99 nM.s−1, respectively when the initial oxalate concentration was saturating (Fig. 5B). Any interpretation of these oxygen consumption rates, however, must reflect the fact that the T165V OxDC variant breaks down oxalate with a turnover number (kcat/Mn = 2.11 s−1) that is approximately 10-fold less than that of WT OxDC (kcat/Mn = 21.4 s−1). For the enzyme concentrations used in these experiments, we therefore computed the level of oxalate consumption per dioxygen molecule, obtaining ratios of 126 and 54 for WT OxDC and the T165V OxDC variant, respectively. Hence, removal of the Arg-92/Thr-165 hydrogen bond does increase dioxygen consumption per oxalate molecule consumed, as expected on the basis of the spin-trapping rates of the formate radical anion.
Fig. 5.
(A) Time-dependence of the EPR signal associated with the radical obtained by reaction of α-phenyl-t-butylnitrone with the formate radical anion generated during catalytic turnover of the T165V OxDC variant. The reaction mixture consisted of enzyme (320 μg), 20 mM PBN, and 100 mM oxalate dissolved in 300 mM NaOAc buffer, pH 4.1, containing 100 mM NaCl. Color code: 105 s, black; 626 s, red; 522 s, green; 712 s, blue; 892 s, yellow; 1079 s, violet; 1265 s, grey; 2090 s, purple. (B) Time-dependence of dioxygen consumption under initial rate conditions for the T165V OxDC variant (purple) and WT OxDC (cyan). The reaction mixtures consisted of WT OxDC or the T165V OxDC variant (at a final concentration of 1.0–1.4 μM) and 100 mM potassium oxalate dissolved in 50 mM acetate buffer, pH 4.2, containing 0.2% Triton X-100.
DISCUSSION
There is general agreement that the initial step of activating oxalate for subsequent C-C bond cleavage involves bidirectional PCET (1, 11, 28) using the nomenclature of Nocera and Stubbe (32, 33). Sequence alignment studies (33) and X-ray crystallography (4) suggest that this PCET step is mediated by the side chain of Glu-162, which is located within a conformationally flexible loop segment. A similar role for an aspartate side chain in “gating” bidirectional PCET has also been proposed for the enzyme ribonucleotide reductase (32). Residues adjacent to the conformationally flexible SENS loop segment form an extensive series of hydrogen bonds (Fig. 1), which likely have functional importance given our demonstration that removal of the interaction between the side chains of Arg-92 and Thr-165 does impact catalytic activity, reducing kcat/KM by an order of magnitude without disrupting metal loading to a significant extent. The X-ray crystal structure of the T165V OxDC variant shows, somewhat unexpectedly, that removing the hydrogen bond does not cause the Arg-92 side chain to adopt a new position within the active site, or perturb the ability of the loop backbone to adopt the “closed” conformation that is observed in WT OxDC. Instead, we observe that the Glu-162 side chain adopts a previously unobserved conformation that incorrectly positions the carboxylate for proton abstraction given the geometrical requirements for PCET (32, 35).
This conclusion is supported by the heavy-atom IE measurements outlined above, which reveal that (i) 18k3 is 3-fold smaller, and (ii) k5/k4 is increased 4-fold for the reaction catalyzed by the T165V OxDC variant when compared to WT OxDC. First, the reduced value of 18k3 shows that the deprotonation step takes place in the T65V variant via an earlier transition state than that employed by the WT enzyme. Second, given that an increase in k5 is unlikely because the amount of positive charge is lower on the central carbon that becomes formate, the larger k5/k4 ratio is a consequence of a decrease in the rate at which the oxalate radical anion intermediate 1 reverts to the Michaelis complex. Both findings can be explained by a model in which removal of the Arg-92/Thr-165 hydrogen bond results in mis-positioning of the Glu-162 side chain. Assuming that Glu-162 only rarely adopts a conformation in which it can abstract a proton from a Mn(II)-bound oxalate monoanion, which would slow the overall turnover of the T165V OxDC mutant, then the neutral side chain will quickly revert to its alternate, mis-positioned conformation after substrate deprotonation, thereby preventing reversal of PCET (reduced k4) and committing the oxalate radical anion 1 to decarboxylation. In addition, some effect on bond orders in the transition state for decarboxylation might be expected if the electrostatic properties of the active site during catalysis were perturbed by mis-positioning the Glu-162 side chain.
When the reaction catalyzed by the T165V OxDC variant was performed at pH 4.2, a substantial external commitment (k3/k2 = 3.2) was calculated, but this assumed that the internal commitment (k5/k4) is unchanged at pH 4.2 and 5.7. The fact that the T165V OxDC variant is an order of magnitude slower than WT OxDC argues, however, against the variant exhibiting an appreciable k3/k2 ratio. We therefore interpret our IE data to mean that k5/k4 is not pH-independent but is larger at pH 4.2. This is most likely the result of k4 being even smaller at pH 4.2 than at pH 5.7. The decreased value of k4 presumably results from an increased tendency at low pH for the Glu-162 side chain to adopt an unproductive conformation for protonation of the oxalate radical anion intermediate.
The nature of the reversible electron sink that must also be present for PCET to take place in OxDC during catalysis remains elusive. Although there is ample precedent for transferring a sigma-bond electron into a d-orbital of an oxidized metal center, as is the case for lipoxygenase (36, 37), the failure of ongoing efforts to observe catalytically competent Mn(III) or Mn(IV) species in WT OxDC (or any of the OxDC variants reported in the literature to date) has stimulated consideration of alternate locations for the electron. In this regard, we note that the tyrosine radical observed during catalytic turnover of the enzyme is not catalytically competent (38) and is therefore not the reversible electron sink required by current mechanistic proposals (1, 11).
Indirect evidence that enzyme-bound dioxygen is the electron sink for PCET is, however, provided by our observation that the T165V OxDC variant consumes more dioxygen than WT OxDC. Spin-trapping experiments clearly show that the leakage rate of formate radical anion 2 into solution is significantly greater in the T165V OxDC variant (7), although the X-ray crystal structure shows that in the conformation observed there is no direct channel between the active site and bulk solvent as is seen in the crystal structure of the “open,” wild-type enzyme (28). Moreover, if the Glu-162 side chain points away from the active site event when the SENS loop adopts its closed conformation, then one would expect the second PCET step (leading to formate production) to be slower, thereby extending the lifetime of formate radical anion 2.
The radical anion 2 can react with dissolved molecular dioxygen when it is in free solution to form CO2 and superoxide, which in turn can rapidly disproportionate to give oxygen and hydrogen peroxide at a rate that is diffusion-controlled at pH 4.1 (39). Consequently, since this process yields hydrogen peroxide, it is difficult to ascertain the amount of dioxygen that is consumed by any oxidase activity in the T165V OxDC variant. Assuming that oxidase activity of the T165V OxDC variant can be neglected gives an upper bound for the apparent first order rate constant of dioxygen consumption due to radical anion leakage of 3.8 × 10−4 s−1. Although any comparison of this rate constant with that observed for formation of the “spin trapped” adduct (1.1 × 10−3 s−1) is complicated by the different enzyme concentrations used in the spin trapping (80 μM) and dioxygen consumption (1.5 μM) experiments (not to mention the unknown level of oxidase activity in the T165V OxDC variant), our finding that the rate constants for dioxygen consumption and formation of the “spin trapped” adduct differ by only approximately 3-fold is intriguing. More specifically, the similarity of these two rate constants implies that dioxygen is not used merely to generate a protein-based radical or oxidized metal center that could then function as a reversible electron sink. Our working hypothesis therefore remains that the role of electron sink is played by enzyme-bound dioxygen, especially given previous studies showing that decarboxylase activity does not take place under anaerobic conditions (40, 41).
Supplementary Material
Acknowledgments
Dr. Stephen Bornemann (John Innes Centre, Norwich, UK) generously provided a plasmid containing the gene encoding His-tagged, WT Bacillus subtilis OxDC. We also acknowledge Dr. Bornemann for giving permission to include Fig. 1 in the paper, which is based on a similar diagram in the Ph.D. thesis of Matthew R. Burrell (John Innes Centre, Norwich, UK). We thank Mario Moral and Dr. David Silverman for providing unpublished preliminary data for OxDC inhibition by bicarbonate, Ms. Larentia Soerianto for help in performing the heavy-atom isotope effect measurements, and Dr. Witcha Imaram for providing unpublished spin trapping data on the T165V OxDC variant.
Footnotes
This work was supported by grants from the National Institutes of Health (DK061666 to N.G.J.R. and GM018938 to W.W.C.) and the National Science Foundation (CHE-0809725 to A.A.).
The coordinates and structure factors for the T165V variant of oxalate decarboxylase have been deposited in the PDB with accession code 3S0M.
Abbreviations: SENS, Ser161-Glu162-Asn163-Ser164; OxDC, oxalate decarboxylase; PCET, proton-coupled electron transfer; OxOx, oxalate oxidase; SEN-DAS, an OxDC/OxOx chimeric variant in which residues Ser161-Glu162-Asn163 are mutated to Asp161-Ala162-Ser163; WT, wild type; QM/MM, quantum mechanical/molecular mechanical; ICP-MS, inductively-coupled plasma mass spectrometry; IRMS, isotope-ratio mass spectrometry; Ni-NTA, nickel-nitrilotriacetic acid agarose; PBN, α-phenyl-t-butylnitrone
Residue numbering throughout the manuscript refers to the oxalate decarboxylase encoded by the YvrK gene in Bacillus subtilis.
The proton is being taken from the end of the substrate that becomes CO2. At this end of the molecule, the bond orders remain 1.5. The electron is removed from the other carboxylate and the fractionation factor at the carbon that becomes product CO2 is NOT significantly affected by changes in bond order at the other end of the substrate (Fig. S1, see supporting information). Therefore it is very unlikely that the equilibrium 13C isotope effect for proton removal from the carboxyl group hydrogen bonded to Glu-162 will differ from that for deprotonation of a normal carboxyl group.
A justification for this assumption is provided in footnote #38 of reference (11).
The presence of high CO2 concentrations in assay mixtures containing OxDC appears to preclude the use of a traditional Clark electrode for monitoring dioxygen consumption under our reaction conditions. This likely reflects a perturbation of the chemical equilibrium operating at the reference electrode due to bicarbonate or carbonate anion formation (31).
Procedures for obtaining the T165S and T165V OxDC variants by overlap extension mutagenesis and primer sequences, and detailed analyses of heavy-atom IE data and spin-trapping kinetics (Figures S1 and S2) are provided as Supporting Information (9 pages). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Burrell MR, Just VJ, Bowater L, Fairhurst SA, Requena L, Lawson DM, Bornemann S. Oxalate decarboxylase and oxalate oxidase activities can be interchanged with a specificity switch of up to 282 000 by mutating an active site lid. Biochemistry. 2007;46:12327–12336. doi: 10.1021/bi700947s. [DOI] [PubMed] [Google Scholar]
- 2.Shimazono H, Haiyashi O. Enzymatic decarboxylation of oxalic acid. J Biol Chem. 1957;227:151–159. [PubMed] [Google Scholar]
- 3.Svedruzic D, Jonsson S, Toyota CG, Reinhardt LA, Ricagno S, Lindqvist Y, Richards NGJ. The enzymes of oxalate metabolism: unexpected structures and mechanisms. Arch Biochem Biophys. 2005;433:176–192. doi: 10.1016/j.abb.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 4.Just VJ, Stevenson CE, Bowater L, Tanner A, Lawson DM, Bornemann S. A closed conformation of Bacillus subtilis oxalate decarboxylase OxdC provides evidence for the true identity of the active site. J Biol Chem. 2004;279:19867–19874. doi: 10.1074/jbc.M313820200. [DOI] [PubMed] [Google Scholar]
- 5.Moomaw EW, Angerhofer A, Moussatche P, Ozarowski A, Garcia-Rubio I, Richards NGJ. The metal-dependence of oxalate decarboxylase activity. Biochemistry. 2009;48:6116–6125. doi: 10.1021/bi801856k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svedruzic D, Liu Y, Reinhardt LA, Wroclawska E, Cleland WW, Richards NGJ. Investigating the functional roles of active site residues in the oxalate decarboxylase from Bacillus subtilis. Arch Biochem Biophys. 2007;464:36–47. doi: 10.1016/j.abb.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imaram W, Saylor BT, Centonze C, Richards NGJ, Angerhofer A. ESR spin trapping of oxalate-derived free radical in the oxalate decarboxylase reaction. Free Rad Biol Med. 2011;50:1009–1015. doi: 10.1016/j.freeradbiomed.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford MM. A rapid and sensitive method for quantiation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Just VJ, Burrell MR, Bowater L, McRobbie I, Stevenson CEM, Lawson DM, Bornemann S. The identity of the active site of oxalate decarboxylase and the importance of the stability of active-site lid conformations. Biochem J. 2007;407:397–406. doi: 10.1042/BJ20070708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain-reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 11.Reinhardt LA, Svedruzic D, Chang CH, Cleland WW, Richards NGJ. Heavy atom isotope effects on the reaction catalyzed by the oxalate decarboxylase from Bacillus subtilis. J Am Chem Soc. 2003;125:1244–1252. doi: 10.1021/ja0286977. [DOI] [PubMed] [Google Scholar]
- 12.Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;62:151–160. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 13.Northrop DB. In: Isotope effects on Enzyme-Catalyzed Reactions. Cleland WW, O’Leary MH, Northrop DB, editors. University Park Press; Baltimore, MD: 1977. pp. 122–148. [Google Scholar]
- 14.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect D – Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 15.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCCP4 suite and current developments. Acta Crystallogr Sect D – Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D – Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, Afonine PV, Bunkoczj G, chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D – Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D – Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK – A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Kraulis PJ. MOLSCRIPT – A program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 21.Fenn TD, Ringe D, Petsko GA. POVScript+. A program for model and data visualization using persistence of vision ray tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- 22.McEvoy AK, McDonagh CM, MacCraith BD. Dissolved oxygen sensor based on fluorescence quenching of oxygen-sensitive ruthenium complexes immobilized in sol gel-derived porous silica coatings. Analyst. 1996;121:785–788. [Google Scholar]
- 23.Schutte H, Flossdorf J, Sahm H, Kula MR. Purification and properties of formaldehyde dehydrogenase and formate dehydrogenase from Candida boidini. Eur J Biochem. 1976;62:152–160. doi: 10.1111/j.1432-1033.1976.tb10108.x. [DOI] [PubMed] [Google Scholar]
- 24.O’Leary MH. Determination of heavy atom isotope effects on enzyme-catalyzed reactions. Meth Enzymol. 1980;64:83–104. doi: 10.1016/s0076-6879(80)64006-3. [DOI] [PubMed] [Google Scholar]
- 25.O’Leary MH. Multiple isotope effects on enzyme-catalyzed reactions. Annu Rev Biochem. 1989;58:377–401. doi: 10.1146/annurev.bi.58.070189.002113. [DOI] [PubMed] [Google Scholar]
- 26.Perrin CL. Secondary equilibrium isotope effects on acidity. Adv Phys Org Chem. 2010;44:123–171. [Google Scholar]
- 27.Edens WA, Urbauer JL, Cleland WW. Determination of the chemical mechanism of malic enzyme by isotope effects. Biochemistry. 1997;36:1141–1144. doi: 10.1021/bi962128s. [DOI] [PubMed] [Google Scholar]
- 28.Anand R, Dorrestein PC, Kinsland C, Begley TP, Ealick SE. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry. 2002;41:7659–7669. doi: 10.1021/bi0200965. [DOI] [PubMed] [Google Scholar]
- 29.Moral MEG, Tu CK, Imaram W, Angerhofer A, Silverman DN, Richards NGJ. Nitric oxide reversibly inhibits Bacillus subtilis oxalate decarboxylase. Chem Commun. 2011;47:3111–3113. doi: 10.1039/c0cc04946h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu CK, Swensen ER, Silverman DN. Membrane inlet for mass spectrometric measurements of nitric oxide. Free Rad Biol Med. 2007;43:1453–1457. doi: 10.1016/j.freeradbiomed.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Clark LC, Wolf R, Granger D, Taylor Z. Continuous recording of blood oxygen tensions by polarography. J Appl Physiol. 1953;6:189–193. doi: 10.1152/jappl.1953.6.3.189. [DOI] [PubMed] [Google Scholar]
- 32.Reece SY, Nocera DG. Proton-coupled electron transfer in biology: Results from synergistic studies in natural and model systems. Annu Rev Biochem. 2009;78:673–699. doi: 10.1146/annurev.biochem.78.080207.092132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the Class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 34.Dunwell JM, Khuri S, Gane JP. Microbial relatives of the seed storage proteins of higher plants: Conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammes-Schiffer S. Theoretical perspectives on proton-coupled electron transfer reactions. Acc Chem Res. 2001;34:273–281. doi: 10.1021/ar9901117. [DOI] [PubMed] [Google Scholar]
- 36.Hatcher E, Soudackov AV, Hammes-Schiffer S. Proton-coupled electron transfer in soybean lipoxygenase: Dynamical behavior and temperature dependence of kinetic isotope effects. J Am Chem Soc. 2007;129:187–196. doi: 10.1021/ja0667211. [DOI] [PubMed] [Google Scholar]
- 37.Rickert KW, Klinman JP. Nature of hydrogen transfer in soybean lipoxygenase1: Separation of primary and secondary isotope effects. Biochemistry. 1999;28:12218–12228. doi: 10.1021/bi990834y. [DOI] [PubMed] [Google Scholar]
- 38.Chang CH, Svedruzic D, Ozarowski A, Walker L, Yeagle G, Britt RD, Angerhofer A, Richards NGJ. Detection of a tyrosyl radical and characterization of the manganese center in the oxalate decarboxylase reaction by electron paramagnetic resonance spectroscopy. J Biol Chem. 2004;279:52840–52849. doi: 10.1074/jbc.M402345200. [DOI] [PubMed] [Google Scholar]
- 39.Bielski BHJ, Allen AO. Mechanism of the disproportionation of superoxide radicals. J Phys Chem. 1977;81:1048–1050. [Google Scholar]
- 40.Tanner A, Bowater L, Fairhurst SA, Bornemann S. Oxalate decarboxylase requires manganese and dioxygen for activity. J Biol Chem. 2001;276:14627–14634. doi: 10.1074/jbc.M107202200. [DOI] [PubMed] [Google Scholar]
- 41.Emiliani E, Bekes P. Enzymatic oxalate decarboxylation in Aspergillus niger. Arch Biochem Biophys. 1964;105:488–493. doi: 10.1016/0003-9861(64)90040-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.