Abstract
Mycobacterium tuberculosis complex (MTBC) genomes contain 2 large gene families termed pe and ppe. The function of pe/ppe proteins remains enigmatic but studies suggest that they are secreted or cell surface associated and are involved in bacterial virulence. Previous studies have also shown that some pe/ppe genes are polymorphic, a finding that suggests involvement in antigenic variation. Using comparative sequence analysis of 18 publicly available MTBC whole genome sequences, we have performed alignments of 33 pe (excluding pe_pgrs) and 66 ppe genes in order to detect the frequency and nature of genetic variation. This work has been supplemented by whole gene sequencing of 14 pe/ppe (including 5 pe_pgrs) genes in a cohort of 40 diverse and well defined clinical isolates covering all the main lineages of the M. tuberculosis phylogenetic tree. We show that nsSNP's in pe (excluding pgrs) and ppe genes are 3.0 and 3.3 times higher than in non-pe/ppe genes respectively and that numerous other mutation types are also present at a high frequency. It has previously been shown that non-pe/ppe M. tuberculosis genes display a remarkably low level of purifying selection. Here, we also show that compared to these genes those of the pe/ppe families show a further reduction of selection pressure that suggests neutral evolution. This is inconsistent with the positive selection pressure of “classical” antigenic variation. Finally, by analyzing such a large number of genes we were able to detect large differences in mutation type and frequency between both individual genes and gene sub-families. The high variation rates and absence of selective constraints provides valuable insights into potential pe/ppe function. Since pe/ppe proteins are highly antigenic and have been studied as potential vaccine components these results should also prove informative for aspects of M. tuberculosis vaccine design.
Introduction
Mycobacterium tuberculosis, the main causative agent of tuberculosis in humans, is a member of the M. tuberculosis complex (MTBC), a closely related group of slow-growing pathogenic mycobacteria. Recent studies of MTBC evolution have revealed that the M. tuberculosis genome appears to be a composite genome created by frequent horizontal gene transfer events in a broad, genetically diverse, progenitor species prior to an evolutionary bottleneck or selective sweep around 35,000 years ago [1]. Divergence of the rare, smooth colony forming tubercle bacilli M. canetti seems to immediately predate this bottleneck/selective sweep while all other members of the MTBC are the result of the clonal expansion of a small number of surviving bacteria. This recent clonal expansion with the concurrent absence of horizontal gene transfer explains the relatively high degree of genetic homogeneity (99.9%) observed between MTBC members despite differences in their phenotypic characteristics and host ranges [2], [3], [4]. Whole genome sequencing of several M. tuberculosis strains, along with M. bovis and M. africanum, has confirmed this genetic homogeneity and revealed many other interesting biological aspects [5], [6], [7].
One of the surprises emerging from the analysis of the first sequenced M. tuberculosis genome (the laboratory strain H37Rv) was the discovery of two large gene families, designated pe and ppe, that in H37Rv comprise 99 and 69 members respectively and together account for around 10% of the organism's genomic coding potential [5]. Pe genes are characterised by the presence of a proline-glutamic acid (PE) motif at positions 8 and 9 within a highly conserved N-terminal domain consisting of around 110 amino acids. Similarly, ppe genes contain a proline-proline-glutamic acid (ppe) at positions 7–9 in a highly conserved N-terminal domain of approximately 180 amino acids. The C-terminal domains of both pe and ppe protein families are highly variable in both size and sequence and often contain repetitive DNA sequences that differ in copy number between genes [5].
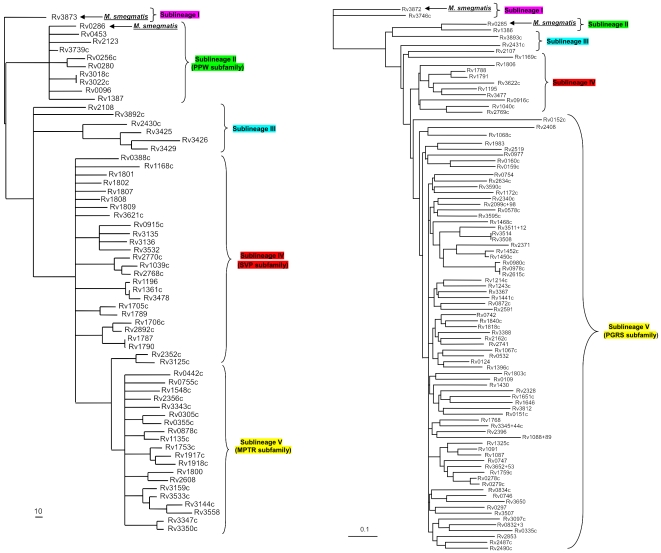
The pe and ppe gene families can be divided into sub-families based on similarities in their N-terminal regions and the phylogenetic relationships between each gene sub-family have been previously described, demonstrating that their evolutionary expansions are linked to the duplications of the ESAT-6 (esx) gene clusters [8]. Ppe genes can be subdivided into 5 subfamilies, the most numerous of which are the ppe_svp (24 members) and the ppe_mptr (major polymorphic tandem repeat) subfamilies (23 members) (Fig. 1a). Pe genes can also be divided into 5 sub-families, the largest of which, the polymorphic GC-rich-repetitive sequence (pe_pgrs), comprises 65 members in H37Rv (Fig. 1b). This sub-family is characterised by a C-terminal domain that contains multiple tandem repeats of a glycine-glycine-alanine (Gly-Gly-Ala) or a glycine-glycine-asparagine (Gly-Gly-Asn) motif. Phylogenetic analysis indicates that the emergence of the large pe_pgrs and ppe_mptr subfamilies is a recent evolutionary event, with their presence being restricted to members of the MTBC and close relatives such as M. marinum and M. ulcerans [8].
Figure 1. Phylogenetic reconstruction of the evolutionary relationships between the members of the pe and ppe protein families.
A. Phylogeny of the ppe protein family. The phylogenetic tree was constructed from the phylogenetic analysis done on the 180 aa N-terminal domains of the ppe proteins. The tree was rooted to the outgroup Rv3873 (ppe68), shown to be the first ppe insertion into the ESAT-6 (esx) gene clusters [8]. Figure reproduced from reference 8 with permission of the authors. B. Phylogeny of the pe protein family. The phylogenetic tree was constructed from the phylogenetic analysis done on the 110 aa N-terminal domains of the pe proteins. The tree was rooted to the outgroup Rv3872 (pe35), shown to be the first pe insertion into the ESAT-6 (esx) gene clusters [8]. Figure reproduced from reference 8 with permission of the authors.
The high pe/ppe gene content of the MTBC suggests an important biological role for their respective proteins. However, their precise function is unknown although recent studies have provided some intriguing clues. In pathogenic organisms it is generally found that proteins that are directly exposed to host immune surveillance show higher levels of polymorphism than that found in general housekeeping proteins [9]. This is thought to reflect their in volvement in antigenic variation and immune evasion. Many pe/ppe proteins have been found to be highly immunogenic and several groups have investigated this aspect of their biology with regard to vaccine production (for example, [10], [11]). Persuasive evidence now exists that many M. tuberculosis pe/ppe proteins are cell surface located [12], [13], [14], [15] and that others are probably secreted [16], [17] and this, in conjunction with their immunogenicity and the well established polymorphic nature of their C-terminal repeats, has led to the suggestion that they may well be involved in antigenic variation and immune evasion [5]. Indeed, several studies have revealed varying degrees of pe/ppe sequence polymorphism between M. tuberculosis clinical isolates. Talarico and colleagues have reported a high degree of polymorphism within the pe_pgrs33, pe_pgrs16 and pe_pgrs26 genes [18], [19], [20]. Similar results have also been found for ppe18 [21]. Less sensitive analysis based on the size of DNA repeats in the C-terminal region of ppe34 and ppe8 have also revealed a high frequency of polymorphism [12], [22]. In addition, some pe/ppe genes have been reported to display elevated levels of IS6110 integration [23], [24], [25], [26], [27], [28] and homologous recombination [26], [29], [30], [31]. However, sequence analysis of 4 pe (pe5, pe11, pe18 and pe31) and 4 ppe (ppe9, ppe27, ppe41 and ppe50) genes found polymorphism to be limited or absent [32]. Along with sequence variation, gene expression alterations may contribute to antigenic variation and these have also been noted in pe/ppe genes from different M. tuberculosis strains [33], [34], [35]. For example, ppe44 shows limited sequence diversity between strains (only isolates of the Beijing genotype were found to be polymorphic) whereas transcript levels of the gene are highly variable [35]. Numerous other reports have documented variation in pe/ppe transcription levels under different environmental and experimental conditions [36], [37], [38], [39]. Furthermore, there does not appear to be a global regulator of pe/ppe expression [36], [40], suggesting a complex regulatory network and a high degree of plasticity in their expression repertoire.
It has been proposed that pe/ppe proteins can aid M. tuberculosis pathogenesis by negatively influencing host immunity [5] and recently Toll-like receptor 2 (TLR2) has assumed a prominent role in this theory. For example, Basu et al showed that pe_pgrs33 is able to enhance the expression of tumour necrosis factor alpha (TNFα) in a TLR2-dependent manner leading to macrophage apoptosis [41]. Interestingly, deletions within the PGRS domain (as is often seen in clinical isolates) inhibited this ability. Ppe proteins have also been shown to function in a TLR2-dependent manner. Nair et al demonstrated that ppe18 binds to TLR2 which stimulates IL-10 production in macrophages [42]. This leads to an anti-inflammatory Th2 type immune response. Evidence also exists to suggest that pe_pgrs proteins may be able to inhibit antigen processing and/or presentation [43] and it has been proposed that the Gly-Ala repeats in the C-terminal PGRS domains are able to inhibit proteasomal degradation of the N-terminal PE domain [44] thus inhibiting antigen processing by CD8+ T cells in a manner similar to that seen in Epstein – Barr virus nuclear antigen 1 [45]. Several other lines of evidence also suggest a major role for pe/ppe proteins in mycobacterial pathogenesis. For example, recent work has shown that pe_pgrs33 localises to host cell mitochondria where it is able to induce apoptosis and primary necrosis [46]. Studies demonstrating increased mycobacterial growth in macrophages and subsequent macrophage necrosis of pe_pgrs33 expressing strains (as oppose to pe_pgrs33 negative strains) have also been reported [47], [48] and other studies have documented an attenuated phenotype with the knockout of specific pe/ppe genes [49], [50] or the upregulation of specific pe/ppe genes upon infection [38], [51].
Evidence for other diverse alternative or additional pe/ppe functions also exists. In silico analysis of PGRS protein sequences reveal that at least 56 pe_pgrs members contain multiple nona-peptide repeats (GGXGXD/NXUX, where X = any amino acid and U = a large non-polar hydrophobic residue) that are predicted to be calcium binding motifs [52]. The authors suggest that these motifs might be involved in the initial attachment of M. tuberculosis to host alveolar macrophages. PGRS domains have also been implicated in cellular structure and colony morphology [14] and in the binding of fibronectin [53]. A possible role in iron uptake has also been proposed for ppe37 following the finding that it is upregulated under low iron conditions [54]. It is also notable that pe/ppe genes are often found paired within operons with the pe gene located upstream of the ppe gene. Pe/ppe genes within these operons are cotranscribed and physically interact with each other and transcription of both is required for correct cellular localization [55], [56]. This is emphasised by the findings of Strong and colleagues who failed in numerous attempts to determine the crystal structures of individual pe and ppe proteins. Coexpression and copurification of the proteins coded by the linked genes Rv2431c (pe25) and Rv2430c (ppe41) was successful, however, and the crystal structure revealed a 1∶1 pe25/ppe41 protein dimer where helices from each protein are predicted to interact and form a stable complex. The structure implies a docking site for an additional protein and suggests a role in signal transduction [56].
Here, we have used recently acquired whole genome sequence data from 18 isolates representing a broad spectrum of the MTBC phylogeny to investigate variation in 33 pe (excluding pe_pgrs) and 66 ppe genes. We have supplemented this data by selecting 14 pe and ppe genes (including 5 pe_pgrs) and performing whole gene sequencing on a cohort of 40 clinical isolates representing a broad and well characterised spectrum of the M. tuberculosis phylogeny. We hypothesise that if pe/ppe proteins are involved in immune evasion and antigenic variation their genes will have undergone rapid evolutionary change, as demonstrated by high levels of DNA sequence polymorphism and evidence for diversifying selection compared to other M. tuberculosis genes. Previous work on this theme [12], [18], [19], [20], [21], [22], [32] has produced conflicting results that may be due to the lack of sensitivity of the analysis technique, the decision to examine genes that belong to a particular pe or ppe sub-family that might show abnormal variation levels, or the decision to examine clinical isolates that are too closely related to reveal polymorphic differences. The resultant comparative gene analysis presented here provides new insights into the variation and evolution of these genes along with their potential role in providing the pathogen with a source of antigenic variation.
Results
Comparative gene analysis using whole genome sequences
A total of 66 ppe and 33 pe genes were analysed. Unfortunately, due to the extensive repetitiveness of their C-terminal regions and the inherent difficulties encountered in sequencing through repetitive regions using the third generation short read sequencing techniques, the pe_pgrs genes of most publicly available whole genome sequences were incomplete or of low sequence quality and could not be included in this analysis. Variability estimates for ppe38/71 and ppe50 could not be determined due to the difficulty in obtaining a reference sequence. Ppe38/71 are completely homologous in most cases and are located in a hypervariable region that is prone to homologous recombination, gene conversion, IS6110 integration and large deletion events [26]. Ppe50 is also highly variable and displays numerous different sequence types due to large deletions and other sequence variations [32]. Due to the exclusion of genes with notations suggesting potential sequence errors, an average of 15.2 and 16.5 genomes (from a possible maximum of 18) were analysed for each ppe and pe gene respectively. Full details of all variations detected can be seen in tables S1, S2, S3.
Confirmation of whole genome sequence accuracy
In order to ascertain the accuracy of the whole genome sequences used in our analysis we obtained the original DNA used in the sequencing process to determine the F11, CPHL_A, K85, T17 and T92 sequences. A total of 40 variations observed in the pe/ppe genes of these 5 isolates were reanalysed by amplifying the surrounding region by PCR and using standard Sanger sequencing methodology to sequence the amplicons. A variety of variations were chosen for analysis and these comprised sSNP's, nsSNP's, frameshifts, and an in-frame deletion. We also ensured that some of the variations detected in the large ppe_mptr genes, ppe5/6 and ppe7/8, were analysed since it could be suggested that mistakes are more likely to be made here due to the highly repetitive nature of their C-terminal domains. Table S4 lists the variations, primer details and results of our analysis. Four of the 40 variations (10%) were found to be erroneous in the publicly available whole genome sequences. One of these (T17, ppe28) appears to be due to an assembly error while another (CPHL_A, ppe13) involves a long poly C region at the 3′ end of the gene. The other 2 errors involve a SNP or single bp deletion. The 10 variations that were checked in the ppe5/6 and 7/8 genes were all confirmed indicating that the large ppe_mptr genes were not more likely to produce sequencing errors than the smaller less complex genes.
Number of structural protein variants
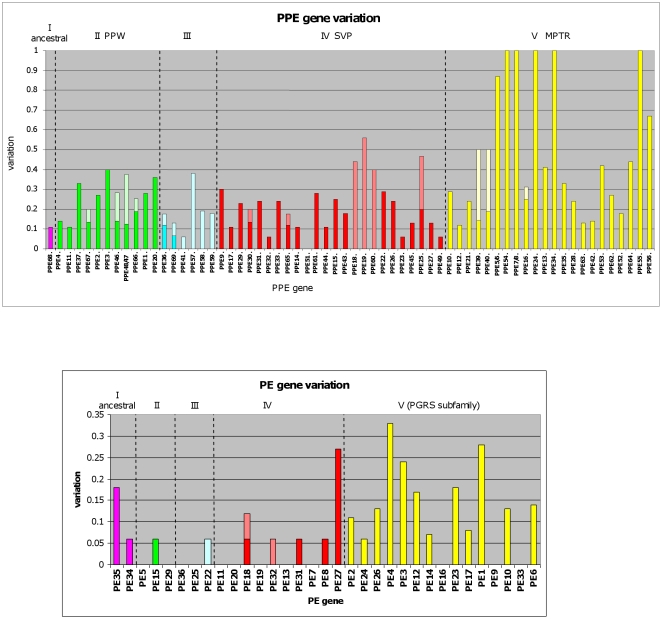
Various aspects of genetic variation between the homologous genes may be analysed. First we wished to determine the number of predicted different structural variants of each pe/ppe protein, based on the observed genetic variations, as a proportion of the total number of isolates analysed. Thus, sSNP's were ignored, variations that were specific to multiple isolates from a single lineage were counted as a single variant and single isolates that contained more than 1 variation were still counted as a single variant. Results for the ppe gene analysis are shown in Fig. 2a. They reveal a high level of variation across all subfamilies, with only one gene (ppe51) showing no variation in all genomes analysed. Subfamily V (the MPTR subfamily) shows many genes with extreme levels of variation. By distinguishing between different types of mutation it is notable that certain genes display alternate mechanisms of variation. For example, homologous recombination events, particularly between closely related ppe genes in close physical proximity, are shown to be responsible for a high degree of variation within certain genes (ppe57/58/59 and ppe18/19/60). Other macromutational events (whole or partial gene deletions and IS6110 integrations) were found to be responsible for a significant proportion of the variation in several genes. Also notable is the finding that macromutational events do not contribute to variation in the most hypervariable genes of the MPTR subfamily. Six genes in particular (ppe5/6, 7/8, 24, 34, 54 and 55) show extreme variation and 5 of these have a variation index of 1 (indicating that each isolate had a unique sequence). These 6 genes are all large (between approximately 3.1 and 10.0 kb) and reveal mutations including nsSNP's, frameshifts and in-frame indels. The sequences of these 6 hypervariable genes were further compared between 3 closely related genomes, KZN 1435, KZN 605 and KZN 4207 [57] in order to ascertain whether they were evolving at a rate that would enable us to distinguish even between extremely closely related isolates. For each gene the sequence in all 3 genomes was identical. The sequences of four of the hypervariable ppe genes (ppe24, 34, 54 and 55) were also compared between the index case and 2 transmission chain endpoint isolates of the Harlingen cluster [58], [59]. An average of 84% of the coding region for each of these genes was available for analysis. No variations were observed. These results indicate that while these ppe genes are hypervariable across the full phylogeny of M. tuberculosis, they do not evolve at a rate fast enough to distinguish between extremely closely related isolates.
Figure 2. Sequence variation levels in ppe and pe genes.
A. Calculations of sequence variation in 64 ppe genes. Synonymous variations have been ignored. The Y axis shows the proportion of sequences that show variation predicted to result in amino acid changes. A value of 1 indicates that all analysed sequences were unique. Average number of genomes analysed per gene = 15.2. Genes have been grouped together according to their subfamily [8] by colour and subfamilies are also separated by dotted lines. Each vertical bar is subdivided into micromutations (nsSNP's, frameshifts, small in-frame indels) in dark shading and macromutations (homologous recombination, IS6110 integration, partial and whole gene deletions) in light shading. Ppe38 and ppe50 were not included due to hypervariability at the macromutational level [26], [30] and the difficulty in establishing a consensus sequence. For details of all variations detected see Tables S1 and S2. B. Calculations of sequence variation in 33 pe (excluding pgrs) genes. Synonymous variations have been ignored. Average number of isolates analysed per gene = 16.5. The genes from subfamily V (pgrs subfamily, yellow) are those which are classified as members of this subfamily by their N-terminal amino acid sequences [8] but that do not include the long PGRS C-terminal region. For details of all variations detected see Tables S1 and S3.
A similar analysis of the ppe (excluding pe_pgrs) genes revealed agenerally lower level of variation with many of the genes showing no variation across the analysed genomes (Fig. 2b). Macro-mutational events, including homologous recombination, were rare.
Pe/ppe variation levels in comparison with other M. tuberculosis genes
In order to ascertain whether the variation levels of the pe and ppe genes differed from other M. tuberculosis genes we compared our results to those obtained by Hershberg and colleagues who identified the SNP's present in 89 non-pe/ppe genes comprising 65,829 bp, from 107 MTBC isolates [60]. This study identified a total of 231 nsSNP's, which when divided by the total number of nucleotides multiplied by the number of isolates gives a nsSNP frequency of 231/(65,829×107) = 0.327×10−4 nsSNP's per nucleotide. Similar calculations using our ppe data required the exclusion of several genes. Ppe38/71 and ppe50, which exhibit extreme levels of macro-mutational variation, were excluded. Ppe18, 19, 24, 34, 54, 55, 57, 58, 59 and 60 were also excluded because of extreme variability or frequent homologous recombination events which resulted in difficulty in determining the consensus sequence of the gene. The remaining 54 ppe genes comprise 100,657 bp and contain 163 nsSNP's and the average number of isolates analysed per gene was 15.17. This results in a nsSNP frequency of 163/(100,657×15.17) = 1.067×10−4 nsSNP's per nucleotide. This value is approximately 3.3-fold greater than that found in the non-pe/ppe MTBC genes [60] despite the exclusion of the most variable ppe genes. Similarly, the 33 pe (excluding pgrs) genes comprised 21,726 bp and contained 35 nsSNP's with an average isolate number per gene of 16.5, resulting in a nsSNP frequency of 35/(21,726×16.5) = 0.976×10−4 nsSNP's per nucleotide. This value is approximately 3.0-fold higher than that found in non-pe/ppe MTBC genes [60]. These results confirm that nsSNP's occur at a far higher frequency in pe/ppe genes than in non-pe/ppe genes.
Whole gene sequencing results of 14 pe and ppe genes
Complete results for all variations found from whole gene sequencing of 14 pe and ppe genes from 40 phylogenetically diverse clinical isolates covering the whole M. tuberculosis phylogenetic tree (PGG1, 2 and 3 strains including members of all the main lineages EAI, CAS, Beijing, LAM, Haarlem, LCC and T) are shown in Table S5. Our sequencing of 3 pe genes (pe35, 11 and 3) and 6 ppe genes (ppe68, 2, 44, 10, 42 and 62) confirmed the results found for these genes in our in silico gene analysis. In each case lineage specific variations were consistent between the 2 different analyses. Interestingly, our 3 EAI samples failed to show the ppe62 G1690A SNP which was present in 2 (T17 and T46) of the 4 EAI samples analysed in silico. This suggests that our EAI isolates are relatively closely related and do not reflect the large genetic diversity observed within this group [60].
Particular interest lies in our analysis of the 5 pe_pgrs genes since the pgrs subfamily could not be analysed using in silico methods. Three of these genes (pe_pgrs16, 26 and 33) have previously been analysed for their variation [18], [19], [20]. Our replication of this work (using better defined M. tuberculosis lineages) confirms that all 3 of these genes display extremely high variation with in-frame indels within the pgrs repetitive region comprising a large proportion of the mutations in each case. These indels were often large. For example, in the pe_pgrs16 gene the 2 EAI isolates SAWC1659 and SAWC2493 both possess 2 deletions of 66 bp and 600 bp and all CAS family isolates possess 2 deletions of 45 bp and 42 bp in pe_pgrs33 (Table S5). Pe_pgrs18 has previously been reported as being part of a duplicated gene pair (with pe_pgrs17) that shows evidence of homologous recombination and gene conversion events [30]. Our results for pe_pgrs18 confirmed a high level of homologous recombination with pe_pgrs17. We were also able to confirm the presence of the 12/40 polymorphism in the Haarlem and LCC groups that appears to result from gene conversion with pe_pgrs17 [30]. Details of the variability characteristics of pe_pgrs62 have not previously been reported. Results for this gene were surprising because despite being the same size as most of the other analysed pe_pgrs genes it showed very little variation and no in-frame indels were observed (Table S5). Additional in silico analysis of this gene in M. bovis confirmed its invariant nature. A closer inspection of the gene's predicted amino acid sequence revealed that it does not possess the C-terminal multiple tandem repeats of Gly-Gly-Ala and Gly-Gly-Asn typical of pgrs proteins. Taken together, our results suggest that pe_pgrs genes generally display high variation levels with in-frame indels making a large proportion of mutations. However, mutational mechanisms and levels of variation can differ greatly between individual genes implying functional variation within this subfamily.
The 252 bp deletion identified in the pe_pgrs16 gene of isolate SAWC 2185 (Haarlem, F2) (Table S5) was further analysed in order to determine how variable this mutation was within both the F2 family and other members of the same cluster as SAWC 2185. Isolates from 36 different F2 clusters as well as 4 isolates from the same cluster were examined. All other members of the cluster to which isolate SAWC 2185 belongs, along with 27 of the additional F2 clusters, were found to contain this mutation. However, isolates representing the remaining F2 clusters lacked the mutation confirming the presence of within-family variation for this mutation.
Analysis of selective constraints in pe/ppe genes
One of the major findings of the MTBC genetic diversity study of Hershberg and colleagues [60] was the low level of purifying selection compared to other bacteria, as assessed by the ratio of nonsynonymous to synonymous SNP's (dN/dS) in 89 non-pe/ppe genes. A dN/dS ratio of <1 is considered to indicate purifying selection, dN/dS = 1 suggests an absence of selection (i.e. neutral evolution) and dN/dS>1 indicates positive or diversifying selection. In our analysis of 54 ppe genes (excluding the genes described above) we discovered a total of 220 SNP's, of which 163 (74%) were nonsynonymous (Table S2). The average pairwise dN/dS ratio for the concatenated ppe genes was 1.045. This is substantially higher than the already extremely high value of 0.57 reported for the non-pe/ppe M. tuberculosis genes [60] and suggests an absence of selection pressure. Similarly, in our analysis of 33 pe genes we detected a total of 47 SNPs, of which 35 (74%) were nonsynonymous (Table S3). The average pairwise dN/dS ratio for the concatenated pe genes was 1.000, again far higher than the value previously obtained for non-pe/ppe genes [60] and again suggesting an absence of selection pressure. We also calculated the dN/dS value for the 3 pe_pgrs genes that show a “typical” pgrs variation profile (pe_pgrs16, 26 and 33 – see above) using the SNPs identified in our current study in addition to those detected previously [18], [19], [20]. A total of 63 SNPs were found in these 3 genes of which 43 (68%) were nonsynonymous. The average pairwise dN/dS ratio for the 3 concatenated pe_pgrs genes was 0.869, a value once again close to that indicating neutral evolution.
Discussion
Although polymorphisms in certain M. tuberculosis pe and ppe genes have been previously documented, this study is the first to make use of publicly available MTBC whole genome sequences, as well as a comprehensive set of 40 clinical isolates covering the known M. tuberculosis phylogenetic tree and all major M. tuberculosis strain lineages including EAI, CAS, Beijing, Haarlem, LAM, LCC and T, to produce an extensive analysis of pe/ppe gene variation. Unfortunately, the large pe_pgrs subfamily was not able to be analysed using these methods due to a lack of sequencing accuracy but our own sequencing analysis of selected pe_pgrs genes, in conjunction with those of previous investigators, also provides important insights into genetic variation within this subfamily.
The first important observation made was the confirmation that pe and ppe genes display a high frequency of variation (Fig. 2) and that this variation exceeds that seen in other MTBC genes. Hershberg and colleagues previously analysed MTBC genetic diversity by sequencing 89 non-pe/ppe genes (classified as either “housekeeping”, “virulence” or “surface”) in 107 MTBC isolates [60]. Compared to these genes, the nsSNP frequency in the ppe and pe (excluding pe_pgrs) genes in our analysis was approximately 3.3 and 3.0 times greater, respectively. Several qualifying points should be emphasised when considering these values. Our quality assurance analysis of 40 selected SNP's from the whole genome sequences revealed that 4 were incorrect (Table S4), indicating an overestimation of variation frequency of approximately 10%. However, it should also be noted that our ppe variation values were obtained without the inclusion of the 12 most variable ppe genes which were excluded from the analysis due to the difficulty in determining a consensus sequence. The inclusion of these genes would undoubtedly result in a significant increase in observed ppe variation. In addition, many of the ppe genes that were included in our analysis displayed high levels of variation that were not due to nsSNP's. Indeed, the mutational spectrum seen in ppe genes was extensive. The most common mutations observed were nsSNP's and frameshifts caused by small indels. However, many large ppe genes of the mptr subfamily often display in-frame indels, certain groups of genes undergo frequent homologous recombination events and, as previously reported, ppe39 and ppe40 are particularly susceptible to IS6110 insertions [27]. Figure 2a reveals that members of the ppe_mptr subfamily have a generally higher frequency of mutation but this does not apply consistently to all members of this group.
Interestingly, although the pe (excluding pgrs) genes revealed far lower variation than the ppe's (Fig. 2), the frequency of nsSNP's was similar to that of the ppe's and was found to be approximately triple that of the non-pe/ppe genes analysed previously [60]. Protein changes in these genes were generally due to nsSNP's and small indels leading to frame shifts. In-frame indels and macromutations (whole or partial gene deletions and IS6110 integrations) were rare. The lower variation in these genes probably reflects a strong functional constraint of the pe protein. It has previously been shown that pe proteins and the pe domain of pe_pgrs proteins are responsible for cell wall localisation [15]. This is presumably essential for optimal protein function and mutations that hinder this process would therefore be subject to strong negative selection pressures.
Our own sequencing analysis of 5 pe_pgrs genes showed that, in general, variation within the pe_pgrs subfamily exists at far higher levels than in non-pgrs pe members and that this increase in variation is largely caused by a higher frequency of in-frame indels within the C-terminal pgrs region. These results support the findings of Talarico and colleagues who have previously reported analysis of genetic polymorphism in pe_pgrs33, 16 and 26 [18], [19], [20]. We show that the deletions in these 3 genes are often large (for example, in EAI isolates 666 bp has been deleted from pe_pgrs16, Table S5). The fact that large deletions were often found in multiple isolates from the same lineage suggests that these mutations are not subjected to strong purifying selection forces. The phenotypic consequences of these deletions may include a reduction in macrophage apoptosis caused by a decrease in TNFα production [41] and, at an epidemiological and clinical level, be associated with clustering and a lack of lung cavitations [19]. Our analysis of pe_pgrs18 and 62 has provided additional interesting information relating to pe_pgrs variation since neither of these genes displayed the “typical” variation pattern seen in pe_pgrs33, 16 and 26 (Table S5). Variation in pe_pgrs18 was found to be largely caused by gene conversion with pe_pgrs17. These genes are in close physical proximity, have high sequence homology, and are presumably the result of a recent duplication event. A previous study has documented homologous recombination between these genes and has identified a polymorphism present in one or both of these genes and used it to infer details of the evolution and clonal expansion of the MTBC [30]. Genetic variation in pe_pgrs62 has not been reported previously and we chose this gene for analysis because studies have shown that it is a T cell antigen with vaccine potential [61], [62], [63] and that its PGRS domain is able to elicit a strong antibody response [64]. The PGRS domain of pe_pgrs62 is atypical as it lacks the Gly-Gly-Ala or Gly-Gly-Asn repeats found in most members of this subfamily. Interestingly, the amount of variation seen in this gene was far less than in the more typical pe_pgrs genes with only 4 SNPs (of which only 2 were non-synonymous) seen in our 40 clinical isolates. This lack of genetic variation is especially interesting since pe_pgrs62 can stimulate both cell-mediated and humoral based host immunity and might therefore be expected to undergo significant levels of antigenic variation. Taken together, these results reveal that while variation in pe_pgrs genes is generally very high, this variation, along with the dominant type of mutational mechanism, can differ greatly between genes. The finding of low variation in a highly immunogenic pe_pgrs member lacking the typical PGRS domain also implies functional variation in certain members of this sub-family.
Another major finding of this study was that selection appears to be absent in pe/ppe genes. Most genomic regions in all organisms are subjected to strong purifying selection pressures. Within the Actinobacteria, for example, pairwise genome-wide comparisons result in a general dN/dS value of 0.15–0.20 [60]. This value appears to be fairly typical of both prokaryotic and eukaryotic organisms [65]. The recent comparative sequence analysis of 89 genes in 107 MTBC isolates [60] found an average pairwise dN/dS ratio of 0.57, a value far higher than that found in other bacteria and an indication that purifying selection is severely reduced in the MTBC on a general genomic level. In pe/ppe genes specifically, a high ratio of nonsynonymous to synonymous SNP's has previously been noted [6], [20] and it has also been shown that these genes are under greater selection for amino acid substitutions than other M. tuberculosis genes [66]. Our pairwise dN/dS ratio calculations for 54 ppe and 33 pe genes were 1.045 and 1.000 respectively, suggesting that selection pressure on these genes is extremely limited or altogether absent. Although our analysis of pe_pgrs genes was numerically limited, the pairwise dN/dS ratio was also close to 1 (0.869), again indicating a selection pressure close to neutral. This result is surprising because pe/ppe proteins are thought to provide antigenic variation and therefore be subjected to positive, rather than neutral, selection pressure. Thus, our results indicate that variation in these proteins is inconsistent with “classical” antigenic variation. It should be noted, however, that these results are an average of the gene families as a whole and that individual genes might be subjected to greater or lesser selective pressures. Evidence that pe/ppe genes are the major targets of positive selection in M. tuberculosis comes from a recent paper that examined the genomes of H37Rv and H37Ra [67]. Of the 12 genes that were found to be positively selected in these strains 6 were from the pe or pe_pgrs families. Our results also need to be interpreted in light of the report of Comas and colleagues [68] who found that the antigenic epitopes (excluding pe and ppe proteins) of M. tuberculosis are highly conserved and that there appears to be a strong selection pressure against sequence diversity in these regions. This finding was unexpected and is also inconsistent with the classical model of an evolutionary immunological arms race between pathogen and host and the authors favour the explanation that the host immune response is, paradoxically, beneficial to the pathogen. Despite the fact that our pe and ppe dN/dS values were far higher than those found for M. tuberculosis antigens in the Comas study, this explanation may also apply (to a lesser extent) to pe and ppe proteins and explain why their dN/dS values were less than those of typical antigens in other organisms.
Although our results suggest that pe/ppe proteins do not act as typical antigenic variants it is also important to consider the impact of population genetics on dN/dS values. The dN/dS ratio is a popular measure of selection pressure not only because it is simple and robust but also because of the simple interpretation of dN/dS<1 as negative selection, dN/dS = 1 as neutral selection and dN/dS>1 as positive selection. This analysis was originally designed for comparisons between sequences from divergent lineages or species and it has recently been shown that the standard signature of positive selection (dN/dS>1) does not hold for comparisons within a population [69]. It can sometimes be difficult to determine the appropriate evolutionary time-scale (distinct lineages/species versus numerous isolates from a single population) associated with a dataset of microbial sequences and it is possible that some of the more closely related sequences in our dataset have not diverged sufficiently for this analysis to be appropriate. If this is the case it is unlikely that our dN/dS values would alter drastically. We would, however, predict that our values are underestimates and therefore conclude that a mild positive selection pressure is acting upon these genes. Many of the pe/ppe genes present within the MTBC have homologues in the closely related, but phylogenetically distinct, species M. marinum [70] and we suggest that a comparison between these genes could provide a more accurate estimate of the evolutionary pressures they have been subjected to.
We hope that our results will allow for a more directed approach towards the use of pe/ppe proteins as vaccine components since it is possible that the high levels of polymorphism observed in certain members of these protein families could limit their effectiveness in some cases. This has been highlighted in a recent mathematical modelling analysis that has predicted the negative impact on vaccine efficacy that may occur when mycobacterial strain diversity is not considered [71]. For example, the Mtb72F vaccine comprises the 2 recombinant proteins pepA (Rv0125) and ppe18 (Rv1196). Mtb72F has been shown to have a protective effect against challenges with two M. tuberculosis laboratory reference strains (H37Rv and Erdmann) in numerous animal models, including a primate model [72], [73], [74], [75]. However, a recent study has shown that over 20% of M. tuberculosis strains taken from 2 geographical regions contain mutations that alter at least 1 amino acid in the ppe18 protein, many of which are in regions predicted to be T cell epitopes [21]. Our study confirms a high rate of ppe18 variation and shows that it is predominantly due to homologous recombination between ppe18, ppe19 and ppe60, which have extremely high sequence similarity. These results suggest that the Mtb72F vaccine could have limitations in a clinical setting and that, in hindsight, a pe/ppe protein that displays higher sequence conservation across many strains may have been a more effective vaccine candidate. An example of this is pe_pgrs62 which has also been investigated for its vaccine potential with promising results [61], [62]. This highly immunogenic, atypical pgrs protein showed extremely limited sequence variation across our cohort of isolates (Table S5) and might be expected to provide more consistent protection against a variety of M. tuberculosis strains. The data available for immunogenicity at the pe/ppe epitope level is limited however and it should be noted that variable regions of pe/ppe genes may be less immunogenic and less important for an immune response. It should also be noted that pe/ppe proteins probably have functional variation and that some may have a limited role in immune function.
The exact nature of pe/ppe function in the host cell is yet to be determined. However, our results also provide some additional insights and allow us to speculate on potential mechanisms of action for these proteins. When the high levels of pe/ppe sequence variation are considered in conjunction with the high inter-strain expressional variation [33], [34], [35] it is apparent that there is likely to be a huge diversity of pe/ppe expressional and functional variation across the MTBC. This would lead to a situation where only extremely closely related isolates have identical functional and expressional profiles across the entire pe/ppe spectrum. We note that this situation has parallels to the classical MHC class I and II systems where highly polymorphic MHC loci produce multiple alleles which, despite their structural and functional similarities, are distinct with regards to the antigenic peptides they present to CD4+ and 8+ T cells. It may be speculated that the large number of polymorphic pe/ppe proteins have evolved in response to the multiple MHC alleles expressed by host populations and that specific pe/ppe proteins are adapted to preferentially coexist alongside specific MHC alleles. The absence of selection exerted on pe/ppe genes may be interpreted as both a result of immune pressure selecting for antigenic variants and an adaptation for these proteins to function alongside new or rare MHC alleles that have not previously been encountered in the bacteria's evolutionary history. Although purely speculative, this theory is consistent with the large pe/ppe expansion within the MTBC (and in the closely related species M. marinum which is a natural pathogen of fish), it's functional and expressional variability, and the finding that some pe/ppe proteins appear to interfere with antigen processing [43], [44]. The true nature of pe/ppe function remains one of the great mysteries of M. tuberculosis pathogenesis however and many additional functional studies will probably be required before we are able to gain a more complete understanding of their role.
Materials and Methods
Ethics statement
We recovered sputum specimens from the National Health Laboratory Service (NHLS) after routine processing. None of the authors were directly involved in sputum collection. This study was approved by the Stellenbosch University Health Research Ethics Committee (approval reference number N10/04/126). Informed consent was not required as we received samples from the NHLS after routine processing. This was approved by the Stellenbosch University IRB.
In silico whole genome sequence analysis
Sequence selection details
Analysis of pe and ppe genes from the following 18 fully sequenced MTBC genomes was conducted: M. bovis strain AF2122/97 [76], H37RV [77], CDC1551 [6], CPHL_A (M. africanum), K85 (M. africanum), T92, T46, T48, EAS054, 94_M4241A, 02_1987, T85, C strain, Haarlem, F11, GM1503, KZN 1435 and 98-R604_INH-RIF-EM [57]. Details of the phylogenetic placements of each isolate are shown in Table 1. At least 1 representative from all 7 major MTBC lineages (including the animal lineage) [78] are included in this study apart from lineage 3 (CAS lineage). Ppe genes from the fully sequenced isolates KZN 605 and KZN 4207 [57] as well as the Harlingen transmission chain [58] were also analysed in specific instances. Orthologues of each gene were located by BLAST searches using the H37Rv gene sequence as the type standard. Gene sequences obtained from the Broad institute [57] were not used if they contained the following messages suggesting possible low sequence quality: “At least one base has a quality score <10”, “EST-based feature contains predicted/unverified ORF” or “Frame Shift: Sequence Error”. Sequence alignments were done using CLUSTALW [79].
Table 1. Details of 18 whole genome sequence isolates used for in silico comparative gene analysis.
| Isolate | Lineage | Reference |
| T92 | Lineage 1. PGG1, EAI family | [57] |
| T17 | Lineage 1. PGG1, EAI family | [57] |
| T46 | Lineage 1. PGG1, EAI family | [57] |
| EAS054 | Lineage 1. PGG1, EAI family | [57] |
| 94_M4241A | Lineage 2. PGG1, Beijing family | [57] |
| 02_1987 | Lineage 2. PGG1, Beijing family | [57] |
| T85 | Lineage 2. PGG1, Beijing family | [57] |
| C strain | Lineage 4. PGG2, low copy clade | [57] |
| CDC1551 | Lineage 4. PGG2, low copy clade | [6] |
| Haarlem | Lineage 4. PGG2, Haarlem family | [57] |
| F11 | Lineage 4. PGG2, LAM family | [57] |
| GM1503 | Lineage 4. PGG2, LAM family | [57] |
| KZN1435 | Lineage 4. PGG2, LAM family | [57] |
| 98-R604_INH-RIF-EM | Lineage 4. PGG2, LAM family | [57] |
| H37Rv | Lineage 4. PGG3 | [5], [77] |
| CPHL_A | Lineage 5. PGG1, West Africa-1 (M. africanum) | [57] |
| K85 | Lineage 6. PGG1, West Africa-2 (M. africanum) | [57] |
| M. bovis AF2122/97 | Animal lineage | [76] |
Confirmation of whole genome sequence accuracy
Genomic DNA from 5 of the whole genome sequenced isolates (F11, CPHL_A, K85, T17 and T92) was used to check the accuracy of 40 variations that were found in various pe and ppe genes (Table S4). Primers were designed to amplify a region surrounding the variation point and PCRs and sequencing of the amplicons were performed as described below. Recently, a number of nucleotides in the H37Rv sequence, including some within pe and ppe genes, were found to be incorrect [78]. These SNPs were corrected before analysis.
dN/dS values
Due to the general low level of SNPs present when analysing individual genes, a concatenated alignment for each gene category (ppe, pe and pe_pgrs) was generated combining all individual genes. Prior to concatenation the consensus sequence of each gene was aligned with the equivalent sequence containing all SNP's identified using CLUSTALW [79]. Other variations (eg frameshifts or in-frame indels) that had been identified were ignored. The resultant alignment files for each gene were concatenated using DnaSP [80] and pairwise dN/dS values were determined by subjecting the alignment to the DnaSP program package.
DNA sequencing of clinical isolates
Bacterial culture conditions, molecular typing and strain selection
Sputum samples were obtained from primary health care clinics in metropolitan Cape Town, South Africa. This region has a very high tuberculosis incidence and has been used extensively in an ongoing, prospective epidemiological study [81]. According to the National Tuberculosis Control Program in line with the Directly Observed Therapy Short-course strategy, diagnosis of tuberculosis is made by sputum smear microscopy in new cases, and by smear microscopy and culture in retreatment cases. We recovered these sputum specimens for our study area of interest from the National Health Laboratory Service (NHLS) after routine processing. M. tuberculosis strains present in sputum culture were genotyped using IS6110 RFLP [82] and spoligotyping [83], [84]. DNA fingerprints were analysed with GelCompar software using the unweighted-pair group method, average linkages and Dice coefficients [85]. Isolates with an IS6110 similarity index of ≥65% were grouped into strain lineages [86]. Fourty isolates of divergent lineages were selected for analysis. Table 2 shows phylogenetic details of these clinical isolates.
Table 2. Details of clinical isolates used in this study.
| Isolate | Lineage | South African IS6110 Lineage [84] |
| SAWC 1659 | 1, PGG1, EAI | - |
| SAWC 2493 | 1, PGG1, EAI | - |
| SAWC 4981 | 1, PGG1, EAI | - |
| SAWC 2803 | 3, PGG1, CAS | F34 |
| SAWC 2240 | 3, PGG1, CAS | F20 |
| SAWC 2666 | 3, PGG1, CAS | F33 |
| SAWC 974 | 3, PGG1, CAS | F25 |
| SAWC 2088 | 2, PGG1, Atypical Beijing | F31 |
| SAWC 2701 | 2, PGG1, Atypical Beijing | F27 |
| SAWC 2076 | 2, PGG1, Typical Beijing | F29 |
| SAWC 1430 | 4, PGG2 | F3 |
| SAWC 3656 | 4, PGG2, LAM | F26 |
| SAWC 2576 | 4, PGG2, LAM | F15 |
| SAWC 2525 | 4, PGG2, LAM | F9 |
| SAWC 1815 | 4, PGG2, LAM | F11 |
| SAWC 1733 | 4, PGG2, LAM | F13 |
| SAWC 3100 | 4, PGG2, LAM | F14 |
| SAWC 1595 | 4, PGG2, Quebec/S | F28 |
| SAWC 198 | 4, PGG2, “1 bander” | F110 |
| SAWC 2073 | 4, PGG2, LCC – “2 bander” | F120 |
| SAWC 233 | 4, PGG2, LCC – “3 bander” | F130 |
| SAWC 861 | 4, PGG2, LCC – “4 bander” | F140 |
| SAWC 1162 | 4, PGG2, LCC – “5 bander” | F150 |
| SAWC 716 | 4, PGG2, Pre-Haarlem | F19 |
| SAWC 1748 | 4, PGG2, Pre-Haarlem | F24 |
| SAWC 1127 | 4, PGG2, Haarlem-like | F6 |
| SAWC 103 | 4, PGG2, Haarlem-like | F7 |
| SAWC 386 | 4, PGG2, Haarlem | F1 |
| SAWC 1645 | 4, PGG2, Haarlem | F10 |
| SAWC 1841 | 4, PGG2, Haarlem | F4 |
| SAWC 2185 | 4, PGG2, Haarlem | F2 |
| SAWC 239 | 4, PGG3, T | F22 |
| SAWC 2901 | 4, PGG3, T | F16 |
| SAWC 1608 | 4, PGG3, T | F5 |
| SAWC 1109 | 4, PGG3, T | F23 |
| SAWC 4302 | 4, PGG3, T | F18 |
| SAWC 1956 | 4, PGG3, T | F17 |
| SAWC 1290 | 4, PGG3, T | F21 |
| SAWC 300 | 4, PGG3, T | F12 |
| SAWC 1870 | 4, PGG3, T | F8 |
Selection of pe/ppe genes for whole gene sequence analysis
A phylogenetic analysis of both the pe and ppe gene families has previously been reported [8]. This has demonstrated that each gene family can be divided into several subfamilies (Fig. 1). In order to maximise the scope of our analysis we selected genes representative of several different sub-families in each case. Where possible, genes for which some aspect of their biology (such as antigenicity) had been previously reported were chosen. A total of 14 pe/ppe genes were selected. These included the ancestral member of each family as well as 5 pe_pgrs genes. Details of the selected genes are listed in Table 3.
Table 3. Details of the pe and ppe genes examined by whole gene sequencing.
| Gene | Rv number | Size in H37Rv (bp) | Sublineage* | Variability in literature | Comments |
| pe35 | Rv3872 | 300 | I | No data. | Ancestral pe protein. Present in RD1 region. Highly immunogenic, eg [89]. |
| pe11, lipX | Rv1169c | 303 | IV | Invariable [32]. | B cell responses in subgroups of patients [90]. Putative lipase [77]. |
| pe3 | Rv0159c | 1407 | V (PGRS subfamily) | No data. | Atypical sublineage V protein. Not pgrs. |
| pe_pgrs16 | Rv0977 | 2772 | V (PGRS subfamily) | Highly variable [20]. | Upregulated in mouse model [39], [91]. |
| pe_pgrs18 | Rv0980c | 1374 | V (PGRS subfamily) | Known to undergo homologous recombination with pe_pgrs17 [30]. | Highly upregulated during the early stages of M. tuberculosis invasion of the blood-brain barrier [91]. High sequence identity to pe_pgrs17 implying recent duplication event [30]. |
| pe_pgrs26 | Rv1441c | 1476 | V (PGRS subfamily) | Highly variable [20]. | Downregulated in mouse model [39]. |
| pe_pgrs33 | Rv1818c | 1497 | V (PGRS subfamily) | Highly variable [18], [19]. | Localised in cell wall [14] and surface exposed [15]. SigA-mediated transcription downregulated during stationary phase and under stress conditions [92]. Implicated in pathogenicity and host immune responses [43], [47], [48], [61], [62], [90], [93], [94]. Possible inhibitor of antigen processing [44]. |
| pe_pgrs62 | Rv3812 | 1515 | V (PGRS subfamily) | No data. | Elicits strong antibody response [64]. T cell antigen [61], [62]. |
| ppe68 | Rv3873 | 1107 | I | No data. | Ancestral ppe protein. |
| ppe2 | Rv0256c | 1671 | II (PPW subfamily) | No data. | PPW subfamily. |
| ppe44 | Rv2770c | 1149 | IV (SVP subfamily) | Limited diversity. Alteration in Beijing isolates [35]. | Variable expression in clinical isolates [35]. Expressed during subcutaneous and intravenous infection by M. bovis BCG in BALB/c mice [95]. |
| ppe10 | Rv0442c | 1464 | V (MPTR subfamily) | No data. | Ancestral ppe MPTR protein. |
| ppe42 | Rv2608 | 1743 | V (MPTR subfamily) | Variable in clinical isolates [96]. | Elicits a high humoral and low T cell response [96]. |
| ppe62 | Rv3533c | 1749 | V (MPTR subfamily) | No data. | MPTR protein. |
As defined in reference [8] .
Each gene sequenced in this study is listed along with its phylogenetic position within its family and any additional information regarding its protein's function available in the literature.
PCR and sequencing
Primer sequences for the 14 selected genes are listed in Table S6. PCRs were done in a reaction mixture containing 0.1 µg template DNA, 3 µl GC-rich solution, 1.5 µl 10× buffer containing MgCl2, 2.4 µl 10 mM dNTP's, 0.6 µl each primer (5 pmol/µl) and 0.12 µl FastStart Taq (Roche, Germany) made up to 15 µl with H2O. Amplification comprised an initial 6 min template denaturation followed by 35 cycles using the appropriate annealing temperature (listed in Table S6) and an extension time of 30 s to 1 min 30 s depending on the length of the amplicon. PCR product was checked by electrophoresis through an agarose gel and an aliquot was treated with ExoSAP-IT (USB). Sequencing was performed using an ABI 3100 automated DNA sequencer. Sequence editing and manipulation was done using the BioEdit Sequence Alignment Editor [87].
Supporting Information
(DOCX)
2 homologous genes (PPE38 & PPE71) in most isolates. Hotspot for IS6110 integration and homologous recombination [26] .
(XLS)
(XLS)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We acknowledge the Wellcome Trust, the James Craig Venter Institute, the Broad Institute, and the Pasteur Institute for making DNA sequences available.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was provided by the South African National Research Foundation through the Centre of Excellence in Biomedical Tuberculosis Research - http://www.tuberculosis.org.za/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, et al. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002;162:1533–1543. doi: 10.1093/genetics/162.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes AL, Friedman R, Murray M. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg Infect Dis. 2002;8:1342–1346. doi: 10.3201/eid0811.020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, et al. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, et al. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunham RC, Plummer FA, Stephens RS. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–2276. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campuzano J, Aguilar D, Arriaga K, Leon JC, Salas-Rangel LP, et al. The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine. 2007;25:3722–3729. doi: 10.1016/j.vaccine.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 11.Chaitra MG, Hariharaputran S, Chandra NR, Shaila MS, Nayak R. Defining putative T cell epitopes from PE and PPE families of proteins of Mycobacterium tuberculosis with vaccine potential. Vaccine. 2005;23:1265–1272. doi: 10.1016/j.vaccine.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 12.Sampson SL, Lukey P, Warren RM, van Helden PD, Richardson M, et al. Expression, characterization and subcellular localization of the Mycobacterium tuberculosis PPE gene Rv1917c. Tuberculosis (Edinb) 2001;81:305–317. doi: 10.1054/tube.2001.0304. [DOI] [PubMed] [Google Scholar]
- 13.Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, et al. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 14.Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, et al. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol Microbiol. 2004;52:725–733. doi: 10.1111/j.1365-2958.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 15.Cascioferro A, Delogu G, Colone M, Sali M, Stringaro A, et al. PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol Microbiol. 2007;66:1536–1547. doi: 10.1111/j.1365-2958.2007.06023.x. [DOI] [PubMed] [Google Scholar]
- 16.Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- 17.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 18.Talarico S, Cave MD, Marrs CF, Foxman B, Zhang L, et al. Variation of the Mycobacterium tuberculosis PE_PGRS 33 gene among clinical isolates. J Clin Microbiol. 2005;43:4954–4960. doi: 10.1128/JCM.43.10.4954-4960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talarico S, Cave MD, Foxman B, Marrs CF, Zhang L, et al. Association of Mycobacterium tuberculosis PE PGRS33 polymorphism with clinical and epidemiological characteristics. Tuberculosis (Edinb) 2007;87:338–346. doi: 10.1016/j.tube.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talarico S, Zhang L, Marrs CF, Foxman B, Cave MD, et al. Mycobacterium tuberculosis PE_PGRS16 and PE_PGRS26 genetic polymorphism among clinical isolates. Tuberculosis (Edinb) 2008;88:283–294. doi: 10.1016/j.tube.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebert AM, Talarico S, Yang D, Durmaz R, Marrs CF, et al. DNA polymorphisms in the pepA and PPE18 genes among clinical strains of Mycobacterium tuberculosis: implications for vaccine efficacy. Infect Immun. 2007;75:5798–5805. doi: 10.1128/IAI.00335-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava R, Kumar D, Waskar MN, Sharma M, Katoch VM, et al. Identification of a repetitive sequence belonging to a PPE gene of Mycobacterium tuberculosis and its use in diagnosis of tuberculosis. J Med Microbiol. 2006;55:1071–1077. doi: 10.1099/jmm.0.46379-0. [DOI] [PubMed] [Google Scholar]
- 23.Sampson SL, Warren RM, Richardson M, van der Spuy GD, van Helden PD. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:349–359. doi: 10.1054/tuld.1999.0218. [DOI] [PubMed] [Google Scholar]
- 24.Warren RM, Sampson SL, Richardson M, Van Der Spuy GD, Lombard CJ, et al. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol Microbiol. 2000;37:1405–1416. doi: 10.1046/j.1365-2958.2000.02090.x. [DOI] [PubMed] [Google Scholar]
- 25.Yesilkaya H, Dale JW, Strachan NJ, Forbes KJ. Natural transposon mutagenesis of clinical isolates of Mycobacterium tuberculosis: how many genes does a pathogen need? J Bacteriol. 2005;187:6726–6732. doi: 10.1128/JB.187.19.6726-6732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEvoy CR, van Helden PD, Warren RM, Gey van Pittius NC. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol Biol. 2009;9:237. doi: 10.1186/1471-2148-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEvoy CR, Warren RM, van Helden PD, Gey van Pittius NC. Multiple, independent, identical IS6110 insertions in Mycobacterium tuberculosis PPE genes. Tuberculosis (Edinb) 2009;89:439–442. doi: 10.1016/j.tube.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Namouchi A, Mardassi H. A genomic library-based amplification approach (GL-PCR) for the mapping of multiple IS6110 insertion sites and strain differentiation of Mycobacterium tuberculosis. J Microbiol Methods. 2006;67:202–211. doi: 10.1016/j.mimet.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Gutacker MM, Musser JM, Fu YX. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol. 2006;188:8169–8177. doi: 10.1128/JB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karboul A, Gey van Pittius NC, Namouchi A, Vincent V, Sola C, et al. Insights into the evolutionary history of tubercle bacilli as disclosed by genetic rearrangements within a PE_PGRS duplicated gene pair. BMC Evol Biol. 2006;6:107. doi: 10.1186/1471-2148-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karboul A, Mazza A, Gey van Pittius NC, Ho JL, Brousseau R, et al. Frequent homologous recombination events in Mycobacterium tuberculosis PE/PPE multigene families: potential role in antigenic variability. J Bacteriol. 2008;190:7838–7846. doi: 10.1128/JB.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musser JM, Amin A, Ramaswamy S. Negligible genetic diversity of mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics. 2000;155:7–16. doi: 10.1093/genetics/155.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flores J, Espitia C. Differential expression of PE and PE_PGRS genes in Mycobacterium tuberculosis strains. Gene. 2003;318:75–81. doi: 10.1016/s0378-1119(03)00751-0. [DOI] [PubMed] [Google Scholar]
- 34.Gao Q, Kripke KE, Saldanha AJ, Yan W, Holmes S, et al. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology. 2005;151:5–14. doi: 10.1099/mic.0.27539-0. [DOI] [PubMed] [Google Scholar]
- 35.Rindi L, Peroni I, Lari N, Bonanni D, Tortoli E, et al. Variation of the expression of Mycobacterium tuberculosis ppe44 gene among clinical isolates. FEMS Immunol Med Microbiol. 2007;51:381–387. doi: 10.1111/j.1574-695X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 36.Voskuil MI, Schnappinger D, Rutherford R, Liu Y, Schoolnik GK. Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis (Edinb) 2004;84:256–262. doi: 10.1016/j.tube.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Fu LM, Tai SC. The Differential Gene Expression Pattern of Mycobacterium tuberculosis in Response to Capreomycin and PA-824 versus First-Line TB Drugs Reveals Stress- and PE/PPE-Related Drug Targets. Int J Microbiol. 2009;2009:879621. doi: 10.1155/2009/879621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava V, Jain A, Srivastava BS, Srivastava R. Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. Tuberculosis (Edinb) 2008;88:171–177. doi: 10.1016/j.tube.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, Brennan MJ. Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol. 2006;188:3721–3725. doi: 10.1128/JB.188.10.3721-3725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstone RM, Goonesekera SD, Bloom BR, Sampson SL. The transcriptional regulator Rv0485 modulates the expression of a pe and ppe gene pair and is required for Mycobacterium tuberculosis virulence. Infect Immun. 2009;77:4654–4667. doi: 10.1128/IAI.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basu S, Pathak SK, Banerjee A, Pathak S, Bhattacharyya A, et al. Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem. 2007;282:1039–1050. doi: 10.1074/jbc.M604379200. [DOI] [PubMed] [Google Scholar]
- 42.Nair S, Ramaswamy PA, Ghosh S, Joshi DC, Pathak N, et al. The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J Immunol. 2009;183:6269–6281. doi: 10.4049/jimmunol.0901367. [DOI] [PubMed] [Google Scholar]
- 43.Delogu G, Brennan MJ. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun. 2001;69:5606–5611. doi: 10.1128/IAI.69.9.5606-5611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh KW, Lehming N, Seah GT. Degradation-resistant protein domains limit host cell processing and immune detection of mycobacteria. Mol Immunol. 2009;46:1312–1318. doi: 10.1016/j.molimm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, et al. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 46.Cadieux N, Parra M, Cohen H, Maric D, Morris SL, et al. Induction of cell death after localization to the host cell mitochondria by the Mycobacterium tuberculosis PE_PGRS33 protein. Microbiology. 2011;157:793–804. doi: 10.1099/mic.0.041996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan MJ, Delogu G, Chen Y, Bardarov S, Kriakov J, et al. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect Immun. 2001;69:7326–7333. doi: 10.1128/IAI.69.12.7326-7333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dheenadhayalan V, Delogu G, Brennan MJ. Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect. 2006;8:262–272. doi: 10.1016/j.micinf.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 49.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 50.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachhawat N, Singh B. Mycobacterial PE_PGRS proteins contain calcium-binding motifs with parallel beta-roll folds. Genomics Proteomics Bioinformatics. 2007;5:236–241. doi: 10.1016/S1672-0229(08)60010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Espitia C, Laclette JP, Mondragon-Palomino M, Amador A, Campuzano J, et al. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology. 1999;145(Pt 12):3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tundup S, Akhter Y, Thiagarajan D, Hasnain SE. Clusters of PE and PPE genes of Mycobacterium tuberculosis are organized in operons: evidence that PE Rv2431c is co-transcribed with PPE Rv2430c and their gene products interact with each other. FEBS Lett. 2006;580:1285–1293. doi: 10.1016/j.febslet.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 56.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broad Institute website. Available ([ www.broadinstitute.org/]). Accessed 2010 January.
- 58.Schurch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, et al. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect Genet Evol. 2010;10:108–114. doi: 10.1016/j.meegid.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Schurch AC, Kremer K, Daviena O, Kiers A, Boeree MJ, et al. High resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J Clin Microbiol. 2010 doi: 10.1128/JCM.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaitra MG, Nayak R, Shaila MS. Modulation of immune responses in mice to recombinant antigens from PE and PPE families of proteins of Mycobacterium tuberculosis by the Ribi adjuvant. Vaccine. 2007;25:7168–7176. doi: 10.1016/j.vaccine.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 62.Chaitra MG, Shaila MS, Nayak R. Characterization of T-cell immunogenicity of two PE/PPE proteins of Mycobacterium tuberculosis. J Med Microbiol. 2008;57:1079–1086. doi: 10.1099/jmm.0.47565-0. [DOI] [PubMed] [Google Scholar]
- 63.Vipond J, Vipond R, Allen-Vercoe E, Clark SO, Hatch GJ, et al. Selection of novel TB vaccine candidates and their evaluation as DNA vaccines against aerosol challenge. Vaccine. 2006;24:6340–6350. doi: 10.1016/j.vaccine.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 64.Koh KW, Soh SE, Seah GT. Strong antibody responses to Mycobacterium tuberculosis PE-PGRS62 protein are associated with latent and active tuberculosis. Infect Immun. 2009;77:3337–3343. doi: 10.1128/IAI.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daubin V, Moran NA. Comment on “The origins of genome complexity”. Science. 2004;306:978; author reply 978. doi: 10.1126/science.1100559. [DOI] [PubMed] [Google Scholar]
- 66.Plotkin JB, Dushoff J, Fraser HB. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature. 2004;428:942–945. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Zhang H, Zhou T, Zhong Y, Jin Q. Genes under positive selection in Mycobacterium tuberculosis. Comput Biol Chem. 2011;35:319–322. doi: 10.1016/j.compbiolchem.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen T, Colijn C, Murray M. Modeling the effects of strain diversity and mechanisms of strain competition on the potential performance of new tuberculosis vaccines. Proc Natl Acad Sci U S A. 2008;105:16302–16307. doi: 10.1073/pnas.0808746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skeiky YA, Alderson MR, Ovendale PJ, Guderian JA, Brandt L, et al. Differential immune responses and protective efficacy induced by components of a tuberculosis polyprotein vaccine, Mtb72F, delivered as naked DNA or recombinant protein. J Immunol. 2004;172:7618–7628. doi: 10.4049/jimmunol.172.12.7618. [DOI] [PubMed] [Google Scholar]
- 73.Brandt L, Skeiky YA, Alderson MR, Lobet Y, Dalemans W, et al. The protective effect of the Mycobacterium bovis BCG vaccine is increased by coadministration with the Mycobacterium tuberculosis 72-kilodalton fusion polyprotein Mtb72F in M. tuberculosis-infected guinea pigs. Infect Immun. 2004;72:6622–6632. doi: 10.1128/IAI.72.11.6622-6632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsenova L, Harbacheuski R, Moreira AL, Ellison E, Dalemans W, et al. Evaluation of the Mtb72F polyprotein vaccine in a rabbit model of tuberculous meningitis. Infect Immun. 2006;74:2392–2401. doi: 10.1128/IAI.74.4.2392-2401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reed SG, Coler RN, Dalemans W, Tan EV, DeLa Cruz EC, et al. Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proc Natl Acad Sci U S A. 2009;106:2301–2306. doi: 10.1073/pnas.0712077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bovilist website. Available: ([ http://genolist.pasteur.fr/BoviList/]). Accessed 2010 January.
- 77.Tuberculist website. Available: ([ http://genolist.pasteur.fr/TubercuList/]). Accessed 2010 January.
- 78.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 81.Verver S, Warren RM, Munch Z, Vynnycky E, van Helden PD, et al. Transmission of tuberculosis in a high incidence urban community in South Africa. Int J Epidemiol. 2004;33:351–357. doi: 10.1093/ije/dyh021. [DOI] [PubMed] [Google Scholar]
- 82.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Streicher EM, Victor TC, van der Spuy G, Sola C, Rastogi N, et al. Spoligotype signatures in the Mycobacterium tuberculosis complex. J Clin Microbiol. 2007;45:237–240. doi: 10.1128/JCM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hermans PW, Messadi F, Guebrexabher H, van Soolingen D, de Haas PE, et al. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 86.Richardson M, Carroll NM, Engelke E, Van Der Spuy GD, Salker F, et al. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J Clin Microbiol. 2002;40:2750–2754. doi: 10.1128/JCM.40.8.2750-2754.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 88.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mukherjee P, Dutta M, Datta P, Dasgupta A, Pradhan R, et al. The RD1-encoded antigen Rv3872 of Mycobacterium tuberculosis as a potential candidate for serodiagnosis of tuberculosis. Clin Microbiol Infect. 2007;13:146–152. doi: 10.1111/j.1469-0691.2006.01660.x. [DOI] [PubMed] [Google Scholar]
- 90.Narayana Y, Joshi B, Katoch VM, Mishra KC, Balaji KN. Differential B-cell responses are induced by Mycobacterium tuberculosis PE antigens Rv1169c, Rv0978c, and Rv1818c. Clin Vaccine Immunol. 2007;14:1334–1341. doi: 10.1128/CVI.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jain SK, Paul-Satyaseela M, Lamichhane G, Kim KS, Bishai WR. Mycobacterium tuberculosis invasion and traversal across an in vitro human blood-brain barrier as a pathogenic mechanism for central nervous system tuberculosis. J Infect Dis. 2006;193:1287–1295. doi: 10.1086/502631. [DOI] [PubMed] [Google Scholar]
- 92.Vallecillo AJ, Espitia C. Expression of Mycobacterium tuberculosis pe_pgrs33 is repressed during stationary phase and stress conditions, and its transcription is mediated by sigma factor A. Microb Pathog. 2009;46:119–127. doi: 10.1016/j.micpath.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 93.Balaji KN, Goyal G, Narayana Y, Srinivas M, Chaturvedi R, et al. Apoptosis triggered by Rv1818c, a PE family gene from Mycobacterium tuberculosis is regulated by mitochondrial intermediates in T cells. Microbes Infect. 2007;9:271–281. doi: 10.1016/j.micinf.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 94.Singh PP, Parra M, Cadieux N, Brennan MJ. A comparative study of host response to three Mycobacterium tuberculosis PE_PGRS proteins. Microbiology. 2008;154:3469–3479. doi: 10.1099/mic.0.2008/019968-0. [DOI] [PubMed] [Google Scholar]
- 95.Bonanni D, Rindi L, Lari N, Garzelli C. Immunogenicity of mycobacterial PPE44 (Rv2770c) in Mycobacterium bovis BCG-infected mice. J Med Microbiol. 2005;54:443–448. doi: 10.1099/jmm.0.45960-0. [DOI] [PubMed] [Google Scholar]
- 96.Chakhaiyar P, Nagalakshmi Y, Aruna B, Murthy KJ, Katoch VM, et al. Regions of high antigenicity within the hypothetical PPE major polymorphic tandem repeat open-reading frame, Rv2608, show a differential humoral response and a low T cell response in various categories of patients with tuberculosis. J Infect Dis. 2004;190:1237–1244. doi: 10.1086/423938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
2 homologous genes (PPE38 & PPE71) in most isolates. Hotspot for IS6110 integration and homologous recombination [26] .
(XLS)
(XLS)
(DOCX)
(DOCX)
(DOCX)