Abstract
The cilium is a microtubule-based organelle that contains a unique complement of proteins for cell motility and signaling functions. Entry into the ciliary compartment is proposed to be regulated at the base of the cilium 1. Recent work demonstrated that components of the nuclear import machinery, including the RanGTPase and importins, regulate ciliary entry 2–4. We hypothesized that the ciliary base contains a ciliary pore complex (CPC) whose molecular nature and selective mechanism are similar to the nuclear pore complex (NPC). By microinjecting fluorescently-labeled dextrans and recombinant proteins of various sizes, we characterize a size-dependent diffusion barrier for the entry of cytoplasmic molecules into primary cilia in mammalian cells. We demonstrate that nucleoporins localize to the base of primary and motile cilia and that microinjection of nucleoporin function-blocking reagents blocks the ciliary entry of kinesin-2 KIF17 motors. Together, this work demonstrates that the physical and molecular nature of the CPC is similar to the NPC, and further extends functional parallels between nuclear and ciliary import.
Cilia are evolutionarily conserved organelles important for normal cellular development, motility and sensory functions 5,6. Disruption of proteins that normally localize to and function within primary cilia leads to a wide range of human diseases collectively termed the ciliopathies, with phenotypes including retinal degeneration, cystic kidney diseases, skeletal defects, and obesity 7–9. As an organelle with a unique protein and lipid composition, cilia must utilize specific mechanisms to ensure the accurate targeting to and retention of proteins within this compartment 10.
The mechanisms regulating entry into the ciliary compartment are unclear at first glance as the cilium lacks a limiting membrane separating it from the cytoplasm. The restriction point is believed to be a specialized region at the base of the cilium called the transition zone where Y-shaped structures and transition fibers can be seen by electron microscopy to link the microtubule core (the axoneme) and basal body structures to the ciliary membrane 11–13. Recent work has uncovered a network of protein components that localize to the transition zone and basal body region, and whose defects lead to various ciliopathies 14–18.
Are cytoplasmic proteins restricted from entering from the ciliary compartment, and if so, what mechanisms prevent diffusive entry? We recently demonstrated that ciliary entry of a cytoplasmic protein, the kinesin-2 motor KIF17, requires an import signal similar to a nuclear localization signal (NLS), the nuclear transport factor importin-β2 (transportin-1), and a RanGTP/GDP gradient between ciliary and cytoplasmic compartments 2. Thus, we hypothesized that the mechanisms that regulate ciliary entry of soluble proteins may be mechanistically similar to those that regulate nuclear entry. Consistent with this hypothesis, ciliary import of X-linked Retina Pigmentosa protein RP2 utilizes importin-β2 and NLS-like sequences 4. In addition, the transition zone region has been compared to the NPC and proposed to serve as a flagellar/ciliary pore complex (CPC) 1,10. Thus, we set out to determine whether primary cilia utilize mechanisms similar to nuclei to regulate the entry of cytoplasmic components.
Nuclear-cytoplasmic shuttling is controlled by the NPC, a large multi-protein complex embedded in the double membrane of the nuclear envelope to form a pore-like structure 19. Transport of cytoplasmic components through NPCs is regulated in two ways 20,21. First, a size exclusion mechanism limits molecules >30 kDa from freely diffusing between compartments. Second, proteins above this size limit utilize NLS import signals, nuclear transport receptors and the small GTPase Ran to cross the physical barrier formed by the NPC.
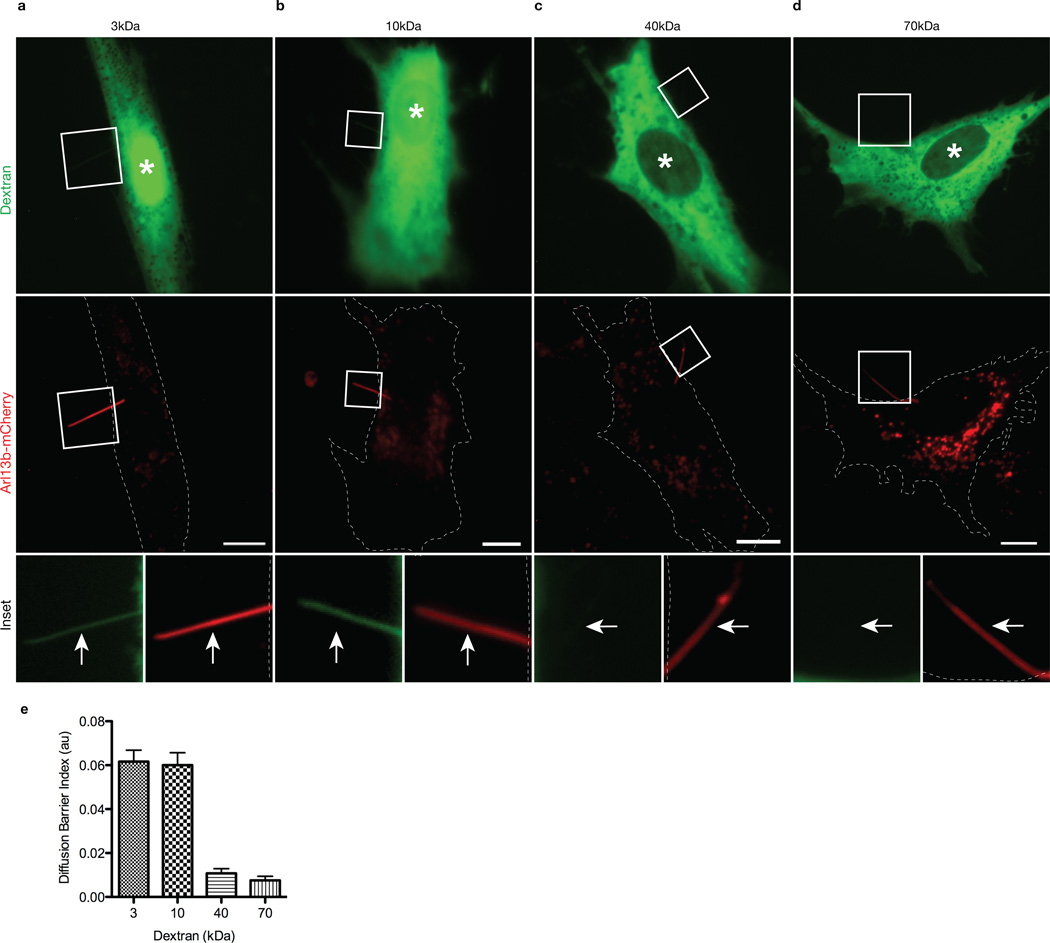
We first tested whether there is a size-dependent barrier that restricts ciliary entry of cytoplasmic molecules by microinjecting ciliated mammalian cells with fluorescently-labeled dextrans of various sizes, analogous to previous work defining a diffusion barrier for the NPC 22,23. hTERT-RPE cells were transfected with mCherry-tagged Arl13b as a live-cell marker of primary cilia (Supplementary Fig. 1) and observed 48 hours later by wide field fluorescence microscopy. Only cells with Arl13b-mCherry-labeled cilia that projected off the cell body were microinjected with fluorescent dextrans to clearly distinguish ciliary from cytoplasmic fluorescence. Dextran localization to the nuclear and ciliary compartments was assessed 20 minutes after microinjection. Small dextrans (3 kDa and 10 kDa) freely diffused into both ciliary and nuclear compartments (Fig. 1a,b). In contrast, larger dextrans (40 kDa and 70 kDa) were excluded from both ciliary and nuclear compartments (Fig. 1c,d). Quantification of the fluorescence intensities in primary cilia demonstrates that the 40 kDa and 70 kDa dextrans were significantly restricted from entering the cilium (Fig. 1e). Ciliary entry of 3 kDa and 10 kDa dextrans was rapid and could be detected within 5 minutes post-injection (Supplementary Fig. 1c), consistent with the rapid diffusion of small molecules into and within sea urchin spermatozoa 24.
Figure 1. The ciliary base acts as a size-dependent barrier for entry of cytoplasmic dextran molecules.
hTERT-RPE cells expressing Arl13b-mCherry to mark the primary cilium in live cells were microinjected with (a) 3 kDa (b) 10 kDa (c) 40 kDa or (d) 70 kDa fluorescent dextrans. Shown are representative images taken 20 min post-injection. White asterisks indicate the nuclei. Dashed white lines represent cell periphery. The bottom row shows higher magnifications of primary cilia in the boxed regions. White arrows point to primary cilia. Scale bars, 10 µm. (e) Quantification of ciliary localization of fluorescent dextrans. The data are expressed as a Diffusion Barrier Index, which represents the ratio of the mean fluorescence intensity in the cilium versus the cytoplasm. Error bars represent SEM. N = 11 (a), 12 (b), 11 (c), 10 (d) cells.
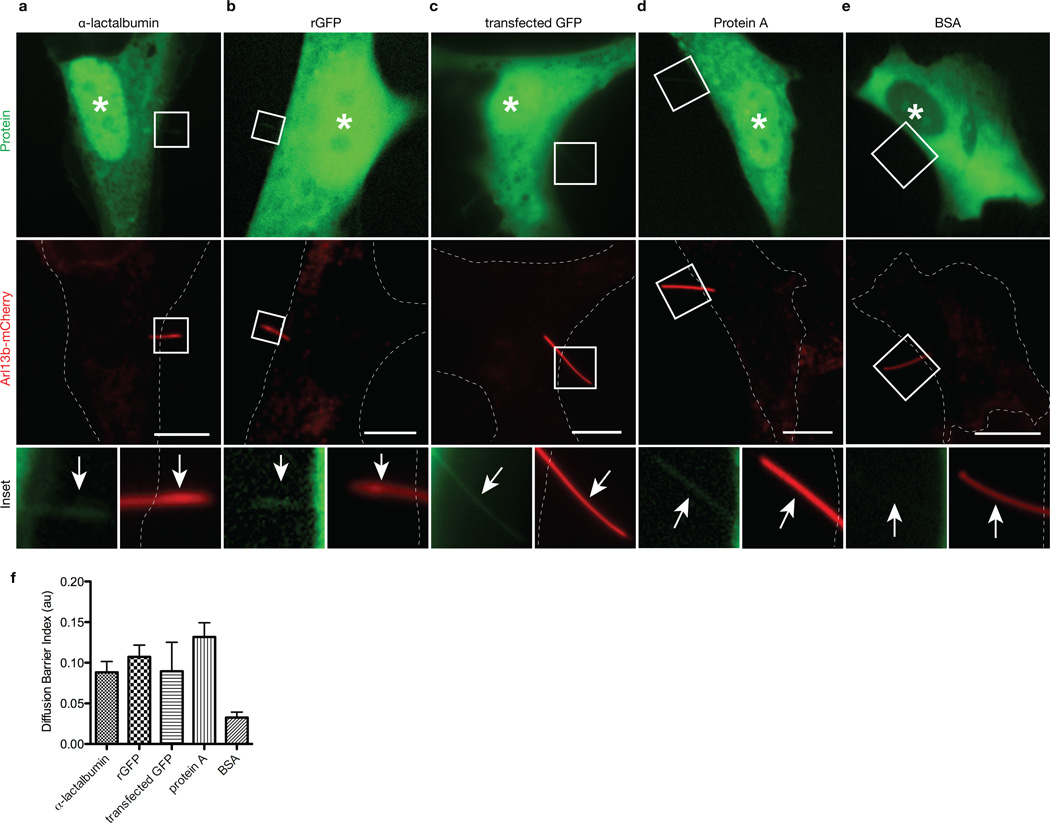
To further investigate the size-dependent diffusion barrier for ciliary entry, we microinjected fluorescently-labeled soluble proteins of nearly spherical shape that act as inert probes 25. Again, only cells with Arl13b-mCherry-labeled cilia projecting off the cell body were selected for microinjection and analysis. We found that α-lactalbumin (14 kDa), recombinant green fluorescent protein (rGFP, 27 kDa) and protein A (41 kDa) could enter both ciliary and nuclear compartments (Fig 2. a,b,d,f). Analysis in live cells is a critical component of these experiments as ciliary localization of expressed GFP could be reproducibly seen in live cells that were transfected (Fig. 2c) but was difficult to determine after fixation and/or permeabilization (data not shown), consistent with recent reports 26,27. In contrast to the ciliary localization of these smaller proteins, bovine serum albumin (BSA, 67 kDa) was restricted from both ciliary and nuclear compartments (Fig 2 e,f). Collectively, these experiments characterize a barrier for diffusion of cytoplasmic molecules into the ciliary compartment. Thus, like nuclei, cilia restrict the diffusional entry of cytoplasmic molecules based on size and utilize active transport mechanisms to facilitate entry of large proteins.
Figure 2. The ciliary base acts as a size-dependent barrier for entry of inert cytoplasmic proteins.
hTERT-RPE cells expressing Arl13b-mCherry to mark the primary cilium in live cells were either microinjected with Alexa448-labeled purified proteins (a) α-lactalbumin (14 kDa), (b) recombinant GFP (rGFP, 27 kDa), (d) Protein A (41 kDa), (e) BSA (67 kDa) or were (c) transfected with GFP-expressing plasmid. Shown are representative images taken 5 min post-injection. White asterisks indicate the nuclei. Dashed white lines indicate the cell periphery. The bottom row shows higher magnifications of primary cilia in the boxed regions. White arrows point to primary cilia. Scale bars, 10 µm. (f) Quantification of ciliary localization of microinjected fluorescent proteins. The data are expressed as a Diffusion Barrier Index. Error bars represent SEM. N = 11 (a), 10 (b), 7 (c), 8 (d), 11 (e) cells.
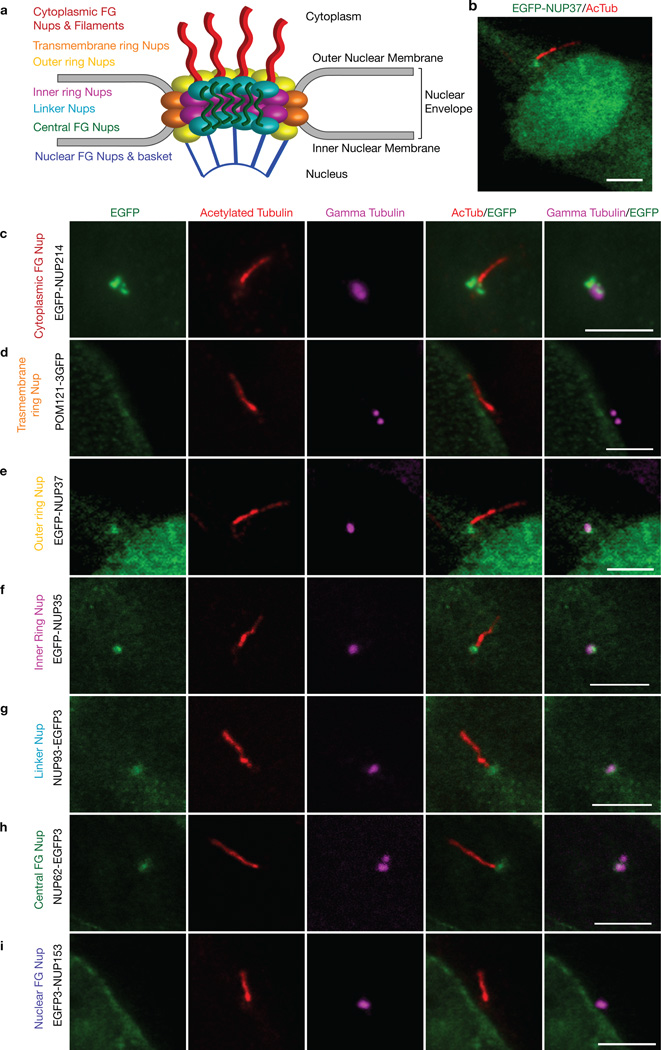
What are the molecular mechanisms by which ciliary entry of cytoplasmic molecules is restricted? One possibility is that ciliary-specific proteins create a diffusional barrier resembling that of the nuclear pore. Alternatively, NPC components could localize to the ciliary base and create a permeability barrier. To test this latter possibility, we tested whether NPC components localize to primary cilia in mammalian cells. The NPC is a large proteinaceous structure composed of multiple copies of 30 different nucleoporin proteins that assemble into subcomplexes (Fig. 3a) 28,29. EGFP-nucleoporins have been used to study the kinetics and organizational dynamics of the NPC in live cells 30,31. We expressed a fluorescently-tagged nucleoporin from each subclass in Odora cells, an immortalized cell line derived from rat olfactory sensory neurons that generates primary cilia 32. All nucleoporins localized to the nuclear membrane as expected (Supplementary Fig. 2). Localization at the ciliary base was observed for EGFP-NUP37 (outer ring nucleoporin, 12/15 cells, Fig. 3e), EGFP-NUP35 (inner ring nucleoporin, 18/18 cells, Fig. 3f), NUP93-EGFP3 (linker nucleoporin, 19/19 cells, Fig. 3g), and NUP62-EGFP3 (central FG nucleoporin, 16/17 cells, Fig. 3h). Ciliary base localization was also observed when fluorescently-tagged nucleoporins were expressed in hTERT-RPE cells (Supplementary Fig. 3). In addition, EGFP-NUP214, a member of the cytoplasmic phenylalanine-glycine (FG)-containing nucleoporin and filament subcomplex, localized in distinct puncta near the γ-tubulin-labeled basal body (Fig. 3c).
Figure 3. Fluorescently-tagged nucleoporins localize to the base of primary cilia.
(a) Diagram depicting the overall structure and subcomplexes of the NPC, based on the model in 37. Odora cells expressing (c) EGFP-NUP214, (d) POM121-3GFP, (b, e) EGFP-NUP37, (f) EGFP-NUP35, (g) NUP93-EGFP3, (h) NUP62-EGFP3, (i) EGFP-NUP153 were fixed and stained with antibodies to acetylated α-tubulin (red) and γ-tubulin (magenta) to mark the primary cilium and basal bodies, respectively. Merged images are shown in the right panels. Shown are representative images of the ciliary region of the cells; whole cell views are shown in Supplementary Figure 2. Scale bars, 5 µm.
Interestingly, not all nucleoporins could be localized at the ciliary base. POM121-3GFP, GFP-GP210 and NDC1-GFP, transmembrane nucleoporins that anchor the NPC to the nuclear envelope, did not localize to the base of the primary cilium (Fig. 3d, Supplementary Fig. 2h, i). Ciliary localization was also not observed for EGFP-NUP153, a nucleoporin that projects into the nucleoplasm (Fig. 3i). These EGFP-nucleoporins localized correctly to the nuclear envelope (Supplementary Fig. 2), suggesting that the GFP tag does not hinder their ability to assemble into nucleoporin complexes. Collectively, these experiments demonstrate that specific nucleoporins localize to the ciliary base where they could regulate passive and active transport into the ciliary compartment.
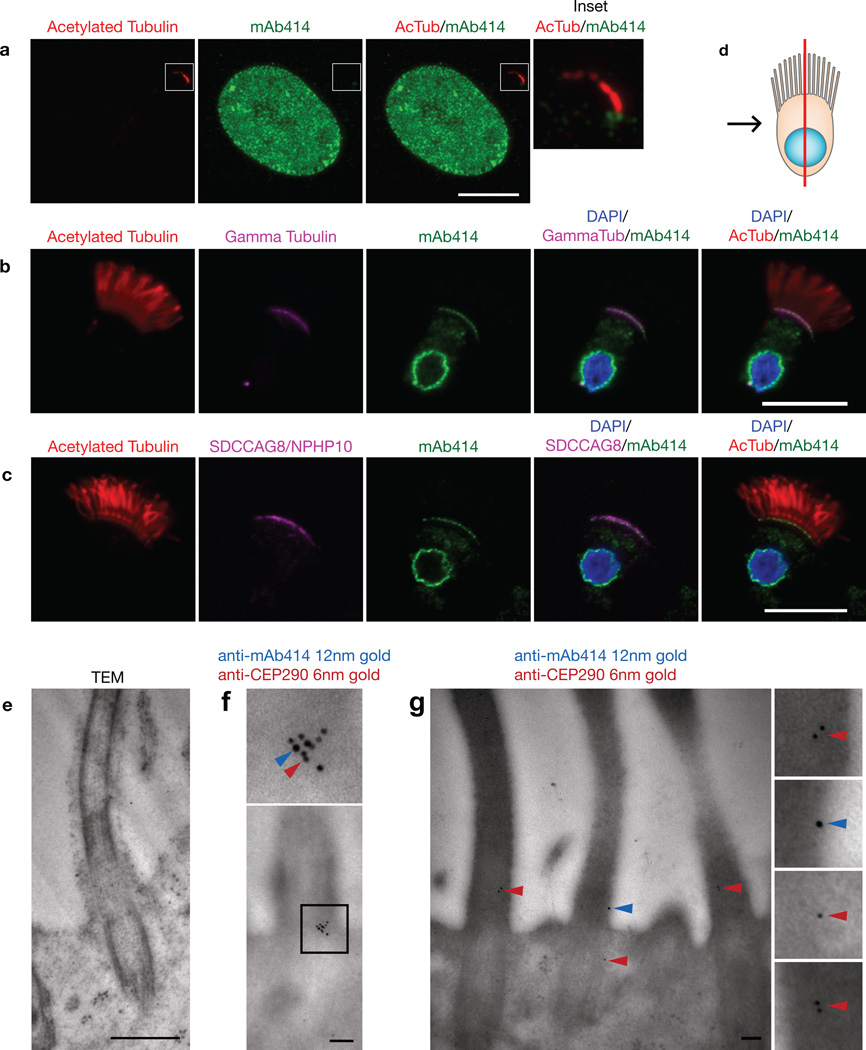
To determine if endogenous nucleoporins localize to the base of the cilium, we immunostained cells with a monoclonal antibody, mAb414, that recognizes several FG-containing nucleoporins (NUP358, NUP214, NUP62 and NUP153) 33. The FG repeats on these nucleoporins function to allow cargo-importin complexes to associate with and shuttle through the NPC meshwork. In NIH3T3 cells, mAb414 staining showed discrete puncta present at the base of the cilium, in addition to the nuclear envelope (Fig. 4a). To more clearly visualize mAb414 staining at the base of cilia, we extracted and performed immunofluorescence on trachea epithelial cells that project multiple motile cilia from their apical surface. In these cells, mAb414 stained not only the nuclear envelope but also the cilia-basal body border as shown by co-localization with γ-tubulin (Fig. 4b, Supplementary Movie 1). mAb414 staining also co-localized with basal body-associated SDCCAG8/NPHP10 protein, whose mutation causes the ciliopathy nephronophthisis 34 (Fig. 4c). In a confocal section along the edge of epithelial cells, punctate mAb414 staining could be observed on the nuclear envelope and at the base of the cilia (Supplementary Fig. 4a). Punctate staining under the cilia was also evident in a confocal bird’s eye projection down on the apical surface (Supplementary Fig. 4b). Using antibodies specific to individual nucleoporins, NUP62 and NUP133 could be detected at the ciliary base of epithelial cells (Supplementary Fig. 4 c, d).
Figure 4. Endogenous nucleoporins localize to the base of cilia.
(a) Representative image of an NIH3T3 cell fixed and stained with antibodies to nucleoporins (mAb414, green) and acetylated α-tubulin (red) to mark the primary cilium. The right panels show higher magnifications of primary cilium in the boxed region. (b) Rat trachea cells were fixed and stained with antibodies to nucleoporins (mAb414, green), acetylated α-tubulin (red) and γ-tubulin (magenta). (c) Rat trachea cells were fixed and stained with antibodies to nucleoporins (mAb414, green), acetylated α-tubulin (red) and SDCCAG8/NPHP10 (magenta). DAPI (blue) indicates the nucleus. For (b) and (c), (d) depicts a schematic representation of the epithelial cells in which the red line indicates the confocal section and the black arrow represents the point of view. Scale bars, 5 µm. (e-g) Rat trachea tissue was fixed and processed for EM. (e) Addition of osmium tetraoxide during EM processing resulted in higher contrast. Scale bar, 500nm. (f, g) Dual-label immunogold EM of sections of tracheal tissue using mAb414 and antibodies to CEP290/NPHP6. Blue arrowheads, 12nm gold particles (mAb414); red arrowheads, 6nm gold particles (anti-CEP290/NPHP6). Scale bars, 100nm.
To more precisely define the localization of nucleoporins at the base of cilia, we performed immuno-electron microscopy (EM) of rat trachea. Using mAb414 in single-label immunogold EM, gold particles labeled the ciliary transition zone and basal body in addition to the expected localization at NPCs on the nuclear envelope (Supplementary Fig. 5b, d). In double-label immunogold EM with mAb414 and antibodies to CEP290/NPHP6, co-staining was observed in clusters (Fig. 4f) and as single gold particles (Fig. 4g) at the transition zone. Thus, while mAb414 staining appears to primarily colocalize with the basal body by fluorescence microscopy, mAb414 antigens localize at the transition zone and basal body by EM, perhaps due to differences in antigen accessibility. Further work is required to define the localization of specific nucleoporins at the ciliary base. Collectively, fluorescence and electron microscopy experiments demonstrate that endogenous nucleoporins localize not only in the NPCs of the nuclear envelope but also in CPCs at the ciliary base.
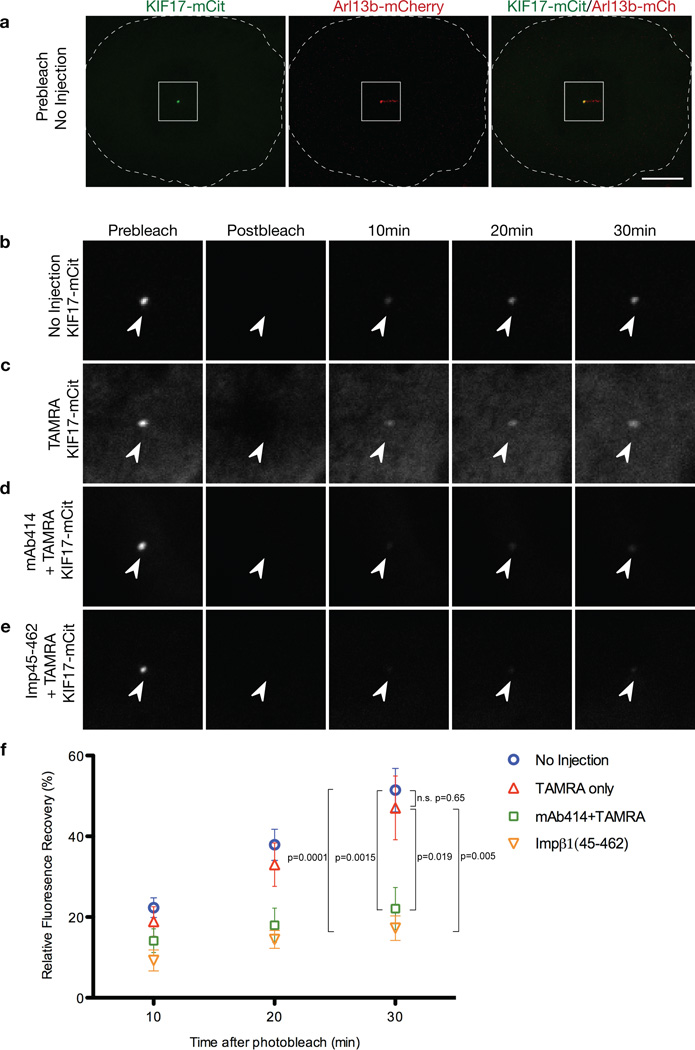
To determine whether nucleoporins function to regulate import of ciliary proteins, we microinjected two different NPC function-blocking reagents into ciliated cells, mAb414 35 and a truncated version of importin-β1 36. NIH3T3 cells co-expressing KIF17-mCitrine and Arl13b-mCherry were microinjected with mAb414 or importin-β1(45-462) together with TAMRA dye to mark injected cells (Fig. 5a). As controls, cells were injected with TAMRA dye alone or not injected. KIF17-mCitrine fluorescence at the tips of cilia was photobleached and the subsequent fluorescence recovery (Fig. 5b-d) was taken as a measure of ciliary entry of new KIF17-mCitrine molecules as described 2. Microinjection of mAb414 or importin-1(45-462) resulted in a significant reduction in KIF17-mCitrine fluorescence recovery when compared to control cells (Fig. 5d, e). We conclude that functionally inhibiting nucleoporins restricts KIF17-mCitrine entry into the ciliary compartment.
Figure 5. Microinjection of nucleoporin function-blocking reagents into cells restricts the ciliary entry of KIF17 motors.
Odora cells coexpressing KIF17-mCitrine and Arl13b-mCherry were (a, b) uninjected or were injected with (c) fluorescent TAMRA dye alone, (d) mAb414 antibodies and TAMRA or (e) Importin-β1(45-462) and TAMRA. Expressing cells were imaged [(a) prebleach] and then the KIF17-mCitrine fluorescence at the distal tip of the cilium was bleached with high laser power. Dashed white line indicates the edge of the cell. Scale bar, 10 µm. Following the bleach, the cells were imaged (postbleach) and the fluorescence recovery of KIF17 was measured over time. (b-e) show prebleach, postbleach and recovery images of KIF17-mCitrine in the cilium. Arrowheads point to the distal tip of the cilium. (f) Quantification of the fluorescence recovery of KIF17-mCitrine in the distal tips of cilia. The data are represented as mean ± SEM of fluorescence recovery after photobleach. N=8 (a), 8 (b), 8 (c) and 7 (d). Statistical significance was assessed by student’s t-test. n.s. denotes no significant difference.
In conclusion, we propose a model in which ciliary import displays selective and molecular features characteristic of nuclear import. First, we demonstrate that soluble molecules above a specific size threshold are restricted from passively entering the ciliary compartment, analogous to the manner in which the NPC acts as a sieve to prevent spurious entry of molecules. This CPC permeability barrier thus allows the enrichment of specific proteins destined for the cilium through active transport mechanisms. Furthermore, we show that several nucleoporins localize to and function at the base of the cilium in a similar manner to their functional roles at the NPC. Nucleoporins are found in several cilia proteomes (http://v3.ciliaproteome.org/cgi-bin/index.php). Thus, we propose that nucleoporins not only form the molecular components that regulate transport across the NPC, but also transport across the CPC. Collectively with our previous work 2,4, we demonstrate that ciliary import utilizes the three defining functional components that regulate nucleocytoplasmic transport – nuclear transport factors like importins, the Ran GTPase system, and nucleoporins.
Interestingly, not all fluorescent nucleoporins localize to the base of the cilium. The absence of EGFP3-NUP153 at the cilia base may be due to the nuclear-specific functions of this subcomplex, which forms a basket structure that serves as a platform for transcriptional regulation and chromatin stability in the nucleoplasm 37. NUP153 was demonstrated not be required for transportin-mediated nuclear import in Xenopus nuclei 38. The absence of POM121-3GFP, GFP-GP210, and NDC1-GFP at the ciliary base may be due to their nuclear-specific roles in anchoring components of the NPC in the nuclear envelope 19. Indeed, the structural differences between nuclear and ciliary membranes suggest that a different membrane-associated protein complex anchors the CPC to the surrounding ciliary membrane. It is tempting to speculate that the Nephronopthisis (NPHP) and Meckel-Gruber Syndrome (MKS) disease-related gene networks may play a role in anchoring nucleoporins at the CPC. Recent work demonstrated that disruption of NPHP and/or MKS genes caused defects in anchoring transition zone structures to the ciliary membrane and abnormal ciliary protein composition 14–18. Clearly, further studies are required to determine whether NPHP, MKS and nucleoporin components are functionally integrated together at the base of the cilium.
As ciliary and nuclear import share several molecular and mechanistic features, how cargoes are distinguished from entering the cilium versus the nucleus is unclear. Cilia have evolved distinct mechanisms for ciliary trafficking that may be important in defining ciliary versus nuclear transport, such as the Intraflagellar Transport (IFT) complex that drives transport along axonemal microtubules, the BBSome coat complex, and small GTPases of the Arf and Rab families (reviewed in 1,10). Future work is clearly required to uncover the complex molecular network at the base of the cilium and to delineate the mechanisms by which these components establish the primary cilium as a complex signaling center and their implications in ciliopathies.
METHODS
Antibodies and Plasmids
Commercial antibodies include: acetylated α-tubulin (1:10,000; clone 6-11B-1, Sigma), gamma-tubulin (1:500; T6557, Sigma), mAb414 (1:400 for immunofluorescence, 1:50 for immunoEM; ab24609, Abcam), NUP62 (1:250; sc-48373, Santa Cruz), polyglutamylated tubulin (1:1000; GT335, Enzo Life Sciences), and CEP290/NPHP6 (1:50 for immunoEM, IHC-00365, Bethyl Laboratories). Rabbit polyclonal anti-acetylated tubulin antibody (1:1000) was raised against the synthetic peptide: CGQMPSD(AcK)TIGGGDD. NUP133 (1:750) and SDCCAG8/NPHP10 (1:1000) antibodies were gifts from Martin Hetzer (Salk Institute) and Friedhelm Hildebrandt (University of Michigan), respectively. Secondary fluorescence-conjugated antibodies were from Invitrogen and Jackson ImmunoResearch.
The KIF17-mCitrine plasmid has been described 39. The Arl13b-mCherry plasmid was constructed by subcloning human Arl13b cDNA from Arl13b-EGFP (gift of Kenji Kontani, University of Tokyo) into the KpnI and AgeI sites of mCherry-N1. The EGFP-Nucleoporin plasmids (EGFP-NUP214, EGFP-NUP37, EGFP-NUP35, NUP93-EGFP3, NUP62-EGFP3, EGFP3-NUP153) were purchased from EUROSCARF. POM121-3GFP, NDC1-GFP and GFP-GP210 were gifts from Richard Wozniak (University of Alberta), Martin Hetzer (Salk Institute), and Bill Dauer (University of Michigan), respectively. Importin-β1(45-462) was a gift from Ulrike Kutay (ETH Zurich) and recombinant protein was purified as described 36.
Cell Culture
hTERT-RPE cells were grown in DMEM/F12 supplemented with 10% fetal bovine serum, 1% Pen/Strep and 0.01 mg/ml Hygromycin B, transfected with Trans-IT (Mirus), and serum starved for 48–60 hr to induce ciliogenesis. Odora cells were grown in DMEM supplemented with 10% fetal calf serum and 1% Pen/Strep/Gluta-MAX I and DNA constructs were transfected cells using Trans-IT LTI (Mirus) transfection reagent.
Microinjection
Fluorescently labeled dextrans of different molecular weights – 3 kDa-FITC, 10 kDa-FITC, 40 kDa-FITC, 70 kDa-FITC (Molecular Probes) - were reconstituted in buffer containing 25 mM Hepes pH7.4, 115 mM KOAc, 5 mM NaOAc, 5 mM MgCl2, 0.5 mM EDTA, 1 mM GTP and 1 mM ATP and microinjected into cells at 10 mg/ml. hTERT-RPE cells were microinjected and visualized using the FemtoJet Microinjector System (Eppendorf) mounted on an inverted epifluorescence microscope (Nikon TE2000-E) with 40×0.75 N.A. (with 1.5×Optivar) and 60×1.40 (oil) objectives and Photometrics CoolSnap ES2 camera. Dextran solutions were centrifuged at 10,000g for 5 min to remove any aggregates before loading into Femtotips (Eppendorf).
Recombinant α-lactalbumin and BSA were purchased from Sigma and recombinant GFP and protein A were purchased from Prospec. All recombinant proteins (except rGFP) were brought to a concentration of 1mg/ml before labeling with Alexa 488 using a microscale protein labeling kit (Invitrogen). rGFP was at 1 mg/ml before injection. All of the proteins were spun down before injection to remove any aggregates.
Quantification of relative mean pixel intensities of the cilia versus cytoplasm in microinjected cells was determined using ImageJ. The ciliary region indicated by Arl13b-mCherry signal projecting off the cell body was used to generate a ciliary region of interest (ROI). The average fluorescence in the ciliary ROI was background subtracted using a ROI off the cell. The average fluorescence intensity in the cytoplasmic region half the distance between the nuclear envelope and cell periphery was measured and background subtracted the same way. The Diffusion Barrier Index was calculated as the ratio of mean fluorescence intensity in the cilia versus the cytoplasm. 10–12 cells from separate experiments were measured for each dextran or protein.
Immunostaining and Microscopy
The trachea was dissected from adult rats and epithelial cells were dissociated with a toothpick, spun onto coverslips, and immediately fixed and immunostained. All experimental procedures were approved by the University of Michigan Committee on the Use and Care of Animals and performed in accordance with the Guide for the Care and Use of Laboratory Animals.
Odora and hTERT-RPE cells were fixed with 4% PFA, permeabilized with 0.1% Triton X-100 or 0.1%SDS in PBS, and blocked with 0.2% fish skin gelatin (FSG, Sigma) in PBS. Samples were incubated in primary antibodies for 1 hr at room temperature and washed three times with 0.2% FSG/PBS. Samples were incubated in fluorescent-conjugated secondary antibodies for 1 hr, washed three times with 0.2% FSG/PBS, and mounted using Prolong Gold (Invitrogen). Immunostaining of NIH3T3 cells were carried out in a similar manner but were washed and incubated with antibodies in a solution of 0.1% Triton X-100, 0.02% SDS, 10 mg/ml BSA in PBS. Immunostaining of rat tracheal cells was carried out in a similar manner except that 0.1% Triton X-100 and 0.05% SDS in PBS was used for permeabilization.
Confocal imaging was performed on Leica SP5X and Olympus Fluoview 500 confocal microscopes with a 60×1.40 numerical aperture (N.A.). Epifluoresence imaging was performed on an inverted epifluorescence microscope (Nikon TE2000-E) with 40×0.75 N.A. and 60×1.40 objectives and Photometrics CoolSnap HQ camera.
Electron Microscopy
Rat tracheas were fixed with 4% paraformaldehyde/0.25% glutaraldehyde in 0.1M cacodylate buffer at 4°C overnight. For TEM only (Figure 4d), tissue was post fixed with 1% Osmium tetraoxide. After dehydration in a graded series of ethanol, the samples were embedded in LR White (EMS; Hatfield, PA) in gelatin capsules and cured at 55°C for 72 hr. Thin sections of 80nm were collected on formvar carbon coated 100 Mesh nickel grids. For immunogold staining, grids were rinsed with PBS with 0.1 % Triton X-100 for 10 min, then rinsed with PBS two more times. After blocking with Aurion blocking buffer at RT for 1 hr, the grids were incubated with primary antibody at 25–50x dilution in 0.2%BSA/PBS overnight at room temperature, followed by 6 nm immunogold conjugated goat anti-rabbit IgG (Aurion, Netherlands) or 12 nm gold conjugated goat anti mouse secondary (Jackson Lab) at 25x dilution for 2 hr. After rinsing with PBS, grids were post fixed with 2.5% glutaraldehyde in PBS for 20 min, and some samples were treated with 4% aqueous uranyl acetate to increase contrast. Images were taken with a Phillip CM-100 transmission electron microscope operated at 60kV.
FRAP experiments
NIH3T3 cells were plated in glass-bottom dishes (MatTek), co-transfected with KIF17-mCitrine and Arl13b-mCherry and serum starved for 24–48 hours. Expressing cells were microinjected with TAMRA dye alone, TAMRA dye (50 µM) + mAb414 antibody (0.95 mg/ml), or TAMRA dye (50 µM) + Importin-β1(45-462) (133 µM) using the FemtoJet Microinjector System (Eppendorf) mounted onto a Nikon Ti Eclipse inverted microscope. The cells were maintained in a live-cell chamber at 37°C and 5% CO2. Using the Nikon A-1 Confocal System with Perfect Focus, microinjected cells were identified due to the presence of the TAMRA dye. A pre-bleach picture was taken and then the KIF17-mCitrine fluorescence at the distal tip of cilia was photobleached using 50% laser power for 1 sec. KIF17-mCitrine fluorescence recovery images were taken postbleach at 10-minute intervals for 30 minutes. Fluorescence signals were quantified using Metamorph software, background subtracted, and the average fluorescence values from 8 cells were plotted using Prism software (Graphpad).
Statistical Analysis
All statistical analysis was performed using Prism software and specific tests are noted in the text. Error bars are ±SEM and significance was assessed as p<0.05.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01GM070862 (to KJV), F32GM089034 (to JFD), and funds from the University of Michigan Center for Organogenesis (to KJV and BM). We thank Ben Allen, Jeff Martens, Toby Hurd, Shuling Fan, Martin Hetzer, and Verhey lab members for discussions of the project. We thank Bill Dauer, Valérie Doye, Martin Hetzer, Richard Wozniak, Ulrike Kutay, Kenji Kontani, and Friedhelm Hildebrandt for reagents and advice. We are grateful to Branch Craige (University of Massachusetts Medical School) for sharing his technique of trachea cell extraction and to Geri Kreitzer (Weill Medical College of Cornell University) for advice on microinjection. We gratefully acknowledge Steve Lentz and the Morphology and Image Analysis Core of the Michigan Diabetes Research and Training Center, funded by NIH Grant 5P60 DK-20572.
Footnotes
AUTHOR CONTRIBUTIONS
H.L.K, J.F.D, T.L.B and C.L performed experiments. H.L.K, J.F.D, T.L.B, C.L, B.M and K.J.V designed experiments. All authors contributed to helpful discussions shaping the investigation. H.L.K and K.J.V wrote the manuscript, with all authors providing comments and suggestions. K.J.V directed the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 2.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan S, et al. Induction of Ran GTP drives ciliogenesis. Mol Biol Cell. 2011;22:4539–4548. doi: 10.1091/mbc.E11-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 7.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 8.Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 9.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54:246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 13.Gilula NB, Satir P. The ciliary necklace. A ciliary membrane specialization. J Cell Biol. 1972;53:494–509. doi: 10.1083/jcb.53.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craige B, et al. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chih B, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. doi: 10.1038/ncb2410. Advance Online Publication. [DOI] [PubMed] [Google Scholar]
- 19.Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 21.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 22.Lang I, Scholz M, Peters R. Molecular mobility and nucleocytoplasmic flux in hepatoma cells. J Cell Biol. 1986;102:1183–1190. doi: 10.1083/jcb.102.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 24.Takao D, Kamimura S. Geometry-specific heterogeneity of the apparent diffusion rate of materials inside sperm cells. Biophys J. 2010;98:1582–1588. doi: 10.1016/j.bpj.2009.12.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvert PD, Schiesser WE, Pugh EN., Jr Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. J Gen Physiol. 2010;135:173–196. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 32.Murrell JR, Hunter DD. An olfactory sensory neuron line, odora, properly targets olfactory proteins and responds to odorants. J Neurosci. 1999;19:8260–8270. doi: 10.1523/JNEUROSCI.19-19-08260.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 34.Otto EA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clever J, Yamada M, Kasamatsu H. Import of simian virus 40 virions through nuclear pore complexes. Proc Natl Acad Sci U S A. 1991;88:7333–7337. doi: 10.1073/pnas.88.16.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 38.Walther TC, et al. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.