Abstract
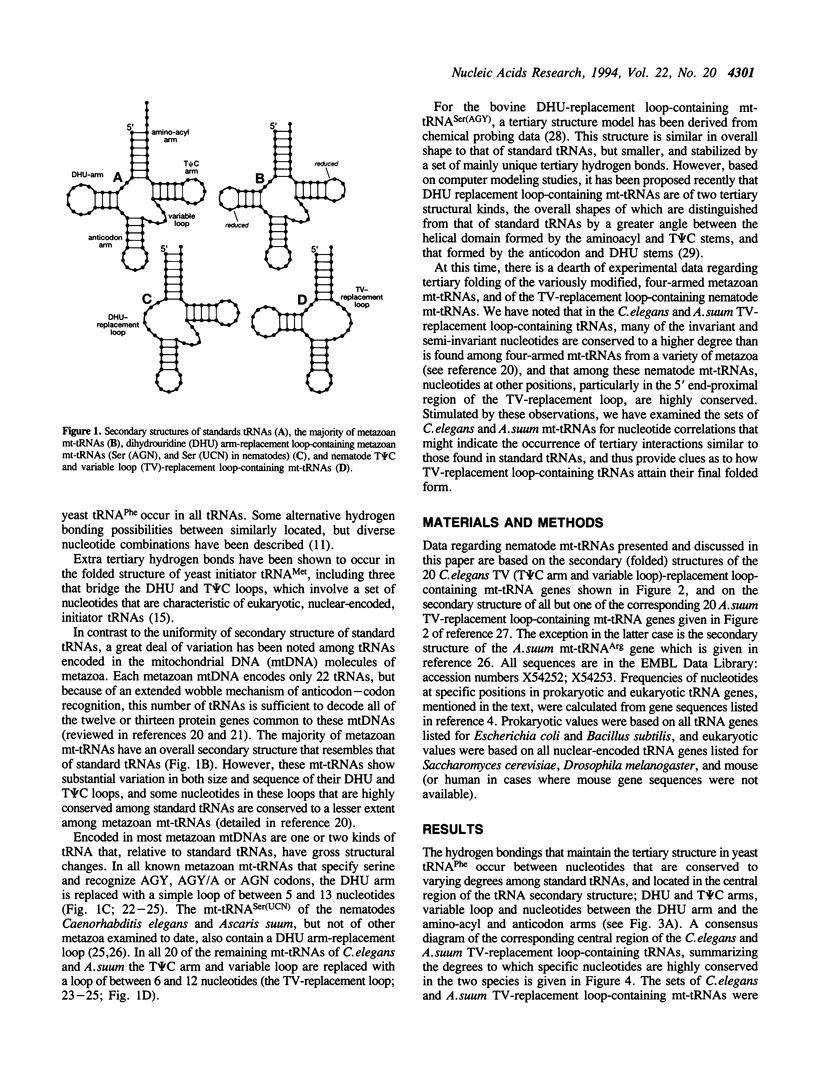
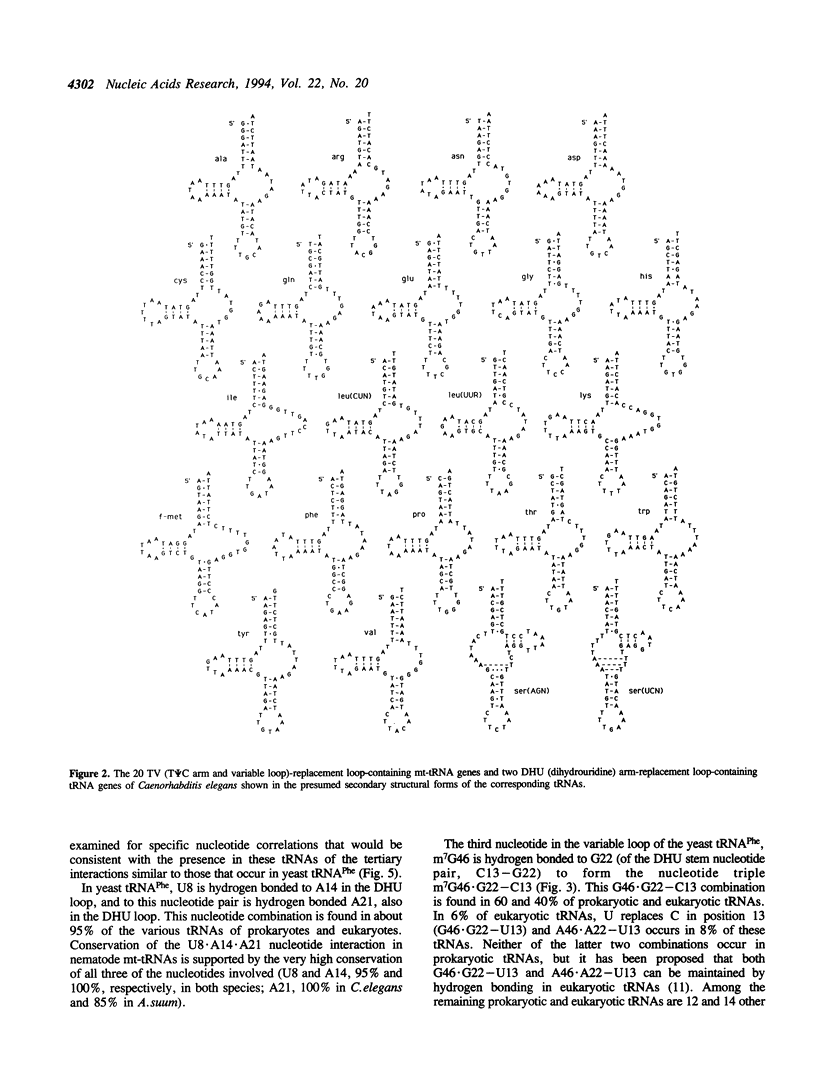
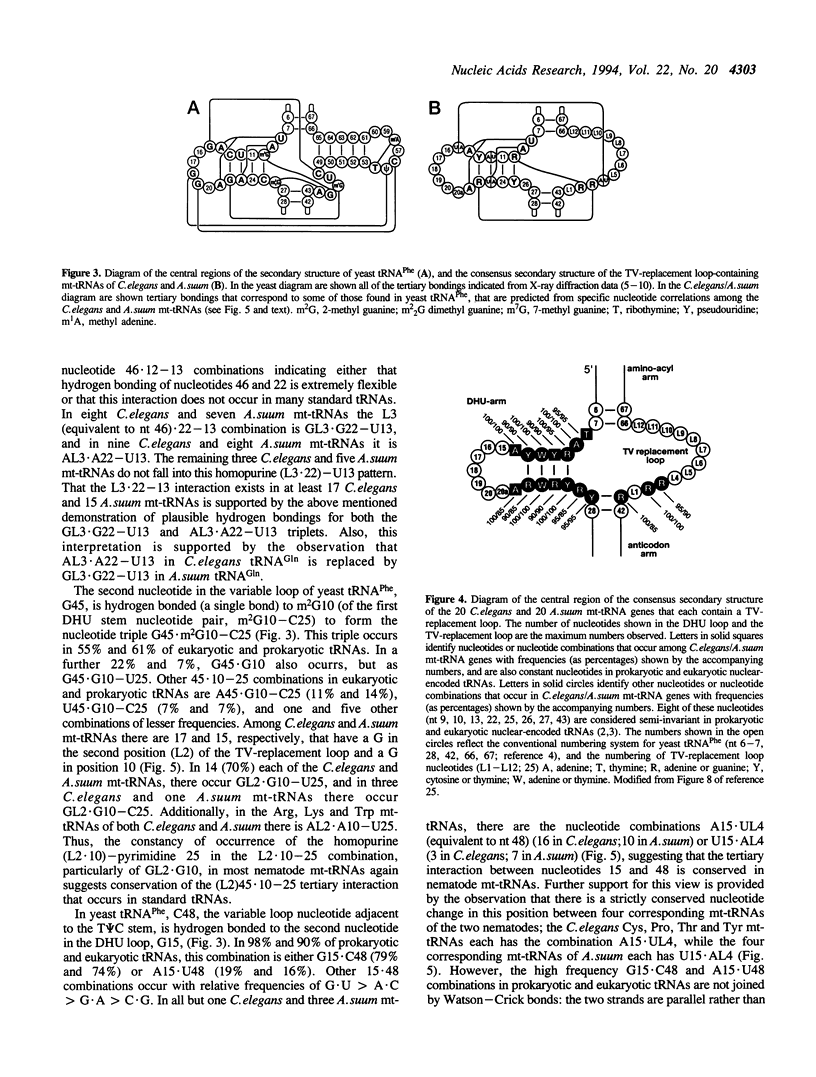
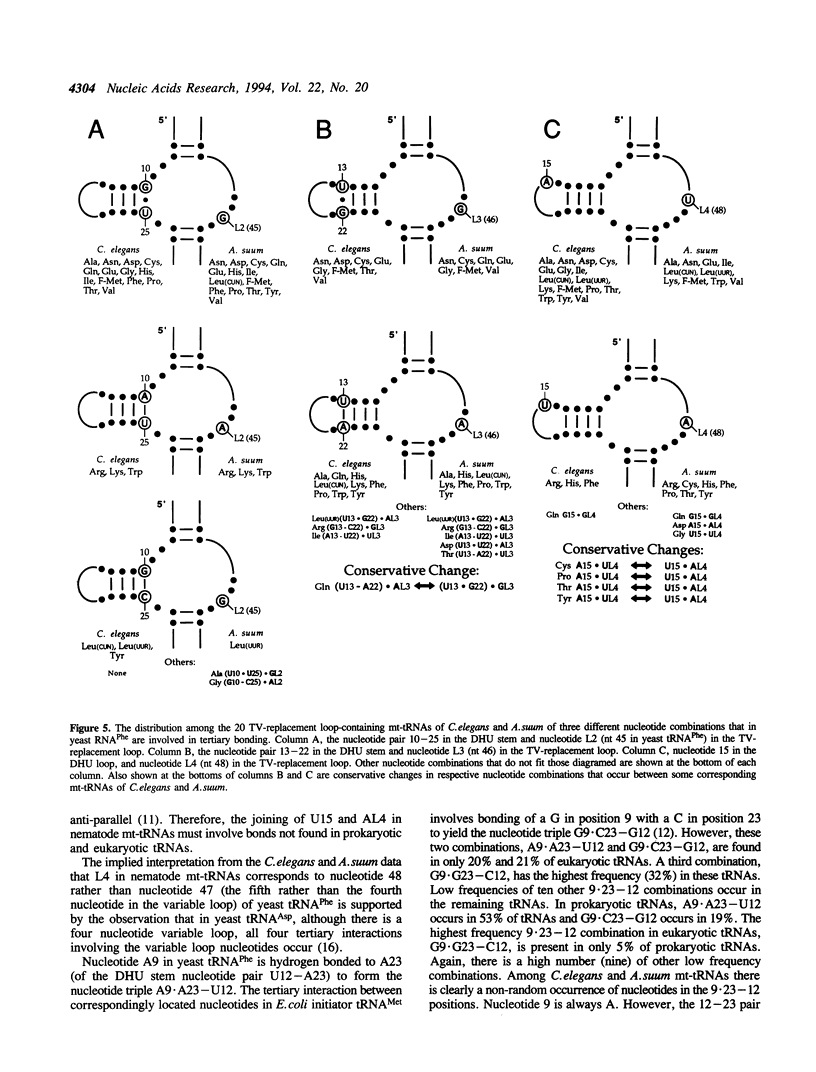
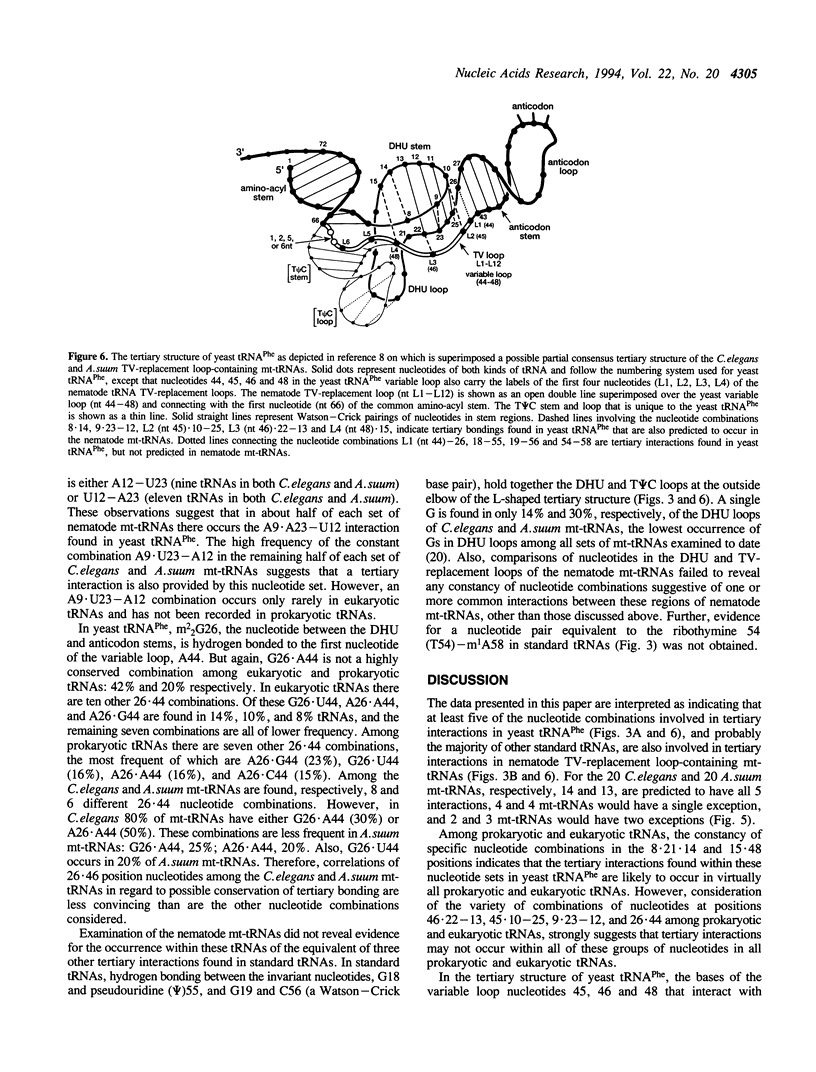
In the predicted secondary structures of 20 of the 22 tRNAs encoded in mitochondrial DNA (mtDNA) molecules of the nematodes, Caenorhabditis elegans and Ascaris suum, the T psi C arm and variable loop are replaced with a loop of 6 to 12 nucleotides: the TV-replacement loop. From considerations of patterns of nucleotide correlations in the central regions of these tRNAs, it seems highly likely that tertiary interactions occur within five sets of binary and ternary combinations of nucleotides that correspond in location to nucleotides known to be involved in tertiary interactions in yeast tRNA(Phe) and other standard tRNAs. These observations are consistent with the nematode TV-replacement loop-containing mt-tRNAs being folded into a similar L-shaped functional form to that demonstrated for standard tRNAs, and for the bovine DHU (dihydrouridine) arm replacement-loop-containing mt-tRNA(Ser(AGY)). However, the apparent occurrence in nematode mt-tRNAs of tertiary bonds common to standard tRNAs contrasts with the situation in bovine mt-tRNA(Ser(AGY)) where the functional form is dependent on an almost unique set of tertiary interactions. Because three of the proposed conserved tertiary interactions in the nematode mt-tRNAs involve nucleotides that occur in the variable loop in standard tRNAs, it seems more likely that in nematode mt-tRNAs it is the T psi C arm rather than the variable loop that has undergone the greatest proportional decrease in nucleotide number.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcari P., Brownlee G. G. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980 Nov 25;8(22):5207–5212. doi: 10.1093/nar/8.22.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R., Sigler P. B. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991 Oct;10(10):3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P., Roberti M., Rainaldi G., Gadaleta M. N., Saccone C. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. J Biol Chem. 1989 Jul 5;264(19):10965–10975. [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Garey J. R., Wolstenholme D. R. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA(serAGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 1989 May;28(5):374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Klug A., Ladner J., Robertus J. D. The structural geometry of co-ordinated base changes in transfer RNA. J Mol Biol. 1974 Nov 5;89(3):511–516. doi: 10.1016/0022-2836(74)90480-x. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Okimoto R., Macfarlane J. L., Clary D. O., Wolstenholme D. R. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics. 1992 Mar;130(3):471–498. doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto R., Wolstenholme D. R. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm: towards a minimal tRNA adaptor. EMBO J. 1990 Oct;9(10):3405–3411. doi: 10.1002/j.1460-2075.1990.tb07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Singhal R. P., Fallis P. A. Structure, function, and evolution of transfer RNAs (with appendix giving complete sequences of 178 tRNAs). Prog Nucleic Acid Res Mol Biol. 1979;23:227–290. doi: 10.1016/s0079-6603(08)60135-x. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Gautheret D., Cedergren R. Fitting the structurally diverse animal mitochondrial tRNAs(Ser) to common three-dimensional constraints. J Mol Biol. 1994 Mar 4;236(4):982–989. doi: 10.1016/0022-2836(94)90004-3. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R. Genetic novelties in mitochondrial genomes of multicellular animals. Curr Opin Genet Dev. 1992 Dec;2(6):918–925. doi: 10.1016/s0959-437x(05)80116-9. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R., Macfarlane J. L., Okimoto R., Clary D. O., Wahleithner J. A. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1324–1328. doi: 10.1073/pnas.84.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- de Bruijn M. H., Klug A. A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire 'dihydrouridine' loop and stem. EMBO J. 1983;2(8):1309–1321. doi: 10.1002/j.1460-2075.1983.tb01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M. H., Schreier P. H., Eperon I. C., Barrell B. G., Chen E. Y., Armstrong P. W., Wong J. F., Roe B. A. A mammalian mitochondrial serine transfer RNA lacking the "dihydrouridine" loop and stem. Nucleic Acids Res. 1980 Nov 25;8(22):5213–5222. doi: 10.1093/nar/8.22.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]



