Abstract
Background
Although the role of microbes in disease pathogenesis is well established, data describing the variability of the vast microbiome in children diagnosed with ulcerative colitis (UC) are lacking. This study characterizes the gut microbiome in hospitalized children with severe UC and determines the relationship between microbiota and response to steroid therapy.
Methods
Fecal samples were collected from 26 healthy controls and 27 children hospitalized with severe UC as part of a prospective multi-center study. DNA extraction, PCR amplification of bacterial 16S rRNA, and microarray hybridization were performed. Results were analyzed in Genespring GX 11.0 comparing healthy controls to children with UC, and steroid responsive (n=17) to non-responsive patients (n=10).
Results
Bacterial signal strength and distribution showed differences between UC and healthy controls (adjusted p<0.05) for Phylum, Class, Order, Family, Genus, and Phylospecies levels with reduction in Clostridia and an increase in Gamma-proteobacteria. The number of microbial phylospecies was reduced in UC (266±69) vs. controls (758±3, p<0.001), as was the Shannon diversity index (6.1±0.23 vs. 6.49±0.04, respectively; p<0.0001). Steroids non-responders harbored less phylospecies than responders (142±49 vs. 338±62, p=0.013).
Conclusions
Richness, evenness, and biodiversity of the gut microbiome were remarkably reduced in children with UC, compared to healthy controls. Children who did not respond to steroids harbored a microbiome that was even less rich than steroid responders. This study is the first to characterize the gut microbiome in a large cohort of pediatric patients with severe ulcerative colitis and describes changes in the gut microbiome as a potential prognostic feature.
Keywords: microbiome, ulcerative colitis, children, steroid therapy, microbial diversity
Introduction
Ulcerative colitis (UC) is a debilitating condition of chronic inflammation of the colon that leads to significant morbidity in both adults and children (1). An understanding of the pathogenesis of UC is incomplete and is vital to the development of new therapeutic approaches. The etiology of inflammatory bowel diseases (IBD), including UC, involves a disruption in the complex balance between microbes and the host, leading to immune activation. An altered host immune response in IBD is demonstrated by genetic predisposition in individuals with numerous susceptibility loci, many of which are involved in microbial handling (2–4). The role of microorganisms is also suspected based on the location of the disease in the distal bowel corresponding to the highest microbial concentration (5). In murine colitis models, chronic inflammation resolves with alteration of the intestinal microbiota, and reappears with reintroduction of microbes (6). In humans, diversion of the fecal stream attenuates inflammation, which returns with re-exposure of the lumen to fecal effluent (7).
Although it is clear that microbes have a crucial role in the pathogenesis of IBD, their exact role is only now being meticulously investigated. Pathogens that have been associated with IBD include Mycobacterium avium subspecies paratuberculosis (MAP), Staphylococcus aureus, and toxigenic Bacteroides fragilis; however, no specific organism has been consistently identified as an etiologic agent (8). Specific bacteria with increased virulence have also been incriminated, including strains of Escherichia coli with a propensity to adhere to and invade mucosal epithelium (9–10). There also appears to be some benefit for antimicrobial or probiotic therapy in IBD (11–13). Recently, an association between infectious gastroenteritis and IBD has been suggested in large population-based analyses (14–16).
Knowledge of intestinal bacterial composition and effects on the host have been limited by a lack of ability to define the microbiota colonizing the host as only a small portion is culturable (17). Recent advances in culture-independent metagenomic approaches have enabled measurement of this immense, diverse, and complex ecosystem (18–19). Detrimental alteration in the balance of the entire intestinal tract community of microorganisms, a phenomenon called dysbiosis, is implicated in the pathogenesis of various disease states, such as obesity, irritable bowel syndrome, and IBD (20–22). Numerous studies have shown changes in the microbiome in IBD patients, including an overall decrease in the diversity of microbial species, as well as a decrease in Firmicutes (23), specifically Clostridium, and a subsequent increase in Enterobacteriaceae (24).
Severe UC represents an extreme clinical setting; recognizing alterations in microbes during UC exacerbation could provide important insight into changes in the intestinal environment and how they relate to disease pathogenesis. Pediatric data have been limited to date, despite the high prevalence of severe exacerbations in this population. Characterizing microbes in children is of additional value since microbial composition is different than in adults (20, 25) and since studies in this population are more likely to identify early changes related to pathogenesis, as opposed to treatment, comorbidities, or environmental exposures that impact adults more frequently.
Detection of bacteria in fecal samples using microarray technology offers a unique approach to a thorough characterization of the microbiome. In this study we used a microarray containing probes to the 16S rRNA of 852 known human commensal species that enables culture-independent characterization of the immense intestinal microbiome to characterize the fecal microbiota of children with severe UC. With this approach, the relative amount of each of the individual phylospecies in the microbiome was assessed simultaneously by quantifying individual signal strengths.
Materials and Methods
Study design and participants
This is a nested case-control study of the Outcome of intravenous Steroid therapy in ulcerative Colitis Individuals (OSCI) study (26) which was a multicenter, prospective cohort of children 2–18 years of age, hospitalized for intravenous corticosteroid therapy for acute UC. The ethical review board of all participating hospitals approved this study. Informed consent and age-appropriate assent were obtained from participants and their caregiver, according to the local policy.
Data were prospectively collected at multiple times during the admission; stool samples were obtained on the third day of intravenous corticosteroid therapy (methylprednisolone 1 mg/kg/day in general). Children receiving antibiotics or probiotics during the month prior to admission were excluded to eliminate the effect of antibiotics as a confounding variable on the gut microbiome (27). Pediatric Ulcerative Colitis Activity Index (PUCAI) was used to assess disease activity (28). Steroid failure was defined as either the need for colectomy or second-line medical therapy with infliximab or cyclosporine prior to discharge.
Stool was also collected from age-matched healthy controls for comparison. None of the controls had a recent or chronic illness and none had been exposed to antibiotics, probiotics, or any other medication during the month prior to stool collection.
The bacterial make up of the gut microbiome was investigated using high-throughput microarray technology as previously reported (25). We have designed, validated, and used a custom GeneChip that is able to identify 16S rRNA of hundreds of species that constitute the human gut microbiome.
Sample collection
All fecal specimens were placed in tightly sealed sterile 50 ml tubes, frozen immediately, and kept at −70°C.
DNA isolation
Fecal DNA was isolated using ZR Fecal DNA kit (Zymo Research Corporation, Orange, CA). Frozen fecal samples (150 mg) were used following the manufacturer’s protocol. Eluted DNA was checked for quality by 1% agarose gel electrophoresis. The eluted DNA was then quantified on a NanoVue Spectrophotometer (GE Health care, NJ). All fecal samples were run individually.
PCR reaction
The genomic DNA (gDNA) was diluted to a concentration of 400 ng/µl and universal primers for 16S rRNA forward (AGA GTT TGA TCC TGG CTC AG) and 16S rRNA reverse (ACG GCT ACC TTG TTA CGA CTT) (Integrated DNA technologies, Coralville, IA) were used to amplify the whole 1.5kb fragment of 16S ribosomal RNA gene.
The reaction was carried out for 30 cycles on a StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA) as a normal thermo-cycling reaction. The amplified product was cleaned using a Qiagen MinElute PCR Purification Kit as per the manufacturer’s suggested protocol and then verified by 1% agarose electrophoresis alongside 100 bp Low Scale DNA Ladder (Fisher Scientific, Pittsburgh, PA) for the presence of a distinct band at 1500 bp position. The amplified DNA was then quantified on a NanoVue Spectrophotometer.
DNA fragmentation
Amplified 16S rDNA (400 ng) was used as target for the microarrays as previously described (25, 29). DNase I enzyme (New England Biolabs, Ipswich, MA) was diluted to a concentration of 0.05 units/µl in DNase reaction buffer. Ribosomal DNA fragmentation was carried out using working concentration of enzyme at 0.05 units DNase I/µg of amplified DNA as this concentration has been shown to produce ideal fragment sizes for microarray hybridization. The DNA was incubated at 37°C along with the enzyme for 10 minutes; thereafter, the enzyme was heat-inactivated at 75°C for another 10 minutes.
DNA labeling reaction
The single color protocol from Agilent Technologies (version 5.0, June 2007) was adopted for DNA labeling reaction using an Agilent Genomic DNA Enzymatic Labeling Kit (Agilent Technologies, Santa Clara, CA) and a single dye. Primers (2.5 µl) were added to 13 µl of the fragmented DNA, and the mixture was incubated at 95°C for 3 minutes and then cooled to 4°C. The samples were briefly spun in a microcentrifuge to collect the contents at the bottom. Reaction buffer, dNTPs, single dye, and Exo-Klenow fragment were added to the annealed primer-template mix and incubated at 37°C for 2 hours followed by heat inactivation of the enzyme at 65°C for 10 minutes. The labeled DNA was mixed in 1X TE buffer (pH 8.0) and passed through Microcon YM-30 filters to clean the DNA.
Microarray hybridization
The following components were added in the order indicated: labeled DNA, Cot-1 DNA, Agilent 10X blocking agent, and Agilent hybridization buffer. Components were mixed thoroughly, quickly spun, then incubated at 95°C for 3 minutes and 37°C for 30 minutes. The reaction mixture (40 µl) was loaded onto a gasket chamber and the printed microarray was placed over the gasket with the active side of the array facing down. The sandwich assembly was thoroughly inspected for any leaks then hybridized for 24 hours. Washing and scanning (Agilent G2565CA Microarray scanner with SureScan High resolution technology; Agilent Technologies, Santa Clara, CA) were then carried out as per the manufacturer’s protocol and the raw data were extracted using the Agilent Feature Extraction software (Version 10.1).
Statistical analysis of microarray data
Identification of microbe phylogeny was systematically achieved using a microarray chip we developed and validated specifically for comprehensive analysis of the human stool microbiome (25). Signals from 852 separate bacterial species were obtained for each patient individually. The bacteria were classified into 119 different genera, 53 families, 25 orders, 15 classes, and 8 phyla. Signal intensities were normalized and the 852 adjusted signals were summed and the percent of the total adjusted signal was determined for all bacteria. Signals were then summed separately by Genus, Family, Order, Class, and Phylum and the percentages of the total signal were determined for each hierarchical classification level.
For statistical analyses of specific signals, all signals were transformed using the log base 2 logarithmic transformation. Comparisons of log transformed signals between the healthy and UC groups were made with two-sample t tests or unequal variances two-sample t tests when group variances were statistically different. P values were adjusted for multiple comparisons with the Benjamini-Hochberg method. All signal analyses were conducted using GeneSpring GX version 11.0 (Agilent Technologies Inc., Santa Clara, CA). The number of phylospecies detected (presented as richness) was compared between the healthy and UC groups, as well as between the responders and non-responders in the UC group, with two-sample t tests. Relative abundance of species, as analyzed by signal intensity, is presented as evenness. Shannon index of biodiversity was calculated using the following formula: H’= −∑(Pi * ln Pi), where Pi is the frequency of each species identified in the array; t test was performed between values in controls and UC patients.
Comparisons between the demographics and clinical features of healthy and UC groups and between responders and non-responders for gender, race, disease onset, disease extent, and pre-admission steroids were made with Fisher’s exact tests. Age, height, BMI, disease duration, PUCAI, and bloody stools/day were compared with two-sample t tests. Demographic comparisons were made using SAS v9.2 (SAS Institute, Inc, Cary, NC). All continuous variables are presented as the mean ± standard deviation (SD) unless otherwise specified.
Results
Baseline characteristics
The OSCI cohort with available stool samples consisted of 101 children, of whom 27 did not receive antibiotics or probiotics for at least one month prior to sample collection; only these individuals were included in this study. Steroid failure was seen in 10 of the 27 eligible children. The remaining 17 children were defined, therefore, as steroid responsive. Fifteen children resided in Canada and 12 in the USA.
Comparisons were made to healthy children (n=26) with similar characteristics to the study population regarding age, gender, and BMI (Table 1A). Since a slight difference in race was noted a subgroup analysis was performed excluding races other than white. Results were similar to the comparisons using all subjects (data not shown). Therefore, only analyses using all subjects are reported. For children with UC, mean (±SD) age at admission was 13.6±3.0 years, median disease duration 3.5 months (IQR 0–13.1 months), and in 12 cases (44%) the admission was associated with first onset of disease. In 89% of patients extensive disease or pancolitis [E3 and E4 Paris Classification (30), respectively] were found; 12% had left sided disease (E2); 63% were males. The average PUCAI score at admission was 70±12. Those who responded to corticosteroid therapy appeared to be more likely to have new-onset of disease and less likely to be admitted on oral steroids, although, in contrast to the larger cohort described in the original OSCI study (26), none of these differences were statistically significant in this limited study (Table 1B). As expected, PUCAI score and the number of bloody stools/day appeared to decline on day 3 of treatment in responders but not in non-responders. All patients had bloody diarrhea on days 1 & 3 of steroid therapy.
Table 1.
Characteristics of healthy children and children with ulcerative colitis (UC).
| A: Healthy controls vs. UC: | |||
|---|---|---|---|
| Variable | Healthy (n=26) |
UC (n=27) |
P Value |
| Gender: | .347 | ||
| Male | 14 (53.8) | 17 (63.0) | |
| Race: | .023 | ||
| White | 26 (100) | 21 (77.8) | |
| Other | 0 (0) | 6 (22.2) | |
| Age (y) | 13.3±2.7 | 13.6±3.0 | .767 |
| Height (cm) | 155.5±16.9 | 159.4±18.0 | .417 |
| BMI (kg/m2) | 19.4±3.7 | 19.9±4.0 | .587 |
| B: Responders vs. non-responders to corticosteroid therapy: | |||
|---|---|---|---|
| Variable | UC responders (n=17) |
UC non- responders (n=10) |
P Value |
| New onset (yes) | 9 (53) | 3 (10) | .167 |
| Disease duration (mo.) | 22±21 | 24±36 | .862 |
| Pancolitis (yes) | 16 (94) | 8 (80) | .261 |
| Pre-admission oral steroids | 2 (12) | 4 (40) | .096 |
| Day 1 PUCAI score | 70±12 | 71±8 | .963 |
| Day 3 PUCAI score | 44±22 | 60±19 | .064 |
| Day 1 bloody stools/day | 8.9±3.9 | 6.8±1.6 | .352 |
| Day 3 bloody stools/day | 4.9±2.8 | 6±3.4 | .368 |
Values in the tables are No. (%) for gender, race, new onset, pancolitis, pre-admission oral steroids; mean±SD for age, height, BMI, disease duration, PUCAI, stools/day. Mean disease duration is calculated only for those who were not new-onset.
Children with severe UC have reduced gut biodiversity
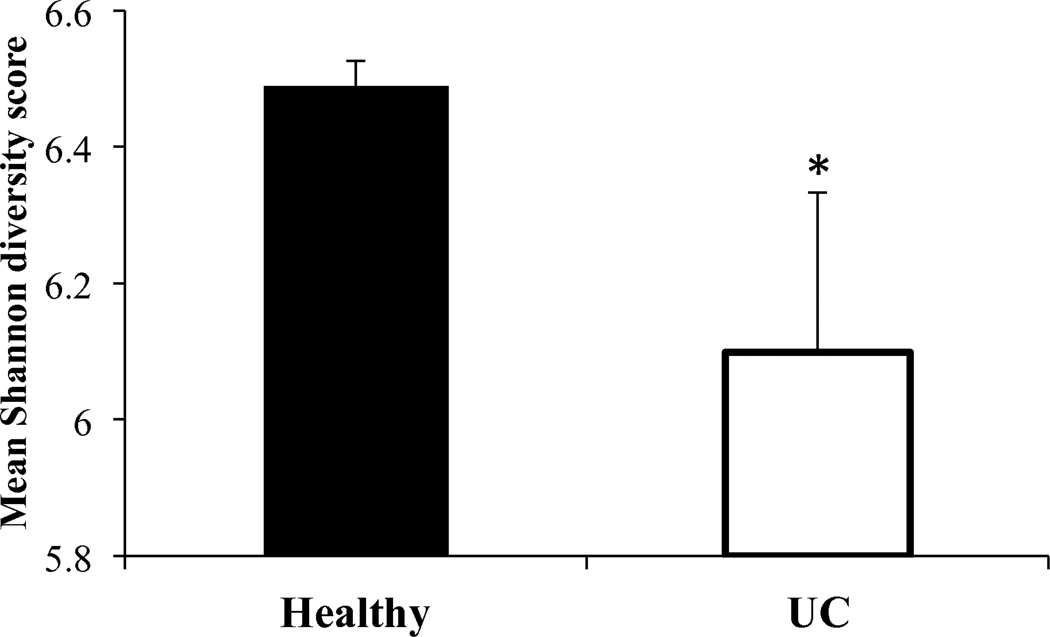
The Shannon diversity index was used as a measure of microbial biodiversity as it accounts for both the number of species (richness) and their relative distribution (evenness). Interestingly, our array showed relatively high biodiversity in both healthy controls and children with severe UC; however, the Shannon index was significantly higher in healthy children (6.488±0.04 vs. 6.098±0.23 in UC; p<0.0001; Figure 1). There were no differences in the Shannon index between UC patients who responded to corticosteroids and those who did not (6.118±0.23 and 6.065±0.25, respectively; p=0.58).
Figure 1. Microbial biodiversity is reduced in children with severe ulcerative colitis.
Mean (±SD) Shannon index of biodiversity in healthy controls (Healthy) and patients with severe ulcerative colitis (UC). * p<0.0001.
Comparisons of signal strength and percent composition of the microbiome (evenness)
In the comparison of both raw and log-transformed signals, microbial intensities were different between the groups at the Phylum, Class, Order, and Family levels. Only 3 of 119 Genera and 42 of 852 phylospecies did not show statistically significant differences between groups (adjusted p>0.05). Furthermore, we found a remarkable overall decrease in signal intensity in 95% of phylospecies in patients with UC; in light of that, we further investigated the percent distribution of individual groups of bacteria that comprise the gut microbiome to identify alterations in relative community composition. Analysis of percent of total signals between groups showed statistically significant differences in 6 of the 8 (75%) studied Phyla, 14 (93%) Classes, 21 (84%) Orders, 35 (66%) Families, 83 (70%) Genera, and 534 (63%) phylospecies (adjusted p<0.05). There was no statistically significant difference in the evenness of microbial signal (log-transformed signal or the percent of total signal) based on steroid response within the UC group of patients; however, as described below, there was a substantial decrease in the richness of phylospecies in children with steroid failure compared to those who were steroid responsive. There was no difference in the richness or evenness of the gut microbiome based on geographic location whether residing in the USA or Canada.
Effects at the Phylum level
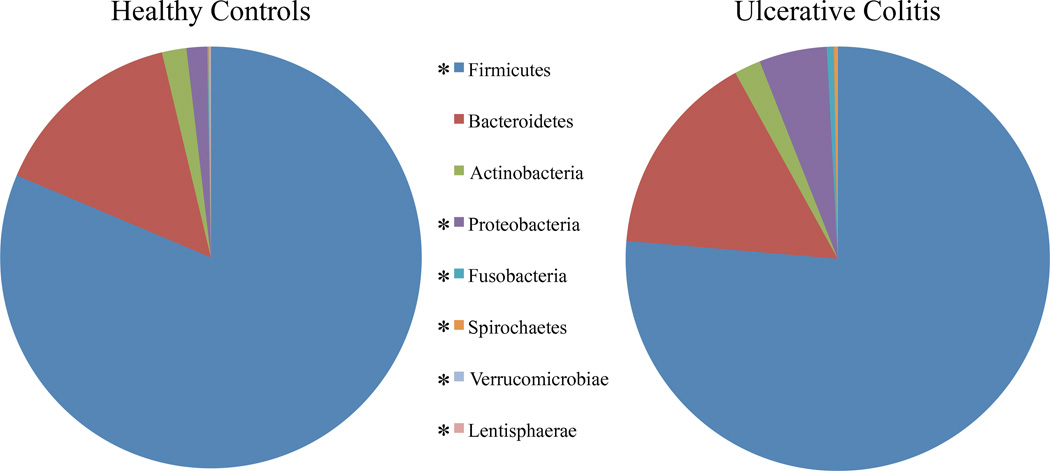
In comparison of both the raw and the log-transformed signals, there was a significant decrease in all 8 Phyla included in the microarray for the fecal microbiome of children with severe UC relative to controls (Table 2). Furthermore, the proportion of the total signal showed a significant difference between groups in the adjusted percentage of the mean signal intensity for six of the eight Phyla, aside from Actinobacteria and Bacteroidetes. In UC, there was a decrease in the percentage of the signal in Firmicutes, Lentisphaerae, and Verrucomicrobia and an increase in Proteobacteria, Fusobacteria, and Spirochaetes (Figure 2 and Table 2). The largest changes were noted as a 5% decrease of relative signal in Firmicutes and a 3.6% increase in Proteobacteria in UC.
Table 2.
Phylum and Class comparisons for healthy children and children with ulcerative colitis (UC).
| Log-transformed signal | Percent of total raw signal | ||||||
|---|---|---|---|---|---|---|---|
| Phylum (Class) | Number of Species |
Healthy (n=26) |
UC (n=27) |
Corrected P value |
Healthy (n=26) |
UC (n=27) |
Corrected P value |
| Actinobacteria | 29 | 19.58±0.76 | 14.11±2.70 | <.0001 | 1.88±0.27 | 2.02±0.80 | .451 |
| (Actinobacteridae) | 20 | 18.42±0.74 | 13.33±2.80 | <.0001 | 0.85±0.14 | 1.21±0.53 | .003 |
| (Coriobacteridae) | 9 | 18.71±0.80 | 12.77±2.60 | <.0001 | 1.03±0.16 | 0.81±0.37 | .009 |
| Bacteroidetes (Bacteroidetes) | 142 | 22.54±0.68 | 16.69±2.77 | <.0001 | 14.90±2.68 | 15.64±11.96 | .760 |
| Firmicutes | 623 | 25.03±0.65 | 19.46±2.51 | <.0001 | 81.38±2.59 | 76.35±11.27 | .041 |
| (Bacilli) | 21 | 19.11±0.80 | 14.37±2.75 | <.0001 | 1.39±0.37 | 2.82±2.22 | .004 |
| (Clostridia) | 563 | 24.97±0.65 | 19.33±2.51 | <.0001 | 78.27±2.55 | 69.98±9.72 | <.001 |
| (Erysipelotrichi) | 39 | 19.43±0.60 | 14.98±2.62 | <.0001 | 1.72±0.41 | 3.55±1.15 | <.0001 |
| Fusobacteria (Fusobacteria) | 3 | 15.08±0.73 | 11.50±2.84 | <.0001 | 0.09±0.02 | 0.53±0.74 | .005 |
| Lentisphaerae (Lentisphaerae) | 1 | 14.15±0.93 | 5.65±3.41 | <.0001 | 0.05±0.02 | 0.02±0.02 | <.001 |
| Proteobacteria | 49 | 19.32±0.66 | 15.33±2.45 | <.0001 | 1.57±0.25 | 5.17±3.93 | <.001 |
| (Alphaproteobacteria) | 9 | 14.87±0.90 | 11.32±3.04 | <.0001 | 0.08±0.03 | 0.39±0.33 | <.0001 |
| (Betaproteobacteria) | 18 | 17.50±0.80 | 12.56±2.85 | <.0001 | 0.47±0.18 | 0.96±0.80 | .005 |
| (Deltaproteobacteria) | 5 | 16.92±0.80 | 12.05±2.59 | <.0001 | 0.30±0.05 | 0.52±0.33 | .003 |
| (Epsilonproteobacteria) | 6 | 16.47±0.61 | 13.07±2.32 | <.0001 | 0.22±0.03 | 0.99±0.52 | <.0001 |
| (Gammaproteobacteria) | 11 | 17.67±0.64 | 13.90±2.39 | <.0001 | 0.51±0.13 | 2.31±2.92 | .005 |
| Spirochaetes (Spirochaetes) | 4 | 15.09±0.69 | 10.58±3.05 | <.0001 | 0.09±0.02 | 0.25±0.28 | .008 |
| Verrucomicrobiae (Verrucomicrobiae) | 1 | 12.60±4.46 | 6.76±3.42 | <.0001 | 0.06±0.06 | 0.03±0.04 | .018 |
Figure 2. Phylum level changes in severe ulcerative colitis.
Percent of raw signals for each of the 9 measured phyla are presented. Patients with severe ulcerative colitis had relative reductions in Firmicutes, Verrucomicrobiae, and Lentisphaerae and increases in Proteobacteria, Fusobacteria, and Spirochaetes. * corrected p<0.05.
Alterations at the Class level
There was a significant decrease in the log transformed signal intensities for each of the 15 examined classes between the groups. In the comparison of the percentage of signal intensity, there was a significant difference for 14 of the 15 Classes. There was a decrease in the percentage of the signal in UC patients in Clostridia, Lentisphaerae, Coriobacteridae, and Verrucomicrobiae and higher signals in Actinobacteridae, Bacilli, Erysipelotrichi, Fusobacteria, Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria, Gammaproteobacteria, and Spirochetes (Table 2).
At the Genus level
In the comparison of log-transformed signal intensities between groups, there was a significant difference in signals for 116 of the 119 Genera. Of the 116 Genera that showed a significant difference, the signal strength was decreased in the UC group in 101 Genera, while it was increased in 15.
In the comparison of the percentage of the total signals between groups with UC and healthy controls, there was a significant difference in the adjusted percentage of the mean signal for 82 of the 119 Genera. Of the 82 Genera that showed a significant difference, the signal strength was decreased in the UC group in 56 and increased in 26, including Escherichia coli.
At the Phylospecies Level
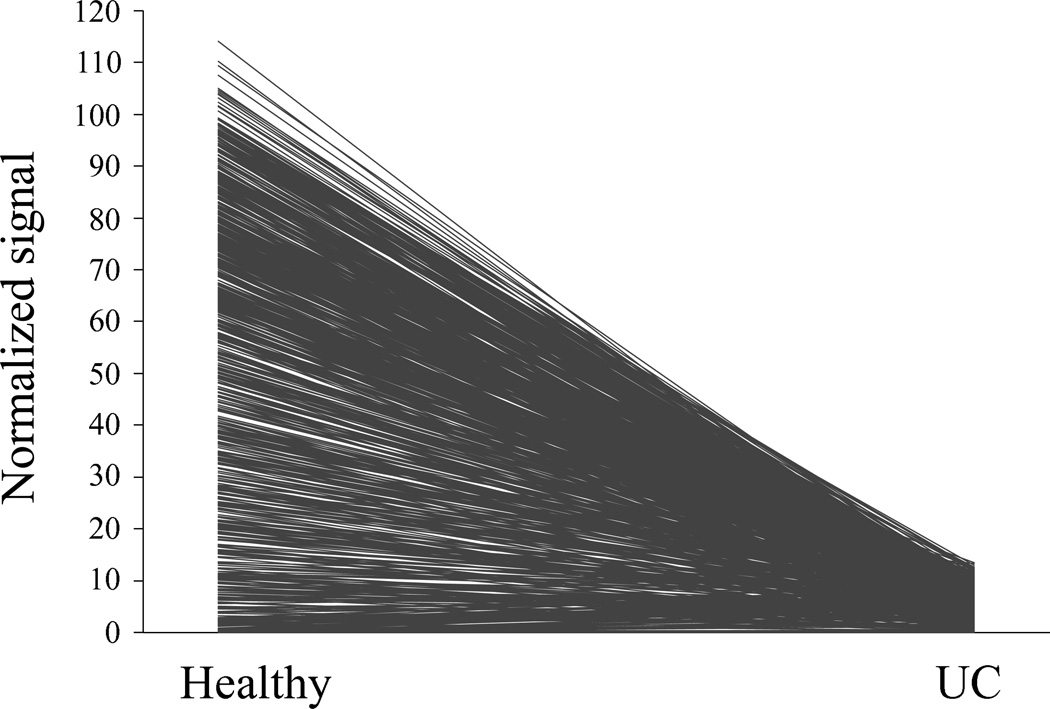
Comparison of the raw and log-transformed signal between the groups demonstrated a significant difference in the raw signal for 810 of the 852 species studied. The signal strength decreased in the UC group in 758 of the species, whereas it increased in 52 of the species (Figure 3).
Figure 3. Reduced microbial intensities in pediatric ulcerative colitis.
Profile plot representation of normalized signal intensities of each of the phylospecies in children with ulcerative colitis (UC) vs. healthy controls (Healthy). Each line represents one of the 852 phylospecies. The figure demonstrates a dramatic decrease in the signal intensity for the majority of the phylospecies.
Richness of the fecal microbiome
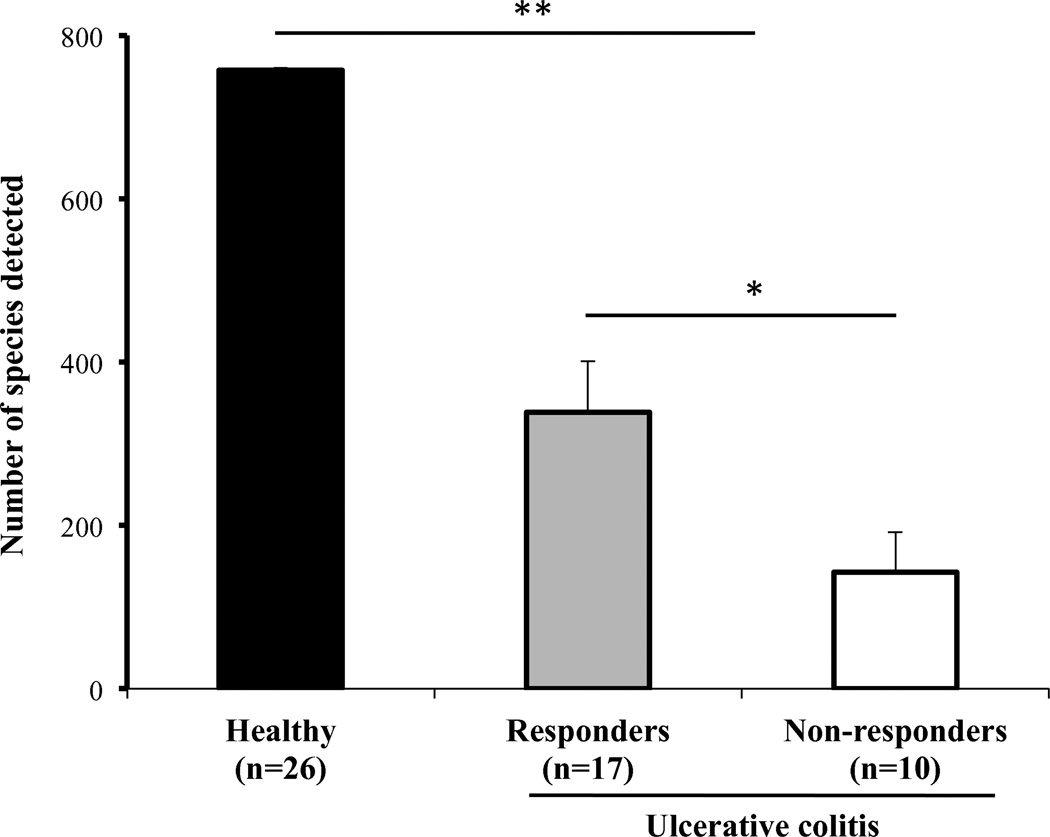
Richness is a reflection of the number of detected phylospecies. Children with UC had severe compromise in gut microbiome diversity with an average number of detected phylospecies of 266±69, while healthy children had 758±3 (p<0.001) phylospecies detected. Interestingly, children who failed steroid therapy had significantly less microbial diversity than children who were steroid responsive (142±49 vs. 338±62 detected phylospecies respectively, p=0.013, Figure 4).
Figure 4. Diminished microbial richness correlates with ulcerative colitis and lack of response to steroids.
Number of species detected in fecal samples of healthy children (Healthy) and children with severe ulcerative colitis showing statistically significant difference (** p<0.001). Differences also noted (* p=0.013) between children with ulcerative colitis who responded to steroids (Responders) and those that failed to respond (Non-responders); values depicted as mean ± SD.
Discussion
This study describes, for the first time, the fecal microbiome profile of a large number of children hospitalized with severe ulcerative colitis using a comprehensive human fecal microbial 16S rRNA microarray. We found substantial reductions in the richness, evenness, and biodiversity of the gut microbiome in these children, supporting a correlation between microbial diversity and bowel health. Although data on disease severity were available for each individual, the number of patients was too small to directly correlate severity with microbial diversity. However, the fact that patients refractory to corticosteroids had further reduction in microbial richness, relative to those who responded to therapy, suggests a ‘dose response’ effect, where more severe illness associates with further reduced richness. Our findings cannot determine whether poor diversity is a cause or result of severe colitis; however, this feature could be used as an important marker of severity, possibly even predicting response to corticosteroids.
In addition, this study demonstrates a decrease in signal in almost all phylospecies (Figure 3), reflecting a reduction in overall abundance of the microbiome in children with severe UC. This is consistent with other studies of the microbiome in adults with UC in which the overall richness is diminished (31–34). Analysis of the relative percent of signals shows a decreased percentage representation of Firmicutes, Lentisphaerae, and Verrucomicrobia phyla and an increase in Fusobacteria, Proteobacteria, and Spirochaetes. At the Class level, we see most notably a decrease in Clostridia by more than 8%, with an increase in Gammaproteobacteria and Erysipelotrichi. These data suggest an overall shift in the entire makeup of the microbiome in severe UC. The most significant Phylum-level changes came with Proteobacteria, which contains E. coli, increasing by 3.6% and Firmicutes decreasing by 5% of the total composition in severe UC (Table 2 and Figure 2).
Immediately after birth the sterile intestine of the neonate is colonized by Enterobacteria and Enterococci. The microbiome continues its dynamic process of change during childhood with reduction in aerobes and facultative anaerobes and an increase in obligate anaerobes (35). The number of microbiota that constitute the gut microbiome is thought to exceed 1014 organisms encompassing vast diversity and complexity (36) with a vital symbiotic role in digestion, development, vitamin and mineral production, modulation of motility, protection from pathogens, and immune function (37). Along with altered host defenses and immune regulation, the intestinal microbiota plays an important role in the pathogenesis of IBD. Recent studies have demonstrated a dysbiosis with alterations in the relative numbers of microorganisms in the microbiome of IBD patients (20, 38–44). The amplification of 16S rRNA from complex microbial communities has been shown to be an effective method for analyzing the complex composition and diversity of the community (25). The use of microarray technology allows scientists to assess the microbiome in a unique manner through measuring the relative composition of a massive number of known inhabitants of the microbiome.
Prior molecular characterization of the microbiota in IBD yielded a number of consistencies. The first is the overall decrease in microbial diversity in the microbiome (32–34, 45–46). Using T-RFLP, Nishikawa et al, showed reduced diversity of mucosa-associated microbes in patients with active UC and that the diversity increased after these patients improved, although the clinical improvement was achieved using antibiotics (42). However, Martinez et al, also found that DGGE patterns changed in UC patients over time, suggesting reduced stability (41). In addition, most studies have found an increase in Enterobacteriaceae and E. coli, and a decrease in Firmicutes, especially Clostridium. More recently, Lepage et al. showed a relative increase in Proteobacteria and reduction in Firmicutes among patients with UC, relative to unaffected twins (34). These results match our own findings. Where studies do not uniformly agree, is the disease-specific composition of the microbiome in Crohn disease and UC, including the abundance vs. depletion of Bacteroides, specifically Faecalibacterium prausnitzii, in disease states, the increased vs. decreased concentration of mucosa-bound bacteria, and the variability in the microbiome during active inflammation. A comprehensive study by Frank et al, (23) showed a decrease in Firmicutes and Bacteroidetes, and an increase in Proteobacteria and Actinobacteria in IBD patients. The decrease in Firmicutes was due to loss of Clostridium, and the increase in Proteobacteria, including E. coli, inflated due to loss of Bacteroidetes and Lachnospiraceae.
The first study in pediatric IBD patients by Conte et al, (47) used culture techniques of biopsy samples from children with IBD and found a higher numbers of mucosa-associated bacteria and a decrease in specific groups of normal anaerobic microbiota, particularly Bacteroides vulgatus. More recently, using qPCR to quantify 9 species/groups, Schwiertz et al, did not identify substantial alterations in the microbiota of children with UC, while changes were observed in individuals with Crohn disease; however, in contrast to our study, most of these children had mild disease at the time of stool sampling (48).
This study is the first to molecularly analyze the microbiome in a relatively large sample size of 27 children investigating a unique cohort of hospitalized children with severe UC. The use of microarray technology with its ability to specifically measure 852 species simultaneously to assess the microbiome, creates a clear picture of the shift in the composition of the microbiome during disease states. The study clearly demonstrates that the richness and evenness of the gut microbiome are severely impacted in children with severe UC. The Shannon diversity index, which encompasses richness and relative abundance of species in a biological system, thus reflecting both increases in the number of different species and in the relative frequency of species, is considered to be an excellent measure of ecosystem health (49). In our study we found that children with severe UC had lower Shannon diversity scores, suggesting that reduced diversity and dysbiosis are linked to severe UC.
Various mechanisms have been proposed for the dysbiosis found in IBD including altered adhesion proteins leading to enhanced binding of microbiota (9). However, it is not yet clear whether changes in bacterial composition are primary disturbances leading to tissue damage or secondary to inflammation occurring in the bowel. Some studies have shown changes in the microbiome during disease quiescence, and in histologically normal sections of bowel, pointing towards a primary disturbance. One of the questions that, therefore, remain unanswered is whether these changes reflect mere inflammation, dysmotility, diet, or a true role of the gut microbiome in the pathogenesis and development of UC. As understanding how luminal microbes relate to inflammation in UC is clearly a research priority (50), future characterization of this community will be important in unraveling the disease pathogenesis as well as potential development of therapeutic regimens tailored to target the microbiome.
In addition, this study is among the first to measure the microbiome in children with IBD. The findings of dysbiosis in this study with a decrease in Firmicutes and an increase in Proteobacteria are similar to changes seen in adult disease. Thus, these changes in the gut microbial composition are evident early in the disease process, and may persist throughout time and course of illness into adulthood, suggesting a potential role in pathogenesis.
Dysbiosis, as exemplified in this study, may lead to inflammation in IBD through a number of mechanisms. Even slight alterations in the balance between protective and aggressive microbiota can favor inflammation. As shown in other molecular analyses of the microbiome, our results indicate a reduction in Gram-positive organisms that constitute the Firmicutes phylum. This phylum of anaerobic cocci possesses specific activities that can alter metabolic and immune function in the intestinal tract (51). Furthermore, Clostridia and Bacteroides are primary producers of butyrate and other short chain fatty acids, which act both as energy substrate for intestinal epithelial cells and as anti-inflammatory agents at the epithelial layer. Specifically, butyrate has been shown to decrease proinflammatory cytokines by inhibiting NF-κB activation in patients with Crohn disease (52–54). Similar to the results shown in other studies, this study shows a dysbiosis with an increase in Gram-negative bacteria such as E. coli. An increase in Gram-negative microbiota on the mucosal surface can be associated with increased concentration of lipopolysaccharide, a component of Gram-negative bacteria cell wall, which triggers inflammation (55). Much remains to be determined in the complex interaction between the immunologic and metabolic function of the microbiome and its role in disease process.
One of the limitations of this report is that the samples analyzed are fecal rather than mucosal. Studies utilizing intestinal biopsies are able to examine bacteria directly interacting with the mucosa; however, biopsies pose additional challenges including the need for surgical prep of the colon that may modify microbiota, sampling of a small and specific portion of the intestinal tract, which itself has shown tremendous variability along the gut (56), and the invasive of this procedure nature leading to small sample sizes, especially in children. Another consideration is the potential effect of diarrhea alone on the stool microbiome, which can lead to a reduction in microbial diversity due to flushing effect (57). In addition, increased water in diarrheal stools could dilute microbial content. While we cannot exclude that flushing alone can explain at least part of our observations, the finding that relative abundance of species and the Shannon diversity index were altered suggests a more global correlation between UC and the microbiome.
Although we have identified changes in microbiota in children with UC, our findings cannot define the cause for these alterations. Since diarrhea and inflammation can both impact the microbiota (58), our findings may represent a secondary effect of disease. For instance, many proteins shift from the blood stream into the lumen of the bowel during active mucosal inflammation (e.g. ferritin) that have anti-microbial properties. Medications, nutrition, or other undefined factors may also have anti-microbial/microbiota altering roles; however, patients were excluded if they received antibiotics one month prior to the study or during the study. Other medications, including corticosteroids that all patients were receiving, could also alter gut microbiota, although no data are currently available on these potential effects. Nevertheless, we cannot exclude that at least part of the observed differences are due to factors indirectly related to the patients’ active disease or pathogenesis.
Another potential limitation of our approach is the lack of ability to identify emergence of microbial species that are not included in the array. This could result in an over-estimation of the relative abundance of known species, as we cannot account for unique species not represented in our array that may appear in specific disease states. Although some studies have suggested that patients with IBD may have higher abundances of specific strains (43–44), many of these would still likely be represented on our array. Deep sequencing of these samples would provide additional valuable data, but the custom array used in this study provides a more manageable and likely reproducible dataset that is advantageous as a practical tool for potential clinical use. In addition, while PCR bias applies to comparisons of abundance of bacterial species to others, rather than comparing the same bacteria among experimental groups (59), this study is focused on comparing the relative abundance of specific bacteria in healthy vs. diseased state, thus circumventing the effect of PCR bias.
It may not be sufficient to understand “who lives” in the intestinal tract of children with UC; there is a need to link transformations of the microbiome with functional changes in the metabolism and immune function profile of these organisms. The pathways and interactions of the gene loci and their role in host epithelial barrier function, bacterial killing, cell signaling, and innate and adaptive immune response should be explored. Advancing our knowledge of the gut microbiome will help develop therapeutic strategies to more accurately target this fascinating microbial community.
This is the first study to extensively measure the microbiome in children hospitalized with severe UC and the first to recognize an association between gut microbial diversity and response to therapy in this fragile pediatric population.
Acknowledgement
This work was supported by NIH; NCCAM AT003400 and the Children’s Medical Center Research Foundation. Partial funding was obtained from Schering-Plough, Canada that allowed patient recruitment and stool collection. E.W. is an Alberta Innovates Health Solutions (AIHS) Clinical Investigator. Infrastructure in E.W.’s laboratory is funded by the Centre for Excellence for Gastrointestinal Inflammation and Immunity Research (CEGIIR) at the University of Alberta and the Alberta Inflammatory Bowel Disease Consortium, which is supported by an AIHS Interdisciplinary Team Grant. We thank all the patients, their families, and health providers that contributed to this study.
References
- 1.Turner D. Severe acute ulcerative colitis: the pediatric perspective. Dig Dis. 2009;27:322–326. doi: 10.1159/000228568. [DOI] [PubMed] [Google Scholar]
- 2.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seksik P, Rigottier-Gois L, Gramet G, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onderdonk AB, Hermos JA, Bartlett JG. The role of the intestinal microflora in experimental colitis. Am J Clin Nutr. 1977;30:1819–1825. doi: 10.1093/ajcn/30.11.1819. [DOI] [PubMed] [Google Scholar]
- 7.Rutgeerts P, Goboes K, Peeters M, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 8.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent-invasive Ecoli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn's disease. Curr Opin Gastroenterol. 2007;23:16–20. doi: 10.1097/MOG.0b013e3280105a38. [DOI] [PubMed] [Google Scholar]
- 11.Prantera C, Scribano ML. Antibiotics and probiotics in inflammatory bowel disease: why, when, and how. Curr Opin Gastroenterol. 2009;25:329–333. doi: 10.1097/MOG.0b013e32832b20bf. [DOI] [PubMed] [Google Scholar]
- 12.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland L, Singleton J, Sessions J, et al. Double blind, placebo controlled trial of metronidazole in Crohn's disease. Gut. 1991;32:1071–1075. doi: 10.1136/gut.32.9.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradel KO, Nielsen HL, Schonheyder HC, et al. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Porter CK, Tribble DR, Aliaga PA, et al. Infectious gastroenteritis and risk of developing inflammatory bowel disease. Gastroenterology. 2008;135:781–786. doi: 10.1053/j.gastro.2008.05.081. [DOI] [PubMed] [Google Scholar]
- 16.Jess T, Simonsen J, Nielsen NM, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut. 2011;60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 17.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondon MR, August PR, Bettermann AD, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cucchiara S, Iebba V, Conte MP, et al. The microbiota in inflammatory bowel disease in different age groups. Dig Dis. 2009;27:252–258. doi: 10.1159/000228558. [DOI] [PubMed] [Google Scholar]
- 21.Codling C, O'Mahony L, Shanahan F, et al. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 55:392–397. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swidsinski A, Loening-Baucke V, Vaneechoutte M, et al. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 25.Paliy O, Kenche H, Abernathy F, et al. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner D, Mack D, Leleiko N, et al. Severe pediatric ulcerative colitis: a prospective multicenter study of outcomes and predictors of response. Gastroenterology. 2010;138:2282–2291. doi: 10.1053/j.gastro.2010.02.047. [DOI] [PubMed] [Google Scholar]
- 27.Jakobsson HE, Jernberg C, Andersson AF, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423–432. doi: 10.1053/j.gastro.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Agans R, Rigsbee L, Kenche H, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: The Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 31.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 32.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lepage P, Hasler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 36.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 38.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 39.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. e1841. [DOI] [PubMed] [Google Scholar]
- 40.Sepehri S, Kotlowski R, Bernstein CN, et al. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675–683. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- 41.Martinez C, Antolin M, Santos J, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–648. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 42.Nishikawa J, Kudo T, Sakata S, et al. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44:180–186. doi: 10.1080/00365520802433231. [DOI] [PubMed] [Google Scholar]
- 43.Sokol H, Seksik P, Rigottier-Gois L, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 44.Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamboli CP, Neut C, Desreumaux P, et al. Dysbiosis as a prerequisite for IBD. Gut. 2004;53:1057. [PMC free article] [PubMed] [Google Scholar]
- 46.Tamboli CP, Neut C, Desreumaux P, et al. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conte MP, Schippa S, Zamboni I, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55:1760–1767. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwiertz A, Jacobi M, Frick JS, et al. Microbiota in pediatric inflammatory bowel disease. J Pediatr. 2010;157:240–244. doi: 10.1016/j.jpeds.2010.02.046. e241. [DOI] [PubMed] [Google Scholar]
- 49.Camp JG, Kanther M, Semova I, et al. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 51.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segain JP, Raingeard de la Bletiere D, Bourreille A, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan SH, Barcenilla A, Stewart CS, et al. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Netea MG, Kullberg BJ, de Jong DJ, et al. NOD2 mediates anti-inflammatory signals induced by TLR2 ligands: implications for Crohn's disease. Eur J Immunol. 2004;34:2052–2059. doi: 10.1002/eji.200425229. [DOI] [PubMed] [Google Scholar]
- 56.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 58.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Rigsbee L, Agans R, Foy BD, et al. Optimizing the analysis of human intestinal microbiota with phylogenetic microarray. FEMS Microbiol Ecol. 2011;75:332–342. doi: 10.1111/j.1574-6941.2010.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]