Abstract
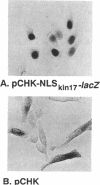
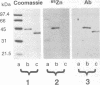
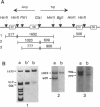
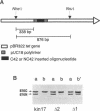
Kin17 is a 45 kDa protein encoded by the KIN17 gene located on mouse chromosome 2, band A. The kin17 amino acid sequence predicts two domains, which were shown to be functional: (i) a bipartite nuclear localization signal (NLS) that can drive the protein to the cell nucleus, (ii) a bona fide zinc finger of the C2H2 type. The zinc finger is involved in kin17 binding to double-stranded DNA since a mutant deleted of the zinc finger, kin17 delta 1, showed reduced binding. Single-stranded DNA was bound poorly by kin17. Interestingly, we found that kin17 protein showed preferential binding to curved DNA from either pBR322 or synthetic oligonucleotides. Binding of kin17 to a non-curved DNA segment increased after we had inserted into it a short curved synthetic oligonucleotide. Kin17 delta 2, a mutant deleted of 110 amino acids at the C-terminal end, still exhibited preferential binding to curved DNA and so did kin17 delta 1, suggesting that a domain recognizing curved DNA is located in the protein core.
Full text
PDF


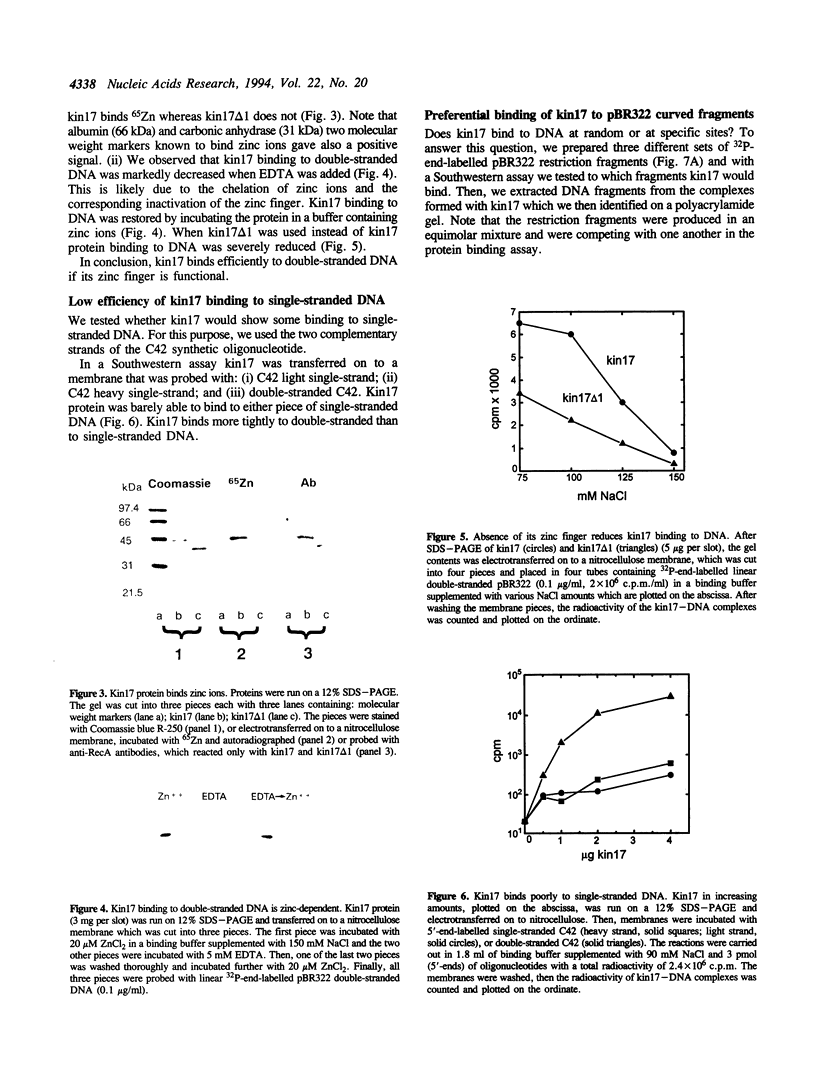
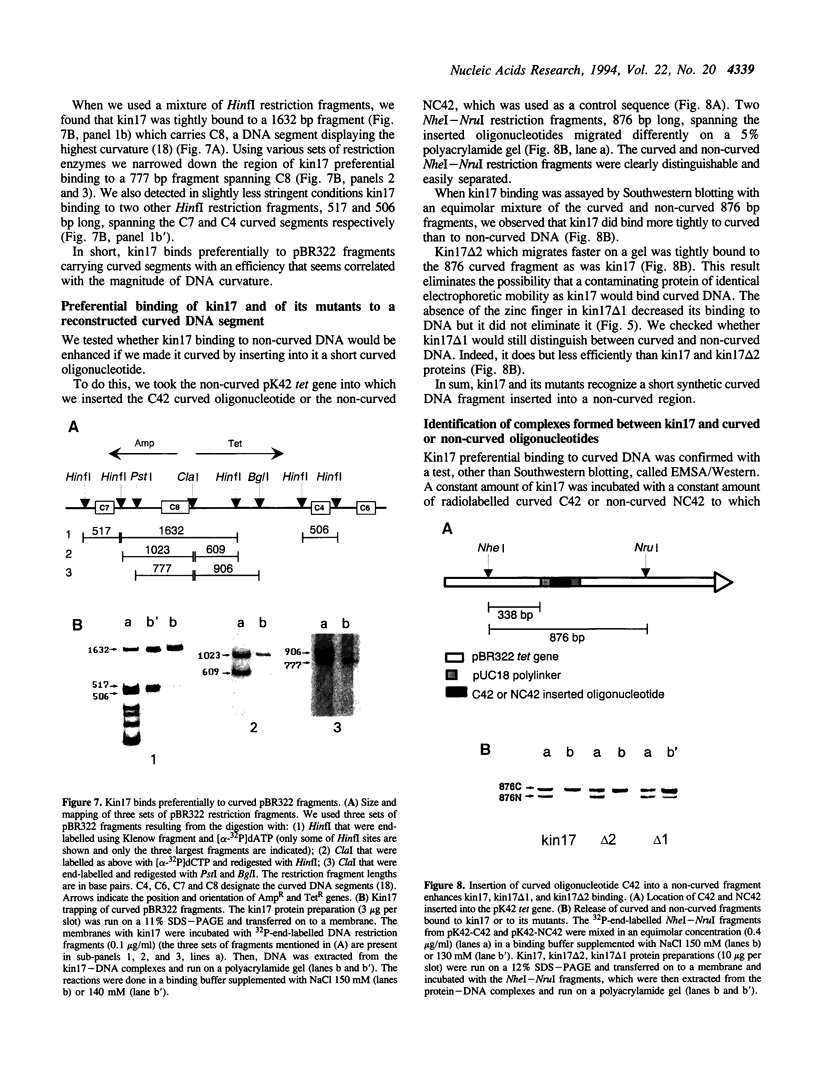


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angulo J. F., Moreau P. L., Maunoury R., Laporte J., Hill A. M., Bertolotti R., Devoret R. KIN, a mammalian nuclear protein immunologically related to E. coli RecA protein. Mutat Res. 1989 Mar;217(2):123–134. doi: 10.1016/0921-8777(89)90064-5. [DOI] [PubMed] [Google Scholar]
- Angulo J. F., Rouer E., Mazin A., Mattei M. G., Tissier A., Horellou P., Benarous R., Devoret R. Identification and expression of the cDNA of KIN17, a zinc-finger gene located on mouse chromosome 2, encoding a new DNA-binding protein. Nucleic Acids Res. 1991 Oct 11;19(19):5117–5123. doi: 10.1093/nar/19.19.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antalis T. M., Godbolt D., Donnan K. D., Stringer B. W. Southwestern blot mapping of potential regulatory proteins binding to the DNA encoding plasminogen activator inhibitor type 2. Gene. 1993 Dec 8;134(2):201–208. doi: 10.1016/0378-1119(93)90094-j. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E., Rouvière-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991 Mar;10(3):687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Coleman J. E. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Analyzing DNA curvature in polyacrylamide gels. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-a. [DOI] [PubMed] [Google Scholar]
- Dutreix M., Burnett B., Bailone A., Radding C. M., Devoret R. A partially deficient mutant, recA1730, that fails to form normal nucleoprotein filaments. Mol Gen Genet. 1992 Apr;232(3):489–497. doi: 10.1007/BF00266254. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Ménissier de Murcia J. M., Molinete M., Simonin F., Koken M., Hoeijmakers J. H., de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury Christianson A. M., Kafatos F. C. Antibody detection of protein complexes bound to DNA. Nucleic Acids Res. 1993 Sep 11;21(18):4416–4417. doi: 10.1093/nar/21.18.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mazen A., Gradwohl G., de Murcia G. Zinc-binding proteins detected by protein blotting. Anal Biochem. 1988 Jul;172(1):39–42. doi: 10.1016/0003-2697(88)90408-3. [DOI] [PubMed] [Google Scholar]
- Mazen A., Menissier-de Murcia J., Molinete M., Simonin F., Gradwohl G., Poirier G., de Murcia G. Poly(ADP-ribose)polymerase: a novel finger protein. Nucleic Acids Res. 1989 Jun 26;17(12):4689–4698. doi: 10.1093/nar/17.12.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A. V., Kuzminov A. V., Dianov G. L., Salganik R. I. Mechanisms of deletion formation in Escherichia coli plasmids. II. Deletions mediated by short direct repeats. Mol Gen Genet. 1991 Aug;228(1-2):209–214. doi: 10.1007/BF00282467. [DOI] [PubMed] [Google Scholar]
- Mazin A., Milot E., Devoret R., Chartrand P. KIN17, a mouse nuclear protein, binds to bent DNA fragments that are found at illegitimate recombination junctions in mammalian cells. Mol Gen Genet. 1994 Aug 15;244(4):435–438. doi: 10.1007/BF00286696. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., Bird A. P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992 Oct 11;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milot E., Belmaaza A., Wallenburg J. C., Gusew N., Bradley W. E., Chartrand P. Chromosomal illegitimate recombination in mammalian cells is associated with intrinsically bent DNA elements. EMBO J. 1992 Dec;11(13):5063–5070. doi: 10.1002/j.1460-2075.1992.tb05613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta T. R., Fukita Y., Miyoshi T., Shimizu A., Honjo T. Isolation of cDNA encoding a binding protein specific to 5'-phosphorylated single-stranded DNA with G-rich sequences. Nucleic Acids Res. 1993 Apr 25;21(8):1761–1766. doi: 10.1093/nar/21.8.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinete M., Vermeulen W., Bürkle A., Ménissier-de Murcia J., Küpper J. H., Hoeijmakers J. H., de Murcia G. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J. 1993 May;12(5):2109–2117. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzard G., Théveny B., Révet B. Electron microscopy mapping of pBR322 DNA curvature. Comparison with theoretical models. EMBO J. 1990 Apr;9(4):1289–1298. doi: 10.1002/j.1460-2075.1990.tb08238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménissier-de Murcia J., Molinete M., Gradwohl G., Simonin F., de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol. 1989 Nov 5;210(1):229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- Pasero P., Sjakste N., Blettry C., Got C., Marilley M. Long-range organization and sequence-directed curvature of Xenopus laevis satellite 1 DNA. Nucleic Acids Res. 1993 Oct 11;21(20):4703–4710. doi: 10.1093/nar/21.20.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Salinovich O., Montelaro R. C. Reversible staining and peptide mapping of proteins transferred to nitrocellulose after separation by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1986 Aug 1;156(2):341–347. doi: 10.1016/0003-2697(86)90263-0. [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986 Dec 1;5(12):3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M. S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992 Mar 26;356(6367):356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Schreiber V., Molinete M., Boeuf H., de Murcia G., Ménissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992 Sep;11(9):3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary A., Sarasin A. Molecular analysis of DNA junctions produced by illegitimate recombination in human cells. Nucleic Acids Res. 1992 Aug 25;20(16):4269–4274. doi: 10.1093/nar/20.16.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J. D., Bronson E. C., Anderson J. N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991 Oct 11;19(19):5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. K., Uhlin B. E., Amini K. S., Clark A. J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183(3):497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- de Murcia G., Ménissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994 Apr;19(4):172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- von Kries J. P., Buck F., Strätling W. H. Chicken MAR binding protein p120 is identical to human heterogeneous nuclear ribonucleoprotein (hnRNP) U. Nucleic Acids Res. 1994 Apr 11;22(7):1215–1220. doi: 10.1093/nar/22.7.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]