Abstract
Bone loss is caused by the dysregulated activity of osteoclasts which degrade the extracellular bone matrix. The tyrosine kinase Pyk2 is highly expressed in osteoclasts, and mice lacking Pyk2 exhibit an increase in bone mass, in part due to impairment of osteoclast function. Pyk2 is activated by phosphorylation at Y402 following integrin activation, but the mechanisms leading to Pyk2 dephosphorylation are poorly understood. In the current study, we examined the mechanism of action of the dynamin GTPase on Pyk2 dephosphorylation. Our studies reveal a novel mechanism for the interaction of Pyk2 with dynamin, which involves the binding of Pyk2’s FERM domain with dynamin’s plextrin homology domain. In addition, we demonstrate that the dephosphorylation of Pyk2 requires dynamin’s GTPase activity and is mediated by the tyrosine phosphatase PTP-PEST. The dephosphorylation of Pyk2 by dynamin and PTP-PEST may be critical for terminating outside-in integrin signaling, and for stabilizing cytoskeletal reorganization during osteoclast bone resorption.
Keywords: signaling, tyrosine kinase, phosphorylation, phosphatase, osteoclasts
1. Introduction
Osteoclasts (OCs) are bone-resorbing multinuclear cells derived from hematopoietic stem cells that play a key role in bone remodeling (Canalis, 2005, Suda et al., 1997). OCs are activated upon adhesion to the bone surface. This results in cytoskeletal rearrangement and formation of the podosome belt/sealing zone which contains highly dynamic actin-rich structures known as podosomes. The podosome belt is important for cell attachment and for circumscribing the ruffled border membrane through which resorption occurs (Vaananen and Horton, 1995, Novack and Faccio, 2009). However, the proteins and cellular mechanisms that regulate podosome belt formation are not well understood.
Proline-rich tyrosine kinase 2 (Pyk2) and the focal adhesion kinase (FAK) are known to play an essential role in cell adhesion (Parsons, 2003, Richardson and Parsons, 1995). Pyk2 is a multi-domain protein containing an N-terminal FERM domain, a central catalytic core, several proline rich domains (PRD) and a C-terminal focal adhesion targeting (FAT) domain (Avraham et al., 2000, Han et al., 2009, Gil-Henn et al., 2007) (Fig. 1A). The FERM domain is found in a number of proteins and is involved in localizing proteins to the plasma membrane and binding to PIP2 (Chishti et al., 1998, Hamada et al., 2000). In response to integrin engagement and in the presence of intracellular calcium [Ca2+]i, Pyk2 is autophosphorylated at tyrosine residue Y402, which is essential for its activity and for downstream signaling (Mitra et al., 2005, Dikic et al., 1996, Avraham et al., 2000). In OCs, Pyk2 is localized to the podosome belt and deletion of Pyk2 leads to an impairment of OC function, which in part contributes to the osteopetrotic phenotype observed in Pyk2-deficient mice (Gil-Henn et al., 2007, Buckbinder et al., 2007).
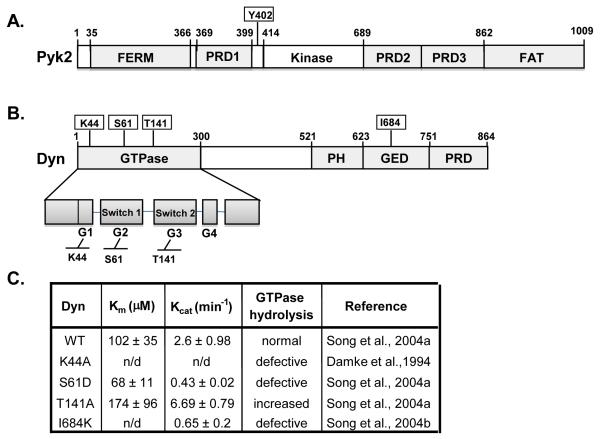
Fig. 1.
Protein structural domains and the effect of point mutations on dynamin’s GTPase activity. (A) Schematic representation of Pyk2 domain structure. Pyk2 contains a central catalytic domain, flanked by non-catalytic N-terminal and C-terminal domains. The C-terminal region has two PRDs and a FAT domain. The Y402 autophosphorylation site is indicated. (B) Schematic representation of dynamin’s domain structure. Dynamin contains a GTPase domain followed by a middle domain, PH, GED and PRD domain. (C) Summary of the effect of dynamin point mutants on dynamin’s Km, catalytic activity and GTP hydrolysis. The reported Km and Kcat values are for human dynamin 1 and are based on the Michaelis-Menten equation (Song et al., 2004b) (n/d; not determined).
Dynamin is a GTP-hydrolyzing enzyme (GTPase) that is involved in membrane fission during clathrin-mediated endocytosis (Schmid et al., 1998). Dynamin has four major domains: an N-terminal GTPase domain, a pleckstrin homology domain (PH) which mediates interaction with membranes lipids, a GTPase effector domain (GED) which is essential for self-assembly and for stimulated GTPase activity, and a C-terminal proline rich domain (PRD) which contains several SH3-binding sites (Hinshaw, 2000, Ahn et al., 2002, Heymann and Hinshaw, 2009). Similar to Pyk2, we found that dynamin localizes to the podosome belt in OCs and we reported that dynamin regulates actin remodeling in the podosomes of OCs (Bruzzaniti et al., 2005, Ochoa et al., 2000). We previously reported that dynamin decreases Pyk2 phosphorylation at Y402 and reduces Src binding to Pyk2 (Bruzzaniti et al., 2009). However, the mechanism of action of dynamin on Pyk2, whether dynamin promotes the dephosphorylation of Pyk2 Y402, or alternatively prevents Pyk2 from being phosphorylated, and the downstream effects on signaling in OCs is unknown. In the current study we demonstrate that Pyk2 associates with dynamin via a novel interaction involving the Pyk2-FERM and dynamin-PH domains. Furthermore, our findings suggest that dynamin acts in a GTPase-dependent manner to regulate Pyk2 dephosphorylation via the tyrosine phosphatase PTP-PEST. The dephosphorylation of Pyk2 is likely to terminate Pyk2 downstream signaling, leading to the reorganization of the actin podosome belt and to a decrease in OC bone resorbing activity.
2. Materials and methods
2.1. Plasmids
Full-length Pyk2 was generated as previously described (Lev et al., 1995, Dikic et al., 1996, Sieg et al., 1998). Pyk2ΔFAT (residues 1-861), Pyk2ΔFERM (residues 367-1009), PRD (residues 690-861), FAT (residues 862-1009) were excised from Pyk2 (wt) and inserted into mammalian vector pEF6/V5-His with in-frame fusion of the V5 epitope at the C-terminal. All Pyk2 cDNA fragments were verified by sequencing and restriction mapping. Dynamin DNA fragments (GTPase-GST, PH-GST, GED-GST, and PRD-GST) were kindly provided by Dr. V. Shah (Minneapolis, MN) and Dr. C.S. Lee (Seoul, S. Korea). A construct of rat dynamin-2 (splice variant aa) (Cao et al., 1998) and the dynamin-2 mutant dynK44A inserted into the pEGFP-N1 vector were generated as previously described (Damke et al., 1994, Altschuler et al., 1998). DynΔPRD was provided by P. De Camilli (Yale University, New Haven, CT). PTP-PEST (wt) and PTP-PEST (R237M) mutant were kindly provided by A. Veillette (Laboratory of Molecular Oncology, Montréal, Québec).
2.2. Preparation of cells
OC precursors were isolated from the bone marrow of 6-8 week old C57Bl/6 mice and differentiated in vitro for 6-8 days with macrophage colony-stimulating factor (M-CSF, 20 ng/ml) and receptor activator of NFκB ligand (RANKL, 80 ng/ml) (Kalliolias et al., 2010).
293VnR cells (HEK293; human embryonic kidney cells stably overexpressing the avβ3 or vitronectin receptor) were previously described and validated (Bruzzaniti et al., 2005). 293VnR cells were transfected using FuGENE 6 according to manufacturer’s protocols (Roche Diagnostics, Indianapolis, IN). For most experiments, a ratio of 2 μg Pyk2 to 1 μg of dynamin was used for these experiments, unless otherwise indicated.
2.3. Immunoprecipitation and GST pull-down assays
OC cultures or transfected 293VnR cells were serum starved for 24 h, washed with phosphate buffered saline (PBS) and lysed in modified RIPA (mRIPA) buffer (50 mM Tris-HCl, pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% NP40, 1% sodium deoxycholate, 0.1% SDS, 50 mM NaF, 1% aprotinin and 0.1mM Na3VO4). Immunoprecipitations (IPs) and Western blotting were performed as previously published (Bruzzaniti et al., 2005). Protein blots were quantified by densitometry analysis using ImageJ software (http://rsb.info.nih.gov/ij/).
GST pull-down assays using anti-GST antibodies were performed as reported (Izumi et al., 2009, Bachand et al., 2002). Briefly, dynamin fragments (GTPase-GST, PH-GST, GED-GST, and PRD-GST) were expressed in bacterial cells induced with IPTG (1mM) for 4 hrs. Bacterial pellets were resuspended with B-PER Protein Extraction Reagents (Pierce, Rockford, IL). Lysates were sonicated, cleared by centrifugation and resuspended in refolding buffer containing 20 mM HEPES-KOH pH 7.5, 150 mM KCl, 1 mM EDTA, 1mM EGTA, 0.5 mM DTT (Leonard et al., 2005). Anti-GST antibody was added to the refolding mixture and incubated for 2 hrs at 4°C. Protein G agarose beads were added and rocked gently for 1hr at 4°C. Cleared lysates from Pyk2-transfected 293VnR cells were added to the dynamin-GST-beads and incubated for 2 hrs at 4°C. Washing and centrifugation were repeated 4 times before proceeding to SDS-PAGE and Western blotting.
3. Results
3.1. Characterization of the effects of dynamin mutants on Pyk2 phosphorylation
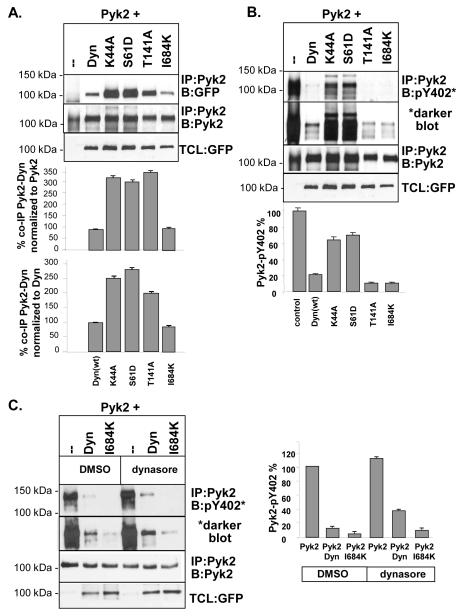
Specific point mutations within dynamin are reported to regulate dynamin’s GTPase activity (Fig. 1C). Therefore, to begin to investigate the mechanism by which dynamin regulates Pyk2 and to determine the role of dynamin’s GTPase activity on Pyk2 Y402 phosphorylation, we used several known dynamin mutants (K44A, S61D, T141A and I684K) which have differential effects on dynamin’s catalytic activity (Fig 1C). Pyk2 was co-expressed in the presence or absence of the dynamin mutants and proteins were immunoprecipitated (IP’d) and subject to SDS-PAGE and Western blotting (Fig. 2A-C). Densitometric analyses were then performed to determine the ability of the dynamin mutants to form a complex with Pyk2, and to regulate Pyk2 Y402 phosphorylation (Fig. 2A-C). We found that Pyk2 co-immunoprecipitated (co-IP’d) with wild-type dynamin (dynWT) as well as all mutants (Fig. 2A). However, dynK44A, dynS61D and dynT141A associated with Pyk2 to a greater extent than dynWT (up to 250-350% increase), whereas the association of Pyk2 with dynI684K was similar to dynWT.
Fig. 2.
Effect of dynamin point mutants on Pyk2 binding and phosphorylation. (A) IPs and Western blotting of Pyk2 co-expressed with dynWT, dynK44A, dynS61D, dyn1T41A or dynI684K in 293VnR cells. The amount of Pyk2 complexed with dynamin mutants was determined by densitometry analysis of IP:Pyk2/B:GFP blots. Results are expressed as a percentage relative to dynWT after normalizing for total Pyk2 in IPs (blot 2) or normalizing for the level of dynamin mutants in TCL blots (blot 3), which gave similar results. As controls, Pyk2 was IP’d with anti-Pyk2 and blotted for Pyk2 and total cell lysates (TCLs) were blotted with GFP to show dynamin expression in cells. (B) IP and densitometry showing the effects of dynamin mutants on Pyk2 phosphorylation. Pyk2 was IP’d and blotted with an antibody to pY402Blots were analyzed by densitometry and normalized for phosphorylated Pyk2 in control IPs (without dynamin). Results are expressed as a percentage of IP’d Pyk2, normalized for total Pyk2 in IPs (blot 3). The expression of dynamin mutants is shown in TCLs blotted with GFP. (C). IP and densitometry showing the effects of the synthetic dynamin inhibitor, dynasore, on Pyk2 phosphorylation in the presence of dynWT or dynI684K. Cells were treated with 90 μM dynasore for 1 hr. Pyk2 was IP’d and blotted with an antibody to pY402 followed by densitometry analysis. Results are expressed as a percentage of IP’d Pyk2. A ratio of 2 μg Pyk2 to 1 μg of dynamin was used for these experiments.
We next determined if Pyk2 phosphorylation was differentially affected by the various dynamin mutants (Fig. 2B). We found that Pyk2 phosphorylation was almost completely abolished in the presence of dynWT, whereas dynK44A and dynS61D only showed a 35-40% decrease. This finding was surprising given that these mutants associated with Pyk2 to a greater extent than dynWT (Fig. 2A). In contrast, Pyk2 phosphorylation was significantly decreased (90% decrease) by dynT141A (Fig. 2B), which was found to strongly associate with Pyk2 (Fig. 2A). Since dynK44A and dynS61D are catalytically-inactive, these findings suggest that Pyk2 dephosphorylation requires dynamin’s catalytic activity. Of further interest, the assembly-defective mutant dynI684K decreased Pyk2 phosphorylation (90% decrease) to a greater extent than dynWT, even though its catalytic activity is reported to be reduced (Fig. 1C). To further examine if dynamin’s GTPase activity was important for its ability to decrease Pyk2 phosphorylation, we performed similar experiments in the presence of the synthetic dynamin GTPase inhibitor, dynasore (Kirchhausen et al., 2008, Nankoe and Sever, 2006). 293VnR cells were transfected with Pyk2 and either dynWT or dynI684K and treated with dynasore for 1 hr. As shown in Fig. 2C, densitometric analyses revealed increased Pyk2 phosphorylation in the presence of dynasore. In addition, dynasore increased Pyk2 phosphorylation in cells expressing Pyk2 and dynWT or dynI684K. This finding was consistent with the increase in Pyk2 phosphorylation we observed in cells co-expressing the GTPase-inactive mutants, dynK444A and dynS61D, compared with dynWT (Fig. 2B). Therefore, our findings indicate that the catalytic activity of dynamin, as well as conformational changes in dynamin, is important for regulating dynamin’s effects on Pyk2 phosphorylation.
3.2. Pyk2-dynamin association involves the Pyk2 FERM and the dynamin PH domains
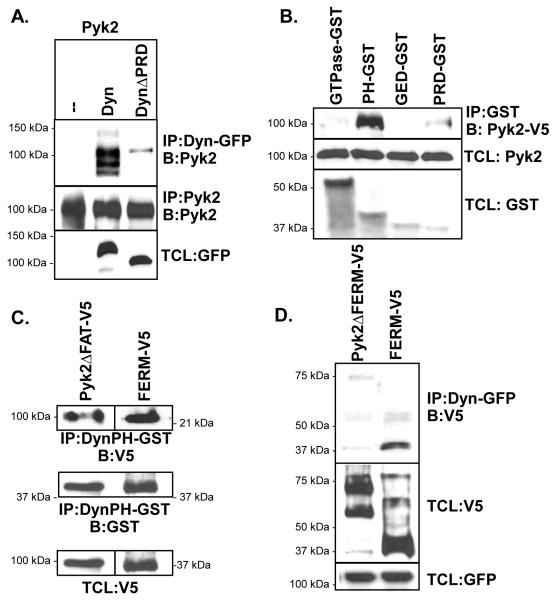
Dynamin and Pyk2 lack obvious complementary motifs through which the direct binding of these proteins may occur. Therefore, to identify the molecular domains involved in the association of dynamin and Pyk2 complexes, we examined the interaction of several dynamin and Pyk2 truncated proteins. First, we compared whether the PRD domains in dynamin or Pyk2 are critical for their mutual association. We found by co-IP that Pyk2 associated with both dynWT and dynΔPRD (lacking the PRD), although its association with dynΔPRD was significantly less than that observed with dynWT (Fig. 3A). This suggested that non-canonical molecular domains were involved in mediating protein complexes. To further elucidate the intramolecular domains involved, we performed GST pull-down assays using truncated dynamin domains. As shown in Fig. 3B, Pyk2 was not detected in pull-down assays using the isolated GTPase or GED domains. However, Pyk2 associated strongly with the dynamin PH domain, while the PRD exhibited reduced association. We next performed GST pull-down experiments to examine which domain of Pyk2 participated in the dynamin-Pyk2 complexes. Our findings revealed that Pyk2ΔFAT and the isolated FERM domain of Pyk2 both associated with the PH domain of dynamin (Fig. 3C). In addition, we found that full-length dynamin strongly co-IP’d with the isolated FERM domain, but not with Pyk2 lacking the FERM domain (Pyk2ΔFERM) (Fig. 3D). Together, these findings indicate that the Pyk2 FERM and dynamin PH domains are likely to be the major domains involved in the association of these proteins.
Fig. 3.
Association of dynamin and Pyk2 domains. (A) IP of dynWT and dynΔPRD, blotted for Pyk2. Proteins were all expressed in 293VnR cells. (B) Bacterially-expressed dynamin domains tagged with GST (GTPase, PH, GED, PRD) were used in GST-pull down assays to capture V5-tagged full-length Pyk2. Proteins were detected with V5 or GST antibodies. (C) Bacterially-expressed dynamin PH domain was used in GST-pull down assays using lysates from 293VnR cells expressing Pyk2ΔFAT or the isolated FERM domain. (D) 293VnR cells expressing full-length dynamin in combination with either Pyk2ΔFERM or the isolated FERM domain were IP’d and blotted. Total cell lysates (TCLs) were blotted as indicated.
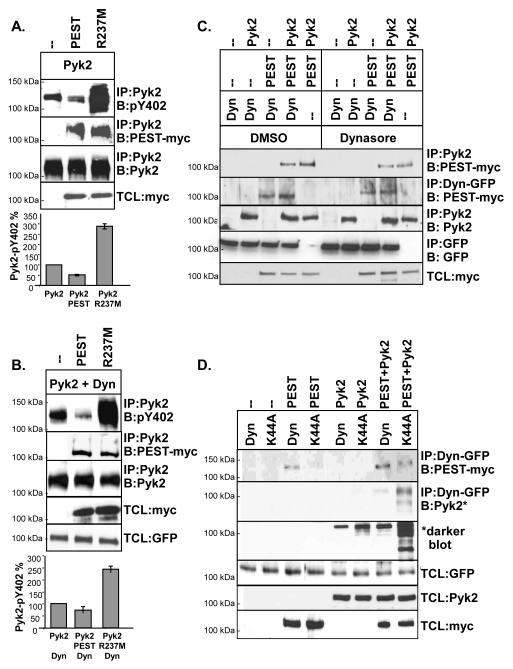
3.3. PTP-PEST regulates the dynamin-induced decrease in Pyk2 phosphorylation
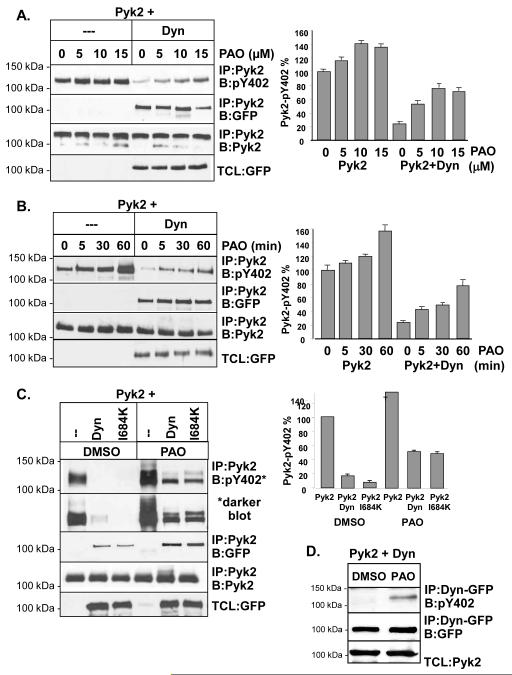
The decrease in Pyk2 phosphorylation that we observed could be the result of a block in the initial autophosphorylation of Pyk2 or the result of Pyk2 dephosphorylation by tyrosine phosphatases. To examine the mechanism of Pyk2 dephosphorylation, Pyk2 was expressed in 293VnR cells in the presence or absence of dynamin. Cells were then treated with a broad spectrum tyrosine phosphatase inhibitor, phenyl arsine oxide (PAO) using increasing inhibitor concentrations and incubation times (Fig. 4A and 4B). Protein complexes were analyzed by Western blotting to detect Pyk2 Y402 phosphorylation, followed by densitometry. Comparable levels of Pyk2 were IP’d as indicated by the control blots (IP:Pyk2, B:Pyk2). These studies revealed that Pyk2 phosphorylation was increased in the presence of PAO in a dose and time-dependent manner (Fig. 4A and 4B, respectively). Although Pyk2 phosphorylation was dramatically decreased by dynWT (Fig 4A and 4B, compare Pyk2 alone and Pyk2+dyn), PAO significantly reversed the inhibitory effect of dynamin on Pyk2 phosphorylation in a concentration and time-dependent manner. These findings therefore suggested that a specific tyrosine phosphatase was involved in the dynamin-induced dephosphorylation of Pyk2. We next examined whether dynamin’s assembly was important in the ability of PAO to restore Pyk2 phosphorylation. We therefore expressed Pyk2 in the presence of either dynWT or dynI684K, and treated cells with the optimal dose of PAO (15 μM for 60 min) (see Fig 4A and 4B). As shown in Fig. 4C, dynWT and dynI684K both induced Pyk2 dephosphorylation, which was partially reversed by PAO. Interestingly, the effect of PAO on Pyk2 dephosphorylation appeared greater for dynI684K than dynWT, again suggesting that the GTPase activity of dynamin as well as its conformation may be important for Pyk2 dephosphorylation, since dynI684K is defective in its ability to hydrolyze GTP (Fig. 1C). Finally, to determine if PAO led to the dephosphorylation of Pyk2 while it was in complex with dynamin, we expressed Pyk2 and in 293VnR cells and then IP’d dynamin and blotted for the presence of phosphorylated Pyk2 Y402. As shown in Fig. 4D, PAO increased the amount of phosphorylated Pyk2 that IP’d with dynamin. Together, our data suggested that a tyrosine phosphatase was likely to be present in the dynamin-Pyk2 complex, leading to Pyk2 dephosphorylation. However, it is possible that some Pyk2 may be dephosphorylated prior to its association with dynamin. the finding that dynWT still IP’d with Pyk2 in the presence of PAO (Fig. 4A and 4B, blot 2) indicated that dephosphorylated Pyk2 remained in complex with dynamin.
Fig. 4.
Tyrosine phosphatases participate in the dephosphorylation of Pyk2 Y402. (A) IP and Western blotting of 293VnR cells expressing Pyk2 in the presence or absence of dynWT and after treatment with PAO or vehicle (dimethyl sulfoxide; DMSO). Cells were treated with increasing concentrations of PAO 0-15 μM PAO for 30 min. As an IP control, membranes were reblotted for Pyk2. TCL were blotted with dynWT (GFP). Bar graph represents densitometry of pY402 expressed as a percentage of total IP’d Pyk2. Experiments were reproduced three times. (B) 293VnR cells expressing Pyk2 and dynWT were treated with PAO (15 μM) for 0-60 min. IP and blotting was performed as indicated. (C) 293VnR cells expressing Pyk2 in the presence of dynWT or dynI684K were treated with 10 μM PAO for 60 min. Proteins were IP’d and blotted as indicated. (D) 293VnR cells expressing Pyk2 and dynWT were treated with DMSO or PAO (10 μM) for 060 min. Dynamin was IP’d and the blot was probed with an antibody to the pY402 site of Pyk2. Blots were stripped and reblotted for dynamin (GFP). A ratio of 1 μg Pyk2 to 1 μg of dynamin was used for experiments examining the effects of PAO.
The tyrosine phosphatase PTP-PEST is essential in regulating both cell spreading and migration by inducing the dephosphorylation of several focal adhesion-associated proteins (Shen et al., 1998, Angers-Loustau et al., 1999). It has also been reported that PTP-PEST specifically dephosphorylates Pyk2 Y402 (Lyons et al., 2001, Sahu et al., 2007) and is inhibited in the presence of PAO or Na3VO4. We therefore investigated the role of PTP-PEST in the dynamin-regulated dephosphorylation of Pyk2. Dynamin and Pyk2 were cotransfected with PTP-PEST (PEST) or an inactive R237M mutant of PTP-PEST (R237M), which is known to act as a dominant negative mutant (Davidson and Veillette, 2001, Shen et al., 1998, Angers-Loustau et al., 1999, Turner, 2000, Lyons et al., 2001). As shown in Fig. 5A, Pyk2 co-IP’d with active PTP-PEST and inactive PEST-R237M. Pyk2 phosphorylation was decreased in the presence of PTP-PEST, but significantly increased by PEST-R237M. A possible explanation for this is that over-expression of inactive PEST-R237M displaces endogenous PTP-PEST or other phosphatases from Pyk2, leading to an overall increase in Pyk2 phosphorylation. Next, Pyk2 was co-expressed with dynamin and either active PTP-PEST or inactive PEST-R237M. Similarly to our earlier findings, PTP-PEST decreased Pyk2 phosphorylation while PEST-R237M significantly rescued Pyk2 Y402 phosphorylation in the presence of dynamin (Fig. 5B). We then examined whether dynamin’s activity was required for the association of Pyk2 and PTP-PEST (Fig. 5C). 293VnR cells expressing various combinations of dynWT, Pyk2 and PTP-PEST were treated with dynasore or vehicle (DMSO) and then subject to IP and Western blotting. As shown in Fig. 5C, dynasore decreased the amount of Pyk2 that was in complex with PTP-PEST (blot 1). We also found that the association of dynamin with PEST also decreased in the presence of dynasore (blot 2), suggesting that dynamin’s GTPase activity was important for the protein complex to form. Next, we used the GTPase-inactive dynK44A mutant to further examine if dynamin’s association with PTP-PEST requires its GTPase activity, or the presence of Pyk2 (Fig. 5D). Indeed, dynamin co-IP’d with PTP-PEST in the presence or absence of Pyk2 (293VnR cells lack endogenous Pyk2). However, we found more PTP-PEST in complex with dynWT than dynK44A, confirming that dynamin’s GTPase activity is important for its association with PTP-PEST. Of interest, co-expression of Pyk2 increased the formation of both the dynamin-PTP-PEST and dynK44A-PTP-PEST complexes, suggesting cooperative binding of these proteins. Finally, we examined if PTP-PEST enhanced the association of dynWT or dynK44A with Pyk2 (Fig. 5D, blot 2 and 3). Although dynK44A associated more strongly with Pyk2 than dynWT (as previously shown in Fig. 1B), PTP-PEST enhanced the formation of Pyk2-dynamin complexes. These findings further suggested a cooperative association, rather than competitive interaction of these proteins. Our findings also suggest that PTP-PEST is involved in the dynamin-induced dephosphorylation of Pyk2. Furthermore, the association of PTP-PEST to dynamin may be regulated by the GTPase activity of dynamin.
Fig. 5.
PTP-PEST promotes Pyk2 Y402 dephosphorylation. (A) IP and Western blotting of 293VnR cells expressing Pyk2 with either PTP-PEST (PEST) or inactive PTP-PEST-R237M (R237M) (myc tag used). Densitometry of blots was performed and the phosphorylation of Pyk2, expressed as a percentage of total Pyk2, is shown. (B) IP and Western blotting of 293VnR cells expressing Pyk2 plus dynWT with either PTP-PEST or PEST-R237M. Experiments were performed as in B. (C) IP and Western blotting of 293VnR cells expressing various combinations of dynWT, PTP-PEST or Pyk2 and treated with 90 uM dynasore for 1 hr. Western blotting and IPs were performed as indicated. Dyn was IP’d with a GFP antibody. (D) 293VnR cells expressing dynWT or dynK44A and either PTP-PEST, Pyk2 or both proteins were analyzed by IP and Western blotting as indicated. Experiments were reproduced 2-3 times.
3.4. Dynamin, Pyk2 and PTP-PEST form a complex in OCs and regulate podosome belt formation
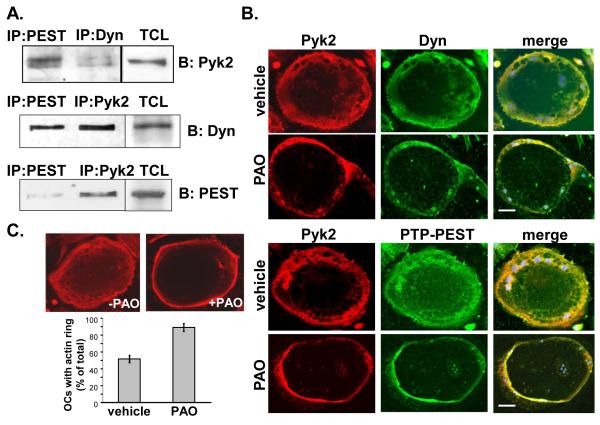
To further elucidate the role of PTP-PEST in the dephosphorylation Pyk2 in OCs, we determined whether these proteins formed a complex in OCs. As shown in Fig. 6A, Pyk2 formed a protein complex with both dynamin and PTP-PEST in OCs. Similarly, PTP-PEST formed a complex with both dynamin and Pyk2, indicating that these three molecules were potentially part of the same molecular complex. In addition, we used microscopic analyses to examine whether podosome belt formation, which is critical for OC function, is affected by inhibition of phosphatase activity. First, we examined if Pyk2 colocalizes with PTP-PEST in the OC podosome belt and whether PAO, which increased Pyk2 phosphorylation in a time and concentration-dependent manner (Fig. 4), affected the cellular localization of these proteins. Consistent with our co-IP studies, Pyk2 colocalized with dynamin as well as with PTP-PEST within the podosome belt in OCs (Fig. 6B). In addition, Pyk2 colocalized with dynamin and PTP-PEST in both untreated and PAO-treated OCs. Of interest, we found that Pyk2, dynamin and PTP-PEST were more concentrated to the cell periphery in PAO-treated cells compared to vehicle-treated cells. We next examined the extent to which PAO affected podosome belt formation in OCs. OCs were treated with or without PAO and the number of cells presenting a peripheral podosome belt were then quantified. As shown in Fig. 6C, in PAO-treated OCs, the podosome belt appeared as a tight band localized to the cell periphery. Furthermore, we found a two-fold increase in the number of OCs containing a peripheral podosome belt following PAO-treatment, suggesting that inhibition of phosphatase activity is likely to decrease OC bone resorption.
Fig. 6.
Dynamin, Pyk2 and PTP-PEST form a complex in OCs. (A) IP of Pyk2, dynamin and PTP-PEST from mature OCs. TCLs were blotted as indicated. (B) Microscopic images of OCs incubated in the presence or absence of PAO (10 μM) for 60 min. Cells were fixed and labeled for Pyk2 (red) and dynamin (green) (upper panels). Alternatively, OCs were labeled for Pyk2 (red) and PTP-PEST (green). Merged images show the colocalization of Pyk2 and dynamin or Pyk2 and PTP-PEST. Scale bar indicates 10 μm. (C) Microscopic analysis of OCs treated with PAO (10 μM) or vehicle for 60 min. OCs were stained with rhodamine phalloidin to detect actin. Bar graph represents the percentage of OCs containing a peripheral podosome belt relative to the total number of OCs present in cultures (>80 OCs counted). Error bars indicate the average of 3 experiments plus SEM.
4. Discussion
OC adhesion and bone resorbing activity requires the assembly of podosomes in the sealing zone/podosome belt which is stabilized by the microtubule network (Gil-Henn et al., 2007). However the signaling proteins involved in the rapid assembly, organization and disassembly of podosomes are not clearly understood. We previously reported that dynamin regulates OC bone resorption in part by decreasing Pyk2 Y402 phosphorylation, which decreases binding to the downstream signaling protein, Src (Bruzzaniti et al., 2009). Our studies now demonstrate the dynamin leads to the dephosphorylation of Pyk2 via recruitment of the tyrosine phosphatase PTP-PEST. Furthermore, we found that dynamin, Pyk2 and PTP-PEST are part of the same molecular complex. In addition, we identified a novel mechanism of interaction of dynamin and Pyk2 which predominantly involves the dynamin PH domain and the Pyk2 FERM domain. Finally, we demonstrated that the inhibition of tyrosine phosphatases stabilizes the podosome belt at the cell periphery, which is likely to significantly affect the bone resorbing activity of OCs.
The phosphotyrosine phosphatase inhibitors PAO and Na3VO4 are both known to inhibit PTP-PEST and we found that PAO (Fig. 4) and Na3VO4 (data not shown) rescued Pyk2 phosphorylation in the presence of dynamin, consistent with the prediction that PTP-PEST is involved in Pyk2 dephosphorylation. In unpublished studies, we also found that Pyk2 co-IPs with PTP-1α, PTP-1β and SHP-2 and that the phosphatase inhibitors PTP-IV (which inhibits SHP-2, PTP1B, PTP-ε, PTP-Meg-2, PTP-σ, PTP-β, and PTP-μ) and okadaic acid led to an increase in Pyk2 phosphorylation. However, the rescue of Pyk2 phosphorylation with these inhibitors was much lower than that results with PAO (data not shown). Therefore, PTP-PEST is likely to be a major phosphatase involved in Pyk2 Y402 dephosphorylation in the presence of dynamin, although other phosphatases may also be involved.
Our studies suggested that dynamin, Pyk2 and PTP-PEST were likely to be present as part of the same molecular complex in OCs and when expressed in 293VnR cells. In addition, the expression of Pyk2 enhanced dynamin’s association with PTP-PEST while over-expression of PTP-PEST enhanced Pyk2’s association with dynamin, suggesting that the association of these molecules within a complex was likely to be cooperative in nature. Furthermore, we found that less dynK44A co-IP’d with PTP-PEST than dynWT, and the expression of PTP-PEST enhanced the association of dynK44A with Pyk2 (Fig. 5D). Consistent with this, we found that in the presence of dynasore or dynK44A, the association of Pyk2 and PTP-PEST was increased, suggesting that dynamin’s GTPase activity regulates the formation of a multi-protein complex. It is possible that the transition of dynamin from GTP-bound to GDP-bound influences the binding of PTP-PEST to dynamin in the complex, which consequently may impact Pyk2 dephosphorylation. It should be noted that dynamin can be tyrosine phosphorylated by Src kinase (Ahn et al., 2002), but the phosphatase involved in dynamin’s dephosphorylation has not yet been determined. Our studies reveal that dynamin forms a complex with PTP-PEST in the presence or absence of Pyk2 (Fig. 5D), indicating that dynamin may be a substrate of PTP-PEST. Together, our findings support the hypothesis that Pyk2 dephosphorylation in the presence of dynamin is mediated by PTP-PEST and that dynamin leads to the dephosphorylation of Pyk2 via PTP-PEST, a step that is likely to occur after Pyk2’s initial autophosphorylation.
The GTPase domain of dynamin contains four distinct structural domains (G1, G2, G3 and G4) that play a major role in binding the guanine nucleotide (GTP) base as well as phosphate/Mg+2 ions (Song et al., 2004a). Residues around G2 and G3 interact with the γ-phosphate of bound GTP, and form the switch 1 and switch 2 regions of the GTPase and are known to alter their conformation upon GTP binding. The critical role of the G1 and G2 domain on dynamin’s GTPase activity was previously demonstrated using the point mutants K44A and S61D (G1 and G2, respectively) (Fig. 1B) (Song et al., 2004a). In addition, these mutants can act as dominant negatives against dynWT in over-expression studies. In contrast, a single point mutation in the G3 domain (T141A) increases dynamin’s catalytic activity. The GTP effector domain (GED) is also reported to bind the GTPase domain and act as an intramolecular GTP-activating protein (GAP), and mutations in this domain significantly decrease dynamin’s catalytic activity (Song et al., 2004b). It was previously reported that dynK44A and dynS61D have decreased GTPase activity, whereas dynT141A has increased GTPase activity compared to dynWT (Damke et al., 1994, Song et al., 2004a, Song et al., 2004b, Soulet et al., 2006, Marks et al., 2001, Lee and De Camilli, 2002) (summarized in Fig 1C). Our findings revealed that GTP hydrolysis is important for the ability of dynamin to promote the dephosphorylation of Pyk2 since dynK44A and dynS61D had only a small effect on Pyk2 dephosphorylation compared to dynWT (Fig. 2). Consistent with this finding, we found that dynT141A, which has increased GTPase activity, associated Pyk2 with the same affinity as dynK44A and dynS61D but reduced Pyk2 phosphorylation by 90% compared to dynWT. Moreover, we found that Pyk2 co-IPs more strongly with dynK44A and dynS61D than dynWT (Fig. 2A. These findings suggested that conformational changes in dynamin’s tertiary structure may contribute to its ability to dephosphorylated Pyk2, perhaps by decreasing the association of dynamin to PTP-PEST (Fig. 5D). In support of this, the crystal structure of the isolated dynamin GTPase domain demonstrates that the G2 and G3 domains, in which the S61D and T141A mutations are located, undergo large conformational changes during their transition from GTP-bound to GDP-bound states (Reubold et al., 2005).
In addition to a basal GTPase activity, the catalytic activity of dynamin can be stimulated by intramolecular assembly and assembly into tetramers, a prerequisite for membrane constriction during endocytosis (Song et al., 2004b, Chappie et al., 2010). The assembly of dynamin is controlled by the GED domain and influences GTP binding and hydrolysis (Damke et al., 1994, Song et al., 2004a, Song et al., 2004b, Soulet et al., 2006, Marks et al., 2001, Lee and De Camilli, 2002). The dynI684K mutant is unable to self-assemble and also exhibits decreased catalytic activity under basal conditions (4-fold lower than dynWT) (Fig. 1C) (Song et al., 2004b, Soulet et al., 2006). We found that dynI684K decreased Pyk2 phosphorylation to a greater extent than dynWT (Fig. 2B and 2C), further supporting the idea that structural changes in dynamin influence the degree the binding to Pyk2 and/or PTP-PEST.
The most commonly reported mechanism for interaction of dynamin with its downstream binding partners involves dynamin’s PRD. Unexpectedly, our experiments demonstrated that the major molecular domain involved in mediating the interaction of dynamin and Pyk2 was not the PRD, but the PH domain. The dynamin PH domain has been reported to be involved in protein-lipid and protein-protein interactions (Shajahan et al., 2004) and mutations in this domain affect the ability of lipids to stimulate dynamin GTPase activity (Achiriloaie et al., 1999, Vallis et al., 1999). In the context of our findings, we speculate that the association of Pyk2 with dynamin may potentially affect the ability of dynamin to be stimulated by lipids. Of clinical interest, mutations in dynamin’s PH domain are associated with dominant intermediate Charcot-Marie-Tooth disease (Zuchner et al., 2005). Moreover, it has been demonstrated that alendronate, a nitrogen-containing bisphosphonate commonly used to treat bone loss associated with osteoporosis, binds dynamin’s PH domain and inhibits dynamin’s catalytic activity (Masaike et al., 2010). These reports further support the idea that the interaction of the dynamin PH domain with Pyk2 potentially inhibits the GTPase activity of dynamin.
Interestingly, our findings demonstrate that Pyk2 interacts with the dynamin PH domain via a unique mechanism involving its N-terminal Pyk2 FERM. The interaction of FERM-domain-containing proteins with PH-domain-containing proteins is not without precedence. For example, the FERM-containing protein FIR can bind directly with the PH domain of the Dbl Rho-GEFs (Kubo et al., 2002). In addition, the activation of Etk is regulated by FAK, which is highly homologous to Pyk2, through the direct association of the Etk-PH and the FAK-FERM domains (Chen et al., 2001). Structural studies have also shown that the FAK FERM domain regulates FAK autophosphorylation when it binds directly to its kinase domain, thus inhibiting access to the activation loop containing Y397 (equivalent to Y402 in Pyk2) (Lietha et al., 2007). Moreover, Pyk2 mutants harboring amino acid changes within the FERM domain have decreased phosphorylation compared to Pyk2, and have reduced capacity to stimulate glioma cell migration (Loftus et al., 2009, Lipinski et al., 2006). Given the amino acid homology between FAK and Pyk2, it is likely that the FERM domain of Pyk2 also regulates its kinase activity by regulating access to Y402.
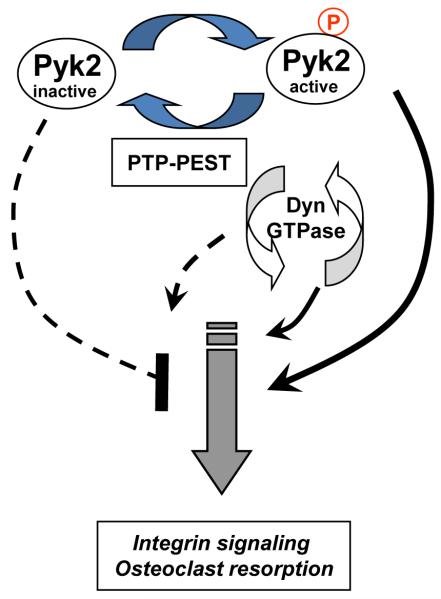
Based on our findings and published data, we propose the following working model (Fig. 7). We propose that integrin activation leads to release of the auto-inhibitory FERM domain, enabling the activation of Pyk2. This results in the autophosphorylation and activation of Pyk2, which promotes OC resorption. Dynamin is also activated and promotes OC resorption. After an appropriate stimulus, PTP-PEST is recruited to dynamin and potentially leads to the dephosphorylation of dynamin, which would in turn decrease its GTPase activity and block OC bone resorption. PTP-PEST also cooperatively enhances dynamin PH domain binding to Pyk2-FERM domain, which permits access to the Y402 site and enables the dephosphorylation of Pyk2 by PTP-PEST. This would subsequently lead to the inactivation of Pyk2 signaling cascades and to the disengagement of integrins. Consistent with cooperative interaction of these proteins, we found that PAO treatment of OCs increased the colocalization of Pyk2, dynamin and PTP-PEST at the cell periphery, where podosome disassembly is likely to occur.
Fig. 7.
Proposed mechanism of interaction of Pyk2, dynamin and PTP-PEST. Pyk2 is activated following integrin engagement which involves release of the auto-inhibitory FERM and the phosphorylation of Y402. Activated Pyk2 and downstream proteins, including dynamin, promote OC resorption. Dynamin potentially recruits PTP-PEST in a GTPase-dependent manner, which subsequently leads to the dephosphorylation of Pyk2 Y402, and inhibits Pyk2 signaling cascades. The cyclic activation/inhibition of Pyk2 and dynamin potentially regulates the attachment, resorption and detachment cycle of OCs.
In conclusion, our data confirm an important role of dynamin GTPase activity in the regulation of Pyk2 phosphorylation. Our studies further demonstrate that dynamin and PTP-PEST control the dephosphorylation of Pyk2 which is critical for the organization of the podosome belt and for OC bone resorbing activity. These studies also raise the possibility that therapeutic strategies that target dynamin’s PH domain or Pyk2’s FERM domain, which are predicted to inhibit bone resorption, may be useful for the treatment of bone loss associated with osteoporosis or related bone diseases.
Acknowledgements
We thank Dr. W. Horne and Dr. R. Baron for important reagents and helpful discussions. We thank Drs S. Huang and H. Largura for technical assistance. We also thank Dr. V. Shah and Dr. C.S. Lee for providing important dynamin contructs, and Dr. Veillette for providing PTP-PEST constructs. This work was funded in part by a Research Support Funds Grant from Indiana University and funds from the Indiana University School of Dentistry. This study was also funded in part by a grant from a Collaborative Research Grant fund of the Office of the Vice President for Research and by a CTSI Collaboration in Biomedical/Translational Research (CBR/CTR) Pilot Program Grant (NIH #RR025761) and by an NIH, R01 grant (#AR060332).
Abbreviations
- PRD
proline rich domain
- WT
wild-type
- PH
plextrin homology domain
- WB
western blot
- IP
immunoprecipitation
Footnotes
Present address for Dr. Shivanna is Indiana University Purdue University Indianapolis, Department of Biology, Center for Regenerative Biology and Medicine, Indiana, USA
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Pierre P. Eleniste, Indiana University School of Dentistry, Department of Oral Biology, Indianapolis, Indiana, USA, peleniste@iupui.edu
Liping Du, Indiana University School of Dentistry, Department of Oral Biology, Indianapolis, Indiana, USA, lipingdu@yahoo.com.
Mahesh Shivanna, Indiana University School of Dentistry, Department of Oral Biology, Indianapolis, Indiana, USA, mshivann@iupui.edu.
Angela Bruzzaniti, Indiana University School of Dentistry, Department of Oral Biology, Indianapolis, Indiana, USA, abruzzan@iupui.edu.
References
- Achiriloaie M, Barylko B, Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol Cell Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Kim J, Lucaveche CL, Reedy MC, Luttrell LM, Lefkowitz RJ, Daaka Y. Src-dependent tyrosine phosphorylation regulates dynamin self-assembly and ligand-induced endocytosis of the epidermal growth factor receptor. J Biol Chem. 2002;277:26642–26651. doi: 10.1074/jbc.M201499200. [DOI] [PubMed] [Google Scholar]
- Altschuler Y, Barbas SM, Terlecky LJ, Tang K, Hardy S, Mostov KE, Schmid SL. Redundant and distinct functions for dynamin-1 and dynamin-2 isoforms. J Cell Biol. 1998;143:1871–1881. doi: 10.1083/jcb.143.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers-Loustau A, Cote JF, Charest A, Dowbenko D, Spencer S, Lasky LA, Tremblay ML. Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J Cell Biol. 1999;144:1019–1031. doi: 10.1083/jcb.144.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham H, Park SY, Schinkmann K, Avraham S. RAFTK/Pyk2-mediated cellular signalling. Cell Signal. 2000;12:123–133. doi: 10.1016/s0898-6568(99)00076-5. [DOI] [PubMed] [Google Scholar]
- Bachand F, Boisvert FM, Cote J, Richard S, Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol Biol Cell. 2002;13:3192–3202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A, Neff L, Sandoval A, Du L, Horne WC, Baron R. Dynamin reduces Pyk2 Y402 phosphorylation and SRC binding in osteoclasts. Mol Cell Biol. 2009;29:3644–3656. doi: 10.1128/MCB.00851-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A, Neff L, Sanjay A, Horne WC, De Camilli P, Baron R. Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol Biol Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckbinder L, Crawford DT, Qi H, Ke HZ, Olson LM, Long KR, Bonnette PC, Baumann AP, Hambor JE, Grasser WA, 3rd, Pan LC, Owen TA, Luzzio MJ, Hulford CA, Gebhard DF, Paralkar VM, Simmons HA, Kath JC, Roberts WG, Smock SL, Guzman-Perez A, Brown TA, Li M. Proline-rich tyrosine kinase 2 regulates osteoprogenitor cells and bone formation, and offers an anabolic treatment approach for osteoporosis. Proc Natl Acad Sci U S A. 2007;104:10619–10624. doi: 10.1073/pnas.0701421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E. The fate of circulating osteoblasts. N Engl J Med. 2005;352:2014–2016. doi: 10.1056/NEJMe058080. [DOI] [PubMed] [Google Scholar]
- Cao H, Garcia F, McNiven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Kim O, Li M, Xiong X, Guan JL, Kung HJ, Chen H, Shimizu Y, Qiu Y. Regulation of the PH-domain-containing tyrosine kinase Etk by focal adhesion kinase through the FERM domain. Nat Cell Biol. 2001;3:439–444. doi: 10.1038/35074500. [DOI] [PubMed] [Google Scholar]
- Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr., Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Hoover KB, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D, Veillette A. PTP-PEST, a scaffold protein tyrosine phosphatase, negatively regulates lymphocyte activation by targeting a unique set of substrates. EMBO J. 2001;20:3414–3426. doi: 10.1093/emboj/20.13.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Tokiwa G, Lev S, Courtneidge SA, Schlessinger J. A role for Pyk2 and Src in linking G-protein-coupled receptors with MAP kinase activation. Nature. 1996;383:547–550. doi: 10.1038/383547a0. [DOI] [PubMed] [Google Scholar]
- Gil-Henn H, Destaing O, Sims NA, Aoki K, Alles N, Neff L, Sanjay A, Bruzzaniti A, De Camilli P, Baron R, Schlessinger J. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2(-/-) mice. J Cell Biol. 2007;178:1053–1064. doi: 10.1083/jcb.200701148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19:4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Mistry A, Chang JS, Cunningham D, Griffor M, Bonnette PC, Wang H, Chrunyk BA, Aspnes GE, Walker DP, Brosius AD, Buckbinder L. Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J Biol Chem. 2009;284:13193–13201. doi: 10.1074/jbc.M809038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JA, Hinshaw JE. Dynamins at a glance. J Cell Sci. 2009;122:3427–3431. doi: 10.1242/jcs.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Matsumoto Y, Ikeuchi T, Saya H, Kajii T, Matsuura S. BubR1 localizes to centrosomes and suppresses centrosome amplification via regulating Plk1 activity in interphase cells. Oncogene. 2009;28:2806–2820. doi: 10.1038/onc.2009.141. [DOI] [PubMed] [Google Scholar]
- Kalliolias GD, Zhao B, Triantafyllopoulou A, Park-Min KH, Ivashkiv LB. Interleukin-27 inhibits human osteoclastogenesis by abrogating RANKL-mediated induction of nuclear factor of activated T cells c1 and suppressing proximal RANK signaling. Arthritis Rheum. 2010;62:402–413. doi: 10.1002/art.27200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77–93. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yamashita T, Yamaguchi A, Sumimoto H, Hosokawa K, Tohyama M. A novel FERM domain including guanine nucleotide exchange factor is involved in Rac signaling and regulates neurite remodeling. J Neurosci. 2002;22:8504–8513. doi: 10.1523/JNEUROSCI.22-19-08504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci U S A. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M, Song BD, Ramachandran R, Schmid SL. Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
- Lev S, Moreno H, Martinez R, Canoll P, Peles E, Musacchio JM, Plowman GD, Rudy B, Schlessinger J. Protein tyrosine kinase PYK2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature. 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Tran NL, Dooley A, Pang YP, Rohl C, Kloss J, Yang Z, McDonough W, Craig D, Berens ME, Loftus JC. Critical role of the FERM domain in Pyk2 stimulated glioma cell migration. Biochem Biophys Res Commun. 2006;349:939–947. doi: 10.1016/j.bbrc.2006.08.134. [DOI] [PubMed] [Google Scholar]
- Loftus JC, Yang Z, Tran NL, Kloss J, Viso C, Berens ME, Lipinski CA. The Pyk2 FERM domain as a target to inhibit glioma migration. Mol Cancer Ther. 2009;8:1505–1514. doi: 10.1158/1535-7163.MCT-08-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons PD, Dunty JM, Schaefer EM, Schaller MD. Inhibition of the catalytic activity of cell adhesion kinase beta by protein-tyrosine phosphatase-PEST-mediated dephosphorylation. J Biol Chem. 2001;276:24422–24431. doi: 10.1074/jbc.M011080200. [DOI] [PubMed] [Google Scholar]
- Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- Masaike Y, Takagi T, Hirota M, Yamada J, Ishihara S, Yung TM, Inoue T, Sawa C, Sagara H, Sakamoto S, Kabe Y, Takahashi Y, Yamaguchi Y, Handa H. Identification of dynamin-2-mediated endocytosis as a new target of osteoporosis drugs, bisphosphonates. Mol Pharmacol. 2010;77:262–269. doi: 10.1124/mol.109.059006. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Nankoe SR, Sever S. Dynasore puts a new spin on dynamin: a surprising dual role during vesicle formation. Trends Cell Biol. 2006;16:607–609. doi: 10.1016/j.tcb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Novack DV, Faccio R. Osteoclast motility: Putting the brakes on bone resorption. Ageing Res Rev. 2009 doi: 10.1016/j.arr.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa GC, Slepnev VI, Neff L, Ringstad N, Takei K, Daniell L, Kim W, Cao H, McNiven M, Baron R, De Camilli P. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Leonard M, Schmid SL, Vallee RB, Kull FJ, Manstein DJ. Crystal structure of the GTPase domain of rat dynamin 1. Proc Natl Acad Sci U S A. 2005;102:13093–13098. doi: 10.1073/pnas.0506491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A, Parsons JT. Signal transduction through integrins: a central role for focal adhesion kinase? Bioessays. 1995;17:229–236. doi: 10.1002/bies.950170309. [DOI] [PubMed] [Google Scholar]
- Sahu SN, Nunez S, Bai G, Gupta A. Interaction of Pyk2 and PTP-PEST with leupaxin in prostate cancer cells. Am J Physiol Cell Physiol. 2007;292:C2288–2296. doi: 10.1152/ajpcell.00503.2006. [DOI] [PubMed] [Google Scholar]
- Schmid SL, McNiven MA, De Camilli P. Dynamin and its partners: a progress report. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- Shajahan AN, Timblin BK, Sandoval R, Tiruppathi C, Malik AB, Minshall RD. Role of Src-induced dynamin-2 phosphorylation in caveolae-mediated endocytosis in endothelial cells. J Biol Chem. 2004;279:20392–20400. doi: 10.1074/jbc.M308710200. [DOI] [PubMed] [Google Scholar]
- Shen Y, Schneider G, Cloutier JF, Veillette A, Schaller MD. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J Biol Chem. 1998;273:6474–6481. doi: 10.1074/jbc.273.11.6474. [DOI] [PubMed] [Google Scholar]
- Sieg DJ, Ilic D, Jones KC, Damsky CH, Hunter T, Schlaepfer DD. Pyk2 and Src-family protein-tyrosine kinases compensate for the loss of FAK in fibronectin-stimulated signaling events but Pyk2 does not fully function to enhance FAK-cell migration. EMBO J. 1998;17:5933–5947. doi: 10.1093/emboj/17.20.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BD, Leonard M, Schmid SL. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J Biol Chem. 2004a;279:40431–40436. doi: 10.1074/jbc.M407007200. [DOI] [PubMed] [Google Scholar]
- Song BD, Yarar D, Schmid SL. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol Biol Cell. 2004b;15:2243–2252. doi: 10.1091/mbc.E04-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet F, Schmid SL, Damke H. Domain requirements for an endocytosis-independent, isoform-specific function of dynamin-2. Exp Cell Res. 2006;312:3539–3545. doi: 10.1016/j.yexcr.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Suda T, Nakamura I, Jimi E, Takahashi N. Regulation of osteoclast function. J Bone Miner Res. 1997;12:869–879. doi: 10.1359/jbmr.1997.12.6.869. [DOI] [PubMed] [Google Scholar]
- Turner CE. Paxillin interactions. J Cell Sci. 2000;113(Pt 23):4139–4140. doi: 10.1242/jcs.113.23.4139. [DOI] [PubMed] [Google Scholar]
- Vaananen HK, Horton M. The osteoclast clear zone is a specialized cell-extracellular matrix adhesion structure. J Cell Sci. 1995;108(Pt 8):2729–2732. doi: 10.1242/jcs.108.8.2729. [DOI] [PubMed] [Google Scholar]
- Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, Merory J, Oliveira SA, Speer MC, Stenger JE, Walizada G, Zhu D, Pericak-Vance MA, Nicholson G, Timmerman V, Vance JM. Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet. 2005;37:289–294. doi: 10.1038/ng1514. [DOI] [PubMed] [Google Scholar]