Abstract
A key aspect of the reaction mechanism of type IB topoisomerases is the controlled unwinding of DNA supercoils while the enzyme is transiently bound to one strand of the DNA duplex via a phosphotyrosyl linkage. In this complex, the mobile segment of the bound DNA downstream from the site of cleavage must rotate around the helical axis, requiring that interactions with the enzyme must break and reform multiple times during the course of removing supercoils. A crystal structure of variola virus type IB topoisomerase (vTopo) bound to DNA shows several positively charged side chains that interact with the downstream mobile and upstream rigid segments, suggesting that these groups may play a role in catalysis, including the processive unwinding of supercoils. We have mutated three such residues, R67, K35 and K271 to Ala and Glu and determined the energetic effects of these mutations at each point along the reaction coordinate of vTopo. R67 interacts with a phosphate group in the rigid DNA segment across from the site of DNA strand cleavage. The ~30-fold damaging effects of the R67A and R67E mutations were primarily on the phosphoryl transfer step, with little effect on enzyme-DNA binding, or the processivity of supercoil unwinding. Removal of the K35 interaction shows similar mutational effects as R67, even though this residue interacts with the mobile segment three base pairs away from the cleavage site. The two mutations of K271, which interacts with mobile region even further from the site of covalent linkage, show significant effects not only on phosphoryl transfer, but also downstream DNA strand positioning. Moreover, supercoil unwinding measurements indicate that the K271A and K271E mutations increase the average number of supercoils that are removed during the lifetime of the covalent complex, enhancing the processivity of supercoil unwinding. These measurements support the proposal that the processivity of supercoil unwinding can be regulated by electrostatic interactions between the enzyme and the mobile DNA phosphate backbone.
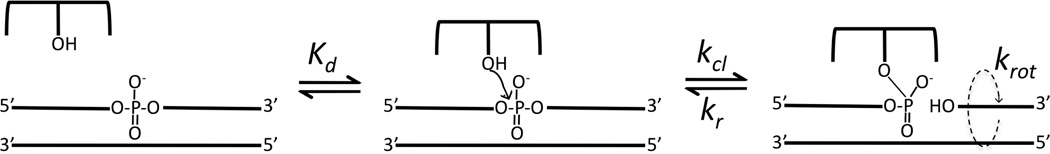
Many essential transactions involving genomic DNA, such as transcription and replication, result in the accumulation of either negative or positive DNA supercoils that represent high-energy topological forms of duplex DNA that must be enzymatically removed to allow progression of replication or transcription forks (1, 2, 3). In addition, a steady-state level of negative supercoiling is a critical feature of genomic DNA that drives local spontaneous unwinding of the DNA strands to allow access of proteins. In higher eukaryotes and mammals the accumulation of these high-energy topological states is regulated type IB topoisomerases (1, 3). These enzymes share a catalytic mechanism that involves the attack of an active site tyrosine nucleophile at a single phosphodiester linkage in duplex DNA to generate a reversible nick in one DNA strand, forming a swivel point for removal of positive or negative supercoils (Scheme 1).
Scheme 1.
The process of supercoil unwinding may be usefully viewed as kinetic competition between strand ligation (kr, Scheme 1), which locks in the number of supercoils, and rotation of the downstream segment (krot) (4–7). Thus, the number of supercoils that may be removed during the lifetime of the covalent complex is given by the ratio krot (in cycles per second)/kr (s−1). The transient formation of a covalent attachment point with the DNA provides a nicely engineered environment for controlling the release of supercoils because the enzyme remains in firm contact with the immobile region of the DNA, and can form tuned interactions with the mobile region downstream of the nick so as to restrict the number of supercoils that are removed during a cleavage event. Both bulk solution and single molecule experiments have established that type IB topoisomerases remove multiple (~5–14) supercoils during the lifetime of the covalent complex (5, 6). A key question is how do these enzymes regulate krot such that strand religation (kr) is competitive with rotation? This is a significant question because genomic DNA cannot exist with a steady-state negative superhelical density if the rotation rate vastly outpaces the ability of the enzyme to lock in DNA supercoils by ligating the cleaved strand.
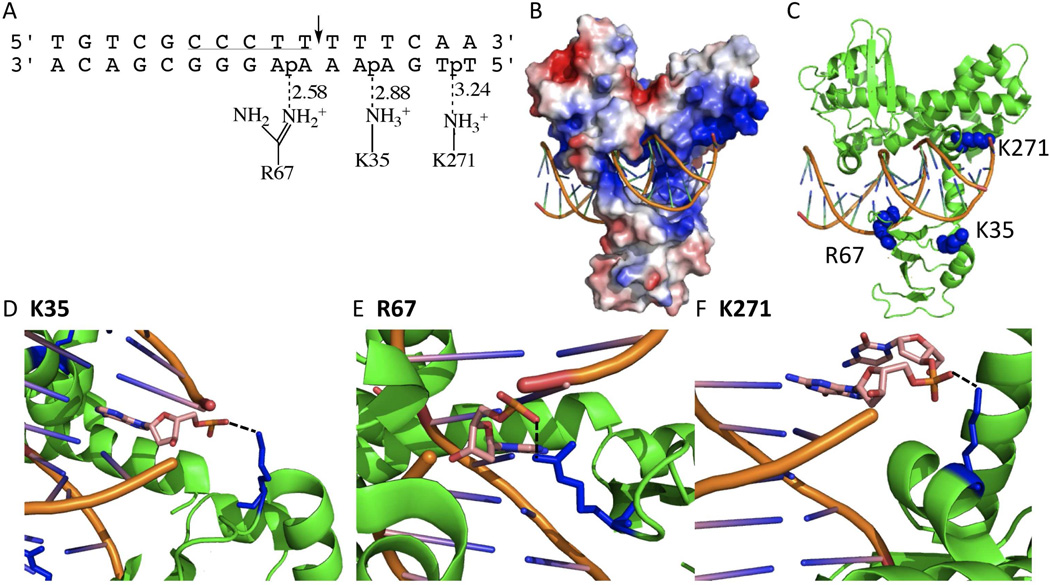
A very extensively studied type IB topoisomerase is that from vaccinia or the closely related variola virus (4). This enzyme has been a useful model for understanding mechanistic aspects of type IB enzymes because of its favorable properties that includes a strong preference for site-specific cleavage at 5′-CCCTT↓X-3′ pentapyrimidine sequences in duplex DNA (Fig. 1A) (8, 9). A recent crystal structure of this enzyme in complex with DNA containing a covalent vanadate transition state mimic is of special interest because it reveals enzyme side chains that may form functionally relevant interactions with the downstream mobile region of the DNA after strand cleavage (Fig. 1B, PDB ID 3IGC) (10). Two conserved charged residues, K35 from the amino terminal domain and K271 from the C-terminal domain, are observed to interact with phosphates of the uncleaved strand three and six base pairs downstream from the cleavage site, respectively (Fig. 1B). A third charged group, R67, interacts with a phosphate group of the uncleaved strand directly across from the cleavage site. With respect to strand rotation kinetics, R67 would be expected to have a distinct functional role from the two downstream groups because it is located within the rigid DNA region of the covalent adduct.
Figure 1.
Variola topoisomerase-DNA interactions (PDB ID 3IGC). (A) Schematic of K35, R67, and K271 interactions with duplex DNA. Dashed lines indicate hydrogen bonds with the distance in angstroms. The vTopo DNA recognition sequence is underlined, and cleavage occurs at the phosphate indicated by the arrow. (B) Global electrostatic map of vTopo complexed with DNA, red and blue indicates negative and postive electrostatic potential, respectively. DNA is shown in stick mode (orange). (C) Same structure from (B) with the mutated residues shown as blue spheres and the protein and DNA shown in green and orange. (C) Interaction of Lys35 with the +3 phosphate on the uncleaved strand, (D) Interaction of Ag67 with the −1 phosphate on the uncleaved strand, (C) interaction of Lys271 with the +6 phosphate on the uncleaved strand. Panels (B–F) were produced using PyMol (17).
Here we examine the functional roles of K35, K271 and R67 at each discrete step of the topoisomerase reaction coordinate using DNA oligonucleotides and a supercoiled plasmid substrate. The functional outcomes of deletion and charge reversal mutagenesis of these side chains indicate that K35 and R67 are important for the chemical steps of cleavage and religation, but not for DNA binding or controlling supercoil unwinding. In contrast, deletion and charge reversal of K271 has a large effect on DNA binding, cleavage, and the downstream strand interactions important for regulating supercoil unwinding. Accordingly, supercoil unwinding measurements indicate that K271E removes significantly more supercoils for each cleavage event than the wild-type enzyme.
Materials and Methods1
Enzymes
The wild-type variola topoisomerase was overexpressed in T7 Express cells that were transformed with the pET21-Topo expression plasmid. The protein was purified using phosphocellulose chromatography as previously described (11). The mutants were created using the QuikChange II Mutagenesis Kit from Stratagene (La Jolla, CA). The mutations were confirmed on both strands by sequencing, and the expressed proteins were purified in a manner similar to wild-type.
DNA Substrates
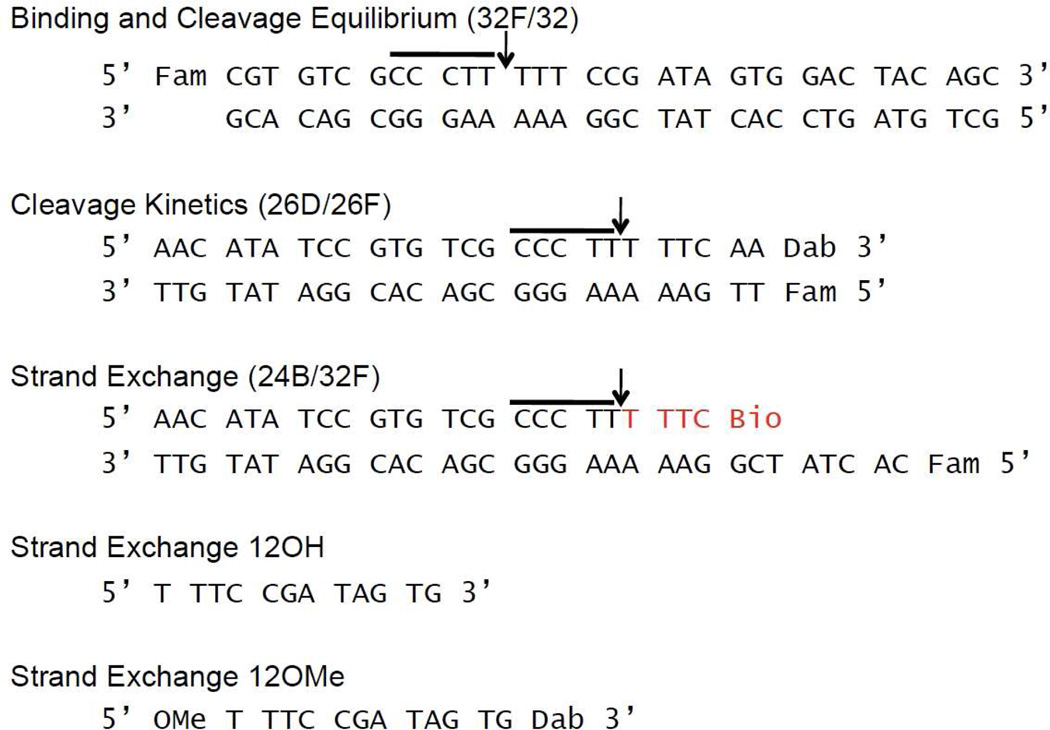
The sequences for the DNA oligonucleotide substrates are shown in Figure 2, where Fam is 6-carboxy-fluoroscein, Dab is 3’-dabcyl-dT, and Bio is biotin connected to the oligonucleotide with a 15-atom tetra-ethyleneglycol spacer. There were two forms of the 12mer used in the assays. One contained a 5’-O-methyl group and a 3’-dabcyl-dT modification (12OMe); the second 12mer had an identical sequence but with no end modifications (12OH). The doubly substituted 12mer and the Fam labeled 32mer used in the binding assay were synthesized on a ABI 394 synthesizer, using nucleoside phosphoramidites purchased from Glen Research (Sterling, VA). The oligonucleotides were purified using an anion-exchange column and desalted using C18 reverse phase chromatography. The purity of the oligonucleotides was confirmed using electrophoresis through a 20% polyacrylamide gel containing 7M urea and MALDI-TOF analysis. The remaining oligonucleotides were ordered from Integrated DNA Technologies (IDT).
Figure 2.
Sequences of the DNA duplexes used in this study. The conserved pentapyrimidine sequence is highlighted by the horizontal line. For the 24B/34F duplex used for formation of the covalent complex (12), the sequence shown in red is the biotinylated portion that is removed when applied to streptavidin beads. The 12mer sequence overhang is complimentary to the strand exchange 12mer.
The binding and cleavage duplexes were made by mixing the complementary strands using a 10% excess of the non-fluorescent strand. The duplex used to prepare the covalent complex (24B/32F) had the 24Bio strand in 50% excess (12). The binding and cleavage duplexes were prepared in Buffer A [20 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 1 mM dithiothreitol (DTT)]; the covalent complex duplex was made in Buffer B [20 mM Tris 8.0, pH 8.0, 30 mM NaCl, and 1 mM DTT. The duplexes were formed by mixing the individual strands at the indicated concentrations, heating to 95°C, and then slowly cooling to room temperature. The DNA sequences used in the current study contain a T at the −1 position relative to the cleavage site. Although previous work has shown that an A at this position accelerates cleavage by about 10-fold relative to T, this substitution does not change the cleavage equilibrium and it facilitates singleturnover kinetic measurements by hand mixing (9, 13). The nature of the base at the -1 position has no bearing on the interpretations of this study.
Noncovalent Binding and Cleavage Equilibrium Measurements
Equilibrium cleavage measurements were made by titrating 60 or 90 nM 32F/32 duplex with increasing concentrations of enzyme (40–1000 nM) in Buffer A. The covalent complexes at equilibrium were trapped by the addition of 4% SDS. Running the samples on a 10% SDS-PAGE allowed for separation of the free duplex and covalent complex. The fraction of the total fluorescence counts migrating as the covalent complex at each enzyme concentration was determined by dividing the counts in the covalent complex by the sum of the counts in the covalent complex and free duplex using Quantity One imaging software. By plotting the fraction covalent complex as a function of enzyme concentration, the binding constant (Kd) and the reversible cleavage-ligation equilibrium constant (Kcl) can be obtained using the following equation:
| (1) |
where a = 1 + 1/Kcl; b = ax + a[S] + Kd/Kcl; Kcl = kcl/kl; and [E] and [S] are enzyme and substrate concentrations, respectively (11).
Single Turnover Cleavage Kinetics
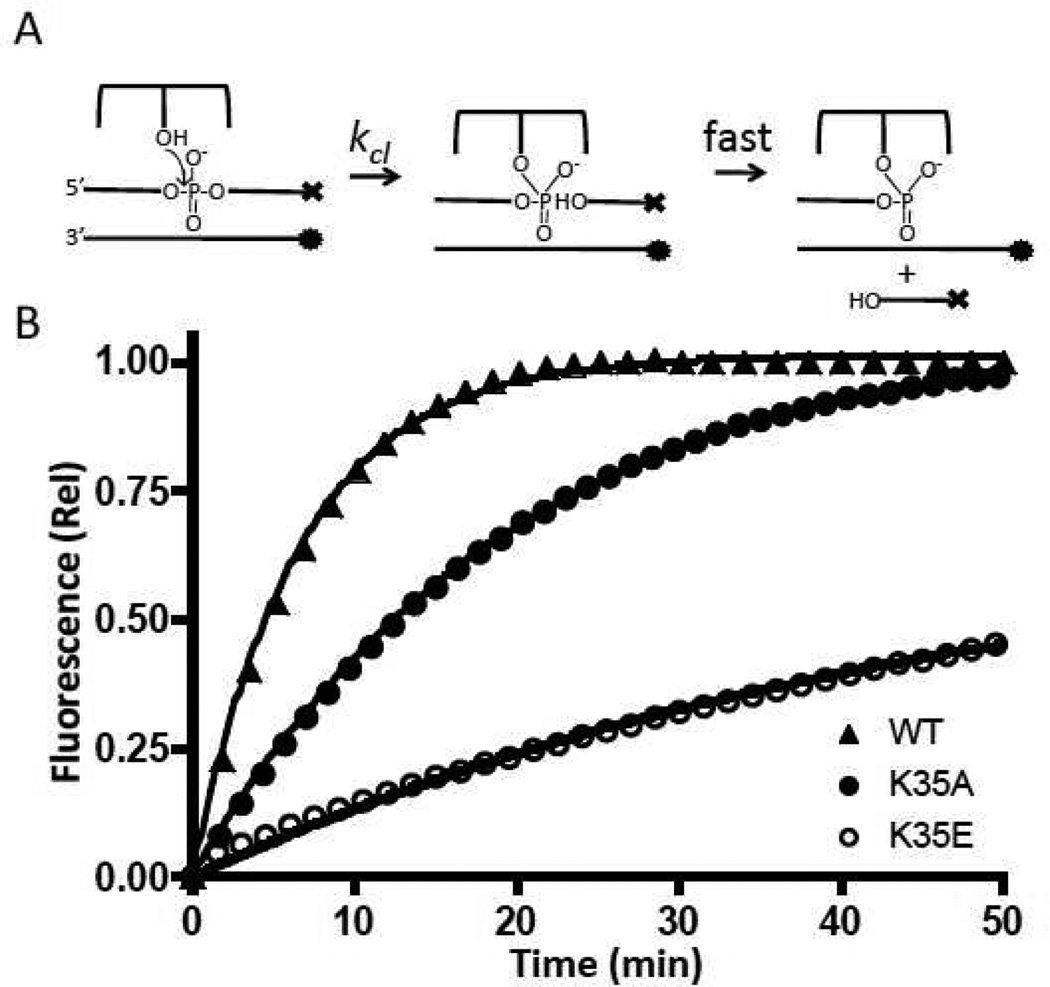
Cleavage measurements were made with a 26D/26F “suicide” substrate that contains a sufficiently short strand downstream of the cleavage site in order to ensure rapid dissociation from the covalent complex (14). The enzyme is in 20-fold excess over the substrate so that the observed kinetics are pseudo-first order and independent of the concentration of the enzyme or DNA. The reactions containing 50 nM duplex and 1 µM vTopo in Buffer A were carried out in a Spex Fluoromax 3 fluorimeter, and the increase in Fam fluorescence was followed at 515 nm with excitation at 495 nm (excitation and emission slits of 3 nm). Raw fluorescence measurements were normalized using the following equation:
| (2) |
where Ft is the fluorescence at t, F0 is the initial fluorescence, and Ff is the final fluorescence signal. It has been previously shown that cleavage is the rate-limiting step for these substrates, therefore, the fluorescence signal reports the rate of cleavage and not strand dissociation. Plots of Ft against time were fitted to a single exponential equation to obtain kcl, the cleavage rate.
Preparation of Covalent Complex for Strand Exchange Measurements
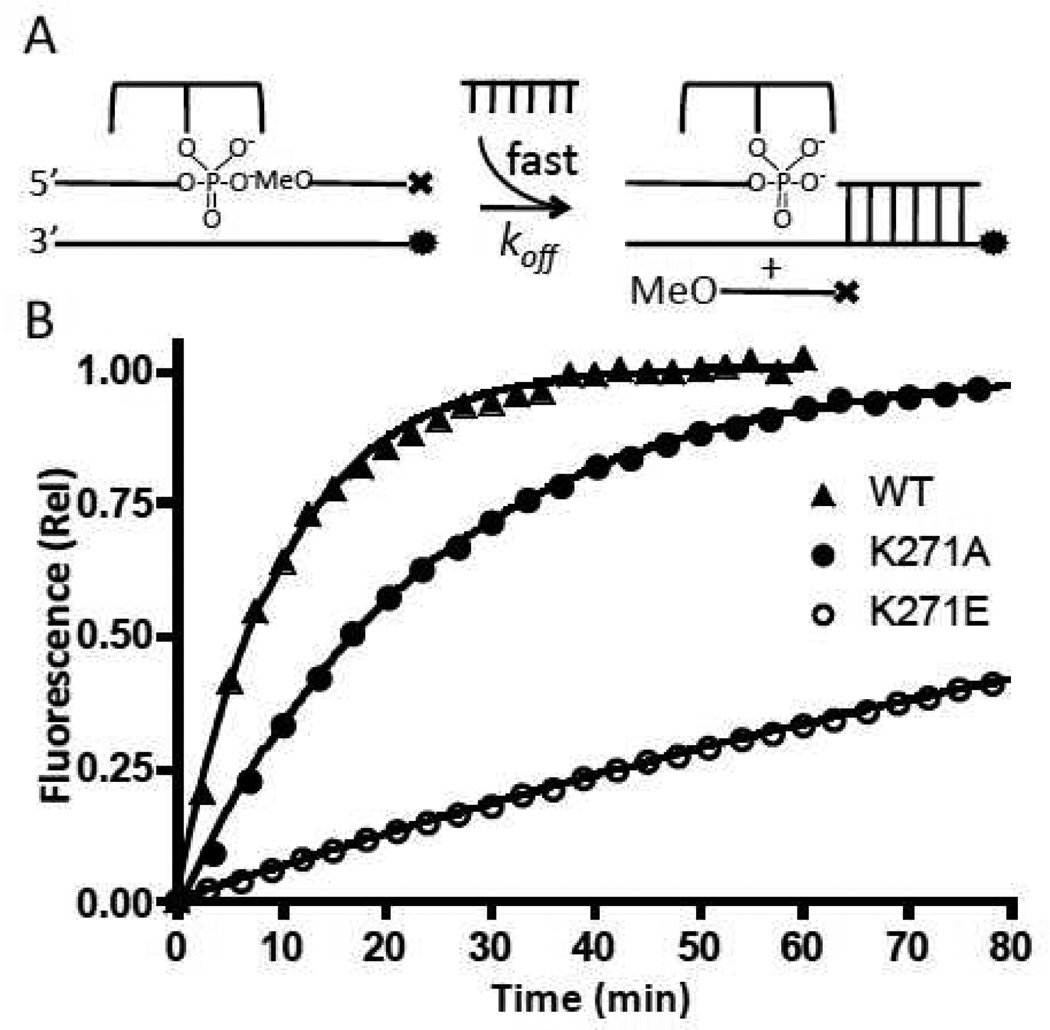
Recently, a method for creating and purifying a vTopo covalent complex with overhang DNA was described (12). By reacting the enzyme with the 24B/32F duplex that has a Fam label on the uncleaved strand and is biotinylated on the cleaved strand (Fig. 2), we are able to purify the covalent complex using a streptavidin-agarose slurry in Buffer B (see reference (12) for further details). This purification of the covalent complex allows us to examine the effects of mutations on the association and dissociation kinetics of DNA strands to the downstream single strand region of the covalent complex.
Strand Dissociation
In order to examine the rate of strand dissociation from the covalent complex, we took 30 nM covalent complex (CC) derived from reaction of each enzyme with 24B/32F, and pre-bound it with 90 nM 12OMeDab, which resulted in quenching of the Fam fluorescence of the overhang strand. Reactions were initiated by adding 3 µM unlabeled chase (12OH), which serves to quickly and irreversibly trap the free covalent complex after the 12OMeDab strand dissociates. The fluorescence increase was plotted as a function of time, and the dissociation rate (koff) was determined from a single exponential fit to the data.
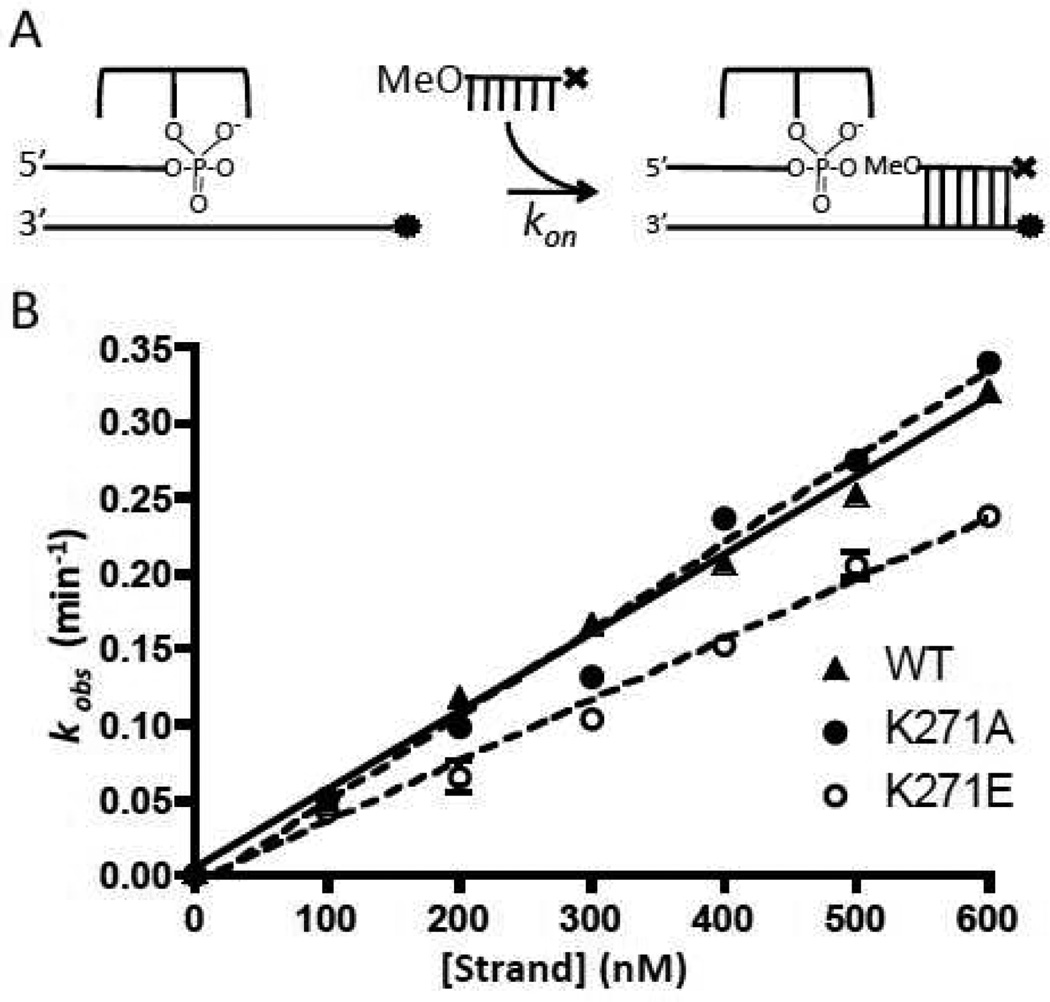
Strand Association
Increasing concentrations of the 12OMeDab strand was added to 10 nM CC in Buffer B to obtain strand association rates. The incoming strand concentration was at least 10-fold greater than the [CC] to maintain pseudo-first-order conditions. The Dab quenching of the Fam signal was monitored using an Applied Photophysics stopped flow instrument, using an excitation wavelength of 495 nm and a >510 nm cutoff optical emission filter. The time dependent decrease in fluorescence was fitted to a single exponential function to obtain kobs. The observed rate constants were then plotted as a function of 12OMeDab concentration and fitted to eq 3, where kon is the strand association rate and koff is obtained from the strand dissociation measurements.
| (3) |
Supercoil Relaxation Measurements
In order to study the processivity of supercoil unwinding by the various constructs (i.e. the number of supercoils unwound per cleavage event), we reacted 40 nM negatively supercoiled pUC19 plasmid with 80 nM vTopo in a 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 20 µg/mL bovine serum albumin (BSA), 0.01% Brij-35 and 5 mM MgCl2 buffer. By using an [E]/[DNA] ratio of two, we statistically favor only one vTopo per DNA molecule, and the DNA molecules that are bound by vTopo react with zero-order kinetics (14). Reactions were initiated by manually pipetting an equal volume of vTopo in reaction buffer into a PCR strip tube containing DNA, also in reaction buffer. The reactions were quenched at appropriate times with 1% SDS (w/v), 10% glycerol, and 0.5% bromophenol blue delivered from a second pipettor. Accurate quench timing for times in the range 1–15s was achieved through the use of a metronome. After quenching, 15 µL of each sample was electrophoresed through a 25 cm horizontal 1% agarose gel in 1X TAE (40 mM Tris-acetate, pH 8.5, 2 mM EDTA) buffer containing 15 µg/mL chloroquine at 80V for 6 hours. (The presence of chloroquine enhances separation of individual topoisomers.) The gel was destained with deionized water for two hours to remove the chloroquine, stained for 10 min with a 1L solution containing 0.5 µg/mL of ethidium bromide, and destained again with deionized water for 30 minutes. Using a transilluminator, the stained gels were preexposed to UV light for 30 min prior to image analysis to insure that strand breaks were introduced into all of the DNA topoisomers, negating the possibility of differential amounts of dye intercalation for individual topoisomers. Background corrected ethidium bromide fluorescence intensities of supercoiled substrate, topoisomer intermediates and relaxed product were quantified using a Typhoon imager.
Analysis of Supercoil Unwinding Data
The time dependent changes in the topoisomer distributions were analyzed based on a previous “topoisomer pooling” approach that simplifies the kinetic analysis (6), with several modifications as follows (see also Supplemental Materials). Although the chloroquine gels provide resolution of nine topoisomers as well as relaxed product, it is not necessary to quantify the appearance and disappearance of each individual topoisomer band to satisfactorily characterize the processivity of the reaction. Instead, an easy to appreciate simplification may be introduced if multiple supercoils are released for each cleavage event. For example, if the starting substrate distribution contains on average 9 ± 2 negative supercoils, and the enzyme removes 5 supercoils for each cleavage event, then this population of four substrate topoisomers (S) will be on average converted into a new distribution (“pool”) of less negatively supercoiled intermediate topoisomers (I) that will not significantly overlap with the substrate pool. Similarly, the I pool will relax directly to product (P), without significant interconversion within the I pool. Thus, a simplified kinetic model may be used that accounts for the conversion of S→I, S→P, and I→P. Such a minimal model may be experimentally justified by carrying out kinetic simulations of the relaxation data to discover the minimum number of pools required (see Supplemental Methods). Data were quantified by determining fluorescence counts in the S, I and P pools at each time point and expressing these counts as a fraction of the total counts (S + I + P) in each lane to account for loading differences between lanes. In the final optimized model, the top four bands constituted S, the next four bands constituted I, and the final band at the bottom of the gel was P. In the results below we extend this approach by introducing two simple parameters to characterize the processivity of the mutant enzymes relative to the wild-type.
Results
Approach
In principle, a given mutation of vTopo could affect many discrete ground states and transition states along the reaction coordinate, making it important that different measurements are performed to ascertain the function of a given residue (11). We have previously developed methods and partial substrates that allow measurement of discrete steps of the vTopo reaction chemistry that include site-specific noncovalent DNA binding (Kd) (15), the cleavage-religation equilibrium (Kcl = kcl/kr), and the single-turnover cleavage rate (kcl) (11, 14). Recently, strand exchange kinetic measurements have been developed that measure potential interactions of the enzyme with the downstream DNA region that is mobile during supercoil unwinding (12). These assays involve measuring the rate of association and dissociation of the downstream portion of the cleaved DNA strand, which can be measured when the region of downstream base pairing is less than 10 bp. Finally, here we introduce a new method for evaluating the effects of mutations on supercoil unwinding. We should point out that because each of these measurements are, by necessity, made using different types of substrates, it is not possible to compare rates or equilibria between these classes of substrates. However, the goal of this work is to compare the effects of mutations on each of these individual substrates and then develop a consistent picture of their roles at discrete steps of the reaction.
Noncovalent DNA Binding and Equilibrium Cleavage
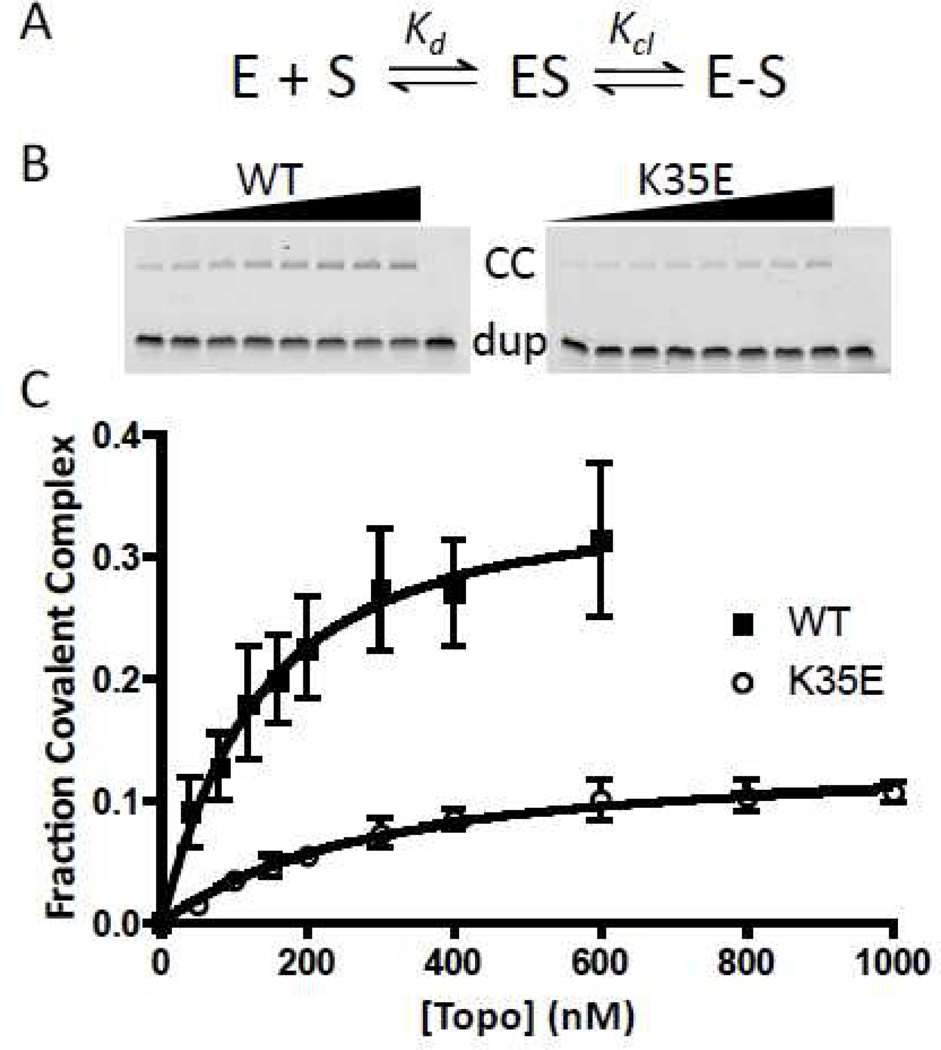
The binding (Kd) and cleavage equilibrium constants (Kcl) of the wild-type and mutant enzymes were measured using the 32F/32 duplex (Fig. 2) and a gel shift assay that follows the formation of the fluorescent covalent complex by resolving it from the free DNA using gel electrophoresis under denaturing conditions (10% SDS-PAGE) (Fig 3a, b). The Kd is determined by the increasing amount of covalent complex formed as the enzyme concentration is increased, while the Kcl is calculated from the endpoint of the curve at saturating enzyme (Fig. 3c, and eq 1). Within error limits of the measurements, the K35A, K35E, R67A, and K271A charge neutralization and reversal mutations had little effect on Kd (Table 1)2, indicating that these mutations do not affect the stability of the ground state noncovalent complex prior to cleavage. In contrast, we were unable to detect any covalent complex with R67E and K271E using this assay, even employing enzyme concentrations as high as 1 µM, which indicates that these charge reversal mutations have caused a defect in binding and/or cleavage. As shown below, since both of these mutations also cause a large decrease in the cleavage rate, this could largely explain the difficulty in detecting the covalent complexes in these measurements. As observed with binding, the K35A and K35E mutations had little effect on the cleavage equilibrium (Kcl), but R67A and K271A showed ~8- and 3-fold decreases in Kcl showing that the native R67 and K271 side chain interactions are used to stabilize the cleaved state over the uncleaved form of the complex.
Figure 3.
DNA binding and cleavage equilibrium assay. (A) Equilibria to be determined. (B) Representative SDS-PAGE gels for separation of the covalent complex (CC) from the free and noncovalently bound duplex DNA. Wild type and K35E enzyme concentrations increase from left to right. (C) Fraction covalent complex plotted as a function of vTopo concentration with error bars (standard deviation of three replicate measurements). Data were fitted to eq 1.
Table 1.
Thermodynamic and kinetic parameters for wild-type and mutant variants of vTopoa
| Construct |
Kd (nM) |
Kcl |
kcl (min−1) |
koff (min−1) |
kon (M−1min−1) × 10−2 |
τ0.18 (s) |
MEI | MEP |
|---|---|---|---|---|---|---|---|---|
| WT | 139 ± 55 | 0.25 ± 0.03 | 0.14 ± 0.01 | 0.104 ± 0.003 | 3.12 ± 0.04 | 3 | 1.00 | 1.00 |
| K35A | 132 ± 35 | 0.32 ± 0.03 | 0.05 ± 0.001 | 0.093 ± 0.013 | 2.11 ± 0.13 | 5 | 0.41 ± 0.33 | 1.07 ± 0.04 |
| K35E | 272 ± 88 | 0.16 ± 0.02 | 0.016 ± 0.004 | 0.142 ± 0.002 | 2.70 ± 0.20 | 10 | 1.34 ± 0.42 | 0.96 ± 0.05 |
| R67A | 148 ± 80 | 0.03 ± 0.01 | 0.005 ± 0.001 | 0.147 ± 0.013 | 2.75 ± 0.09 | 22 | 0.34 ± 0.81 | 1.08 ± 0.10 |
| R67E | ND | ND | 0.004 ± 0.001 | ND | ND | 255 | 0.94 ± 0.43 | 1.01 ± 0.05 |
| K271A | 282 ± 118 | 0.09 ± 0.02 | 0.010 ± 0.0002 | 0.030 ± 0.015 | 3.44 ± 0.04 | 12 | −1.04 ± 0.67 | 1.25 ± 0.08 |
| K271E | ND | ND | 0.006 ± 0.0004 | 0.006 ± 0.0003 | 2.41 ± 0.08 | 10 | −2.23 ± 0.36 | 1.39 ± 0.04 |
Kd is the dissociation constant for noncovalent DNA binding (Scheme 1), Kcl = kcl/kr is the equilibrium constant for reversible strand cleavage (Scheme 1), kcl is the rate constant for single turnover irreversible strand cleavage, koff and kon are the rate constants for 12mer strand dissociation and association (eq 3), τ0.18 is the time for 18% substrate reaction in the supercoil unwinding assay, MEI is the mutational effect on processivity based on intermediate topoisomer accumulation (eq 5), and MEP is the mutational effect on processivity based on product accumulation (eq 5).
Single-turnover Strand Cleavage Kinetics
In order to measure the mutational effects on strand cleavage, a fluorescent substrate was used (26D/26F, Fig. 2). This substrate has its cleaved strand labeled with a 3′-dabcyl fluorescence quench, and its complementary non-scissile strand labeled with a 5′-Fam fluorophore. When vTopo cleaves the scissile strand, a dabcyl labeled 6mer will rapidly dissociate to solution, increasing the fluorescence of the 5′ Fam group on the uncleaved strand (Fig 4a). In order for this assay to report solely on the cleavage rate, strand dissociation must be rapid compared to cleavage, which has been previously established for the substrates used here (11). Except for the R67 position, the charge reversal mutations each showed a 2–3 fold greater damaging effect than the alanine charge ablation mutations (Table 1). The total magnitude of the damaging effects for K35E and K271E mutations were in the range of 10 to 20-fold (Table 1). Although the R67E charge reversal showed one of the largest damaging effects on cleavage (28-fold), it was distinct from the other charge reversals in that its damaging effect on the cleavage rate was similar to the simple charge ablation mutation (Table 1). This would suggest that the negatively charged glutamate side chain does not introduce an unfavorable charge repulsion with the phosphate backbone at this position.
Figure 4.
DNA cleavage kinetic measurements. (A) Reaction to be measured; ⊤ represents dabcyl and • represents fluorescein. (B) Normalized fluorescence intensities as a function of time. The curves were fitted to a single exponential equation. For clarity, only one of every ten data points is plotted.
DNA Strand Dissociation and Association
During supercoil unwinding, interactions between the enzyme and the downstream region of the cleaved DNA must be broken and reformed in order for strand rotation to occur (10). An indirect method to investigate the interactions of enzyme residues with the downstream region of the DNA in the context of the covalent complex (CC) is to measure mutational effects on strand dissociation and association. In the strand dissociation assay, the Fam labeled CC is pre-annealed with the 5′-O-methyl and 3′-dabcylated 12 mer downstream strand (12OMe) to prevent ligation from occurring. By having an excess of unlabeled 12OH trap present, dissociation of the 12OMe strand is irreversible, resulting in a single exponential increase in fluorescence that provides koff (Table 1). Despite having effects on other steps of the reaction (see above), mutations K35A and K35E had no effect on strand dissociation (not shown). Neither did R67A, as would be expected because it is observed to interact with the opposite strand upstream of the cleavage position (Fig. 1). In contrast, K271A and K271E show ~3- and 20-fold decreases in the strand dissociation rate, consistent with the observed interaction of K271 with the phosphate backbone (Fig 4b), but not consistent with the most naïve energetic expectation that removing a potentially favorable charge interaction with the phosphate backbone should increase the off-rate (see Discussion).
The second-order rate constants for strand association were obtained from the slopes of the concentration dependence of the pseudo-first order rate constants for association of a 12OMe to the Fam labeled covalent complex (Figure 5). For all of the mutants, these rates were within ~70% of the value for wild-type vTopo (Table 1). Since strand association and dissociation likely occur through the same rate-limiting transition state, the large effects of the K271 mutations on strand dissociation most likely result from increasing the barrier to dissociation by stabilizing a ground state (or states) that precede the dissociation transition state. In any event, these measurements support the structural observation that K271 forms an important interaction with the downstream strand in the covalent complex (Fig. 1).
Figure 5.
DNA strand dissociation kinetics. (A) Experimental scheme; ⊤ represents dabcyl and • represents fluorescein. (B) Normalized fluorescence intensity as a function of time (eq 2). For clarity, only one in every ten data points is plotted. Curves are best fits to a single exponential equation.
Supercoil Unwinding
A steady-state supercoil unwinding assay can potentially measure every step between enzyme encounter with the plasmid DNA and product dissociation. However, the kinetic steps of cleavage, ligation and supercoil unwinding may be examined in isolation by using excess (single-turnover) enzyme conditions, where both [E] and [S] are well above the Kd for binding to plasmid DNA (<4 nM) (6). The average number of supercoils that are removed for each cleavage event in the ensemble of plasmid molecules will be determined not by the rate of cleavage, but instead, by competition between the ligation rate (kr) and the rate of supercoil unwinding (krot). If a mutation decreases (or increases) the ratio krot/kr, fewer (or more) supercoils will be removed per cleavage event as compared to the wild-type enzyme. It is important to note that in this context, cleavage merely serves as the rate-limiting gateway that allows this kinetic competition to occur. Accordingly, although the magnitude of kcl will determine the overall rate of supercoil removal, a mutational change in kcl alone should have no effect on the important ratio that determines the processivity of supercoil removal. The analysis of supercoil unwinding that follows uses the simplified S→I→P and S′P kinetic pathways described in the Methods section.
We conceptualized two simple and independent empirical measures to describe the processivity for supercoil unwinding which we call processivity factors (Pf, eq 4). These measures simply compare how much of the original substrate pool (S) is converted into intermediate and/or product at a constant fractional reaction (18 ± 2%). This approach takes advantage of the general property that a processive enzyme will convert more substrate to product (P) at a given fractional reaction, and a less processive enzyme will generate more intermediates (I). This concept is expressed algebraically in eq 4, where So is the initial substrate concentration, and St is the remaining substrate concentration at time t after the start of the unwinding reaction. The difference Nt–No (where N = I or P) is the change in the intermediate pool (I) or the product (P) over the same time period.
| (4) |
The mutational effect on processivity (MEN) is defined as the ratio of the processivity factors for the mutant and wild-type enzymes (eq 5), where N designates whether the Pf values for I or P are being compared (see eq 4).
| (5) |
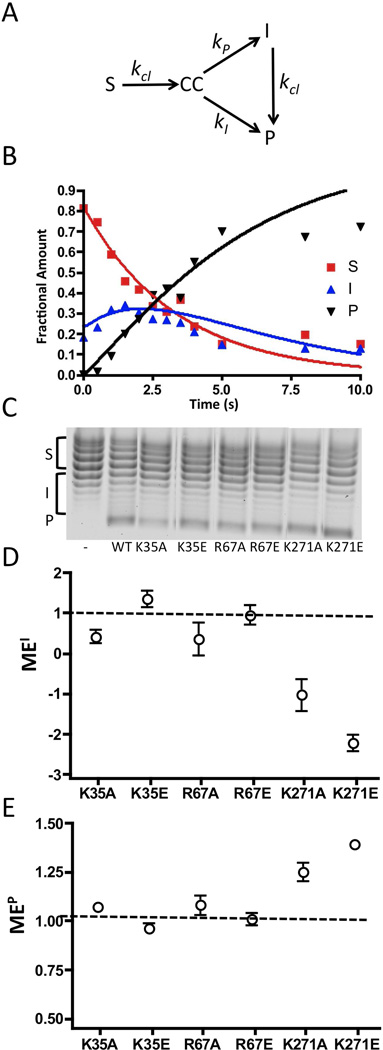
Before applying the above approach for assessing processivity, we confirmed its validity using kinetic simulations (Figure 7a). The time course for reaction of 80 nM wild-type vTopo with 40 nM pUC19 was fitted using a kinetic model employing a substrate pool (S) that is cleaved with a rate constant kcl to generate a cleaved DNA intermediate (CC) that can then partition into an intermediate pool (I) or product (P), with the rate constants kI and kP, respectively (see legend to Fig. 7a, b). This simulation also included a step where the intermediate pool is converted to product with rate-limiting cleavage (kcl). For wild-type vTopo, kcl = 0.3, kI = 5.0, and kP =2.3. In the Supplemental Materials we further show that the ratio kI/kP is a measure of supercoil unwinding processivity that provides qualitatively similar information as the simple processivity factor (eq 4).
Figure 7.
Supercoil unwinding measurements. (A) Kinetic model for analysis of supercoil unwinding (see Supplemental Materials), and (B) the time course for supercoil unwinding by wild-type vTopo. The fraction of S, I and P at each time point were modeled using the numerical integration program Dynafit (16). The rates for S→CC, CC→I, CC→P and I→P are kcl, kI, kP, and kcl respectively. The fitted kinetic parameters for the wild-type enzyme were kcl = 0.3 s−1, kI = 5.0 s−1 kP = 2.3 s−1 (see Supplemental Materials for an analysis of K271A). The ratio kI/kP = 2 is a measure of how much of the covalent complex generated from rate limiting cleavage of S partitions to I as opposed to P. Thus, this ratio is a quantitative measure of processivity of supercoil unwinding that is analogous to the simple processivity factor (eq 4). (C) Representative chloroquine gel analysis for supercoil unwinding by wild-type and mutant topoisomerases as indicated. In this system, negatively supercoiled substrate topoisomers (S) migrate at the top of the gel and relaxed products (P) migrate at the bottom. Each reaction was terminated after 18 ± 2% of substrate had been consumed (see text). The left most lane is the pUC19 substrate in the absence of topoisomerase. The bands corresponding to S, I and P are shown by brackets along side the gel image (see Methods). (E) Mutational effect on the processivity factor (Pf) based on accumulation of intermediates MEI (see text). (E) Mutational effect on the processivity factor (Pf) based on appearance of product MEP (see text). The dashed line at one indicates no mutational effect.
The measurement of Pf for the wild-type enzyme was made at a fractional conversion of 18% which corresponds to the reaction time point of 3s in Fig. 7b. This fractional conversion is optimal to detect changes in intermediate and product pools required for reliable Pf and MEN measurements (eqs 4 and 5). The Pf values for each mutant enzyme were measured in triplicate at fractional conversions of 18 ± 2% (Fig. 7c), and the times to achieve 18% substrate conversion ("τ0.18) for each mutant are listed in Table 1. The processivity factors for most mutants were not significantly different than the Pf (I) = 0.11 and Pf (P) = 1.07 values measured for wild-type vTopo. However, the values for K271A and K271E did differ significantly, with Pf (I) = −.18 and Pf (P) = 1.32.
Calculation of the mutational effects on processivity (MEI and MEP) indicated that K35A, K35E, R67A, and R67E do not differ significantly from wild-type vTopo even though these mutations had substantial effects on cleavage and the cleavage equilibrium (Fig. 7d, e). In contrast, both MEI and MEP were significant for K271A and K271E, with both mutational effects being consistent with an increase in the number of supercoils removed per cleavage event (i.e. negative MEI values, indicating that fewer intermediates accumulate; and positive MEP values, indicating that more substrate goes directly to product rather than passing through the intermediate pool).
Discussion
In order to function properly in a cell, type IB topoisomerases must unwind supercoils in a controlled fashion. If this optimal steady-state condition is not achieved, cellular DNA would either exist in a completely relaxed state, or alternatively, replicative or transcriptional processes would grind to a halt because of the accumulation of excessive supercoils. Previous ensemble and single molecule kinetic measurements have converged on a mechanism by which this optimal steady-state condition is achieved: type IB topoisomerases unwind multiple supercoils during the lifetime of the covalent complex (i.e. krot/kr ≫ 1) (5, 6). This high processivity of supercoil unwinding is balanced by a moderate cleavage rate that serves to limit the time-averaged amount of covalent complex that is present (i.e. Kcl = kcl/kr ≪ 1). A key question for this mechanism is how these enzymes regulate the number of supercoils that are released during the lifetime of the covalent complex. One reasonable proposal is that the enzyme contacts with the downstream segment of the DNA dynamically break and reform in order to allow strand rotation, and also to provide pausing events of long enough duration to allow strand religation to occur. We evaluate the observed properties of the six mutations studied here in the context of such a model.
Lysine-35
K35 forms a 2.9Å hydrogen bond with a phosphate group on the uncleaved strand, 3 base pairs downstream from the cleavage site (Fig. 1). Despite its proximity to the cleavage site, mutating this residue to either alanine or glutamate has minimal effects on DNA binding or the cleavage equilibrium constant. These two measurements alone allow us to conclude that the difference in the free energy levels between the ground state noncovalent (ES) and covalent (E-S) complexes for K35A and K35E are the nearly the same as wild-type vTopo (Scheme 1). Therefore, the 3- and 10-fold decreases in the cleavage rate observed with K35A and K35E must arise from increases in the transition state free energy for cleavage and ligation, rather than ground state stabilization of either the ES or E-S complexes. Thus, K35 is a modestly important catalytic residue by virtue of its selective stabilization of the transition state. The lack of involvement of this group in DNA binding, or in stabilization of the covalent E-S complex, once again indicates that the interactions observed in the crystal structure PDB ID 3IGC approximate the transition-state conformation of the enzyme-DNA complex (10). Consistent with this interpretation, removal of the K35 charged side chain has no effect on the rate of DNA strand association or dissociation, both of which occur from a distinct state from that of strand cleavage and ligation (12). The modest increase in strand association and dissociation with the charge reversal mutant K35E, is consistent with a modest unfavorable electrostatic interaction of the glutamate side chain with the phosphate backbone. This effect may be attenuated by intervening water molecules or even bound monovalent cations.
The effects of the K35 mutations on supercoil unwinding are partially consistent with the effects on the individual steps measured with the oligonucleotide substrates. A significant result is that the time for 18% conversion is about 2 to 3-fold longer than for wild-type vTopo, which are similar damaging effects to those observed on cleavage of the oligonucleotide substrate (Table 1). This is not surprising, because cleavage has been suggested to be the rate-limiting step for supercoil unwinding (11). It is more difficult to reconcile the negligible effects of the K35A and K35E mutations on supercoil unwinding processivity (MEI and MEP, Table 1) because these mutations selectively raise the energy of the cleavage/ligation transition state (i.e decrease kr), which would be expected to increase the ratio krot/kr and lead to an increase in the number of supercoils removed for each cleavage event. This discrepancy may in part arise from the uncertainty in using fairly small changes in kinetic and thermodynamic parameters obtained using oligonucleotide substrates towards the understanding of supercoil unwinding. Nevertheless, the negligible effects of these mutations on supercoil unwinding are consistent with general view that this group interacts primarily in the transition state conformation, and relatively small compensating changes in kr or krot may lead to the similar processivity factors observed with K35A, K35E and wild-type vTopo.
Arginine-67
R67 forms a charged 2.9Å hydrogen bond with a phosphate on the uncleaved strand one base pair upstream from the cleavage site (Fig. 1). Unlike the other residues, the location of R67 upstream relative to the cleavage site would obviate any direct role for this group in modulating the dynamics of strand rotation, although mutations at this site could increase processivity by destabilizing the transition state for strand ligation (i.e decreasing kr). Although we found that R67A and R67E had no effect on DNA binding or strand exchange, both mutations had a significant ~30-fold damaging effect on oligonucleotide strand cleavage, and correspondingly large 7- and 85-fold damaging effects on the time constant for supercoiled substrate unwinding (Table 1). As with K35A and K35E, these comparable mutational effects on strand cleavage and the overall rate for supercoil unwinding are consistent with the conclusion that supercoil unwinding is limited by DNA cleavage. Despite the large decreases in the cleavage rates observed with R67A and R67E, neither mutation had a significant effect on the processivity of supercoil unwinding. Within the context of the kinetic model for supercoil unwinding, where processivity depends on krot/kr rather than kcl, the negligible mutational effects R67A and R67E on processivity could suggest that the ligation rate constant (kr) for these mutants may not be as damaged as kcl. This possibility is supported by the observation that the R67A mutation only decreases the cleavage equilibrium by eight-fold, as compared to its 28-fold effect on the cleavage rate. Since Kcl = kcl/kr, a 3-fold reduction in the mutational effect on Kcl as compared to kcl would require that kr is only 3-fold slower than the wild-type value. A similar or smaller damaging effect on kr in the context of the supercoiled substrate could account for the absence of a measurable effect of the R67A and R67E mutations on the processivity factors.
Lysine-271
K271 forms a 3.2Å charged hydrogen bond with the phosphate backbone of the uncleaved strand six base pairs downstream of the cleavage site and is a clear candidate residue for modulating the dynamics of strand rotation (Fig. 1). The K271A charge ablation mutation has a small effect on the DNA binding affinity, which may be increased in the K271E mutant, because we were unable to measure Kd or Kcl for K271E (Table 1). Despite the large distance between K271 and the site of covalent chemistry, both K271A and K271E have substantial effects on the activation barrier for cleavage. For K271A and K271E, kcl is diminished by 10 and 20-fold compared to wild type vTopo, respectively (Table 1). Since Kcl is only diminished by about 2-fold for K271A, we also infer a significant 5-fold decrease in kr for this mutant based on its effect on kcl and the relationship Kcl = kcl/kr. Such a decrease in kr would serve to increase processivity of unwinding, consistent with the mutational effects on processivity (Fig. 7d, e).
Interestingly, the K271 mutations were the only mutations investigated here that had an effect on both strand dissociation and processivity (Table 1). Our initial hypothesis was that removing or reversing a charged interaction with the phosphate backbone might accelerate strand dissociation because a stabilizing interaction should increase the activation barrier for dissociation3. However, the opposite effect is observed with K271A and K271E. Such an outcome could arise if the binding energy of K271 is used to destabilize the DNA strand, and accordingly, the mutations eliminate this effect, resulting in a slower strand off-rate. Alternatively, K271 could facilitate the exit of the bound strand by interacting with a partially dissociated state, thereby pulling the system toward complete dissociation. The absence of this interaction would therefore result in abortive strand dissociation events and a slower observed koff. Finally, the absence of a mutational effect on strand association is most simply explained by a multistep pathway for strand association/dissociation, in which the interaction with K271 is formed after the rate-limiting transition state for strand association4.
The K271 mutants were the only ones to show a significant change in MEI and MEP values. For both mutants, the MEI value was negative, which means that compared to wild type, these mutants had a decreased accumulation of topoisomers in the I pool. In addition, the MEP values for the K271 mutants were greater than 1, which shows an increase in product formation relative to wild-type. These two values show that as the K271 mutants cleave substrate, they release more supercoils such that topoisomers in the S pool bypass the I pool and move directly to P. This increase in the processivity of the enzyme arises regardless of whether the charge at this position is ablated or reversed, although K271E does show a greater increase in processivity as compared to K271A. It thus appears that this enzyme-DNA interaction plays a role in controlling the number of supercoils released per cleavage event by increasing the lifetime of the covalent complex, but we cannot exclude the additional possibility that the K271 mutants might also increase krot. In summary, the data provide strong evidence for an interaction of K271 with the phosphate backbone that regulates both processivity and strand exchange.
Conclusions
The findings reported here reveal diverse roles for electrostatic interactions between side chains of vTopo and the DNA phosphate backbone, and shed new light on the mechanism for supercoil unwinding. First, mutations that have a large effect on the cleavage rate do not necessarily lead to changes in the processivity of supercoil relaxation. Such effects are consistent with the “free” or “controlled” rotation model for supercoil unwinding (6, 7), and affirm that cleavage merely serves as the gateway to the covalent complex, whose lifetime and dynamic behavior determine the number of supercoils that are removed after each cleavage event. Second, interactions with the downstream strands that are far away from the site of chemistry can have significant effects on the transition state for strand cleavage and/or ligation indicating long-range connectivity effects in the reaction. Finally, the findings reveal how removal of an electrostatic interaction with the downstream mobile duplex region can increase the lifetime of the covalent complex and result in enhanced processivity of supercoil unwinding.
Supplementary Material
Figure 6.
DNA strand association kinetics. (A) Experimental scheme; ⊤ represents the dabcyl quench and • represents the fluorescein dye. (B) Plots of kobs as a function of 12mer concentration were fitted by linear regression to obtain kon (eq 3).
Footnotes
This work was supported by NIH grant GM068626 (J.T.S).
Abbreviations: vTopo, variola type I topoisomerase; Fam, 6-carboxy-fluoroscein; Dab, 3’-dabcyldT; Bio, biotin connected to the oligonucleotide with a 15-atom tetra-ethyleneglycol spacer; OMe, 5’-O-methyl group; OH, hydroxyl group; MALDI, matrix-assisted laser desorptionionization mass spectrometry; Tris, tris(hydroxymethyl)aminomethane; SDS, sodium dodecyl sulfate; SDS-PAGE, polyacrylamide gel electrophoresis in the presence of the denaturant SDS; CC, covalent complex of vTopo with DNA.
Two previous studies have investigated some kinetic properties of the R67A and K271A mutations (18, 19). The present studies extend these investigations to include DNA binding, cleavage equilbrium, single-turnover kinetic measurements, and supercoil processivity measurements.
Previous comparisons of 12mer strand association and dissociation kinetics using identical vTopo-DNA complexes as employed here revealed no effect of the enzyme on either kinetic constant. We interpreted these results as being indicative of a “passive” role of the enzyme in strand positioning. The current mutational findings with K271 suggest that select interactions of the enzyme with the downstream region can have modest effects on strand dissociation. This requires modification of the previous conclusion to include the finding that the identical overall rates of strand dissociation for free DNA and the wild-type enzyme complex occur by different mechanisms.
Although a multistep pathway for strand dissociation would seem to violate the simple two-state approximation for strand exchange (eq 3), such an empirical analysis still holds if the intermediates prior to the rate-limiting step for dissociation are in rapid equilibrium.
Supporting Information Available. Analysis of the purity of vTopo enzyme variants and kinetic simulations of supercoil unwinding reactions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Champoux JJ. DNA Topoisomerases: Structure, Function, and Mechanism. Annual Reviews. 2001 doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Schoeffler AJ, Berger JM. DNA Topoisomerases: Harnessing and Constraining Energy to Govern Chromosome Topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. DNA Topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Shuman S. Vaccinia Virus DNA Topoisomerase: A Model Eukaryotic Type IB Enzyme. Biochimica et Biophysica Acta (BBA)- Gene Structure and Expression. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 5.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and Torque Govern the Relaxation of DNA Supercoils by Eukaryotic Topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 6.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA Topoisomerase I: Evidence Supporting a Free Rotation Mechanism for DNA Supercoil Relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 7.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A Model for the Mechanism of Human Topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 8.Shuman S. Site-Specific DNA Cleavage by Vaccinia Virus DNA Topoisomerase I. Role of Nucleotide Sequence and DNA Secondary Structure. Journal of Biological Chemistry. 1991;266:1796–1803. [PubMed] [Google Scholar]
- 9.Shuman S, Prescott J. Specific DNA Cleavage and Binding by Vaccinia Virus DNA Topoisomerase I. Journal of Biological Chemistry. 1990;265:17826–17836. [PubMed] [Google Scholar]
- 10.Perry K, Hwang Y, Bushman FD, Van Duyne G. Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic. Structure. 2010;18:127–137. doi: 10.1016/j.str.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon K, Stivers JT. Fluorescence Spectroscopy Studies of Vaccinia Type IB DNA Topoisomerase. Closing of the Enzyme Clamp is Faster than DNA Cleavage. J. Biol. Chem. 2002;277:345–352. doi: 10.1074/jbc.M109449200. [DOI] [PubMed] [Google Scholar]
- 12.Stahley MR, Stivers JT. Mechanism and Specificity of DNA Strand Exchange Catalyzed by Vaccinia DNA Topoisomerase Type I. Biochemistry. 2010;49:2786–2795. doi: 10.1021/bi902204v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minkah N, Hwang Y, Perry K, Van Duyne G. Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes. Virology. 2007;365:60–69. doi: 10.1016/j.virol.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stivers JT, Shuman S. Vaccinia DNA Topoisomerase I: Single-Turnover and Steady-State Kinetic Analysis of the DNA Strand Cleavage and Ligation Reactions. Biochemistry. 1994;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 15.Nagarajan R, Stivers JT. Unmasking Anticooperative DNA-Binding Interactions of Vaccinia DNA Topoisomerase I. Biochemistry. 2007;46:192–199. doi: 10.1021/bi061706u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzmic P. Program DYNAFIT for the Analysis of Enzyme Kinetic Data: Application to HIV Proteinase. Analytical Biochemistry. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 17.The PyMOL Molecular Graphics System, Version 1.2r3pre. Schrödinger, LLC; [Google Scholar]
- 18.Wittschieben J, Shuman S. Mutational Analysis of Vaccinia Topoisomerase Defines Amino Acid Residues Essential for Covalent Catalysis. J. Biol. Chem. 1994;269:29978–29983. [PubMed] [Google Scholar]
- 19.Sekiguchi J, Shuman S. Mutational Analysis of Vaccinia Virus Topoisomerase Identifies Residues Involved in DNA Binding. Nucleic Acids Res. 1997;25:3649–3656. doi: 10.1093/nar/25.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.