Summary
Most proteins are glycosylated. Mass spectrometry methods are used for mapping glycoprotein glycosylation and detailed glycan structural determination. This technology enables precise characterization of recombinant glycoproteins in the pharmaceutical industry and academic biomedicine.
1. Introduction to glycoscience
This feature article is intended as a tutorial for scientists wishing to learn more about mass spectrometry applied to glycan and glycopeptide structure. It is not intended as a comprehensive review, and apologies are made to those whose work could not be referenced.
Despite the fact that glycosylation is energetically costly, all cells, in all kingdoms of life, are coated with glycoconjugates1. Not only are glycosylated molecules required for organismal success in an evolutionary context2, but they are required for myriad physiological functions. The functions of glycoconjugates cannot be understood fully using a linear model for information flow in biological systems; it is not possible to predict glycoconjugate structure and function from genomic information. Yet, the functions of glycoconjugates are a direct consequence of their non-template driven biosynthesis. Because the biosynthetic reactions that form glycoconjugate glycans are mediated by enzymatic reactions governed by complex factors including substrate availability and kinetics, glycan structures are heterogeneous as a rule. This results in a distribution of physico-chemical properties in the mature glycoconjugates and concomitant diversity of protein-binding properties. Glycan binding proteins have evolved with glycans and may be viewed as a lens for understanding the functions of glycosylation. Thus, the diversity of glycosylation is a mechanism whereby the binding interactions between a glycosylated protein and its ligands and/or receptors are modulated. In addition, glycosylation is dynamically regulated according to spatial and temporal factors in tissues; this drives the need for effective analytical methods for producing data to inform a clear understanding of glycobiology.
a. N-glycosylation and protein folding
Polypeptide chains destined for the lysosome or for secretion are biosynthesized into the lumen of the endoplasmic reticulum. Many of these proteins are modified co-translationally by N-glycosylation using evolutionarily conserved machinery. These N-glycans help regulate protein folding by binding to calnexin and calreticulin. Through this quality control pathway, only correctly folded proteins are allowed to exit the endoplasmic reticulum.
As an example, consider antibody glycosylation. All antibodies are N-glycosylated at a conserved Asn residue in the Fc region. This glycosylation supports the three dimensional structure of the Fc region and is required for the effector functions whereby antibodies mediate their cytotoxic effects3. Monoclonal antibody therapeutics are a $ multibillion drug market. These biologic drugs are produced in non-human cells but must be shown to produce a human-like glycosylation pattern. As a result, analytical chemistry is essential to establishing lot release criteria for antibody drugs. As the first generation of antibody drugs moves off-patent, there is impetus for the development of biogenerics. Establishment of the correct N-glycosylation structure is a key challenge to demonstration of generic monoclonal antibody equivalence. In the past few years, researchers have discovered how the N-glycosylation fine structure influences monoclonal antibody efficacy3. Specifically, afucosylated antibodies have increased antibody dependent cellular cytotoxicity. Those carrying high mannose N-glycans exhibit increased clearance rates relative to those with complex N-glycans.
b. Carbohydrate-Protein interactions
Numerous carbohydrate binding proteins exist, including antibodies, lectins, receptors, toxins, and microbial adhesins4. Carbohydrate binding domains present in numerous mammalian proteins dictate their interactions with glycosylated partner molecules. These protein binding interactions are mediated by carbohydrate epitopes that may be expressed on one or more glycan classes in a given biological context. Such determinants are made up of 2–10 monosaccharide residues. The blood group antigens are classic examples of glycan epitopes that are expressed in high abundances on the surfaces of red blood cells. In all, there are thousands of glycan epitopes and more to be discovered. The types of such epitopes present on the antennae of glycoconjugate glycans determine the lectin domains to which the molecule will interact. It is therefore important to keep in mind the types of epitopes that may be present when designing a glycomics or glycoproteomics workflow.
c. Host-pathogen interactions
Pathogens evolve to recognize host cell surface glycans in the gastrointestinal tract and in the airways2. Hosts immune systems have evolved to neutralize pathogens by recognizing non-self glycosylation. As an example, the virulence of the influenza A virus depends on the ability of the hemagglutinin glycoprotein to recognize sialic acid containing glycans on host airway surfaces. Alteration of N-glycosylation is a mechanism whereby the influenza A virus maintains virulence in the presence of the selective pressure of the host innate and antibody mediated immune systems. Thus, the longer an influenza A virus variant exists in the human population, the greater the extent of N-glycosylation on the head domain of hemagglutinin5, 6. It is thought that the increasing degree of N-glycosylation enables the virus to evade antibody neutralization. Although the presence of a consensus amino acid sequence for N-glycosylation (NXT or NXS) is readily determined from genomic sequences, it is not possible to predict the degree of site occupancy or the glycan structures expressed. As a result, it is not possible to predict how the glycosylation influences the interactions between hemagglutinin and binding partners. Therefore, analysis of glycosylation of influenza proteins is necessary in order to understand influenza virulence.
2. MS methods for analysis of glycans
It is essential to understand the structural information produced in each type of MS experiment7–9. To begin this process, the reader is referred to Table 1 for a glossary of glycoscience terms used in this review. Mass measurement produces information on the composition of biomolecules. In a glycomics or glycoproteomics experiment, an accurate mass may be used to calculate the general monosaccharide composition of the detected ions, i.e. Hex, HexNAc, dHex, NeuAc, etc. For this purpose, the more accurate the mass measurement, the greater the certainty of the interpretation of the glycosylation pattern. A single stage of MS is often used to profile glycoconjugate mixtures based on extracted mass and abundance information. If one makes assumptions regarding the glycan compound class (for example N-glycans), then this information may be elaborated to include the presence of structures, for example the chitobiose core, that are common to all N-glycans. It is important to recognize that such assumptions derive from information not produced directly in the MS experiment.
Table 1.
A glossary of glycoscience terms used in this article
| Monosaccharide nomenclature | Glc | Glucose |
| GlcNAc | 2-N-acetylglucosamine | |
| Gal | Galactose | |
| GalNAc | 2-N-acetylgalactosamine | |
| Man | Mannose | |
| Fuc | Fucose | |
| NeuAc | 5-N-acetylneuraminic acid | |
| Sialic acid | A general term for neuraminic acids | |
| Hex | Hexose (Glc or Gal or Man) | |
| HexNAc | N-acetylhexosamine (GlcNAc or GalNAc) | |
| dHex | Deoxyhexose (Fuc) | |
| Glycan structure | Anomeric position | Epimers of the chiral carbons in a monosaccharide ring. The C-1 carbon of a given monosaccharide may be present as either of two anomers, α or β. |
| Glycan epitopes | Structural elements of 2–10 monosaccharides that are recognized by a glycan binding protein4 | |
| Chitobiose core | Conserved core of all N-glycans (GlcNAc2Man3) | |
| Glycan derivatization reactions | Reductive amination | Reaction of the reducing end aldyhyde of a glycan with a primary amine and reduction of the newly formed bond to create a glycan alkylated with a secondary amine56. |
| Permethylation | Conversion of all OH to O-methyl and NH to N- methyl31 | |
| Tandem MS terms | Backbone cleavage | Dissociation of a glycosidic bond |
| Cross-ring cleavage | Dissociation across a monosaccharide ring | |
| Scar | A non-methylated site revealed by tandem MS dissociaton of a glycosidic bond in a permethylated glycan |
One may perform tandem MS on some of the ions observed in the profiling experiment. It is very important to understand, however, that such ions will likely be present as isomeric mixtures; the tandem mass spectra will reflect the mixtures present. If desired, one may purify the glycans and perform additional tandem MS experiments. It is possible in some cases to directly interpret the glycan branching and linkage directly from the tandem mass spectra. This task is complicated if more than one glycan positional isomer is present in the sample. As a result, the investigator must judge the purity of the glycan sample. Glycan tandem mass spectra may also be interpreted in reference to glycan standards of known structure. The use of such standards is necessary to validate tandem mass spectral interpretation.
The concept of sequencing is best applied to linear biopolymers. Many glycan classes, however, are branched and as such do not have a linear sequence. The analytical challenge in glycomics is therefore to determine the connectivity of monosaccharides in the glycan and the linkages for each glycosidic bond. Each glycosidic bond has an anomeric position (α or β), but this cannot be determined using tandem MS except under rare circumstances. The monosaccharide linkages present in a purified glycan can be determined using gas chromatography-mass spectrometry linkage analysis10. In the absence of sufficient quantities of purified glycan material for this method, anomeric positions are often assigned using biosynthetic assumptions. The tandem MS experiment determines Hex, HexNAc, dHex, NeuAc, etc based on mass values. The identity of the Hex and HexNAc monosaccharides may be inferred by making assumptions based on the compound class and biosynthetic rules. These assumptions are often made implicitly in publications rather than stated directly.
a. Ionization
There are substantial differences between the two primary mass spectrometric biomolecular ionization methods with respect to analysis of glycans and glycoconjugates. Under typical vacuum source conditions, matrix assisted laser desorption/ionization (MALDI) results in dissociation of labile glycosidic bonds in glycan and glycoconjugate analytes7, 11, 12. This is of greatest concerns for acidic monosaccharide and substituents (sialic acids, uronic acids, sulfate, phosphate), and for fucose residues. As a result, use of vacuum MALDI for profiling of native glycans or glycoconjugates will typically underestimate the abundances of these labile monosaccharides in the ions detected. Dissociation resulting from the ionization process limits the usefulness of vacuum MALDI tandem MS of native and reductively aminated glycans and glycoconjugates. Permethylated glycans are considerably more stable than their native and reductively aminated counterparts, and may be profiled effectively using vacuum MALDI MS and tandem MS methods.
Because it is considerably gentler than MALDI, electrospray ionization (ESI) MS may be used to profile intact native glycans without dissociation of fragile acidic groups. To be successful in the application of ESI MS, the investigator is advised to purchase glycan standards to verify the performance of the instrument. In addition, it is typically necessary to use source desolvation settings that are gentler than those used for peptides and other analytes.
The peak capacity of MS experiments is increased by the addition of chromatographic separations, and ESI is directly compatible with on-line chromatography8, 13, 14. On-line liquid chromatography/mass spectrometry (LC/MS) improves the ability to detect low abundance glycans/glycoconjugates relative to analysis of unseparated mixtures. For this purpose, hydrophilic interaction chromatography (HILIC) and porous graphitized carbon chromatography (PGC) may be used for analysis of native and reductively aminated glycans. PGC and reversed phase (RP) chromatography may be used for reductively aminated glycans. RP, PGC and HILIC may be used for glycopeptides15. Permethylated glycans may be analyzed using RP and PGC16.
3. Glycan tandem MS
a. Glycan dissociation nomenclature
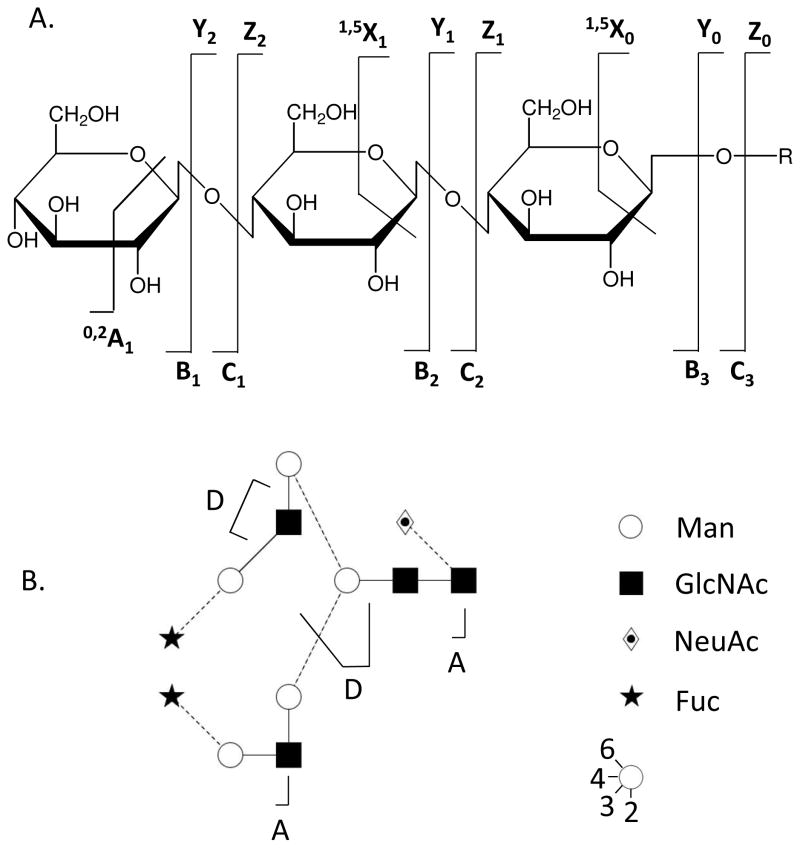
Except where indicated, the nomenclature used in this review follows that developed in 1988 by Domon and Costello17 as shown in Figure 1. Note that C- and Y-type ions correspond in mass to intact glycans with the same monosaccharide composition. The masses of cross-ring cleavages (A and X ions) help determine positions of glycosidic bond attachment, referred to as linkages. Note also that in some cases a double glycosidic bond cleavages occur on either side of a monosaccharides. These product ions, known as D ions, occur to Hex and HexNAc residues that have are substituted in the 3-position7. Analogous residues unsubstituted at the 3-position are likely to undergo A-type cross ring cleavage.
Figure 1.
Nomenclature for glycoconjugate tandem mass spectrometry17.
b. Ion isolation
Tandem mass spectra on glycans and glycoconjugates may be interpreted directly or in reference to standard compounds. Unlike with proteomics, genomics databases are not used for the spectral interpretation. As with all tandem MS experiments, however, precursor ion isolation is a very important consideration for glycomics. Typically precursor ions are isolated with a window of 3–5 u in ESI instruments; for MALDI time-of-flight instruments, the isolation window is considerably wider. As a result, the possibility of co-isolation of background or contaminant ions must be considered. The formation of product ions from such co-isolated ions has the potential to cause false interpretation of the data. Because the likelihood of such co-isolation increases with the complexity of the sample entering the MS analyzer at any given time, chromatographic separation steps are recommended to increase confidence in the data. The use of on-line LC/MS has the advantage that the number of glycan ions entering the source at any given time is fewer than with direct MS analysis of unseparated mixtures. In addition, the abundance of each precursor and product ion may be plotted versus time. Such extracted ion abundance data are useful for demonstrating that observed product ions derive from the target precursor ion18.
c. Dissociation of native and reductively aminated glycans
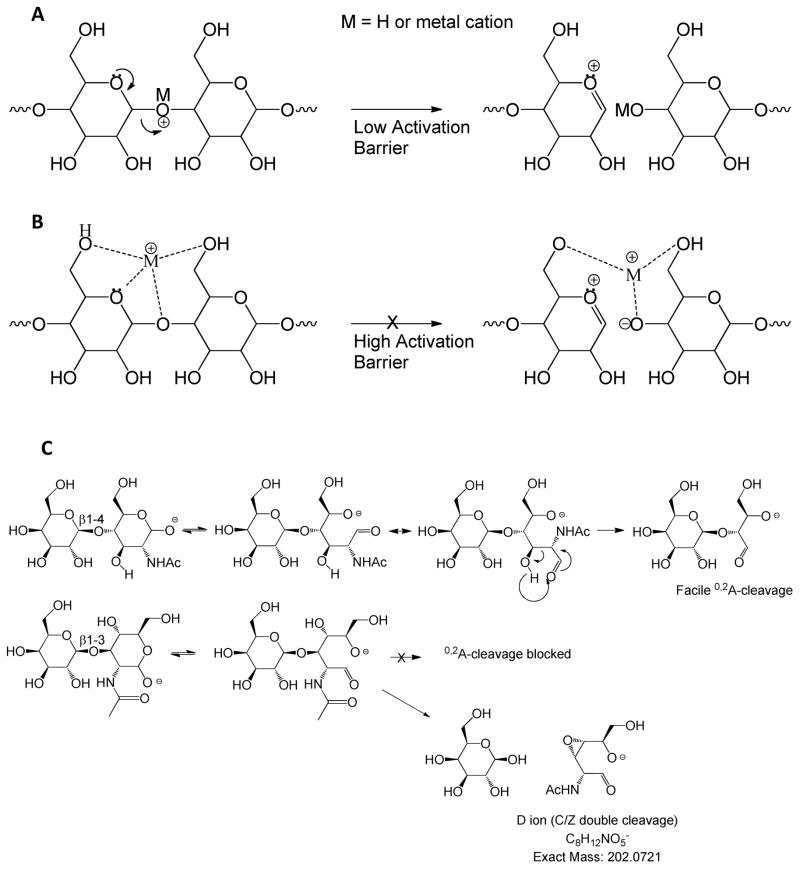
Dissociation induced by collisional heating includes the collisional activated dissociation (CAD) and infrared multiphoton dissociation (IRMPD) methods. In positive ion mode, glycosidic bond dissociation occurs when a cation associates with an electron lone pair of a glycosidic oxygen atom19, 20 (see Figure 2). Protons that associate with the glycosidic oxygen weaken the glycosidic bond, giving rise to a low activation energy barrier to dissociation (Figure 2A). So low is the activation energy for protonated ions that monosaccharide residues attached to glycans and glycopeptides have been observed to rearrange during the collisional heating process7, 21, 22. As a result, it is not recommended to use such tandem mass spectra for interpretation of glycan fine structure. The extent of glycosidic bond destabilization decreases with increasing cation radius and/or affinity for sites other than the glycosidic oxygen atom (Figure 2B). Rearrangements of sodiated glycan or glycopeptide ions have not been reported. As a result, the use of sodium (or other metal) cationized ions is recommended for instances when monosaccharide rearrangements of native and reductively aminated glycans and glycopeptides must be eliminated. Typical LC/MS conditions, however, result in formation of protonated ions. Despite the fact that tandem mass spectra are generated readily using LC/MS/MS, it is risky to interpret detailed glycan structures from the tandem mass spectra or protonated precursor ions. As shown in Figure 2C, the most commonly observed cross-ring cleavages for CAD and IRMPD of glycans are those formed by facile rearrangement mechanisms. Examples include the 0,2A ion observed for the reducing end of native N-glycans. This ion is formed at the reducing end of glycans via retro-aldol condensation23. Formation of this ion is observed for 4- and 6-linked monosaccharide residues, but not those substituted in the three position. The 0,2A ions often undergo further dissociations to form 2,4A ions. Although cross-ring cleavages occur to native and reductively aminated glycans, their abundances are relatively low for cationized precursor ions. The formation of A ions is most facile for the precursor ion and following C-type glycosidic bond cleavage ions.
Figure 2.
Glycan and glycoconjugate tandem MS mechanisms. (A) Proton mediated glycosidic bond dissociation, (B) higher activation energy barrier as cation radius increases, (C) A-type cross-ring cleavages resulting from dissociation of intact glycan precursor ions or C-type ions.
Glycans form negative ions either through deprotonation or anion adduction24. Glycans lacking acidic groups undergo negative polarity CAD to form a series of Cn ions, resulting from consecutive losses of monosaccharide residues from the reducing end25, 26. This is in contrast to cation mediated dissociation in which B- and Y-type ions are formed. Each Cn ion may undergo retro-aldol rearrangement in the gas phase to form a corresponding 0,2A ion, as shown in Figure 2C. The formation of such ions is blocked for 3-linked monosaccharides. For such residues a second glycosidic cleavage occurs, known as a D-type ion. These mechanisms are reliable for determination of linkages for glycans lacking acidic groups.
The situation is more complicated for acidic glycans. Such glycans ionize by dissociation of the acidic protons. For sialylated glycans, the sialic acid residues typically dissociate to produce abundant B1 ion, reducing the abundances of structurally informative Cn ions27. For sulfated glycans, the most acidic functional group is the sulfate itself. Thus, charge will tend to reside on the sulfate. A protonated sulfate group undergoes rapid and undesirable loss of SO328. Such losses are minimized for precursors in which all sulfate groups are deprotonated. In practice, it is not possible to produce such charge states for highly sulfated glycans due to repulsion between adjacent negative charges. Thus, a degree of SO3 losses is observed with tandem MS of sulfated glycans, regardless of the type of dissociation used.
d. Dissociation of permethylated glycans
When a glycosidic bond cleavage occurs for a native glycan ion, the product ion masses do not determine which bond was cleaved. Such dissociation for permethylated glycans results in the formation of product ions with unique masses by virtue of the lack of methyl or methoxy groups. Such scars, as they are known, provide high value for determination of the topology and linkages for glycans29. The permethylation chemistry30, 31 is very robust, and new variants have been published to increase sample throughput and decrease consumption32. Permethylated glycans are typically observed as sodiated ions, unless reversed phase LC/MS is used, under which conditions protonated ions are formed. Multistage tandem MS methods have been developed for gas phase dissection of permethylated glycans using ion trap instruments33–35. This approach is clearly the most powerful mass spectrometric method for glycan structural analysis. However, as with all tandem MS methods, confidence in the interpretation depends on clear understanding of the limitations of the method. There is the possibility of ion co-isolation leading to incorrect interpretations. Such concerns are reduced by chromatographic steps to reduce mixture complexity. There are also possibilities for derivatization side reactions, and separations help increase confidence in data interpretation. Also, application of high accuracy mass spectrometry helps to increase confidences in the mass assignments of permethylated glycans.
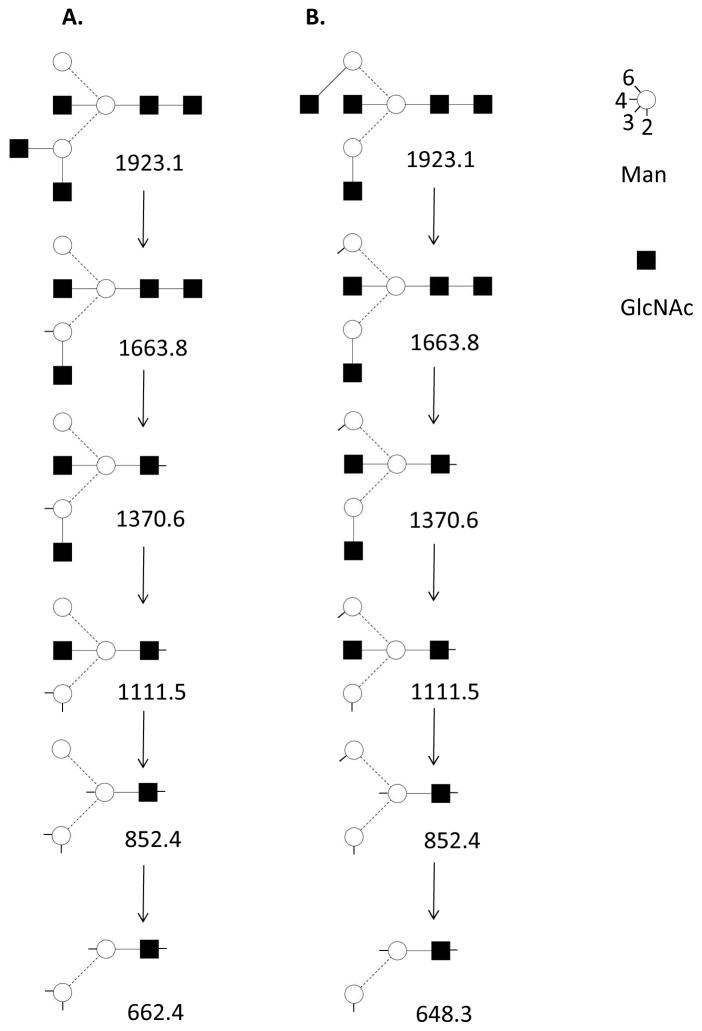
An example of the use of multistage tandem MS to determine ovalbumin N-glycan connectivity is diagrammed in Figure 3 based on published data36. The data were acquired using an ion trap mass spectrometer by selecting successive ions for tandem MS up to MS6. For such data, glycosidic bond dissociation produced abundant ions with scars, each of which corresponded to the absence of a methyl or methoxy group. The data were consistent with the presence of the two structural isomers shown. As is often the case with MSn datasets, the isomeric differences became apparent when the structure was dissected in the gas phase down to the core branching residues, the patterns of which are indicated by mass, due to the scars present. The order of losses of non-reducing end HexNAc residues cannot be determined from the data; the order shown is arbitrary. The antennae HexNAc residues were labeled as GlcNAc based on glycan biosynthetic principles and prior knowledge of ovalbumin glycans. This example illustrates the use of multistage tandem for analysis of glycans present as isomeric mixtures.
Figure 3.
Diagram of multistage tandem MS of permethylated N-glycan GlcNAc5Man3 based on published data36. The diagram is based on Figure 1 and Table 1 from the referenced publication. A symbolic key is shown, based on the Oxford system for glycan representation57.
e. Activated electron dissociation of glycans
Collisional excitation of ions results from slow (on the molecular scale) vibrational heating of ions, resulting in bond dissociation. IRMPD also results from vibrational excitation of ions. Typically, the most labile bonds produce the most abundant ions using these dissociation mechanisms. For glycans, this means that the abundances of cross-ring cleavage ions that provide the information for assigning monosaccharide linkages are often low in abundance. As a result, not all linkages can be assigned in a typical CAD or IRMPD tandem mass spectrum. Although multistage tandem MS increases the information produced, the time and sample consumption scales with each additional tandem MS stage. Therefore, in order to maximize the structural information that can be produced on limiting sample quantities, it would be highly desirable to increase the structural information that may be produced on glycans in a single stage of tandem MS.
Towards these ends, researchers have been investigating use of activated electron dissociation methods. Activated electron dissociation (ExD) methods, for the purposes of this review, include electron capture dissociation (ECD)37, electron transfer dissociation (ETD)38. Each of these methods involves transfer of an electron to the precursor ion, turning it into a radical species. As has been described for peptides, such radicals undergo rearrangement to create bond cleavage. Fortunately, the types of bond cleavages observed do not depend on bond lability; rather, they depend on proximity to the site of radical production and subsequent rearrangement mechanisms. Therefore, the bond dissociation patterns observed using ExD are complementary to those from vibrational excitation (CAD and IRMPD) methods.
During ExD, a positively charged precursor ion gains an electron, decreasing the ion charge. This radical cation then undergoes rearrangement to dissociate backbone bonds. ExD, depending on the conditions, has the potential to dissociate glycans based on a mechanism that is complementary to vibrational excitation (the mechanism behind CAD and IRMPD). Studies have shown that ECD of metal cationized native glycans results in comparatively subtle differences compared to CAD tandem mass spectra on native glycans39. ECD produced A-type cross-ring cleavages that were somewhat higher in abundance than in CAD spectra. Abundances of A-type cross-ring cleavages are higher for neutral and sialylated glycans ionized in the negative mode25, 26.
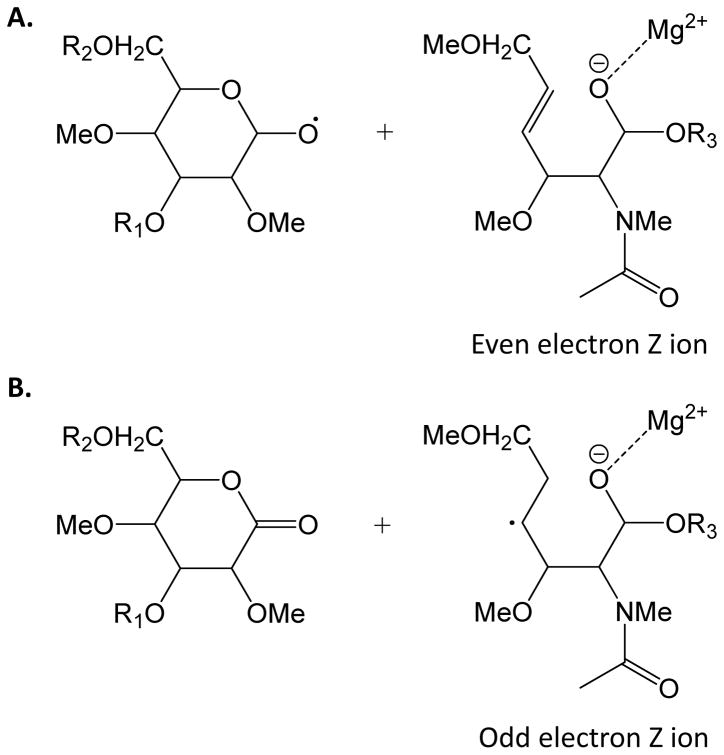
The greatest potential benefit of ExD methods is to increase the dissociation yield of cross-ring cleavage ions in general and X-ions in particular. X-type ions are generally not observed using CAD or IRMPD methods unless there is a facile ene-type rearrangement, such as for unsaturated monosaccharides. The most promising results published thus far are for metal cationized permethylated glycans40, 41. Use of permethylated ions eliminates uncertainties regarding multiple bond dissociation observed with native or reductively aminated glycans. The cation has been observed to direct the product ion pattern. In particular, because the dissociation requires diminishment of positive charge, there is an advantage to the use of multivalent metal cations to maximize charge state41. The formation of a radical cation has the potential to produce both C-type ions, similar to observed using CAD, and Z-type ions. It has been observed that ETD of magnesium cationized permethylated glycans results in abundant A- and X-type cross-ring cleavages. This is consistent with the formation of Z-type ions through a mechanism that results in opening of the glycosidic ring, as shown in Figure 4. The formation of such open ring Z-type ions is significant because they lead to the formation of X-type ions. It will be of interest to see how these new results play out in biochemical applications using ExD methods.
Figure 4.
Generation of (A) even electron Z ions and (B) odd electron Z ions resulting from ETD of permethylated glycans41. These Z ion structures are significant because they undergo subsequent rearrangements to form X-type cross-ring cleavage ions. R1, R2, R3 = monosaccharide residues.
Electron detachment dissociation (EDD) is a technique whereby an electron is removed from a negatively charged precursor ion42. The use of this technique is advantageous for carbohydrate classes that contain acidic groups, owing to their propensities to form negatively charged ions. Despite the fact that acidic glycans form abundant negative ions, the acidic groups tend to undergo facile dissociation during CAD and IRMPD. As a result, in many cases the loss of the acidic group dominates the tandem mass spectrum, and dissociation events occurring to the remainder of the glycan bonds are disfavored. Fortunately, EDD product ion patterns for such glycans complement those generated by CAD. The use of EDD improves the abundances of A-type cross-ring cleavages relative to IRMPD and CAD and produces some X-type ions43. EDD is particularly useful for sialylated glycans44 and for glycosaminoglycans45–47. The EDD method suffers, however, from poor efficiency, requiring extended acquisition times for spectral averaging. As a result, EDD has not been applied to on-line LC/tandem MS of glycans. It is possible to detach an electron from a negatively charged precursors in the gas phase using ion-ion reactions. Such negative ion ETD reactions may be accomplished in a trapped ion mass spectrometer with subsequent high resolution, high mass accuracy mass analysis. This has been demonstrated for glycosaminoglycans, for which product ion patterns are similar to those of EDD48. In summary, methods for the use of ExD for negative ions are developing rapidly and may become applicable to on-line LC/MS experiments.
4. Glycopeptides
Returning to the example of influenza A virus hemagglutinin, the glycosylation of amino acid residues adjacent to the sialic acid binding site strongly influence virulence and recognition by the immune system6. Thus, the understanding of influenza biology depends on the ability to determine the structures of glycans that modify specific amino acid residues. In fact, the ability to determine and exploit the biochemical mechanisms whereby glycosylation on numerous glycoproteins mediate biological function depends on the ability to determine site-specific glycan structure. Analytical methods capable of confident mapping of glycoprotein glycans to specific amino acid residues are now emerging. This section gives an overview of methods that enable understanding of how glycosylation is used as a means of modifying protein properties at specific amino acid sites.
a. Glycopeptide mapping
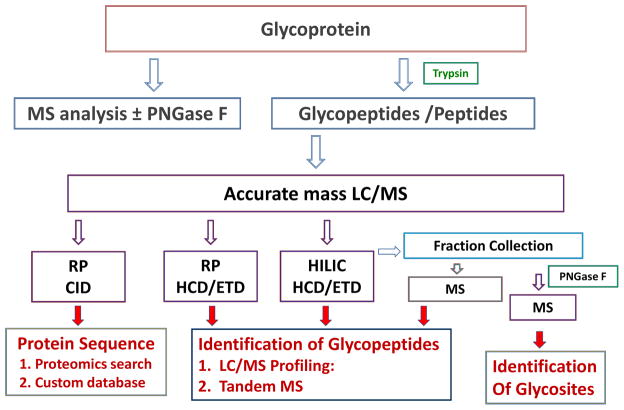
The assignment of glycopeptide glycosylation sites, the compositions of attached glycans, and relative abundances of glycoforms is known collectively as glycopeptide mapping49. Accurate glycopeptide mapping requires a combination of techniques to provide verification of interpretation necessary to minimize false identifications50. A workflow that reflects the use of MS methods available in many proteomics facilities is shown in Figure 5. In order to produce accurate glycopeptide mapping results, it is essential to perform proteomic analysis of the polypeptides in the sample. Real world glycoprotein samples are often contaminated with other glycoproteins and/or proteases. By identifying these contaminants, the investigator will be able to generate a custom proteomics database reflecting the polypeptides detected in the sample. This custom database is then the basis for interpreting glycopeptide mass data so as to minimize false positive identifications. The value of accurate mass measurement cannot be overstated for glycopeptide profiling; the more accurate the measurement, the greater the certainty of the interpretation. Reversed phase chromatography and HILIC provide complementary LC/MS results for glycopeptides, and there is value for combining data from both methods to maximize coverage of the target glycoprotein sequence51. It is useful to conduct proteomics MS before and after deglycosylation of the glycoprotein using peptide N-glycosidase F (PNGase F). In the presence of H2180, the formerly glycosylated Asn residue is converted into 18O-Asp, and peptides containing this residue may be identified using the custom proteomics database.
Figure 5.
Workflow for N-linked glycopeptide mapping
b. Glycopeptide tandem MS
The CAD and ExD methods provide complementary information on glycopeptide structure52. CAD dissociation produces abundant cleavages of the glycan with peptide bond cleavages in low abundance. The extent of glycan dissociation in ion traps is considerably less than observed in instruments with beam type dissociation cells. The latter such instruments include quadrupole time-of-flight, triple quadrupole, and Fourier Transform mass spectrometers with external dissociation cells. Dissociation of protonated glycan ions is not recommended because monosaccharide rearrangements may result. Glycopeptides, however, are ionized typically as protonated ions in MS and LC/MS workflows. As a result, it is risky to attempt to derive detailed glycan structures from tandem MS of these protonated ions. It is more appropriate to use the CAD tandem MS data to verify the composition of the glycopeptide glycan. ExD dissociation occurs primarily to the peptide backbone, leaving the glycan intact. Thus, ExD is useful for confirming the identification of the peptide. In favorable cases, the site of glycosylation may be identified.
The ability to confidently map glycoprotein glycosylation depends on the sensitivity and mass accuracy of the MS instrument. Due to the increased spectral summation times, the ability to detect low abundance glycopeptides is greater for collected LC fractions than obtainable using on-line LC/MS53. As a result, there is value in use of automated nano-ESI robots for acquiring glycopeptide mapping data.
c. Mucin-type O-glycopeptides
Mucin type O-glycans are built on GalNAc residues that modify Ser/Thr amino acid side chains. This type of glycosylation occurs typically clustered into domains with many modified Ser/Thr residues. There is a substantial degree of heterogeneity of the O-glycan structure on each modified amino acid residue. As a result, analysis of mucin type O-glycopeptides poses a considerable analytical challenge54. Unlike for N-glycosylation, there is no consensus sequence for predicting the location of amino acid residues glycosylation by mucin O-glyans. In addition, there is no enzyme for universal release of O-glycans. In the face of these difficulties, there is a strong need for determining the patterns of O-glycan modification and the glycan structures at each modified amino acid residue. The most promising approach to emerge in recent years to address this problem is the application of ECD/ETD methods. Similar to N-linked glycopeptides, ECD/ETD dissociates the peptide backbone preferentially, leaving the O-glycans intact. Thus, it is possible to assign the pattern of O-glycan substitution using ECD/ETD.
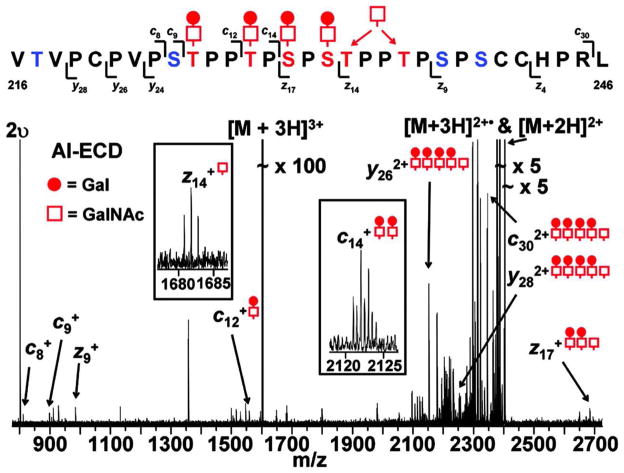
A particularly impressive example of O-glycopeptide analysis is shown in Figure 655. Immunoglobulin A (IgA) molecules have a hinge region that carries a mucin-like domain with a cluster of O-glycans. In IgA nephropathy, IgA molecules are present with incomplete galacotsylation in the hinge region, meaning that some of the O-glycans are present as GalNAc with no other monosaccharides present. The research question involved determination of the pattern of expression of deficiently galactosylated O-glycans of the IgA hinge region. It was possible to determine the pattern of glycosylation for a 31 amino acid hinge region peptide that contained five sites of O-glycosylation. The ECD tandem mass spectrum shown shows a series of product ions, the m/z values of which determine the masses of the corresponding peptide fragments and attached glycans. Assuming that glycosylation occurs only at Ser and Thr residues, the m/z values of the series of product ions suffice to determine the amino acids modified by four Gal-GalNAc disaccharide groups. It was also possible to narrow the site of incomplete galactosylation (i.e. modification with a single GalNAc) to one of two Ser residues, as shown in the figure. This example demonstrates the utility of the ExD approach for determining patterns of glycosylaton in mucin-like domains. The ability to apply ExD to biological samples is improving rapidly with increases in instrumentation sensitivity and efficiency.
Figure 6.
ESI ECD tandem mass spectrum of an O-glycosylated peptide from immunoglobulin type A1 hinge region peptide55. The spectrum shows that 11 of the 30 peptide bonds are cleaved. The sites of mucin-type O-glycosylation on Ser/Thr residues as determined from the tandem mass spectrum are shown. Open squares = GalNAc, shaded circles = Gal. The m/z values of the product ions identified four glycosylation positions unequivocally. One GalNA modifies either of the two positions indicated; the data were not sufficient to differentiate the two possibilities. ©2005, American Society for Biochemistry and Molecular Biology, used with permission.
In summary, confident glycopeptide mapping requires acquisition of proteomics, and glycopeptide MS and tandem MS data. The interpretation of such data remains the most time consuming part of the analysis. Today there exist no software programs that integrate the diverse data types needed for glycopeptide mapping.
5. Conclusions and outlook
The structures of glycans determine the carbohydrate binding protein partners to which a glycoprotein binds. Carbohydrate binding domains are common in cell surface and secreted proteins. In order to understand fully the structure-function relationships for glycoproteins, analysis of the glycan structure is unavoidable. Mass spectrometry is an enabling technology in this area.
Accurate mass measurement determines glycan compositions; this information may be used to infer the presence of glycan epitopes based on biosynthetic principles. Although tandem mass spectrometry may be accomplished on native glycans, the quality of the product ion patterns is better for permethylated glycans. Collision based tandem MS of permethylated glycans produces primarily glycosidic bond cleavages that are useful for determining glycan connectivity. The determination of linkage positions, however, requires formation of cross-ring cleavages, the abundances of which are often limiting. Cross-ring cleavage abundances may be improved through the use of multistage tandem MS. Such analyses are not possible using accurate mass measurement at the present. The use of activated electron dissociation results in product ion patterns with more abundant cross ring cleavages than those generated using collisional excitation.
Mapping of glycoprotein glycans is best achieved using a combination of MS based approaches. It is important to acquire proteomics data to generate a custom database of the proteins in the sample. With this database, one can then analyzed MS and tandem MS data produced on glycopeptides, peptides, and released glycans, respectively. The analysis of the data remains time consuming at present. Improvements in the ability to map glycoprotein glycans are likely to come from improvement of mass spectrometer sensitivity, activated electron dissociation methods, and the availability of comprehensive software for analysis of the data.
Acknowledgments
The author is supported by NIH grants R01098050 and P41RR10888
Biography
Author biographies
Nancy Leymarie is a Senior Research Scientist working at the Center for Biomedical Mass Spectrometry (CBMS) at Boston University. Her research is focused on the development and application of advanced mass spectrometry methods in the fields of glycoproteomics and glycomics. Joseph Zaia is Professor of Biochemistry and Associate Director of the CBMS. His research is focused on structural glycosylation phenotypes related to human diseases. He has developed mass spectral methods for glycomics and is applying these to amyloid diseases and cancers. His research group focuses on biochemistry and structural analysis of glycosaminoglycans, glycoproteins, and bacterial polysaccharides.
References
- 1.Varki A. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A. Cell. 2006;126:841–845. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Jefferis R. Biotechnol Prog. 2005;21:11–16. doi: 10.1021/bp040016j. [DOI] [PubMed] [Google Scholar]
- 4.Cummings RD. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 5.Cherry JL, Lipman DJ, Nikolskaya A, Wolf YI. PLoS Curr Influenza. 2009:RRN1001. doi: 10.1371/currents.RRN1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartshorn KL, Webby R, White MR, Tecle T, Pan C, Boucher S, Moreland RJ, Crouch EC, Scheule RK. Respir Res. 2008;9:65. doi: 10.1186/1465-9921-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaia J. Mass Spectrom Reviews. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 8.Zaia J. OMICS. 2010;14:401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaia J. Chem Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geyer R, Geyer H. Methods Enzymol. 1994;230:86–108. doi: 10.1016/0076-6879(94)30009-7. [DOI] [PubMed] [Google Scholar]
- 11.Zaia J. In: Ionization Methods. Gross ML, Caprioli RM, editors. Vol. 6. Elsevier; Amsterdam: 2006. pp. 889–903. [Google Scholar]
- 12.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Glycobiology. 2007;17:411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 13.Mechref Y, Novotny MV. Mass Spectrom Rev. 2009;28:191–191. doi: 10.1002/mas.20209. [DOI] [PubMed] [Google Scholar]
- 14.Zaia J. Mass Spectrom Rev. 2009;28:254–272. doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuhrer M, Deelder AM, Hokke CH. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825:124–133. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 16.Costello CE, Contado-Miller JM, Cipollo JF. J Am Soc Mass Spectrom. 2007;18:1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domon B, Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 18.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Nat Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 19.Orlando R, Bush CA, Fenselau C. Biomed Environ Mass Spectrom. 1990;19:747–754. doi: 10.1002/bms.1200190408. [DOI] [PubMed] [Google Scholar]
- 20.Cancilla MT, Penn SG, Carroll JA, Lebrilla CB. J Am Chem Soc. 1996;118:6736–6745. [Google Scholar]
- 21.Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. RAPID COMMUN MASS SP. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- 22.Wuhrer M, Koeleman CA, Deelder AM. Anal Chem. 2009;81:4422–4432. doi: 10.1021/ac900278q. [DOI] [PubMed] [Google Scholar]
- 23.Spengler B, Dolce JW, Cotter RJ. Anal Chem. 1990;62:1731–1737. doi: 10.1021/ac00207a004. [DOI] [PubMed] [Google Scholar]
- 24.Harvey DJ. J Am Soc Mass Spectrom. 2005;16:622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Pfenninger A, Karas M, Finke B, Stahl B. J Am Soc Mass Spectrom. 2002;13:1331–1340. doi: 10.1016/S1044-0305(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 26.Chai W, Piskarev V, Lawson AM. Anal Chem. 2001;73:631–657. doi: 10.1021/ac0010126. [DOI] [PubMed] [Google Scholar]
- 27.Seymour JL, Costello CE, Zaia J. J Am Soc Mass Spectrom. 2006;17:844–854. doi: 10.1016/j.jasms.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naggar EF, Costello CE, Zaia J. J Am Soc Mass Spectrom. 2004;15:1534–1544. doi: 10.1016/j.jasms.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Reinhold VN, Reinhold BB, Costello CE. Anal Chem. 1995;67:1772–1784. doi: 10.1021/ac00107a005. [DOI] [PubMed] [Google Scholar]
- 30.Ciucanu I, Costello CE. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 31.Ciucanu I, Kerek F. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 32.Mechref Y, Kang P, Novotny MV. Methods Mol Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Singh S, Reinhold VN. Anal Chem. 2005 doi: 10.1021/ac050725r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapadula AJ, Hatcher PJ, Hanneman AJ, Ashline DJ, Zhang H, Reinhold VN. Anal Chem. 2005;77:6271–6279. doi: 10.1021/ac050726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashline D, Singh S, Hanneman A, Reinhold V. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zubarev RA. Curr Opin Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamson JT, Hakansson K. Anal Chem. 2007;79:2901–2910. doi: 10.1021/ac0621423. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C, Xie B, Chan SY, Costello CE, O’Connor PB. J Am Soc Mass Spectrom. 2008;19:138–150. doi: 10.1016/j.jasms.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 41.Han L, Costello C. J Am Soc Mass Spectrom. 2011;22:997–1013. doi: 10.1007/s13361-011-0117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budnik BA, Haselmann KF, Zubarev RA. Chemical Physics Letters. 2001;342:299–302. [Google Scholar]
- 43.Adamson JT, Hakansson K. J Am Soc Mass Spectrom. 2007;18:2162–2172. doi: 10.1016/j.jasms.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Zhou W, Hakansson K. Electrophoresis. 2011;32:3526–3535. doi: 10.1002/elps.201100327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolff JJ, Chi L, Linhardt RJ, Amster IJ. Anal Chem. 2007;79:2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. J Am Soc Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolff JJ, Leach FE, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Anal Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalpathado DS, Desaire H. Analyst. 2008;133:731–738. doi: 10.1039/b713816d. [DOI] [PubMed] [Google Scholar]
- 50.Desaire H, Hua D. Int J Mass Spectrom. 2009;287:21–26. [Google Scholar]
- 51.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849:115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 52.Håkansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 53.Stalnaker SH, Hashmi S, Lim JM, Aoki K, Porterfield M, Gutierrez-Sanchez G, Wheeler J, Ervasti JM, Bergmann C, Tiemeyer M, Wells L. J Biol Chem. 2010;285:24882–24891. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen PH, Kolarich D, Packer NH. FEBS J. 2010;277:81–94. doi: 10.1111/j.1742-4658.2009.07429.x. [DOI] [PubMed] [Google Scholar]
- 55.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, Hiki Y, Toma K, Emmett MR, Mestecky J, Marshall AG, Novak J. J Biol Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 56.Anumula KR. Anal Biochem. 2006;350:1–23. doi: 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 57.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]