Abstract
The T-cell development program is specifically triggered by Notch-Delta signaling, but most transcription factors needed to establish T-cell lineage identity also have crossover roles in other hematopoietic lineages. This factor sharing complicates full definition of the core gene regulatory circuits required for T-cell specification. But new advances illuminate the roles of three of the most T-cell specific transcription factors. Commitment to the T-cell lineage is now shown to depend on Bcl11b, while initiation of the T-cell differentiation program begins earlier with the induction of TCF-1 (Tcf7 gene product) and GATA-3. Several reports now reveal how TCF-1 and GATA-3 are mobilized in early T cells and the pathways for their T-lineage specific effects.
Introduction
Multipotent or lymphoid-biased precursors enter the T-cell developmental pathway in response to thymic microenvironmental signals [1]. The most important trigger is Notch pathway signaling, activated in the precursors by contact with Delta-family Notch ligands expressed by thymic epithelial cells. Pro-T cells then proliferate under continued influence of Notch signaling and remain Notch-dependent through T-lineage commitment, until after successful gene rearrangement enables them to express TCRβ or TCRγδ. However, something more durable and portable than a direct response to Notch pathway signaling must sustain the T-cell gene expression program later, during cell migration through multiple environments and more or less proliferation. The cells establish expression combinations of transcription factors that not only drive T-cell “identity” genes – those encoding TCR/CD3 components, signaling kinases, phosphatases, and adaptors – but also cross-regulate each other to stabilize the T-cell regulatory state.
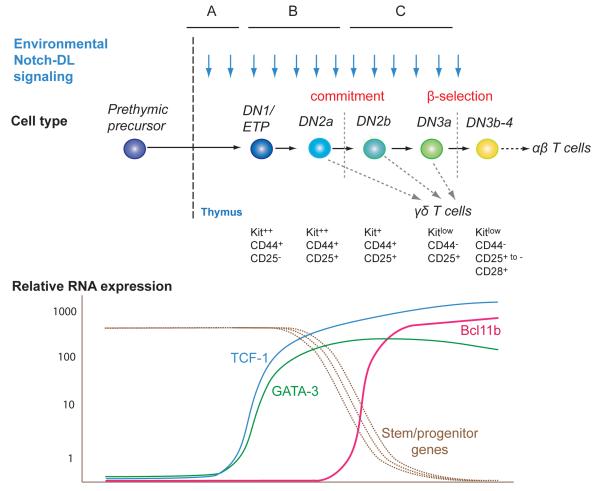
T cell specification has multiple regulatory requirements, but most of the factors needed by T cells are also needed in other hematopoietic differentiation programs (e.g. Ikaros, Gfi1, Myb, Runx1/CBFβ, E2A and its relatives)[2]. Presumably these regulate T-cell specific genes mainly as components of lineage-specific combinations. Three transcription factors are much more T-cell specific in their expression: Bcl11b, GATA-3, and TCF-1 (encoded by Tcf7). These three factors are steeply upregulated in precursors in response to Notch signaling and sustained in T-cell development at varying levels thereafter (Fig. 1). Their sharp profiles of induction during T-cell development offer clear opportunities to reveal how their activities change multipotent progenitors into true pro-T cells.
Figure 1.
Stages in progression through T-lineage commitment and β–selection. Phenotypes and key developmental events are indicated. γδ cells can emerge at several branchpoints. Notch interactions are sustained as shown. RNA expression patterns are approximate.
The T-cell specification process is actually a succession of three distinct regulatory states (Fig. 1A,B,C). Activation of GATA-3 and TCF-1 marks the transition from the first to the second, and activation of Bcl11b marks the transition from the second to the third. By considering how the advent of each factor affects the appropriate transition, the nature of the process is emerging in sharper focus.
Punctuating the T-lineage commitment process: role of Bcl11b
Bcl11b, a bifunctional C2H2 zinc finger factor, was discovered as a potential suppressor of radiation-induced T-cell lymphomas, and independently as a corepressor collaborating with the nuclear receptor COUP-TF (rev. in [3]). Its relative Bcl11a is a proto-oncogene in B lineage cells, essential for B-cell development, and a repressor of fetal-type hemoglobins; but Bcl11b is restricted to T-lineage cells (and at a lower level, NK cells) within hematopoiesis. Bcl11b is crucial for αβ T cell development but dispensable for some γδ T cells, and for some time its main role appeared to be exerted during β-selection [4-6]. It regulates αβ-lineage cell survival, developmental fidelity, and mature T-cell function from the CD4+ CD8+ stage onward [*6,7], restraining the latent NK-like differentiation potential in mature CD8 cells [**8], and supporting both CD8-cell expansion and regulatory T cell differentiation [9,*10]. However, its expression is initially induced in a dramatic upsurge much earlier, within the DN2 stage of T cell development [11]. This event closely coincides with the time frame when most T-lineage precursors become committed to a T cell fate [**12,13].
The linkage is more than a coincidence, as three recent reports have established that unless Bcl11b can be turned on, most T-cell precursors cannot become committed at all [**8,**14,**15]. These studies tracked precursors developing in response to continuous Notch-Delta signaling in vitro, where the lineage commitment impact of Bcl11b was readily observed. Bcl11b was perfectly dispensable for pro-T cell survival and proliferation under these conditions. However, deletion of Bcl11b could extensively prolong the time window in which T-cell precursors retain myeloid potential [**14,**15], and it enabled the developing cells to build up a formidable potential as natural killer (NK) cell precursors [**8,**15].
The role and mode of action of Bcl11b are now under investigation by many groups. It may be a timing factor for T-lineage commitment, since in fetal thymocytes, where differentiation is accelerated as compared to adult thymocytes, Bcl11b RNA begins to be detectable even at the DN1 stage (D.D. Scripture-Adams, M.M. Del Real, K.J. Elihu, and E.V.R., unpublished results). One aspect of the Bcl11b knockout phenotype is interesting from another perspective, however: namely, how much of the T-cell program can be activated without Bcl11b. Multiple gene expression changes normally occur in the DN2 to DN3 transition, since most genes involved in T-cell identity are upregulated immediately after Bcl11b [2], some but not all as direct Notch target genes [16-18]. Concomitantly, multiple “stem/progenitor-associated” genes are profoundly downregulated [19]. The DN2-like Bcl11b deletion phenotype dissects the T-cell specification process into separable modules. Stem/progenitor-associated genes fail to be shut off in the mutant cells, but T-cell transcription factors including GATA-3, and TCF-1 are fully induced to DN3-like levels, GATA-3 perhaps even higher than in normal DN2 cells [**15]. The T-lineage identity genes show split responses, with initial phases of Cd3g and Cd3e upregulation and Rag1 induction occurring, but Zap70 and Ptcra expression remaining weak [**15]. These responses dissect commitment, which depends on Bcl11b, from activation of the T-cell program, which GATA-3, TCF-1, and Notch can initiate even in cells that cannot become committed.
The difficulty with T-cell factors: asymmetric gain and loss of function phenotypes
TCF-1 and GATA-3 have long been recognized as essential for T-cell development [20,21]. Pioneering work using antisense oligonucleotides in fetal thymic organ culture systems showed that these factors were rate-limiting for early T-cell development, and implied additive roles [22]. However, dissecting what they do for T-cell specification has been held back by the peculiarities of their effects when ectopically activated.
GATA-3 plays at least three roles in T cell development: during initial specification, during TCRαβ-dependent positive selection, and in mature T cells where it establishes the Th2 effector program. In the Th2 context the addition of GATA-3 clearly promotes Th2 fate just as loss of GATA-3 inhibits it [23,24]. In TCRαβ-mediated positive selection of CD4+ lineage thymocytes its effects can be more complicated, but again the gain of function of GATA-3 promotes the CD4+ fate relative to other options [25-27]. However, its role has been murkier in the earliest stages. Loss of GATA-3 is profoundly inhibitory to T-cell precursors from the earliest ETP stage (see Fig. 1)[28,29]. GATA-3 binds to genes encoding TCR complex components [30,31], and in acute loss of function at certain stages some of these targets, e.g. the CD3 genes, are downregulated [**32](S. Damle, E.V.R., unpublished). But ectopic expression of GATA-3 in prethymic cells does not turn these genes on without Notch [33]. In early pro-T cells, added GATA-3 does not accelerate T-cell development, but can actually block it, whether or not Notch signaling is provided [34]. Forced GATA-3 expression can activate various inappropriate, non-T programs, depending on the cells into which it is introduced: a nonlymphoid, mast cell program in bone marrow cells or thymocyte progenitors [34], a nonlymphoid, megakaryocyte program in fetal liver precursors [35], and a myeloid program in Pax5-knockout pro-B cells [36]. GATA-3 dose-dependence may account for its problematic behavior in early T cells.
TCF-1 is best known as a signal-dependent transducer of environmental signals from the Wnt pathway via β-catenin [37-39]. In this pathway, without β-catenin, TCF family members act as repressors via recruitment of Groucho/TLE factors, but when Wnt signaling mobilizes β-catenin, β-catenin binding to TCF factors makes them work as activators. By this model the balance of positive and negative regulation of gene targets by TCF-1 should depend on the level of β-catenin. Indeed, ectopic expression of the Wnt antagonist, Dickkopf, could block T-cell development from a very early stage [40]. However, when hematopoietic progenitors have been forced to activate β-catenin, which should optimally activate TCF-1, the cells did not appear to upregulate lineage-specific T-cell genes. Instead, if they were already in the T-cell pathway they upregulated general survival genes [41], and if not, they reverted to a deregulated, multilineage gene expression state [42-44]. Furthermore, in loss of function experiments, mutation of the β-catenin cofactor alone or together with its relatives has had shockingly little effect on T-cell development [45,46], much less than the effect of mutating Tcf7 alone or together with its relative Lef1. Then is TCF-1 mostly important as a repressor? In general, evidence for direct positive regulation of T-cell specification genes by GATA-3 or TCF-1 has been lacking, leaving a major roadblock to understanding the early events in T-cell development.
Three levels of regulation for GATA-3 activity
The essential but complex roles of GATA-3 in T cell development can be explained in part because it does not always regulate the same target genes. Genome-wide maps of GATA-3 binding at different stages now confirm that this factor occupies different genomic sites depending on its developmental context [**32](J.A. Zhang, A. Mortazavi, B. Williams, B.J. Wold, and E.V.R., submitted). Site affinity and factor concentration probably contribute: e.g., the particularly high levels of GATA-3 in Th2 cells enable binding to many more sites than in other T-lineage cells [**32]. However, even in cells with similar levels of GATA-3, occupancy patterns are distinct according to developmental stage. When it is first upregulated in DN1 cells, we find that GATA-3 occupies sites in many stem/progenitor genes, then vacates them during commitment, shifting to bind more “T-cell identity” gene sites in the postcommitment CD4+ CD8+ stage (J.A. Zhang et al., op. cit.). It follows that GATA-3 normally collaborates with other developmentally-restricted factors to help define its targets. Because even the earlier-stage specific sites are genuine GATA-3 sites, however, it is easy to see how experimental overexpression could cause some GATA-3 binding to be inappropriately deployed. The control of GATA-3 levels itself must be extremely precise.
For years, the mechanisms responsible for inducing Gata3 expression at the beginning of T cell development were inaccessible. Two promoters were defined: a proximal one driving most T-cell expression, and a distal Th2-specific one where direct input from Notch was demonstrated in Th2 cells [47,48]. An intronic enhancer and an upstream lineage-specific silencer were also reported [49,50]. However, even a 650 kb Gata3 YAC transgene containing all these elements could not work in vivo to promote expression in T lineage cells [51]. Furthermore, in mature peripheral T cells and DN thymocytes alike, the proposed site of Notch input is buried under repressive H3K27me3 marking in all except Th2 cells, and is probably inaccessible in early stages [52,53](J.A. Zhang et al., op. cit.).
In a tour de force, Engel and coworkers have now found a T-lineage specific enhancer for Gata3 which can mediate activation from the earliest T-cell stages [**54]. The new enhancer is in a gene desert 280 kb downstream (3′) of the Gata3 gene, and it is necessary and sufficient to enable Gata3 transgene expression in T lineage cells. Activity of this enhancer may still need to be modulated by interaction with other cis-regulatory elements. However, this is now the region where to seek the mechanism that Notch signaling first uses to turn on Gata3 expression.
Importantly, GATA-3 is not only regulated through transcriptional control. In thymocyte development there are some mismatches between protein and RNA levels, e.g. during β-selection after successful TCRβ gene rearrangement, when RNA decreases but protein increases [25,55]. The key factor increasing GATA-3 protein seems to be activation via TCR. One reported mechanism is MAP kinase-controlled protein stabilization [56], but this could be more effective to preserve a pool in nondividing cells than to supply new protein during rapid cell division. Cook & Miller have now found another mechanism that enables signaled T cells to produce more GATA-3 protein de novo even from a declining pool of RNA [*57]. Through a PI3K-Akt-mTOR dependent pathway, translational efficiency of GATA-3 is specifically enhanced, possibly by unwinding secondary structure from the Gata3 translational start site. Interestingly, the PI3K-Akt-mTOR pathway can also be activated by IL-7R in DN2 cells and Notch signaling in DN3 cells [58,59]. Thus, GATA-3 protein levels could possibly “measure” integrated transcriptional and survival signals in the cells.
TCF-1: essential driver of T-cell specification downstream of Notch
As a widely used developmental signaling trigger, Notch activation does not embody enough specificity to be the origin of the whole T-cell program. One set of factors that may help to select T-cell specific target genes comprises basic helix-loop-helix E protein genes, including E2A [17]. However only a subset of T-cell genes is turned on by Notch and E2A together, conspicuously omitting Bcl11b, and none of these are sustained by E2A when Notch signals are withdrawn. Something else downstream of Notch must help activate and then sustain the T cell program, something that can act epistatically to Notch signaling in a gain of function assay. Neither GATA-3 nor Bcl11b can do this.
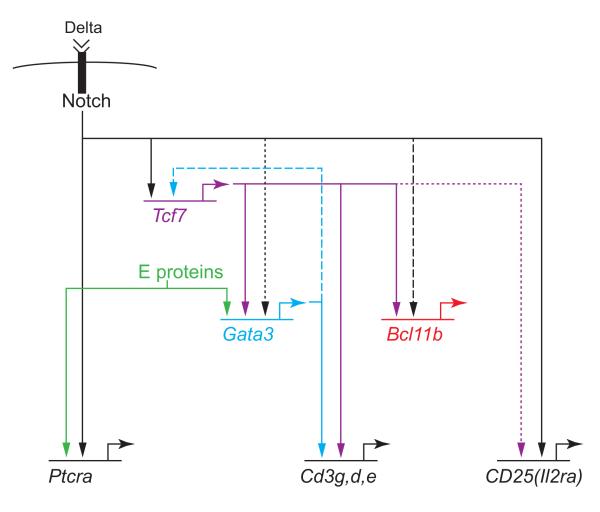
In the past year several groups have returned to TCF-1 as a candidate for initiating T-cell development downstream of Notch. These studies show that Tcf7 is directly activated by Notch through an enhancer 31.5 kb upstream of the promoter [*60,**61], and that TCF-1 becomes essential in adult T-lineage progenitors as soon as it is turned on at the ETP stage. An important role for TCF-1 seems to be antiapoptotic even in ETPs [*60]. However, Weber et al. have also demonstrated that forced expression of TCF-1 can activate most of the T-cell identity program in prethymic precursors, in the absence of Notch signaling [**61]. It is the first T-cell transcription factor shown to do so. TCF-1 need not do everything alone: among the first genes it turns on are Gata3 and Bcl11b, but its effect is impervious to high levels of γ-secretase inhibitors that block Notch signaling [**61]. It fails to turn on the sensitive Notch target gene Ptcra, yet activates many other “DN3-stage” genes that would normally accompany Ptcra expression [16]. Thus, many parts of the T-cell program are dependent on Notch only indirectly, through Notch-activated TCF-1 and its own targets (Fig. 2).
Figure 2.
Gene network relationship among Notch, TCF-1 (Tcf7), GATA-3, Bcl11b, and selected target genes. Solid arrows: support from perturbation and binding evidence. Dashed: binding evidence. Dotted: perturbation evidence. Data from [60-*62]. Bcl11b targets are still being defined.
How TCF-1 works here is a crucial question. Results shown by Weber et al. notably imply that β-catenin and relatives are dispensable for forced expression of TCF-1 to turn on T-cell genes [**61], in harmony with the negative evidence in vivo [45,46]. However, they also show TCF-1 binding directly to genes like Gata3 and Bcl11b that it positively regulates, so it is not acting as a repressor [**61]. This could imply a novel coactivator to replace β-catenin, or even a new mode of TCF-1 action that might have broader developmental significance. Yet there remain questions about the nature of the β-catenin mutation that has been tested in this context [38]. Clarifying the relationship of this new, early positive regulatory role with classic β-catenin mediated signaling pathways will be important.
Initiating the T-cell program: an emerging circuit
These results excitingly open new opportunities and new questions. TCF-1 only appears to be able to bypass Notch when it is introduced at high levels, typical of DN3 rather than DN1 stage cells. Thus at earlier stages, a classic feed-forward network circuit probably operates, in which lower levels of TCF-1 initially induced synergize with Notch signals on first-tier targets (Fig. 2)[*62]. Then with continued Notch signals, TCF-1 autoregulation, and/or cross-regulation by other factors, TCF-1 levels could rise to the point of being able to sustain the T-cell program without Notch. Most interestingly, the interaction between TCF-1 and GATA-3 may be mutual: not only Notch/RBP-Jκ (CSL) but also GATA-3 binds to the Tcf7 upstream enhancer (J.A. Zhang et al., op. cit.). There is much yet to be learned about how dosage regulation is effected in this network circuit, and how these factors intersect with E proteins, Bcl11b, and other regulators. But the outlines of the T-cell specification process are coming into clearer focus.
HIGHLIGHTS.
Notch, TCF-1, GATA-3, and Bcl11b are linked in a T-cell gene network
Bcl11b is needed for commitment but not to initiate the T-cell program
GATA-3 action is regulated via transcription, translation, and binding site selection
TCF-1 (Tcf7) is a direct Notch target required from the Early T-cell Precursor stage
TCF-1 can bypass the requirement for Notch itself to activate Gata3 and Bcl11b
ACKNOWLEDGMENTS
I apologize to authors whose work could not be adequately cited, and thank members of my group for use of our unpublished results. Support was from USPHS grants R01AI083514, R01AI095943, and RC2CA148278, and the Albert Billings Ruddock Professorship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- [2].Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu P, Li P, Burke S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol.Rev. 2010;238:138–149. doi: 10.1111/j.1600-065X.2010.00953.x. [DOI] [PubMed] [Google Scholar]
- [4].Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- [5].Inoue J, Kanefuji T, Okazuka K, Watanabe H, Mishima Y, Kominami R. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in Bcl11b−/− mice. J Immunol. 2006;176:5871–5879. doi: 10.4049/jimmunol.176.10.5871. [DOI] [PubMed] [Google Scholar]
- *[6].Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur.J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. Kastner and colleagues show that Bcl11b deletion enables maturation genes to be activated precociously in CD4+CD8+ thymocytes, seemingly regulators of both CD4 and CD8 cell function. They present selected examples of ChIP-seq data to suggest that the dysregulated genes are direct Bcl11b targets. An unusual, Bcl11b-independent CD8+ cell population in the thymus is also shown to consist of γδ cells, emphasizing the different importance of Bcl11b in αβ and γδ lineage cells.
- [7].Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **[8].Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. Li et al. demonstrate that deletion of Bcl11b at various stages of T cell development rapidly enables T cells to switch to NK-like properties. Notable upregulation of NK-associated genes is found even in CD8+ TCRαβ cells from which Bcl11b is deleted after the cells leave the thymus, and the function switches to NK-like even though the cells retain TCR complex expression. Loss of Bcl11b enables spontaneous “awakening” of genes that are otherwise silent in all mature T cells. A case is made for these cells as potentially valuable for immunotherapy against cancer. The major implication is that Bcl11b is required permanently to restrain NK potential throughout the lifetimes of mature CD8+ T cells. Also see ref. *[10] for another interpretation.
- [9].Vanvalkenburgh J, Albu DI, Bapanpally C, Casanova S, Califano D, Jones DM, Ignatowicz L, Kawamoto S, Fagarasan S, Jenkins NA, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208:2069–81. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[10].Zhang S, Rozell M, Verma RK, Albu DI, Califano D, VanValkenburgh J, Merchant A, Rangel-Moreno J, Randall TD, Jenkins NA, et al. Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J Exp Med. 2010;207:1687–99. doi: 10.1084/jem.20092136. The functions provided by Bcl11b are essential to prevent CD8 cells from differentiating excessively to cytolytic effectors, thus protecting their ability to proliferate. The net impact on antigen-specific cytolysis is positive because of the requirement for clonal expansion in order to achieve a biological impact. Also relevant to the interpretation that Bcl11b restrains “NK-like” or precocious effector function: see refs. *6 and **8.
- [11].Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- **[12].Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. This paper shows that uncommitted DN2a and newly-committed DN2b cells can be separated phenotypically in normal mice, increasing the accessibility of previous insights into the timing of commitment based on use of a transgenic reporter (ref. [13]). The commitment event is also shown to coincide with multiple, specific regulatory gene expression changes, and with sharp changes in the response to Notch signaling. Bcl11b upregulation appears to precede commitment most immediately, with subsequent decreases in the level of progenitor-cell transcription factor PU.1.
- [13].Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- **[14].Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. The authors use an elegant stroma-free in vitro system to show that T-cell specification can be initiated by Notch signaling and cytokines alone; however, under these conditions progression then halts at a stage corresponding to the DN2a stage. Bcl11b is not upregulated under these conditions and can only be induced if the IL-7 concentration is dropped. The implication is that Bcl11b is under negative cytokine-dependent control. In contrast to these results with wildtype cells, Bcl11b-knockout fetal precursor cells are shown to arrest at the same point even under optimal developmental conditions, continuing to grow with persistently elevated PU.1 and myeloid gene expression and subnormal levels of CD3. Transfection of the mutant precursors with exogenous Bcl11b before culture enables them to proceed through T-cell development, confirming the gene specificity of the developmental block.
- **[15].Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. A conditional knockout strategy is used to determine how early T-cell development is affected by the deletion of Bcl11b. The knockout cells differentiate to a DN2a-like stage, but then continue to proliferate extensively with a DN2a-like phenotype rather than progressing further in the T-cell pathway. The cells easily switch to an NK program, especially when Notch-Delta signals become limiting (also see ref. **8). However, they also show enhanced myeloid potential access (cf. ref. **14) and abnormally sustained expression of stem/progenitor-associated regulatory genes, both of which distinguish them from NK cells. Despite failure of TCRαβ lineage progression, Bcl11b-deficient precursors generate some TCRγδ cells, enriched in this system for the fetal-thymus types of Vγ TCR rearrangements. Notably, fetal-type γδ T cells also share with NK cells their expression of high levels of Id2 and IL-2/IL-15Rβ. The failure of stem/progenitor gene repression in the absence of Bcl11b implies that silencing not only of NK-like effector genes but also of primitive self-renewal functions may be crucial for commitment.
- [16].Georgescu C, Longabaugh WJ, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proc.Natl.Acad.Sci.U.S.A. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp. Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol.Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- [21].Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- [22].Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell Mol.Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- [24].Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- [25].Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- [26].Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- [27].Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat.Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- [30].McMillan RE, Sikes ML. Promoter activity 5′ of Dβ2 is coordinated by E47, Runx1, and GATA-3. Mol Immunol. 2009;46:3009–3017. doi: 10.1016/j.molimm.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tripathi RK, Mathieu N, Spicuglia S, Payet D, Verthuy C, Bouvier G, Depetris D, Mattei MG, Hempel WM, Ferrier P. Definition of a T-cell receptor β gene core enhancer of V(D)J recombination by transgenic mapping. Mol.Cell Biol. 2000;20:42–53. doi: 10.1128/mcb.20.1.42-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **[32].Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. The first comprehensively documented genome-wide analysis of GATA-3 binding in different T-lineage cells reveals the high level of context-dependence of GATA-3 in its recruitment and regulatory function. ChIP-seq mapping of GATA-3 binding and conditional GATA-3 knockout effects on RNA expression show that the roles of GATA-3 change from bulk CD4− CD8− cells to CD4+ CD8+ cells to naïve CD4 T cells to Th1, Th2, Th17, and Treg effector and regulatory mature cells. Only a minority of genes with detectable GATA-3 binding sites in a given stage are affected by deletion of GATA-3. GATA-3 is associated with both positive and negative regulatory effects. However, both binding and GATA-3 dependence give intersecting but distinct profiles among the different cell types.
- [33].Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano K, Suzuki D, Yamamoto M, Engel JD, Habu S. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur.J.Immunol. 2008;38:977–985. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- [34].Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen D, Zhang G. Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Exp.Hematol. 2001;29:971–980. doi: 10.1016/s0301-472x(01)00670-1. [DOI] [PubMed] [Google Scholar]
- [36].Heavey B, Charalambous C, Cobaleda C, Busslinger M. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPα and GATA factors. EMBO J. 2003;22:3887–3897. doi: 10.1093/emboj/cdg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–33. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- [38].Staal FJ, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–94. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- [40].Weerkamp F, Baert MR, Naber BA, Koster EE, de Haas EF, Atkuri KR, van Dongen JJ, Herzenberg LA, Staal FJ. Wnt signaling in the thymus is regulated by differential expression of intracellular signaling molecules. Proc.Natl.Acad.Sci.U.S.A. 2006;103:3322–3326. doi: 10.1073/pnas.0511299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Staal FJT, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, van Dongen JJ. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J.Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- [42].Baba Y, Garrett KP, Kincade PW. Constitutively active β-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 2005;23:599–609. doi: 10.1016/j.immuni.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat.Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- [44].Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat.Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- [45].Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of β-catenin and γ-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- [46].Koch U, Wilson A, Cobas M, Kemler R, MacDonald HR, Radtke F. Simultaneous loss of β- and γ-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008;111:160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- [47].Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Grégoire JM, Roméo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage specific silencer. J Biol Chem. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- [50].Hwang ES, Choi A, Ho IC. Transcriptional regulation of GATA-3 by an intronic regulatory region and fetal liver zinc finger protein 1. J.Immunol. 2002;169:248–253. doi: 10.4049/jimmunol.169.1.248. [DOI] [PubMed] [Google Scholar]
- [51].Lakshmanan G, Lieuw KH, Lim KC, Gu Y, Grosveld F, Engel JD, Karis A. Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol.Cell Biol. 1999;19:1558–1568. doi: 10.1128/mcb.19.2.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- [53].Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **[54].Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, Engel JD. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol Cell Biol. 2011;31:1894–904. doi: 10.1128/MCB.05065-11. Engel and colleagues use a BAC-transgenic strategy in mice to explore hundreds of kilobases beyond the Gata3 gene for the presence of a long-sought enhancer that would allow expression in T lineage cells. The region they find that provides this function can confer T-lineage expression based on in vivo assays with gain and loss of function, yet it maps far downstream of the locus in a gene desert. The pattern of expression driven by this enhancer using a GFP reporter does not completely match the endogenous pattern of expression, but provides the first permissive cis-regulatory function for pan-T lineage activation and NK lineage expression that has been found for the murine Gata3 locus. As it is activated in earliest DN stages, it must contain the direct or indirect inputs that mobilize Gata3 in response to early intrathymic Notch signaling.
- [55].David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev.Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yamashita M, Shinnakasu R, Asou H, Kimura M, Hasegawa A, Hashimoto K, Hatano N, Ogata M, Nakayama T. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J.Biol.Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- *[57].Cook KD, Miller J. TCR-dependent translational control of GATA-3 enhances Th2 differentiation. J Immunol. 2010;185:3209–16. doi: 10.4049/jimmunol.0902544. The authors carefully document the disproportionate increase in GATA-3 protein relative to Gata3 RNA that occurs in TCR-signaled T cells. In the case analyzed, protein stability is not detectably increased but translational efficiency of Gata3 RNA is enhanced. Using inhibitor assays they show that the increased protein is the result of an mTOR and PI 3-kinase-dependent pathway. The Discussion considers the possible mechanisms in light of the extensive secondary structure of 5′-untranslated region of Gata3 and the ways access to the initiator codon could be actively controlled.
- [58].Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat.Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- [59].Pallard C, Stegmann AP, van Kleffens T, Smart F, Venkitaraman A, Spits H. Distinct roles of the phosphatidylinositol 3-kinase and STAT5 pathways in IL-7-mediated development of human thymocyte precursors. Immunity. 1999;10:525–535. doi: 10.1016/s1074-7613(00)80052-7. [DOI] [PubMed] [Google Scholar]
- *[60].Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–5. doi: 10.1073/pnas.1110230108. The authors carry out a state-of-the-art analysis of the effects of TCF-1 loss of function in adult bone-marrow-derived T cell precursors. Detailed knockout and cell transfer experiments are shown to pinpoint the stage when TCF-1 loss affects the developmental fitness of T-cell precursors. The authors demonstrate that this does not occur prethymically but affects the population by the ETP (Kithigh DN1) stage and all subsequent stages. The mechanism is probed via RNA analysis and shown to affect DNA damage repair, CD3 expression, IL-7R, Kit, and Id2 and E2A at the ETP stage. Tcf7 is shown to be a Notch-dependent target gene, and ChIP-seq analysis is used to demonstrate strong Notch occupancy in the same putative regulatory region of Tcf7 where TCF-1 itself binds, about 31.5 kb upstream of the promoter, in agreement with ref. **61.
- **[61].Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–8. doi: 10.1038/nature10279. These authors use a gain of function approach to show that forced expression of TCF-1 can bypass most of the requirement for Notch signaling for entry into the T-cell program, provided that the level of TCF-1 is high enough. Levels of Gata3, Bcl11b, Cd3, and Rag1/2 expression are strongly upregulated by TCF-1 gain of function, to levels similar to those in DN2 or even DN3 cells. TCF-1 can do this even under conditions where essentially no Notch pathway input can be invoked. Surprisingly, this powerful effect is intact in β-catenin mutant cells, and insensitive to the effect of adding the catenin inhibitor ICAT to wildtype cells, implying that TCF-1 is not working through the canonical pathway of interaction with β-catenin or one of its relatives. ChIP-seq and direct transfection experiments are used to show that Tcf7 itself is a Notch target, with Notch binding at −31.5 kb (as in *61); thus, part of the requirement for Notch signaling is to induce adequate levels of TCF-1. TCF-1 itself exerts its inductive effects, at least in part, by direct binding to potential cis-regulatory elements in Gata3 and Bcl11b.
- *[62].Kueh HY, Rothenberg EV. Regulatory gene network circuits underlying T cell development from multipotent progenitors. Wiley Interdiscip Rev Syst Biol Med. 2011 doi: 10.1002/wsbm.162. This review includes an updated compilation of data from the literature for effects of transcription factor perturbation and signaling perturbation on T-cell precursor gene expression, and the results are organized in terms of a new T-cell specification gene regulatory network model. Evidence for potential subcircuit architectures that could mediate particular developmental functions like feed-forward amplification or IL-7 dosage sensing are discussed.