Abstract
Fragile X Syndrome (FXS) is the most common form of inherited intellectual disability and autism. The protein (FMRP) encoded by the fragile X mental retardation gene (FMR1), is an RNA-binding protein linked to translational control. Recently, in the Fmr1 knockout mouse model of FXS, dysregulated translation initiation signaling was observed. To investigate whether an altered signaling was also a feature of subjects with FXS compared to typical developing controls, we isolated total RNA and translational control proteins from lymphocytes of subjects from both groups (38 FXS and 14 TD). Although we did not observe any difference in the expression level of mRNAs for translational initiation control proteins isolated from participant with FXS, we found increased phosphorylation of the mammalian target of rapamycin (mTOR) substrate, p70 ribosomal subunit 6 kinase1 (S6K1) and of the mTOR regulator, the serine/threonine protein kinase (Akt), in their protein lysates. In addition, we observed increased phosphorylation of the cap binding protein eukaryotic initiation factor 4E (eIF4E) suggesting that protein synthesis is upregulated in FXS. Similarly to the findings in lymphocytes, we observed increased phosphorylation of S6K1 in brain tissue from patients with FXS (n=6) compared to normal age matched controls (n=4). Finally, we detected increased expression of the cytoplasmic FMR1-interacting protein 2 (CYFIP2), a known FMRP interactor. This data verify and extend previous findings using lymphocytes for studies of neuropsychiatric disorders and provide evidence that misregulation of mTOR signaling observed in a FXS mouse model also occurs in human FXS and may provide useful biomarkers for designing target treatments in FXS.
Keywords: Fragile X, CYFIP1, CYFIP2, mTOR, phosphorylation
Introduction
Fragile X syndrome (FXS) is the most common inherited cause of mental retardation, and the most common single gene mutation associated with autism (Hagerman & Hagerman, 2002, Jacquemont et al., 2007, Loesch et al., 2007). It is caused by a trinucleotide repeat expansion (CGG)n in the 5′ untranslated region of the fragile X mental retardation 1 gene (FMR1) located at Xq27.3. The full mutation, present in individuals having more than 200 CGG repeats, typically involves methylation and subsequent transcriptional silencing of the FMR1 gene, resulting in diminished or absent production of the FMR1 protein, FMRP (Fu et al., 1991, Pieretti et al., 1991, Verkerk et al., 1991, Yu et al., 1991). Loss of FMRP results in aberrant brain development and function (Bagni & Greenough, 2005).
It was recently reported that the Mammalian target of rapamycin (mTOR) signaling is upregulated in a mouse model of FXS (Sharma et al., 2010). The loss of FMRP resulted in enhanced mTOR signaling that was associated with increased formation of the eukaryotic initiation factor complex 4F (eIF4F). These findings suggest that in addition to its RNA binding activity, FMRP also plays a role in the regulation of translation initiation and subsequent protein synthesis. Whether this is the case in individuals with FXS is unknown.
Although FXS is associated with a characteristic phenotype, there is considerable within-syndrome variation in the severity of the phenotype and the profile of impairments, with the most interesting being co-morbidity with autism. From the most recent studies, the prevalence of autism spectrum disorder (ASD) is approximately 60% in individuals with FXS (Harris et al., 2008). A subgroup of patients with FXS also presents with a Prader-Willi-like phenotype (PWP), which includes severe hyperphagia, obesity, hypogonadism, and autism. Prader Willi Syndrome (PWS) is an obesity syndrome resulting from a loss of paternally derived genes at 15q11-13, which regulate metabolism or energy homeostasis. The PWP of FXS was first reported in males with extreme obesity, short stature, short fingers and toes, and hypogonadism (de Vries et al., 1993, de Vries & Niermeijer, 1994, Fryns et al., 1987, Hagerman & Hagerman, 2002, Schrander-Stumpel et al., 1994) but the patients do not show genetic abnormalities at 15q11–13, which normally is associated with PWS. Interestingly, very high rates of autism are observed in patients with FXS and the PWP (Demark et al., 2003, Hatton et al., 2006, Kaufmann et al., 2004, Nowicki et al., 2007, Rogers et al., 2001).
The findings of an up-regulation of the mTOR signaling in the FXS mouse model, combined with the very high rates of autism associated with FXS with and without the PWP (Nowicki et al., 2007), prompted us to investigate mTOR signaling in subjects with FXS and FXS with PWP. Because the molecular signaling effects resulting from FMRP loss are likely causal in the wide-ranging severity of FXS symptoms, including autism, identifying the effects of FMRP loss on molecular signaling pathways, like those governing translation, are key to advancing our ability to treat the disorder.
Methods
Study Participants
52 subjects (49 males, 3 females) were included in the study, except for the cytoplasmic FMR1 interacting protein 1 (CYFIP1) mRNA measurements, which comprised an additional 30 subjects. Participants were recruited through Fragile X Research and Treatment Center at the UC Davis MIND Institute in Sacramento (CA) and included a total of 38 cases with FXS, 10 of which were mosaics (both methylation and size mosaics) (mean 19 ± 2 years, range 4-68 years old). Seven patients had FXS without ASD, while 31 participants presented with both FXS and ASD. 14 subjects also had the PWP and 12 of them had ASD. 14 typically developing (TD) controls (ranging from 21 to 40 CGG repeats) (mean age 26 ± 5 years, range 2-55 years old) were also included in the study. This study was approved by the Institutional Review Board of the University of California.
Clinical evaluation and assessment measures for autism
A complete medical evaluation, including medical history, psychological testing and physical examination was conducted on each subject including controls. Individuals were confirmed to have ASD by a multidisciplinary assessment. This assessment included the Autism Diagnostic Interview-Revised (ADI-R) (Rutter et al., 2003a) which is a standardized, semi-structured, investigator-based interview for caregivers of individuals with autism or pervasive developmental disorders, and using the Autism Diagnostic Observation Schedules (ADOS) (Lord et al., 1999) which is a semi-structured, standardized assessment of the child in which the researcher observes the social interaction, communication, play, and imaginative use of materials for children suspected of having ASD. The DSM IV-TR (APA, 2000) criteria for ASD, was also applied and a team consensus lead to a diagnosis of either autism, pervasive developmental disorders not otherwise specified (PDD-NOS) or no ASD. In this study the autism and PDD-NOS diagnoses were collapsed to ASD.
Cognitive and Adaptive measures
The Wechsler Scale including the WISC-IV (Wechsler, 2003), WPPSI-III (Wechsler, 2002), or WASI (Wechsler, 1999) (where age appropriate) were used for all patients with FXS for assessing IQ. Controls were screened for ASD traits using Social Communication Questionnaire (SCQ) (Rutter et al., 2003b) and for developmental delay with the Vineland Adaptative Behavior Scale (VABS) (Sparrow et al., 2005). Only those who scored within the normal range were utilized in this study.
Molecular measures
DNA analysis
To confirm the presence of FXS, a blood sample was obtained from each subject for the measurement of the CGG repeat number in the FMR1 gene. Genomic DNA was isolated from peripheral blood leukocytes using standard methods (Puregene Kit; Gentra Inc., Minneapolis, MN). For Southern blot analysis, 5-10 μg of isolated genomic DNA was digested with EcoRI and NruI. Probe hybridization used the FMR1-specific dig-labelled StB12.3. Details were as previously described (Tassone et al., 2008). PCR analysis was performed on all the subjects using primer c and f as described in (Filipovic-Sadic et al., 2010, Tassone et al., 2008). Analysis and measurement of trinucleotide allele size, as well as the determination of the methylation status were determined using an Alpha Innotech Fluor Chem 8800 Image Detection System (Alpha Innotech, San Leandro, CA) and the ABI 3730XL 96-Capillary Electrophoresis Genetic Analyzer (Applied Biosystems, Carlsbad, California, USA)
Brain tissues
Brain tissue derived from four males with FXS were from brain autopsies, performed in accordance with University of California, Davis, Institutional Review Board (IRB) approved protocols. One hemisphere of the fresh brain was cut into one cm coronal sections and frozen at −80°C and protein extracts and total RNA were isolated from a frozen 0.5 cm section. Case history and clinical /molecular details of the subjects are as described in (Greco et al., 2011) and (Hunsaker et al., 2011). Age matched control samples were taken from four neurologically normal male cases that were obtained from the autopsy tissue repository at the University of California, Davis Medical Center Department of Pathology and from the Maryland Bank (Table 1).
Table 1.
Summary of the pathology of the postmortem brain tissue of the 8 cases describe
| Subject | Category | Age at death (years) |
PMI (hours) |
Cause of death |
|---|---|---|---|---|
| Case 1 | FXS | 23 | 16 | Cardiac arrest |
| Case 2 | FXS | 57 | 20 | Choking on food |
| Case 3 | FXS | 64 | 12 | Liver neoplasm |
| Case 4 | FXS | 74 | 40 | Pulmonary disease & abdominal complications |
| Control 1 | Normal | 20 | 36 | Gun shot |
| Control 2 | Normal | 57 | 16 | Accident, multiple injuries |
| Control 3 | Normal | 68 | 17 | Cardiac arrest |
| Control 4 | Normal | 88 | 11 | Cardiac arrest |
FXS= Fragile X Syndrome
PMI= Post Mortem Interval
Measurement of gene expression levels
Total RNA was isolated from Tempus tubes using the ABI PRISM™ 6100 Nucleic Acid PrepStation per the manufacturer’s protocol (Applied Biosystems, Carlsbad, California, USA). Total RNA was isolated from brain tissue using standard procedures (Trizol, Invitrogen, Grand Island, NY, USA). Expression levels of the FMR1 gene and of those genes involved in the mTOR pathway cascade were measured by real-time quantitative fluorescence RT-PCR method using primers and probe specific for each single gene (Assay On Demand, Applied Biosystems, Carlsbad, California, USA). Details of the method and its application to the study of FMR1 mRNAs are as described in (Tassone et al., 2000).
Lymphocyte Extraction and Storage
Approximately 6 ml of whole blood from subjects was collected into BD Vacutainer™ CPT™ tubes (Becton-Dickinson, Franklin Lakes, NJ) containing heparin, for isolation of white cells. Lymphocytes were separated, aliquoted (~2×106 cells for cryovial), and stored in liquid nitrogen within 24 hours of collection according to manufacturer’s directions until use. Lymphocytes were maintained under these storage conditions until used for extractions.
Protein extraction and western blotting
Cells were spun down at 17,000g for 10 minutes and washed two times in wash buffer (150 mM NaCl, 50 mM Tris, 2 mM EDTA), then added to homogenization buffer containing protease and phosphatase inhibitors. Cells were briefly sonicated on ice (~10 sec each) in brief pulses (2-3 sec/pulse). Nucleic acids in homogenate was then sheared using sterile 21 gauge syringe three times. Lysed cell slurry was cleared at 17000g at 4°C then quantified using Bradford technique (Pierce, Rockford, IL USA). Protein concentrations were determined by absorbance reading at 562λ (Biotek Synergy 2 Plate reader, Winooski, VT, USA). 30 μg of total protein were combined with 6X SDS/P.A.G.E. buffer (final SDS 1%). Samples were heated at 95°C for five minutes and snap chilled before loading. Proteins were separated on Novex 4-12% gradient Tris-Bis gels (Invitrogen, Grand Island, NY, USA) then transferred to PVDF blots using conventional methodology. Blots were blocked in 0.2% I-Block (Tropix, Carlsbad, California, USA), and then incubated overnight with primaries at 4°C. Bands were resolved using HRP conjugated secondary and visualized using ECL+ (GE-Amersham, Waukesha, WI, USA) on a KODAK 4000MM (Carestream, Rochester, NY) or G.E. LAS4000 (Piscataway, NJ, USA) imaging system. All chemiluminescent signals were obtained in the linear range of detection as confirmed by time course of exposures and saturation detection (G.E. LAS4000 Piscataway, NJ, USA). Blots were subsequently stripped and reprobed with total antibody. Samples that failed to generate western signals that detected Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or generated protein of interest signals less than 10% of background were excluded from analyses.
Antibodies
All antibodies used in this study except for CYFIP1 were commercially obtained. Primaries: total mTOR (Bethyl labs, Montgomery, TX, USA) 1:2000, phospho Ser 2448 mTOR (Cell Signaling, Danvers, MA, USA ) 1:1000, phospho Threonine 389 p70 (Millipore, Billerica, MA, USA) 1:1000, total p70 (Cell Signaling, Danvers, MA, USA), phospho-Ser235/236 S6 (Bethyl labs, Montgomery, TX, USA) 1:2000, phospho Serine 209 eIF4E (Cell Signaling, Danvers, MA, USA) 1:1000, total eIF4E (Bethyl labs, Montgomery, TX, USA) 1:2000, phospho Serine 473 Akt (Cell Signaling, Danvers, MA, USA) 1:1000, pan Akt (Cell Signaling, Danvers, MA, USA) 1:1000, phospho Thr202/Tyr204 p44/42 (Erk1/2) (Cell Signaling, Danvers, MA, USA) 1:3000, p44/42 MAPK (Erk1/2) rabbit monoclonal (Bethyl labs, Montgomery, TX, USA) 1:3000, CYFIP1 1:1000, CYFIP2 (Millipore, Billerica, MA, USA) 1:1000, GAPDH (Novus, St. Charles, MO, USA) 1:10000, Secondaries: Goat anti rabbit-HRP (Promega, Madison, WI, USA) 1:5000, Goat anti mouse-HRP (Promega, Madison, WI, USA) 1:5000.
Statistical Analysis
Statistical analysis comparison of mRNA expression (CYFIP2, FMR1, Janus kinase and microtubule interacting protein 1 (JAKMIP1), RPS6KB1, RPS6KB2, mTOR, Akt, EIF4EBP1, and G protein-coupled receptor 155 (GPR155)) was based on analysis of variance after standard descriptive and graphical analyses. Analysis comparing protein (p389/p70, p209/EIF4E, CYFIP2, and pmTOR/mTOR) collapsed over groups (e.g., FXS vs. control) was based on the standard t-test; if equal variance hypothesis was rejected, then Satterthwaite Two Sample t-test was applied. All statistical tests are two-tailed at significance level 0.05. Statistical analysis was performed in SAS version 9.2.
Results
Measurements of mRNA expression levels in lymphocytes from patients with FXS
Gene expression levels were measured by QRT-PCR on total RNA isolated from peripheral blood leukocytes derived from subjects with FXS with and without the PWP and controls. As expected, we found substantial reduction in the relative amounts of FMR1 mRNA expression levels between FXS (with and without PWP) and normal groups, with a ~75% reduction in total normal signal (FXS=0.368 SD 0.573; normal=1.459 SD 0.244; df=46, t=9.49, p<0.01). Detectable FMR1 mRNA was observed in FXS mosaics, which carry unmethylated, transcriptionally active, expanded alleles.
Consistent with our previous findings we found a reduction in CYFIP1 mRNA expression levels, which encodes an FMRP binding protein and is a repressor of eIF4E activity (Napoli et al., 2008, Schenck et al., 2003), in the blood of patients with FXS and PWP (df=78, t=3.14, p=0.04) compared to normal controls (Nowicki et al., 2007) (Table 2). A recent study reported an increase in CYFIP1 expression in lymphoblastoid cells isolated from patients with autism (Nishimura et al., 2007). They also reported on an increased expression of JAKMIP1 and of GPR155 in both lymphoblastoid cell lines derived from subjects with autism and in the brains of Fmr1 knockout mice (Nishimura et al., 2007). Interestingly, we did not observe a change in the mRNA expression for these two genes in our study (Table 2). Using QRT-PCR, we also measured the mRNA expression levels of CYFIP2, the CYFIP1 paralog, and of the translational control elements, including S6K1, S6K2, mTOR, Akt and eIF4E-binding protein (4E-BP) in patients with FXS compared to typical developing controls. However, no significant differences were observed (Table 2) indicating that in lymphocytes, it is unlikely that FMRP exerts control over the abundance or stability of mRNAs encoding regulators of translational initiation. Gene expression levels were also measured in brain tissue from subjects with FXS and controls (Table 1). Although, as expected, FMR1 mRNA expression levels in the brain were significantly different between FXS and controls (FXS=0.012 SD=0.017; normal=0.631 SD=0.192; df=3, t=6.42, p<0.01 in frontal cortex and FXS=0.015 SD=0.009; normal= 0.463 SD=0.166; df= 3, t= 5.05, p=0.01 in cerebellum) the expression levels of mRNAs for translational initiation control proteins were similar in the two groups.
Table 2.
| Full mutation with PWP (A) | Full mutation w/o PWP (B) | Normal Control (C) | A vs C | A vs B | B vs C | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mRNA | N | Mean * | SD | N | Mean * | SD | N | Mean * | SD | df | t | p | df | t | p | df | t | p |
| FMR1 | 15 | 0.383 | 0.428 | 23 | 0.358 | 0.659 | 13 | 1.459 | 0.244 | 48 | −5.49 | <0.01 | 48 | 0.14 | 0.89 | 48 | −6.13 | <0.01 |
| CYFIP1 | 23 | 0.423 | 0.256 | 37 | 0.532 | 0.162 | 21 | 0.616 | 0.204 | 78 | −3.14 | <0.01 | 78 | −2.02 | 0.05 | 78 | −1.5 | 0.14 |
| CYFIP2 | 9 | 0.912 | 0.134 | 20 | 0.963 | 0.207 | 10 | 0.867 | 0.225 | 36 | 0.49 | 0.62 | 36 | −0.63 | 0.53 | 36 | 1.24 | 0.22 |
| RPS6KB1 | 15 | 1.128 | 0.198 | 23 | 1.151 | 0.194 | 13 | 1.153 | 0.233 | 48 | −0.32 | 0.75 | 48 | −0.33 | 0.74 | 48 | −0.02 | 0.98 |
| RPS6KB2 | 15 | 1.274 | 0.451 | 23 | 1.26 | 0.224 | 13 | 1.074 | 0.374 | 48 | 1.55 | 0.13 | 48 | 0.13 | 0.90 | 48 | 1.57 | 0.12 |
| mTOR | 15 | 1.138 | 0.293 | 23 | 1.206 | 0.285 | 12 | 1.037 | 0.312 | 47 | 0.89 | 0.38 | 47 | −0.7 | 0.49 | 47 | 1.62 | 0.11 |
| Akt | 15 | 1.035 | 0.201 | 23 | 1.103 | 0.122 | 13 | 1.426 | 0.179 | 48 | −0.29 | 0.69 | 48 | −0.3 | 0.73 | 48 | −0.1 | 0.10 |
| EIF4EBP1 | 15 | 0.4 | 0.108 | 23 | 0.377 | 0.082 | 12 | 0.422 | 0.138 | 47 | −0.53 | 0.60 | 47 | 0.66 | 0.51 | 47 | −1.19 | 0.24 |
| GPR155 | 15 | 12.21 | 7.132 | 23 | 13.784 | 4.344 | 13 | 11.408 | 5.238 | 48 | 0.38 | 0.70 | 48 | −0.86 | 0.39 | 48 | 1.24 | 0.22 |
| JAKMIP1 | 15 | 0.597 | 0.293 | 23 | 0.596 | 0.365 | 13 | 0.695 | 0.381 | 48 | −0.75 | 0.46 | 48 | 0.01 | 0.99 | 48 | −0.82 | 0.42 |
Gene expression levels
PWP= Prader Willi-like phenotype
FMR1= fragile X mental retardation 1
CYFIP1 = Cytoplasmic FMR1-interacting protein 1
CYFIP2= Cytoplasmic FMR1-interacting protein 2
RPS6KB1= Ribosomal protein S6 kinase, 70kDa, polypeptide 1
RPS6KB2= Ribosomal protein S6 kinase, 70kDa, polypeptide 2
mTOR= mammalian target of rapamycin
Akt= Protein Kinase B
EIF4EBP1= Eukaryotic translation initiation factor 4E-binding protein 1
GPR155= G protein-coupled receptor 155
JAKMIP1= janus kinase and microtubule interacting protein 1
Phosphorylation of substrates and regulators of mTOR signaling is increased in patients with FXS
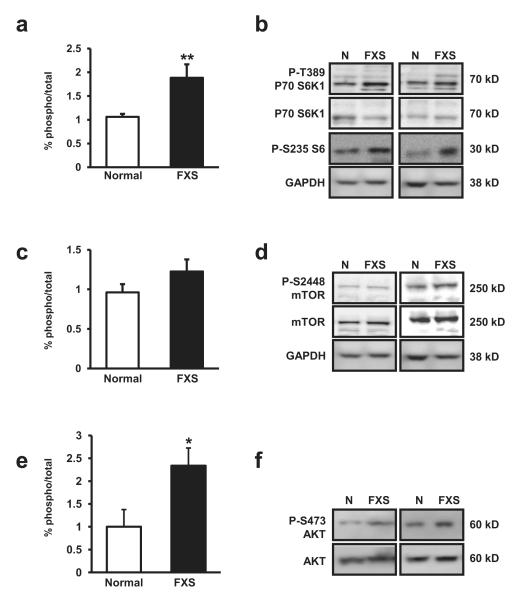
One of the most well described mTOR substrates is p70 S6K1 (S6K1), which regulates a number of activities related to translation initiation including ribosomal maturation and RNA helicase activity. The activity of S6K1 is regulated by phosphorylation at multiple sites (Jacinto & Lorberg, 2008) with threonine 389 (T389) being the site of mTOR-dependent regulation (Burnett et al., 1998, Klann & Dever, 2004). Because mTOR is known to be dysregulated in mouse models of FXS (Sharma et al., 2010) we examined regulation of this site in S6K isolated from lymphocytes of human subjects. We found that compared to normal controls, phosphorylation at T389 (pT389) was enhanced in individuals with FXS. The ratio of levels of pT389 S6K1/Total S6K1 (pS6K1/S6K1) for individuals with FXS compared to normal individuals was (pS6K1/s6K1 ratio: FXS mean 1.885 SD 1.463, Normal 1.062 SD 0.233, df=30, t=2.90, p<0.01) (Figure 1a). A subset of our patients with FXS and normal controls were tested for the activation of S6K1 substrates. Consistent with our S6K1 observations, we also observed increased phosphorylation levels of the S6K1 substrate, ribosomal protein S6 at serine 235/236 (pS6/GAPDH ratio: FXS mean 1.414 SD 0.358, Normal mean 0.638 SD 0.275, df=7, t=4.72, p<0.01) (Figure 1b). No difference in the total amount of protein expression was observed. Surprisingly, when we examined phosphorylation of S2448 on mTOR, a site of multiple kinase action including S6K1 (Sharma et al., 2010) we found no significant difference between FXS and normal individuals in the ratio of levels of phospho/total mTOR phosphorylation (pmTOR/mTOR) (Figure 1c-d). For individuals with FXS compared to normal individuals this ratio was (pmTOR/mTOR ratio: FXS 1.225 SD 0.794, Normal 0.961 pmTOR/mTOR SD 0.390, df=39, t=-1.43, p=0.16). Sharma et al. (2010) also reported increased levels of phosphorylation of Akt, an upstream activator of mTOR, at Serine 473 (S473) in FMRP KO mouse brains. This regulatory site is conserved in humans; therefore we also examined S473 phosphorylation levels in FXS tissues (Figure 1e). Consistent with Sharma et al. (2010) and with our data (Figure 1a, 1b) we also observed elevated S473 expression levels in FXS lymphocytes (Figure 1e, 1f) (pS473 Akt/Tot Akt ratio: FXS mean 0.177 SD 0.030, Normal mean 0.076 SD 0.029 df=13, t=2.34, p=0.04).
Figure 1.
(a) Patients with FXS (n=28) display increased levels of phosphorylated Threonine 389 (pT389) p70 S6 kinase 1 (S6K1) and (pS235/236) S6 compared to normal controls (n=14, p=0.0069). (b) Representative western blot images for pT389 P70 S6K, P70 S6K, pS235/236 S6, GAPDH (loading control) from lysates from four patient sets. (c) Patients with FXS (n=27) show no statistical significant difference in the levels of phosphorylated Serine 2448 (pS2448) mammalian target of rapamycin (mTOR) compared to normal controls (n=14). (d) Representative western blot images for pS2448 mTOR, mTOR, GAPDH (loading control, same blot as above) from lysates from four patient sets. (e) Patients with FXS (n=9) show increased levels of phosphorylated Serine 473 (pS473) AKT (PKB) kinase compared to normal controls (n=7, p=0.0354). (f) Representative western blot images for pS473 AKT, AKT, from lysates from four patient sets. The percent (%) of phospho-signal were normalized to total protein signal for each graph. The error bars represent standard error in each graph. Blots were checked for efficient stripping prior to re-probing.
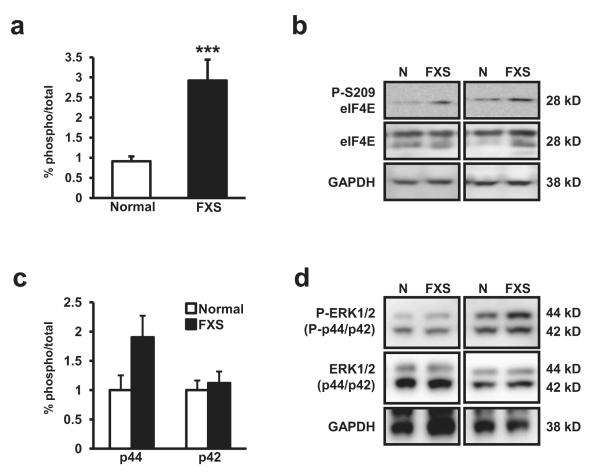
The finding that T389 of S6K was enhanced in patients with FXS prompted us to examine the phosphorylation levels of eIF4E, another critical regulator of cap-dependent translation. eIF4E encodes m7-GTP cap binding activity, providing a physical link between substrate mRNA and the translational initiation machinery. Phosphorylation at serine 209 (pS209) of eIF4E is correlated with increased translation (Flynn & Proud, 1995, Gingras et al., 1999, Mckendrick et al., 1999). Similar to S6K1, we found robust increases in the ratio of levels of pS209 eIF4E/total eIF4E (p4E/4E) in patients with FXS (Figure 2a-b) while the total amount of eIF4E protein was unaltered. The ratio of levels of p4E/4E for individuals with FXS compared to normal individuals was (p4E/4E ratio: FXS mean 2.923 SD 2.690, Normal mean 0.912 SD 0.447, df=30, t=3.85, p<0.01). eIF4E is regulated by the activity of the extracellular-signal regulated kinase (p44)1 and (p42)2 (ERK 1/2) (Waskiewicz et al., 1999) and levels of phosphorylation of ERK1/2 at Threonine 202/Tyrosine 204 (pERK1/2) have been shown to be elevated FXS model mice (Hou et al., 2006). Thus we investigated the possibility that pERK1/2 levels were also elevated in FXS lymphocytes. Although we found a trend for increased levels of pERK in FXS lymphocytes in the larger isoform (ERK1) (Figure 2c, 2d), the difference was not statistically significant (pERK1/ERK1:p=0.10; pERK2/ERK2: p=0.10) (Figure 2c). These results, combined with the increased levels of phosphorylation of T389 on S6K1 (Figure 1a), suggest that the mTOR but not ERK1/2 signaling dysregulation observed in Fmr1 KO mice also is present in individuals with FXS and provides evidence of increased translational activity in lymphocytes of subjects with FXS.
Figure 2.
(a) Patients with FXS (n=20) display increased levels of phosphorylated Serine 209 (pS209) eukaryotic initiation factor 4E (eIF4E) compared to normal controls (n=14, p=0.0006). (b) Representative western blot images for pS2098 eIF4E, eIF4E, GAPDH (loading control) from lysates from four patient sets. (c) Patients with FXS (n=13) show no difference in phospho Threonine 202/Tyrosine 204 ERK1(p44)/ ERK2(p42) levels compared to normal controls (n=10). We did observe a trend for increased pERK1/ERK2 in FXS patients but the difference was not significant (p=.0989). (d) Representative western blot images for pT202/Y204 ERK1/2, Total ERK1/2, and GAPDH (loading control) from lysates from four patient sets. The percent (%) of phospho-signal was normalized to total protein signal for each graph. The error bars represent standard error in each graph. Blots were checked for efficient stripping prior to re-probing.
CYFIP1 protein levels are normal but CYFIP2 protein expression is increased in patients with FXS
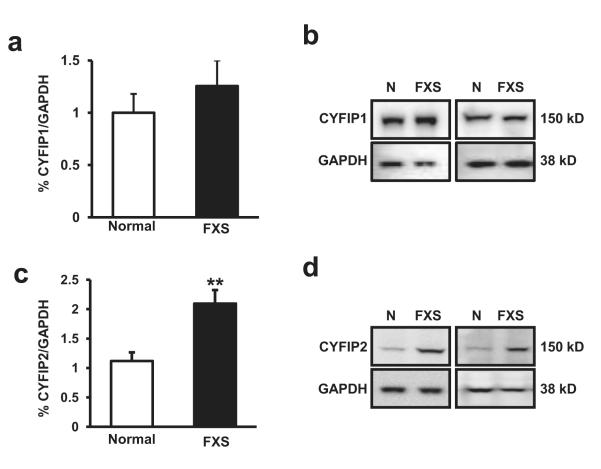
Because we found markers for signaling consistent with enhanced translation and decreased CYFIP1 mRNA levels in FXS patients, we chose to examine the RNA and protein expression levels of CYFIP1 and CYFIP2 (Schenck et al., 2001). Surprisingly, when we examined CYFIP1 protein levels in protein lysates obtained from lymphocytes we detected no difference in the levels of CYFIP1 protein between FXS patients and normal controls (Figure 3a, 3b) (CYFIP1/GAPDH ratio: FXS mean 0.524 SD 0.249 Normal mean 0.386 SD 0.214; df=15, t=1.21, p=0.24).
Figure 3.
(a) Patients with FXS (n=6) display no difference in levels of FMRP interacting protein, CYFIP1, compared to normal controls (n=9). (b) Representative western blot images for CYFIP1, GAPDH (loading control) from lysates from four patient sets. (c) Patients with FXS (n=27) display increased levels of FMRP interacting protein, CYFIP2, compared to normal controls (n=16, p=0.0010). (d) Representative western blot images for CYFIP2, GAPDH (loading control) from lysates from four patient sets. The percent (%) of CYFIP1 or CYFIP2 were normalized to total GAPDH protein signal for each graph. The error bars represent standard error in each graph.
Consistent with the notion that FMRP acts as suppressor of CYFIP2 expression by sequestration of its mRNA (Schenck et al., 2001); we found elevated expression levels of CYFIP2 in the lymphocytes of FXS when compared to normal controls (Figure 3c, 3d). The ratio of CYFIP2/GAPDH for individuals with FXS compared to normal individuals was (CYFIP2/GAPDH: FXS mean 2.094 SD 1.195, Normal mean 1.118 SD 0.553, df=39, t=-3.57, p<0.01). This increase was specific to CYFIP2 protein as total levels of GAPDH, mTOR and S6K1 (Figure 1b, 1d, 3d) were unchanged in FXS. Interestingly, in contrast to CYFIP1 mRNA expression levels, this increase was observed without a detectable change in CYFIP2 mRNA expression levels (Table 2) suggesting that CYFIP2 expression in the blood is normally limited by the availability of its mRNA for translation and not by increases in transcription.
Phosphorylation of p70 S6K1 is increased in brains of patients with FXS
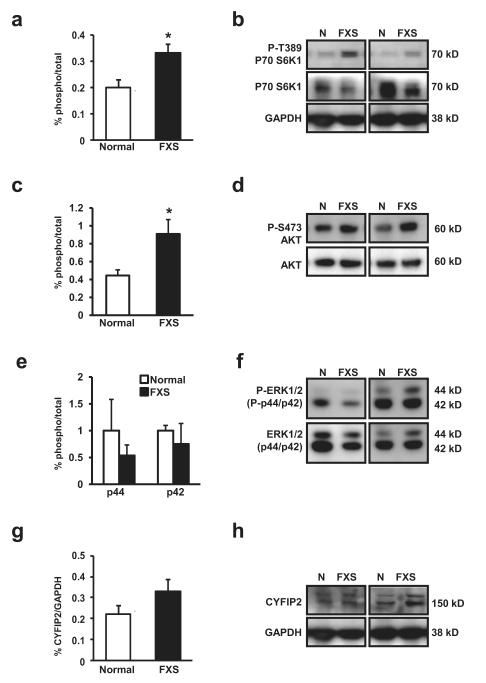
Because FXS defines a series of symptoms that includes intellectual disabilities, increased anxiety, mild to severe cognitive impairment and co-morbidity with autism (Loesch et al., 2007, Rogers et al., 2001) we sought to explore if our findings in fresh lymphocytes could be extended to brain tissue from FXS individuals. We obtained tissue from the cerebellum and frontal lobes of four patients with FXS and normal age matched individuals (Table 1) and isolated proteins from homogenates. Comparable to what we observed in lymphocytes, we detected a ~70% increase in the levels of phospho-T389 S6K1 in the brain (Figure 4a) (pT389 S6K1/Tot S6K1 FXS mean 0.333 SD 0.067 normal mean 0.198 SD 0.064 df=7, t=2.78, p=0.03). Similar to what we observed in lymphocytes, we did not see a change in overall phospho-mTOR/Total mTOR (data not shown). The increased levels of p389 S6K1 was not due to overall difference in protein levels as GAPDH expression was indistinguishable between FXS and normal controls (Figure 4b). We also examined pS473 levels in the brains of FXS patients and again, consistent with what we observed with lymphocytes, we observed increased pS473 levels in the brains of FXS individuals compared to normal controls (Figure 4c, 4d) (pS473 Akt/Tot Akt FXS mean 0.908 SD 0.161, Normal 0.444 SD 0.063, df=7, t=2.72, p=0.03).
Figure 4.
(a) Patients with FXS (n=4) display increased levels of phosphorylated Threonine 389 (T389) p70 S6 kinase 1 (S6K1) compared to normal controls (n=4, p=0.0274). The percent (%) phospho-signal normalized to total protein signal for each graph. (b) Representative western blot images for pT389 P70 S6K, P70 S6K, GAPDH (loading control) from lysates from four patient sets. (c) Patients with FXS show increased pS473 Akt (PKB) levels in the brain compared to normal controls (p=0.0298). The percent (%) of phospho-signal was normalized to total protein signal for graph. (d) Representative western blot images for pS473 AKT and total AKT from lysates from four patient sets. (FXS, n=4; N, n=4) (e) Patients with FXS (n=4) show no difference in pERK1/2 levels compared to normal controls (n=4), for both pERK1 or pERK2. The percent (%) of phospho-signal normalized to total protein signal for graph. (f) Representative western blot images for pERK 1/2 total ERK1/2 from lysates from four patient sets. (g) Levels of CYFIP2 in the frontal lobe are not different than normal controls. CYFIP2 (145 kD band) normalized to total GAPDH protein signal for graph (FXS, n=4; normal, n=4). (h) Representative western blot images for CYFIP2, GAPDH (loading control) from lysates from four patient sets. The presence of the larger band ~150 kD was only seen in brain derived samples. All ECL signal detection was non-saturation (65K bit detection, GE Las400 imager). The error bars represent standard error in each graph. Blots were checked for efficient stripping prior to re-probing.
We also assessed pERK 1/2 levels in brain as they have been reported to be higher in the hippocampus of the mouse models of FXS (Hou et al., 2006). Interestingly, pERK 1/2 levels in the frontal lobe of patients with FXS were no different than normal controls (Figure 4e, 4f) (p>0.5 for both pERK1/ERK1 and pERK2/ERK2). This result is not likely an artifact of post-mortem tissue treatment as pERK 1/2 differences have been reliably detected from other post-mortem samples (Dwivedi et al., 2006).
With respect to CYFIP2, we detected a trend toward increased expression in brain tissue from all four subjects with FXS but the difference did not reach statistical significance (Figure 4g) (CYFIP2/GAPDH FXS mean 0.331 SD 0.109, normal mean 0.221 SD 0.079, df=7, t=1.60, p=0.15). Unlike immunostaining of the protein isolated from the blood (Figure 3d), staining for CYFIP2 in the brain revealed two bands of closely related size, one at the predicted size (~145 kD) and one slightly larger (Figure 4h). Several reports have identified CYFIP2 with a single band (Jackson et al., 2007, Mayne et al., 2004, Mongroo et al., 2011) consistent with what we observe in lymphocytes. However, multiple bands for CYFIP2 have also been reported (Derivery et al., 2009). The presence of the larger band may indicate the presence of brain specific post-translationally modified CYFIP2 isoforms, expression of CYFIP2 paralogs, or the presence of peptides in the brain that are non-specifically recognized by the CYFIP2 antibody used for this study. Levels of CYFIP2 in the cerebellum were too low to quantify reliably (data not shown).
We also examined phosphorylation of eIF4E from brain tissue and found that although the total protein was easily detectable, phosphorylation levels at S209 was extremely low (data not shown). Although it is possible that pS209 signal was degraded by phosphatase activities associated with the post-mortem interval of tissue retrieval, the detectability of pT389 and pERK1/2 argue against this. This low level of frontal lobe pS209 eIF4E may in fact indicate very low steady-state eIF4E activation in the human brain. Because it is known that are important differences in steady-state metabolic rates (Sokoloff, 1981) and thus translational rates in the brain, it is possible that what we observe is specific to the frontal lobe and that other regions may display differences between FXS and normally developing individuals. Regardless, our lymphocyte data, combined with pT389-S6K1 brain results shown in Figure 4a, is consistent with the idea that the loss of FMRP in brains of FXS patients results in dysregulation of mechanisms of translational initiation control rather than transcriptional regulation.
Discussion
Emerging evidence from both human patients with ASD and mouse model of FXS (Hagerman et al., 2010, Sharma et al., 2010) indicate that mTOR signaling cascade dysregulation and eccentric protein synthesis are present and may provide the molecular markers that could enhance diagnostics for the development of potential treatment regimens and will allow evaluation of therapies aimed at ameliorating FXS associated symptoms. The identification of mTOR signaling dysregulation, even in a subset of patients with FXS may also open up new avenues for therapeutic intervention in pathways unrelated to translation. Thus, the present study was undertaken to examine a possible role for mTOR signaling in FXS and our findings indeed implicate dysregulation of mTOR signaling, which may lead to the impaired cognitive functions observed in subjects with FXS.
Our results show a robust activation of the mTOR substrate, S6K1 at the mTOR dependent phosphorylation site, T389, and in the phosphorylation levels of the upstream mTOR regulator, Akt at S473, in both lymphocytes and brains of subjects with FXS. Somewhat surprisingly, we did not observe a statistically significant increase in mTOR phosphorylation levels at S2448 in FXS lymphocytes, although there was a trend for increased phosphorylation at this site. These findings indicate that although the mTOR substrate, S6K1, and mTOR regulator, Akt, both demonstrate enhanced activation in lymphocytes, this enhancement does not likely extend in an S6K1-mediated feedback regulatory loop to S2448 on mTOR. It may also indicate that this mTOR phospho-site does not have the same correlational relevance as it does in other tissue and cell culture types (Reynolds et al., 2002). mTOR phosphorylation is tightly regulated and changes in phosphorylation levels of greater than 50% are rarely observed even under ideal conditions (Avni et al., 1997). Thus, it is also possible that increased phosphorylation is indeed present but our quantitative resolution using whole lysates is not sufficient to resolve small differences in basal activation. Further, the larger sample size and much higher variance for the full mutation group compared to the normal group may have underpowered our ability to analyze results from this phosphorylation site. Finally, although phosphorylation levels at the S2448 site is correlated with increased mTOR activity (Reynolds et al., 2002), it is not required for it. In HEK293 cell culture studies where mTOR serine 2448 was replaced with an alanine, no loss of translational efficiency was observed (Sekulic et al., 2000). So it may be that in lymphocytes this site is not an appropriate marker of mTOR activation.
Although we have demonstrated dysregulation of the mTOR cascade signaling, we did not detect any alteration in the ERK1/2 signaling in either lymphocytes or brain tissue from individuals with FXS. Our findings are consistent with a previous study where using subject lymphocytes, early activation kinetics of ERK were found to be different between patients with FXS and typical developing controls but steady-state (i.e. resting) levels were not (Weng et al., 2008). It is also possible that ERK signaling in brains of FXS patients is perturbed in region or cell-specific manner (i.e. hippocampus) and that our power to detect changes was obfuscated by the gross anatomical level of dissection used for lysate preparation. A more detailed examination of ERK signaling from specific brain areas using sectioning will be needed to examine this question in greater detail.
Consistent with what we observed in earlier study (Nowicki et al., 2007); we saw decreased CYFIP1 mRNA expression levels in lymphocytes from subjects with FXS and more so in those with the PWP (Table 1). However, a similar study reported an increase in expression of CYFIP1 mRNA (Nishimura et al., 2007). This study also reported dysregulation in JAKMIP1 and GPR155 expression from a variety of sources: lymphoblasts from dup (15q) and non-dup (15q) idiopathic ASD patients, FXS model mice, and finally in neuronal cell lines over-expressing CYFIP1. They found cases of both up-regulation and down-regulation of these genes depending on the source of the genetic manipulation. Our study differs from Nishimura et al. (2007) in several important ways. First, our lymphocytes were from patients specifically identified from FXS and FXS with PWP populations rather than from a much larger ASD patient pool and we measured expression levels directly in peripheral blood leukocytes rather than in lymphoblastoid cells lines, which often do not behave as the fresh cells from which they are derived. Second, Nishimura et al. (2007) confirmed their JAKMIP1 expression in Fmr1 KO mice and neuronal cell lines by Western blot analyses rather than mRNA expression. Third, their study used primarily microarray analyses compared to real-time PCR in our study. Finally, it should be noted that earlier microarray studies using pooled FXS lymphoblastoid cells or tissue from FMR1 KO mice did not report CYFIP1 or JAKMIP1 expression differences (Brown et al., 2001, Miyashiro et al., 2003). Thus, our different results may be explained by differences in sample source, microarray type, or statistical analyses used. Although CYFIP1 mRNA levels appear to be clearly reduced in subjects with FXS and the PWP, the correspondent decreased expression level of CYFIP1 protein observed was based on a small sample size analyzed; thus, future studies are required to investigate this possibility and determine if the difference protein expression in this subgroup of individuals with FXS, is statistically significant.
In contrast to what we observed for CYFIP1, we saw no difference in CYFIP2 mRNA expression but rather an increase in CYFIP2 protein levels between FXS, FXS with the PWP and normal control groups (Table1, Figure 3). CYFIP2 is located at 5q33.3 and highly homologous to CYFIP1 and like CYFIP1 binds FMRP (Schenck et al., 2003). Interestingly, CYFIP2 mRNA contains no obvious structure that would be recognized by FMRP (Levanon et al., 2005) although it was identified in a screen aimed at isolating mRNPs associated with FMRP (Darnell et al., 2011). It is possible that RNA secondary structure prediction has important limitations with respect to FMRP function or that CYFIP2 interaction with FMRP mRNPs occurs through the activities of alternative RNA binding proteins that interact with FMRP. Two obvious possibilities are Fragile X related protein 1(FXRP1) and Fragile X related protein 2 (FXRP2), which have been shown to interact with CYFIP2 but not CYFIP1 (Napoli et al., 2008). Our finding of increased expression levels of CYFIP2 in FXS is potentially significant in the context of CYFIP2 relationship to apoptosis (Jackson et al., 2007, Saller et al., 1999). CYFIP2 is an p53-inducible target that may be pro-apoptotic (Jackson et al., 2007), thus it is possible that some symptoms of FXS are mediated by perturbation of apoptotic/cell death. In support of this idea are the aging problems that are associated with FXS including Parkinson symptoms, cognitive decline and MRI changes (Utari et al., 2010). Further examination of CYFIP2 protein expression in tissue of patients with FXS or FXS model mice will be required to address this important question.
Dysregulation of CYFIP1/2 levels at either the protein or mRNA level also have the potential to influence the activities of actin enucleating WASP family verprolin homologous protein (WAVE) complex (Derivery & Gautreau, 2010, Takenawa & Suetsugu, 2007). The multi-protein WAVE complex is composed five core subunits, which include either CYFIP1 (also known as Sra1) or CYFIP2 (also known as Pir121) (Derivery et al., 2009, Takenawa & Suetsugu, 2007). The WAVE complex is critically involved in cell motility and lamellipodium formation and has been shown to play an important role in axon guidance and thus the development of the nervous system (Schenck et al., 2003, Schenck et al., 2004, Schrander-Stumpel et al., 1994, Suetsugu et al., 2003, Tahirovic et al., 2010). Abnormal dendritic spine morphology and maturation are observed in the brains of FXS patients and in FXS model mice (Comery et al., 1997, Irwin et al., 2000). Because these processes critically rely on actin cytoskeletal network functions, it is possible that these FXS associated cellular phenotypes arise from altered WAVE activity resulting from increased FMRP-mediated CYFIP2 expression. This interesting possibility can be investigated in future studies using mouse FXS models and neuronal cell culture where WAVE complex components can be experimentally manipulated and dendritic spine morphology examined along a developmental time course.
Our biochemical data can clearly distinguish full mutation patients with FXS from normal controls. We were, however, unable to further discriminate FXS with and without PWP using molecular markers of translation initiation. Finding that mTOR signaling is dysregulated in patients with FXS (or a subset) may help explain the wide degree of clinical severity presented by FXS. More importantly, the availability of such diagnostic tools may provide insight into the therapeutic course one should take in treating individuals with FXS. Finally, molecular markers of mTOR signaling may also provide outcome measures as a means to assess the long- and short-term therapeutic efficacy of pharmaceutical interventions being used to treat FXS. Additional studies will be needed to better understand the relationship between the loss of FMRP and translational dysregulation in FXS.
Acknowledgements
This work was supported by National Institute of Health grants HD036071 and HD02274 and by Grant RR024146 from the National Center of Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This research was also supported by FRAXA research foundation (E.K.) and NS047834 (E.K.). CYFIP1 antibody was kindly provided by Dr. Sharon Eden. This work is dedicated to the memory of Matteo.
References
- APA . Diagnostic and Statistical Manual of Mental Disorders. Fourth Edition - Text Revision (DSMIV-TR), Fourth Edition, Text Revision American Psychiatric Association; Washington DC: 2000. [Google Scholar]
- Avni D, Biberman Y, Meyuhas O. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Fryns JP, Butler MG, Canziani F, Wesby-van Swaay E, van Hemel JO, Oostra BA, Halley DJ, Niermeijer MF. Clinical and molecular studies in fragile X patients with a Prader-Willi-like phenotype. J Med Genet. 1993;30:761–766. doi: 10.1136/jmg.30.9.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Niermeijer MF. The Prader-Willi-like phenotype in fragile X patients: a designation facilitating clinical (and molecular) differential diagnosis. J Med Genet. 1994;31:820. doi: 10.1136/jmg.31.10.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demark JL, Feldman MA, Holden JJA. Behavioral relationship between autism and fragile X syndrome. Am J Ment Retard. 2003;108:314–326. doi: 10.1352/0895-8017(2003)108<314:BRBAAF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Derivery E, Gautreau A. Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. Bioessays. 2010;32:119–131. doi: 10.1002/bies.200900123. [DOI] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009;66:777–790. doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Pandey GN. ERK MAP kinase signaling in post-mortem brain of suicide subjects: differential regulation of upstream Raf kinases Raf-1 and B-Raf. Mol psychiatry. 2006;11:86–98. doi: 10.1038/sj.mp.4001744. [DOI] [PubMed] [Google Scholar]
- Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, Hagerman PJ, Stenzel TT, Hadd AG, Latham GJ, Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- Fryns JP, Haspeslagh M, Dereymaeker AM, Volcke P, Van den Berghe H. A peculiar subphenotype in the fra(X) syndrome: extreme obesity-short stature-stubby hands and feet-diffuse hyperpigmentation. Further evidence of disturbed hypothalamic function in the fra(X) syndrome? Clin Genet. 1987;32:388–392. doi: 10.1111/j.1399-0004.1987.tb03155.x. [DOI] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr., Warren ST, Oostra BA, Nelson DL, Caskey CT. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Greco CM, Navarro CS, Hunsaker MR, Maezawa I, Shuler JF, Tassone F, Delany M, Au JW, Berman RF, Jin LW, Schumann C, Hagerman PJ, Hagerman RJ. Neuropathologic features in the hippocampus and cerebellum of three older men with fragile X syndrome. Mol Autism. 2011;2:2. doi: 10.1186/2040-2392-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R, Hoem G, Hagerman P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1:12. doi: 10.1186/2040-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ. The fragile X premutation: into the phenotypic fold. Curr Opin Genet Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- Harris SW, Hessl D, Goodlin-Jones B, Ferranti J, Bacalman S, Barabato I, Tassone F, Hagerman PJ, Herman K, Hagerman RJ. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113:427–438. doi: 10.1352/2008.113:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton DD, Sideris J, Skinner M, Mankowski J, Bailey DB, Jr., Roberts JE, Mirrett P. Autistic behavior in children with fragile X syndrome: Prevalence, stability, and the impact of FMRP. Am J Med Genet. 2006;140:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Hunsaker MR, Greco CM, Tassone F, Berman RF, Willemsen R, Hagerman RJ, Hagerman PJ. Rare Intranuclear Inclusions in the Brains of 3 Older Adult Males With Fragile X Syndrome: Implications for the Spectrum of Fragile X-Associated Disorders. J Neuropathol Exp Neurol. 2011;70:462–469. doi: 10.1097/NEN.0b013e31821d3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem J. 2008;410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- Jackson RS, 2nd, Cho YJ, Stein S, Liang P. CYFIP2, a direct p53 target, is leptomycin-B sensitive. Cell Cycle. 2007;6:95–103. doi: 10.4161/cc.6.1.3665. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Hagerman PJ, Leehey MA. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 2007;6:45–55. doi: 10.1016/S1474-4422(06)70676-7. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Cortell R, Kau AS, Bukelis I, Tierney E, Gray RM, Cox C, Capone GT, Stanard P. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. Am J Med Genet. 2004;129A:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch DZ, Bui QM, Dissanayake C, Clifford S, Gould E, Bulhak-Paterson D, Tassone F, Taylor AK, Hessl D, Hagerman R, Huggins RM. Molecular and cognitive predictors of the continuum of autistic behaviours in fragile X. Neurosci Biobehav Rev. 2007;31:315–326. doi: 10.1016/j.neubiorev.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore PC, Risi S. Autism Diagnostic Observation Schedule. Western Psychological Services; Los Angeles, CA: 1999. [Google Scholar]
- Mayne M, Moffatt T, Kong H, McLaren PJ, Fowke KR, Becker KG, Namaka M, Schenck A, Bardoni B, Bernstein CN, Melanson M. CYFIP2 is highly abundant in CD4+ cells from multiple sclerosis patients and is involved in T cell adhesion. Eur J Immunol. 2004;34:1217–1227. doi: 10.1002/eji.200324726. [DOI] [PubMed] [Google Scholar]
- McKendrick L, Pain VM, Morley SJ. Translation initiation factor 4E. Int J Biochem Cell Biol. 1999;31:31–35. doi: 10.1016/s1357-2725(98)00129-0. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, Bowser MJ, Castells A, Spiegelman VS, Rustgi AK. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172–2182. doi: 10.1158/0008-5472.CAN-10-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Steindler C, Pellegrini S, Schanen NC, Warren ST, Geschwind DH. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Nowicki ST, Tassone F, Ono MY, Ferranti J, Croquette MF, Goodlin-Jones B, Hagerman RJ. The Prader-Willi phenotype of fragile X syndrome. J Dev Behav Pediatr. 2007;28:133–138. doi: 10.1097/01.DBP.0000267563.18952.c9. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Reynolds T.H.t., Bodine SC, Lawrence JC., Jr. Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J Biol Chem. 2002;277:17657–17662. doi: 10.1074/jbc.M201142200. [DOI] [PubMed] [Google Scholar]
- Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22:409–417. doi: 10.1097/00004703-200112000-00008. [DOI] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Berument SK, Lord C, Pickles A. Social Communication Questionnaire (SCQ) Western Psychological Services; Los Angeles: 2003b. [Google Scholar]
- Rutter M, Le Couteur A, Lord C. Autism Diagnostic Interview - Revised (ADI-R) Western Psychological Services; Los Angeles, CA: 2003a. [Google Scholar]
- Saller E, Tom E, Brunori M, Otter M, Estreicher A, Mack DH, Iggo R. Increased apoptosis induction by 121F mutant p53. EMBO J. 1999;18:4424–4437. doi: 10.1093/emboj/18.16.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Qurashi A, Carrera P, Bardoni B, Diebold C, Schejter E, Mandel JL, Giangrande A. WAVE/SCAR, a multifunctional complex coordinating different aspects of neuronal connectivity. Dev Biol. 2004;274:260–270. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Schrander-Stumpel C, Meinecke P, Wilson G, Gillessen-Kaesbach G, Tinschert S, Konig R, Philip N, Rizzo R, Schrander J, Pfeiffer L, Maat-Kievit A, van der Burgt L, van Essen T, Latta E, Hillig U, Verloes A, Journel H, Fryns JP. The Kabuki (Niikawa-Kuroki) syndrome: further delineation of the phenotype in 29 non-Japanese patients. Eur J Pediatr. 1994;153:438–445. doi: 10.1007/BF01983409. [DOI] [PubMed] [Google Scholar]
- Sekulic A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000;60:3504–3513. [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff L. Localization of functional activity in the central nervous system by measurement of glucose utilization with radioactive deoxyglucose. J Cereb Blood Flow Metab. 1981;1:7–36. doi: 10.1038/jcbfm.1981.4. [DOI] [PubMed] [Google Scholar]
- Sparrow SS, Cicchetti DV, Balla DA. Vineland Adaptive Behavior Scales. Second edition AGS Publishing; Circle Pines: 2005. [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-Grashoff A, Brakebusch C, Bradke F. Rac1 regulates neuronal polarization through the WAVE complex. J Neurosci. 2010;30:6930–6943. doi: 10.1523/JNEUROSCI.5395-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, Hagerman PJ. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am J Med Genet. 2000;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utari A, Adams E, Berry-Kravis E, Chavez A, Scaggs F, Ngotran L, Boyd A, Hessl D, Gane LW, Tassone F, Tartaglia N, Leehey MA, Hagerman RJ. Aging in fragile X syndrome. J Neurodev Disord. 2010;2:70–76. doi: 10.1007/s11689-010-9047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, Eussen BE, VanOmmen GJB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey TC, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Abbreviated Scale of Intelligence (WASI) Harcourt Assessment, Inc.; San Antonio: 1999. [Google Scholar]
- Wechsler D. Wechsler Preschool and Primary Scale of Intelligence. 3rd Edition The Psychological Corporation; San Antonio: 2002. [Google Scholar]
- Wechsler D. Wechsler Intelligence Scale for Children. Fourth Edition Harcourt Assessment, Inc.; San Antonio: 2003. [Google Scholar]
- Weng N, Weiler IJ, Sumis A, Berry-Kravis E, Greenough WT. Early-phase ERK activation as a biomarker for metabolic status in fragile X syndrome. Am J Med Genet. 2008;147B:1253–1257. doi: 10.1002/ajmg.b.30765. [DOI] [PubMed] [Google Scholar]
- Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, et al. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]