Abstract
Apicomplexan parasites employ gliding motility that depends on the polymerization of parasite actin filaments for host cell entry. Despite this requirement, parasite actin remains almost entirely unpolymerized at steady state; formation of filaments required for motility relies on a small repertoire of actin-binding proteins. Previous studies have shown that apicomplexan formins and profilin exhibit canonical functions on heterologous actins from higher eukaryotes; however, their biochemical properties on parasite actins are unknown. We therefore analyzed the impact of T. gondii profilin (TgPRF) and FH1-FH2 domains of two formin isoforms in T. gondii (TgFRM1 and TgFRM2) on the polymerization of T. gondii actin (TgACTI). Our findings based on in vitro assays demonstrate that TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 dramatically enhanced TgACTI polymerization in the absence of profilin, making them the sole protein factors known to initiate polymerization of this normally unstable actin. In addition, T. gondii formin domains were shown to both initiate polymerization and induce bundling of TgACTI filaments; however, they did not rely on TgPRF for these activities. In contrast, TgPRF sequestered TgACTI monomers, thus inhibiting polymerization even in the presence of formins. Collectively, these findings provide insight into the unusual control mechanisms of actin dynamics within the parasite.
Keywords: actin-binding proteins, cytoskeleton, gliding motility, apicomplexan
Toxoplasma gondii is a protozoan pathogen of the phylum Apicomplexa. T. gondii has an obligate intracellular life cycle and must therefore enter into host cells prior to replication. Along with the other members of the phylum, T. gondii employs a unique form of gliding motility for active invasion of host cells (1). Gliding motility relies on a small myosin, called TgMyoA, anchored within the parasite inner membrane complex to translocate actin filaments toward the posterior of the parasite (2). The coupling of transmembrane adhesins to the myosin motor complex occurs via an interaction between their C-termini and the glycolytic enzyme aldolase, which also serves as an F-actin binding protein (3). Rearward translocation of these adhesin-aldolase-actin complexes facilitates forward motion (1, 2).
Treatment of T. gondii with cytochalasin-D disrupts gliding motility and inhibits host cell invasion (4). Despite the requirement for filamentous actin to support parasite motility and host cell invasion, T. gondii parasites maintain actin in a largely unpolymerized state (5, 6). In exception to this pattern, actin filaments have been visualized beneath the membrane of gliding parasites, as detected by sonication and rapid freezing followed by electron microscopy (6). T. gondii contains one actin isoform, TgACTI, and studies with recombinant baculovirus-expressed parasite actin revealed that it fails to co-polymerize with vertebrate actin and undergoes inefficient polymerization on its own to form short filaments in vitro (7). Additionally, treatment of parasites with jasplakinolide to stabilize actin filaments results in aberrant hyper-motility and disruption of host cell invasion (6). Therefore, it appears that actin filaments are formed only transiently in the parasite in order to control the proper directionality and timing of motility. Invasion of red blood cells by Plasmodium knowlesi (8) merozoites, and motility of Cryptosporidium parvum (9) and Eimeria tenella (10) sporozoites are also sensitive to cytochalasins. Short unstable actin filaments have been described in Plasmodium falciparum(11), and actins from this organism also show unusual polymerization kinetics in vitro (12, 13), suggesting that many of these features are conserved within the phylum. These properties raise the question of how apicomplexan actin polymerization is regulated, especially given the reduced repertoire of actin-binding proteins in these organisms.
Regulation of actin turnover is critical for maintaining proper filamentous networks within cells and therefore, eukaryotic organisms have evolved numerous actin-binding proteins to ensure proper regulation of actin polymerization (14). Searches within the genomes of apicomplexan parasites have revealed a minimal set of actin-binding proteins as compared to other organisms (15, 16). Notably absent is the actin nucleating complex Arp 2/3 (17). One abundant actin-binding protein is actin depolymerizing factor (ADF), which has recently been shown to act primarily by sequestering actin in T. gondii (18, 19). In addition, apicomplexans contain profilin and formins, which normally interact to drive actin polymerization (20).
Profilins are small monomeric actin binding proteins that play multiple roles in regulation of actin polymerization. Profilins were initially shown to sequester G-actin, thereby resulting in filament depolymerization (21). However, profilin also plays a role in promoting polymerization by enhancing nucleotide exchange to convert ADP-actin to ATP-actin, thus creating a polymerization competent state and lowering the critical concentration for polymerization (22). More recently, profilin has been shown to enhance polymerization through interaction with the FH1 domain of yeast formin Bni1 (23), a property subsequently shown for a number of different profilin-formin pairs (24). Profilin has previously been shown to be essential for gliding motility in T. gondii through the use of a conditional knockout (25). Depletion of T. gondii profilin, TgPRF, also results in defects in parasite invasion into and egress from host cells (25). Biochemical assays demonstrate that TgPRF aids in the assembly of skeletal muscle actin filaments at free barbed ends but blocks assembly at the pointed end (25). Despite interacting with heterologous actin in vitro, TgPRF is unable to complement depletion of profilin in yeast (25). Additionally, opposite of conventional profilins, TgPRF was shown to inhibit nucleotide exchange by rabbit actin (26). Finally, profilin in the apicomplexan parasite P. falciparum is essential in the blood-stage of the parasite life cycle (27) and functionally complements the depletion of PRF in T. gondii (25).
Formins contain a formin-homology 2 (FH2) domain that assembles into a homodimer and binds barbed ends of actin filaments (28). N-terminal of the FH2 domain is the formin-homology 1 (FH1) domain that typically contains a number of polyproline stretches involved in recruitment of profilin-actin (28). Formins have been shown to enhance actin polymerization by moving processively along the filament, allowing addition of actin monomers that are donated by profilin (29). In the absence of homologs of other actin nucleating proteins in apicomplexans, such as Arp2/3, formins have become the likely candidate to nucleate parasite actin filament formation. There are three formins in T. gondii and two of them, TgFRM1 and TgFRM2, have been shown to act as nucleators of rabbit actin in vitro and contribute to parasite motility (30). Recently TgFRM3 was shown to bind TgACTI and nucleate rabbit actin assembly in vitro, yet it is not required for parasite survival in culture (31). P. falciparum also encodes three proteins with similarity to formins, and PfFormin1 and PfFormin2 have been shown to act as barbed end nucleators of chicken actin in vitro (32).
The overall sequences of TgACTI as well as those for TgPRF, TgFRM1 and TgFRM2 diverge from their counterparts in higher eukaryotes (25, 30). Hence, these regulatory actin-binding proteins may differ in their interaction with TgACTI compared to what has been observed with heterologous actins in previous studies. Therefore, we tested the functions of TgPRF along with TgFRM1 and TgFRM2 in regulating TgACTI polymerization. We observed that while T. gondii formins are capable of enhancing TgACTI polymerization in vitro, TgPRF acts primarily to sequester actin monomers.
MATERIALS AND METHODS
Recombinant profilin
The profilin gene (TgPRF) (GenBank AAX33672.1) was amplified from T. gondii RH strain cDNA using primers 5′-GCGCGCCCATATGTCCGACTGGGACCCTGTTGT-3′ (forward) and 5′-CGCGGATCCTTAGTACCCAGACTGGTGAA-3′ (reverse) and cloned into the pET16b vector (Novagen, Madison, WI) using the NdeI and BamHI sites to incorporate a His10 tag at the N-terminus. The resulting plasmid was transfected into BL21 E. coli (Novagen) for protein expression. Recombinant protein was purified using ProBond Nickel beads (Invitrogen, Carlsbad, CA). Alternatively, GST-TgPRF described previously (25), was purified on Glutathione sepharose 4 Fast flow (Amersham) using a 10/20 Tricorn column (Amersham). GST was cleaved using the Precision protease (Amersham) and TgPRF was purified and assessed by SDS-PAGE and Coomassie Blue staining (Figure 1A). Proteins were stored at −80°C until use.
Figure 1.
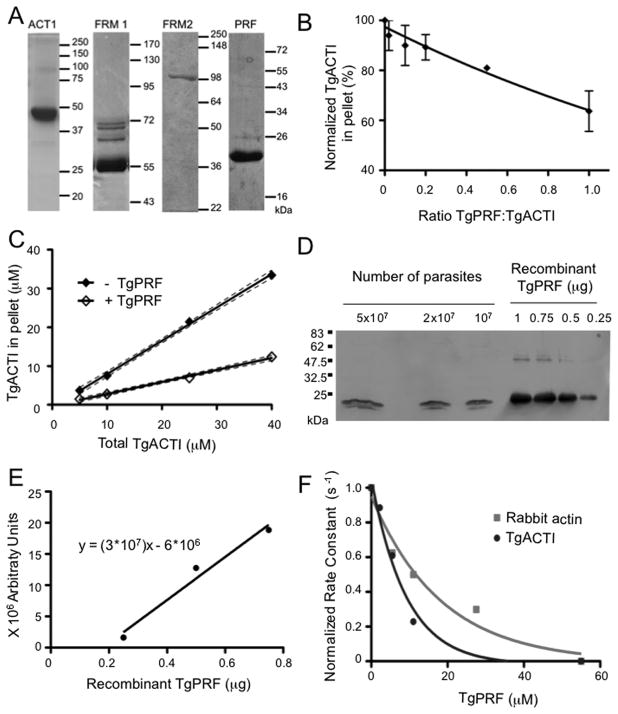
TgPRF acts to sequester TgACTI and prevent polymerization. (A) Purified recombinant T. gondii proteins were assessed by SDS-PAGE, Coomassie blue stained gels. The fainter bands in FRM1 lane reflect contaminants from E. coli that bind to nickel resin. Samples include actin (ACTI), TgFRM1-FH1-FH2 (FRM1), TgFRM2-FH1-FH2 (FRM2) and profilin (PRF). The expected mass of FRM2 is 82 kDa, although it migrates at 100 kDa. Mass ladders in kilodaltons (kDa). (B) Sedimentation analysis of TgACTI polymerized with varying concentrations of TgPRF. TgACTI (25 μM) was incubated with TgPRF (0.1 – 1 molar ratio) for 1.5 h. Reactions were centrifuged at 350,000g for 1 h at room temperature, separated by SDS-PAGE, stained with SYPRO Ruby and visualized using a phosphorimager. Values were normalized to the amount of pelleted TgACTI in the absence of TgPRF. Means ± S.D. from three or more separate experiments are shown. Curve was fitted using a second order polynomial. (C) Steady state sedimentation analysis of varying concentrations of TgACTI ± equimolar TgPRF. Reactions were incubated for ~20 h and then centrifuged at 350,000g for 1 h at room temperature, separated by SDS-PAGE stained with SYPRO Ruby and visualized using a phosphorimager. 95% confidence interval of the linear best-fit line from two independent experiments is shown. (D) Western blot to compare amount of TgPRF in parasite lysates to known concentrations of recombinant TgPRF. Parasite lysates and recombinant protein were resolved on a 12% SDS-PAGE gel, transferred to nitrocellulose and probed with rabbit αTgPRF. (E) Bands from Western in (D) were quantified with a phosphorimager and used to calculate a standard curve from known concentrations of TgPRF (using points for 0.25, 0.5 and 0.75 μM, 1μM was excluded due to saturation of signal) based on a linear regression fit (r2=0.9718). (F) The effect of TgPRF on nucleotide exchange by ATP-rabbit actin (gray squares) or ATP-TgACTI (black circles). Nucleotide exchange was monitored by the loss of fluorescence from ε-ATP labeled actin (1 μM) over time following addition of 1.25 mM unlabeled ATP in the absence or presence of different concentrations of TgPRF (2 μM – 55 μM). The initial rates of fluorescence loss were used to calculate rate constants and are normalized and plotted verses TgPRF concentration to obtain a curve of one phase decay. Representative experiments are shown. Recombinant His-tagged TgPRF was used for experiments shown.
Actin expression and purification
N-terminally His-tagged TgACTI was purified on NiNTA agarose (Invitrogen) from a baculovirus expression system using protocols described previously (13). Purified protein was dialyzed overnight in G buffer containing 0.5 mM DTT with 100 mM sucrose and clarified by centrifugation at 100,000g, 4°C, for 30 min to remove aggregates. Purified TgACTI was resolved on 12% SDS-PAGE gels followed by SYPRO Ruby (Molecular Probes, Eugene, OR) staining (33), visualized using a FLA-5000 phosphorimager (Fuji Film Medical Systems, Stamford, CT), and quantified using Image Gauge v4.23. TgACTI was stored at 4°C and used within 2–3 d. Purity was assessed by SDS-PAGE and Coomassie Blue staining (Figure 1A).
Purification of recombinant formins
The nomenclature for formin domains studied here follows that previously defined (30). In brief, FRM1-FH1-FH2 (amino acid positions 4582–5051) and FRM2-FH1-FH2 (amino acid positions 3317–4043) correspond to the FH2 domains of formin 1 and formin 2, together with N-terminal extensions that constitute putative FH1 domains (30). His-tagged constructs expressing FRM1-FH1-FH2 (56 kDa) and FRM2-FH1-FH2 (82 kDa) were purified on Qiagen Ni-NTA superflow resin under native conditions, as previously described (30). Proteins were stored at −80°C until use in biochemical assays. Purity was assessed by SDS-PAGE and Coomassie Blue staining (Figure 1A).
Quantitation of intracellular profilin concentration in T. gondii
To estimate the intracellular concentration of TgPRF, freshly isolated parasites were lysed in actin stabilization buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 125 mM KCl) containing 1% Triton-X-100 and 1X protease inhibitor cocktail (1 μg/ml E-64, 10 μg/ml AEBSF, 10 μg/ml TLCK and 1 μg/ml leupeptin) for 30 min followed by centrifugation at 21,000g for 10 min to remove insoluble material. Cell lysate was resuspended in 1X Laemmli sample buffer and resolved on 12% SDS-PAGE gels along with a range of recombinant TgPRF standards (0.25–1 μg). Gels were transferred to nitrocellulose, western blotted with anti-TgPRF antibody, visualized using a FLA-5000 phosphorimager (Fuji Film Medical Systems), and quantified using Image Gauge v4.23. The volume of a single T. gondii cell has previously been estimated by biochemical means (34), and this was used to determine the approximate intracellular protein concentration.
90° light scattering
Purified recombinant actin was centrifuged at 100,000g, 4°C, for 30 min using a TL100 rotor and a Beckman Optima TL ultracentrifuge (Beckman Coulter) to remove aggregates. TgACTI was diluted to 5 μM in G buffer and preincubated with 1 mM EGTA and 50 μM MgCl2 for 10 min to replace bound Ca2+ with Mg2+, as described previously (35). Samples were placed in a 100 μl cuvette (Submicro Quartz Fluorometer cell, Starna Cells, Atascadero, CA) and light scattering was monitored with the PTI Quantmaster spectrofluorometer (Photon Technology International, Santa Clara, CA): excitation 310 nm (1 nm bandpass), emission 310 nm (1 nm bandpass) at room temperature. Once a steady reading was obtained, the acquisition was paused and 1/10th volume of 10X F buffer (500 mM KCl, 20mM MgCl2, 10 mM ATP) was added to induce polymerization. Purified recombinant FRM1-FH1-FH2, FRM2-FH1-FH2, or TgPRF were added along with F buffer to induce polymerization. The acquisition was restarted and counts collected until the readings reached a plateau. Light scattering curves were normalized by subtracting the values of TgFRM1-FH1FH2, TgFRM2-FH1FH2 or TgPRF added to G buffer in the absence of TgACTI.
Sedimentation Analysis
Samples were prepared for light scattering analysis as described above and following reading for 1.5 h, samples were centrifuged at 100,000g or 350,000g for 1 h at room temperature. For steady state experiments, samples were incubated for 20 hrs (determined by testing various time intervals) prior to centrifugation at 350,000g. Protein in the supernatant was precipitated in 2 volumes acetone overnight, centrifuged at 21,000g for 30 min, washed with 70% ethanol followed by centrifugation at 21,000g for 10 min. All pellets were resuspended in 1X Laemmli sample buffer. Proteins were resolved on a 12% SDS-PAGE gels, stained with SYPRO Ruby (Molecular Probes), visualized using a FLA-5000 phosphorimager (Fuji Film Medical System), and quantified using Image Gauge v4.23.
Fluorescence Microscopy
Purified recombinant TgACTI was clarified as described above and various concentrations were diluted to final molarity in F buffer. To analyze the affects of formins, 25 μM TgACTI was incubated with 500 nM TgFRM1-FH1-FH2 or 60 nM TgFRM2-FH1-FH2. Alexa-488 phalloidin (0.33 μM; Molecular Probes) was added to each sample to allow visualization of actin filaments. Polymerization was allowed to proceed for 1 h followed by examination using a Zeiss Axioskop (Carl Zeiss, Thornwood, NY) microscope equipped with a 63X Plan-NeoFluar oil immersion lens (1.30 NA). Images were collected using a Zeiss Axiocam with Axiovision v3.1. All Images were processed by linear adjustment in the same manner using Adobe Photoshop v8.0.
Nucleotide Exchange
Nucleotide exchange by actin was monitored as previously described (18). Briefly, 40–60 μM of purified recombinant TgACTI was clarified as described above and treated with 10% volume of 50% slurry 1×8 Cl (200–400 mesh) Dowex beads (Sigma Aldrich, St. Louis, MO) to remove ATP. Actin was then incubated with 500 μM 1,N6-ethenoadenosine 5′triphosphate (ε-ATP) (Molecular Probes) for 1 h at 4°C. Dowex beads were added to remove unbound ε-ATP followed by addition of 20μM ε-ATP to stabilize the actin. Labeled ε-ATP TgACTI was diluted to 1 μM, preincubated with varying concentrations of TgPRF for 10 min, and Mg2+ was exchanged for Ca2+ by incubation with 1 mM EGTA and 50 μM MgCl2 for 5 min. The sample was placed in a submicrocuvette and analyzed using a PTI Quantmaster spectrofluorometer (Photon Technology International) at 25°C using 360 nm excitation and 410 nm emission. Following stabilization of the signal, unlabeled ATP (1.25 mM) was added to compete with the ε-ATP and measurements were continued. The rates of ε-ATP exchange were calculated by plotting the initial change in fluorescence vs. time. The affinity of TgPRF for TgACTI was estimated by comparing rate constants for ATP exchange vs. TgPRF concentration using nonlinear regression analysis based on single-phase decay in Prism (GraphPad, San Diego, CA).
Electron microscopy
Purified recombinant TgACTI was clarified as described above and diluted to 25 μM and polymerization initiated by addition of F buffer with or without 500 nM TgFRM1-FH1-FH2 or 60 nM TgFRM2-FH1-FH2 and incubated for 1 h at room temperature. Quick–freeze, deep-etch electron microscopy (EM) was performed as described previously (36) with minor modifications. Protein samples were deposited onto an acid cleaned, air dried 3 mm2 glass coverslips for 1 min, rinsed briefly in F buffer and transferred to 2% glutaraldehyde in F buffer for 15 min at room temperature. Prior to freezing, glass coverslips were rinsed with dH2O and then frozen by forceful impact against a pure copper block, cooled to 4K with liquid helium. Frozen samples were mounted in a Balzers 400 vacuum evaporator, etched for 20 min at −80°C and rotary replicated with ~ 3 nm platinum deposited from a 15° angle above the horizontal, followed by an immediate ~10 nm stabilization film of pure carbon deposited from a 85° angle. Replicas were floated onto a dish of concentrated hydrofluoric acid, rinsed in dH20, mounted on formvar coated copper grids, and photographed with on JEOL 1400 microscope with attached AMT digital camera.
Statistical Analysis
Statistics were calculated in Excel or Prism (GraphPad) using an unpaired, two-tailed Student’s t-test for normally distributed data with equal variances. Significant differences were defined as P ≤ 0.05.
RESULTS
TgPRF acts to sequester T. gondii actin
Conventional profilins bind actin monomers and can sequester them to inhibit polymerization of actin filaments (21). To determine how TgPRF affects TgACTI assembly, sedimentation was used to monitor the extent of actin polymerization. We have previously demonstrated that TgACTI filaments do not sediment efficiently at 100,000g (7), and hence we performed sedimentation at 350,000g, which generally results in pelleting of > 90% of actin polymerized in F buffer (i.e. using 25 μM) (13). A dose-dependent decrease in sedimentation was observed with increasing concentrations of TgPRF up to a ratio of 1:1, which reduced the amount of actin in the pellet by ~ 40% (Figure 1B). This assay was conducted following 1.5 h incubation and 1 h centrifugation, hence it is possible that greater effects would be seen with longer incubation times. Therefore, to further examine the effects of TgPRF on TgACTI polymerization over a longer time frame, we examined sedimentation at steady state (i.e. after 20 h of polymerization as described previously (18)) with increasing concentrations of actin and a fixed ratio of TgPRF to actin of 1:1. As expected, increasing amounts of TgACTI in the reaction lead to significantly greater polymerization, as detected by the amount of actin in the pellet following sedimentation at 350,000g (Figure 1C). Addition of TgPRF at 1:1 molar ratio significantly impaired polymerization of TgACTI, even at high concentrations (Figure 1C). Collectively, these results indicate that TgPRF sequesters TgACTI monomers and prevents polymerization.
Previous studies have reported that TgPRF is able to bind to TgACTI in a pull-down assay (25), although the affinity of this interaction was not established. Prior estimates have indicated that TgACTI is present in cells at ~ 40 μM (19). To provide a comparison for the level of TgPRF in T. gondii cells, we performed quantitative Western blotting of parasite cell lysates and compared them to known amounts of recombinant TgPRF (Figure 1D,E). Quantitative comparison of the signals obtained by Western blotting combined with estimates of the volume of the cell (34), revealed that the concentration of TgPRF is ~ 38 μM, or very comparable to that of TgACTI. Hence, evaluation of actin dynamics in the presence of TgPRF at equimolar ratios may approximate conditions found in the cell.
TgPRF weakly inhibits nucleotide exchange by actin
Profilins conventionally enhance actin polymerization by converting ADP-actin monomers to ATP-actin, preparing it for addition to the growing barbed end (22). It was recently demonstrated that when combined with heterologous rabbit actin, TgPRF acts unconventionally and inhibits nucleotide exchange (26). We observed a similar inhibition, based on decreased rate of ε-ATP exchange from rabbit actin in the presence of TgPRF (Figure 1F). TgPRF also showed a modest inhibition of nucleotide exchange from ε-ATP-TgACTI (Figure 1F). The inhibition of nucleotide exchange by TgPRF had an estimated observed affinity (Kobs) of 6.3 μM for TgACTI and 12.7 μM for rabbit actin as calculated using a single-order decay curve fit of the rates of exchange.
TgFormins Enhance TgACTI Polymerization
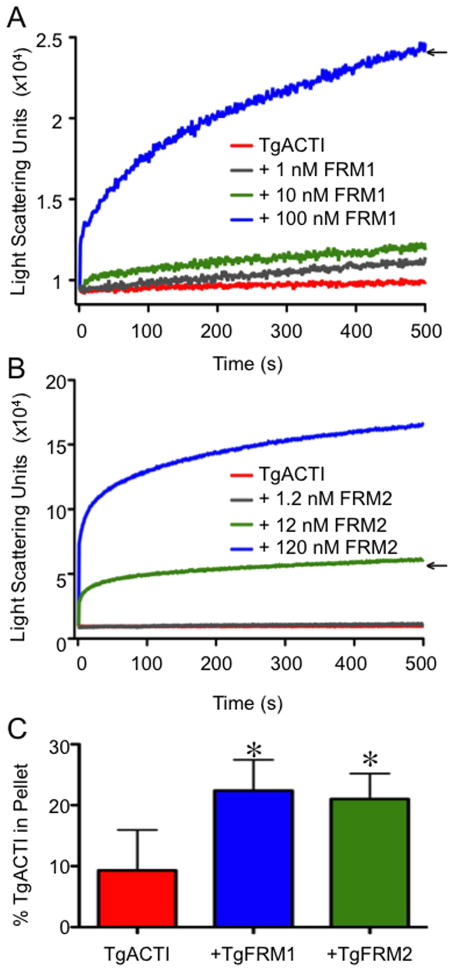
Because formins in T. gondii are quite large (495–555 kDa) we choose to examine the function of FH1-FH2 domains for FRM1 and FRM2, which have previously been shown to be active against of rabbit actin in vitro (30). To evaluate their influence on parasite actin, recombinant T. gondii TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 domains were added to TgACTI and polymerization was monitored by 90° light scattering. Consistent with results from previous studies (13), polymerization of 5 μM TgACTI alone was very modest (Figure 2A,B). Upon addition of 1 nM or 10 nM TgFRM1-FH1-FH2, light scattering increased slightly; however, a substantial increase in light scattering was observed with addition of 100 nM TgFRM1-FH1-FH2 to TgACTI (Figure 2A). A similar range of concentrations was used to test the effects of TgFRM2 domains, and robust light scattering occurred with addition of only 12 nM TgFRM2-FH1-FH2 and an even greater increase was observed at 120 nM (Figure 2B). Concentrations of 100 nM TgFRM1-FH1-FH2 (1:50 molar ratio with TgACTI) and 12 nM TgFRM2-FH1-FH2 (1: ~420 molar ratio with TgACTI) were chosen for use in subsequent experiments due to their ability to similarly increase TgACTI polymerization, as reflected by increased light scattering (Figure 2A,B). These enhanced signals are unlikely to be due to protein aggregation, since incubation of similar concentrations of formin domains alone led to negligible light scattering (data are adjusted for these background levels in Figures 2 and 4). Upon completion of light scattering, the samples were centrifuged at 100,000g for 1 h and the amount of TgACTI in the pellet and supernatant were calculated by imaging SYPRO Ruby stained samples resolved by SDS-PAGE. Consistent with previous reports, these centrifugation conditions did not efficiently pellet parasite actin in F buffer alone (i.e. only ~ 10% was seen in the pellet) due to the small size of filaments (7, 13). However, a significant increase in pelletable actin was observed in the TgACTI samples incubated with TgFRM1-FH1-FH2 (1:50 molar ratio) or TgFRM2-FH1-FH2 (1: ~420 molar ratio) (Figure 2C). These results demonstrate that these domains of TgFRM1 and TgFRM2 substantially enhance TgACTI polymerization even in the absence of profilin. However, the fact that light scattering gave a much higher fold change in the signal when compared to sedimentation, suggests that formins may have additional activities, such as bundling of filaments, which is expected to greatly increase the signal in the light scattering assay.
Figure 2.
TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 enhance polymerization of TgACTI. (A) Comparison of polymerization kinetics of TgACTI in the presence and absence of TgFRM1-FH1-FH2. Polymerization of 5 μM actin in F buffer alone (red) or with the addition of 1 nM (gray), 10 nM (green) or 100 nM (blue) TgFRM1-FH1-FH2 monitored by light scattering. Representative of 2 experiments. (B) Comparison of polymerization kinetics of TgACTI in the presence and absence of TgFRM2-FH1-FH2. Polymerization of 5 μM actin in F buffer alone (red) or with the addition of 1.2 nM (gray), 12 nM (green) or 120 nM (blue) TgFRM2-FH1-FH2 monitored by light scattering. Representative of 2 experiments. Concentrations chosen for subsequent experiments are denoted with arrows in A and B. Data in A and B were adjusted for levels of light scattering observed with FRM domains alone (although these changes were negligible). For comparison please note that the Y axes in A and B are different scales. (C) Upon completion of light scattering, samples of TgACTI alone or in the presence of 100 nM TgFRM1-FH1-FH2 or 12 nM TgFRM2-FH1-FH2 were centrifuged for 1 h at 100,000g to pellet actin filaments. Protein from the pellet or supernatants of all samples was resolved on a 12% SDS-PAGE gel, stained with SYPRO Ruby and quantified by phosphorimager analysis. The average percentage of protein in the pellet fraction from three replicate experiments is shown. Percent in the TgACTI pellet alone was compared to +TgFRM1-FH1-FH2 or +TgFRM2-FH1-FH2 and * denotes significance using Students two tailed t-test. P < 0.05.
Figure 4.
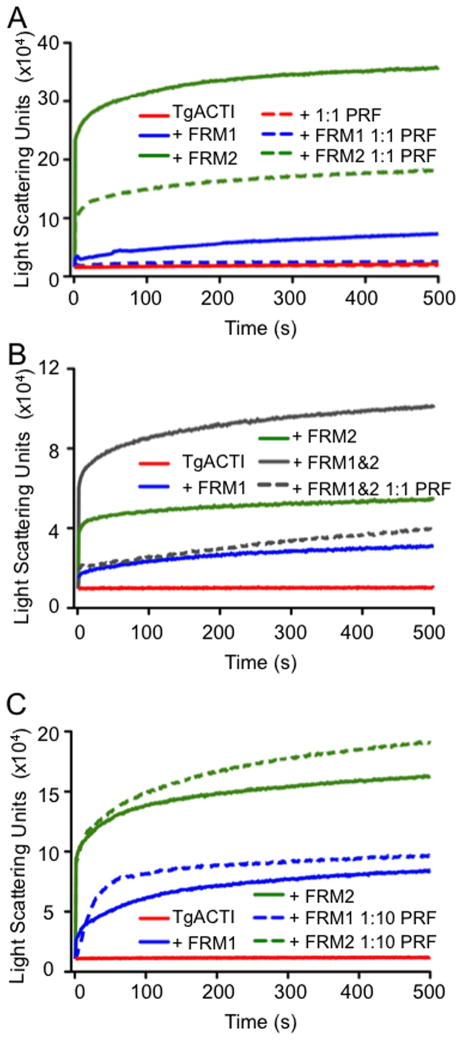
Effect of TgPRF on formin-domain mediated TgACTI polymerization. (A) Comparison of polymerization kinetics of TgACTI in the presence or absence of formin domains with or without addition of equimolar TgPRF. Polymerization of TgACTI (5 μM) alone in F buffer alone (solid red), or TgACTI combined with 5 μM TgPRF (dashed red), 100 nM TgFRM1-FH1-FH2 (solid blue), 100 nM TgFRM1-FH1-FH2 and 5 μM TgPRF (dashed blue), 12 nM TgFRM2-FH1-FH2 (solid green), or 12 nM TgFRM2-FH1-FH2 and 5 μM TgPRF (dashed green) as monitored by light scattering. (B) Comparison of polymerization kinetics of TgACTI in the presence or absence of TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 in combination with and without addition of TgPRF. Polymerization of TgACTI (5 μM) in F buffer alone (solid red), or with 100 nM TgFRM1-FH1-FH2 (solid blue), 12 nM TgFRM2-FH1-FH2 (solid green), 100 nM TgFRM1-FH1-FH2 and 12 nM TgFRM2-FH1-FH2 (solid gray), or 100 nM TgFRM1-FH1-FH2, 12 nM TgFRM2-FH1-FH2 and 5 μM TgPRF (dashed gray) as monitored by light scattering. For comparison please note that the Y axes in A and B are different scales. (C) Comparison of polymerization kinetics of TgACTI in the presence or absence of formin with and without addition of 1:10 concentration of TgPRF. Polymerization of TgACTI (5 μM) in F buffer alone (solid red), or with 100 nM TgFRM1-FH1-FH2 (solid blue), 100 nM TgFRM1-FH1-FH2 and 0.5 μM TgPRF (dashed blue), 12 nM TgFRM2-FH1-FH2 (solid green), or with 12 nM TgFRM2-FH1-FH2 and 0.5 μM TgPRF (dashed green) as monitored by light scattering. Representative data from 2 or 3 experiments are shown. Purified recombinant TgPRF (after GST cleavage) was used for experiments shown.
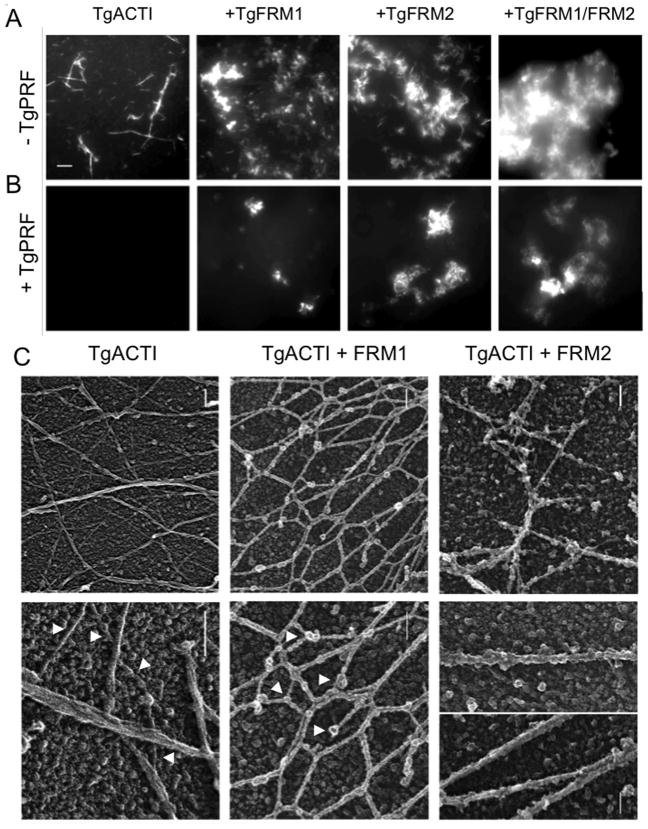
To further examine the extent of polymerization, we visualized TgACTI filaments in the absence and presence of TgFRM1 and TgFRM2 domains using fluorescence microscopy. TgACTI (25 μM) was incubated in F buffer alone, with TgFRM1-FH1-FH2 (1:50 molar ratio), or with TgFRM2-FH1-FH2 (1: ~420 molar ratio), and 0.33 μM Alexa-488 phalloidin was added to all samples to allow for actin filament visualization. We chose a relatively high concentration of actin for this assay because at lower concentrations, TgACTI does not form filaments as detected by phalloidin (13). On its own, TgACTI polymerized into short filament clusters that also formed bundles of longer filaments (Figure 3A). Based on their size and appearance, the more prominently stained structures are likely not single filaments but rather bundles of filaments. In the presence of TgFRM1-FH1-FH2 or TgFRM2-FH1-FH2, there were many more clusters of actin filament bundles, although the length of individual filaments was shorter than with TgACTI alone (Figure 3A). Addition of TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 together in the actin polymerization reactions resulted in a dense cloud (Figure 3A). Collectively, these findings demonstrate that TgACTI is strongly induced to form short filament bundles in the presence of domains from TgFRM1 and TgFRM2.
Figure 3.
Influence of TgFRM1-FH1-FH2, TgFRM2-FH1-FH2, and TgPRF on formation of TgACTI filaments revealed by microscopy. (A) In vitro polymerization of TgACTI with formin domains. TgACTI (25 μM) was incubated in F buffer alone, with 500 nM TgFRM1-FH1-FH2, 60 nM TgFRM2-FH1-FH2, or 500 nM TgFRM1-FH1-FH2 combined with 60 nM TgFRM2-FH1-FH2 for 1 h then visualized by fluorescence microscopy using 0.33 μM Alexa 488-phalloidin. Scale bar, 5 μm. (B) In vitro polymerization of TgACTI with formin domains and profilin. TgACTI (25 μM) was incubated in F buffer alone, with 500 nM TgFRM1-FH1-FH2, 60 nM TgFRM2-FH1-FH2, or 500 nM TgFRM1-FH1-FH2 and 60 nM TgFRM2-FH1-FH2 with 25 μM TgPRF for 1 h then visualized by fluorescence microscopy using 0.33 μM Alexa 488-phalloidin. A representative of 2 similar experiments is shown. Same scale as A. (C) Electron micrographs of replicas of TgACTI polymerized alone or in the presence of TgFRM1-FH1-FH2 or TgFRM2-FH1-FH2. TgACTI (25 μM) was polymerized by addition of F buffer and 500 nM TgFRM1-FH1-FH2 (TgACTI + FRM1) or 60 nM TgFRM2-FH1-FH2 (TgACTI + FRM2) for 1 h then fixed and frozen followed by quick-freeze, deep-etch and platinum replica formation. TgACTI alone (top left panel) formed long straight filaments that often bundled into clusters. Enlarged view (bottom left panel) shows singlet filaments (arrowheads) as well as bundles. In the presence of FRM1 (top central panel), filaments formed interconnected honeycomb networks that were decorated by globular proteins. Enlargement (bottom central panel) of the filaments revealed they were short, formed interconnected networks, and were heavily decorated along their sides as well as containing globular clusters at nodes or intervals along the filaments (arrowheads). In the presence of FRM2 (top right panel), filaments were typically straight and collected into bundles, which on higher magnification show extensive decoration (bottom right panel). Scale bars = 100 nm.
To further visualize the influence of formins on TgACTI filaments, we performed quick-freeze deep-etch electron microscopy to examine filament ultrastructure. Polymerization of TgACTI at high concentrations led to formation of single filaments or bundles of tightly packed filaments (Figure 3C, top left panel). In contrast to previous reports that TgACTI forms unstable filaments (7, 13), long stable filaments of TgACTI were observed, in part due to the use of much higher protein concentrations and the use of rapid freezing techniques. Examination of the filaments at higher magnification revealed the characteristic striations and helical pattern typical of actin filaments (Figure 3C bottom left panel), although these were much more subtle than the patterns seen in conventional actins such as yeast. Addition of TgFRM1-FH1-FH2 to the reaction caused a dramatic shortening of the filaments and resulted in interconnected networks (Figure 3C, top central panel). The actin filaments themselves appear thickened and the striations were less apparent (Figure 3C, bottom central panel), suggesting TgFRM1 domains may coat the filament sides. In addition, large globular proteins occurred at nodes where the filaments connected or at the ends of filaments (Figure 3C, bottom central panel). Formins typically function as dimers (24) and based on the subunit size of TgFRM1-FH1-FH2 (56 kDa) each of these dimers would be expected to be ~112 kDa. The average size of these globular domains is substantially larger than the width of the filament, which is made up of alternating protofilaments of 42 kDa monomers, comprising 5 nm globular subunits (Figure 3C), suggesting that formins may oligomerize in the presence of TgACTI filaments. Addition of TgFRM2-FH1-FH2 to the polymerization reaction also resulted in extensive filament bundling; however, in this case the filaments remained fairly long and straight (Figure 3C, top right panel). Comparison of the diameter of TgACTI filaments in enlarged images revealed that they consisted of bundles of 2 or more filaments that appeared heavily decorated by globular protein domains (Figure 3C, bottom right panel). Although we have not performed specific immunolabeling, these globular proteins are absent from the TgACTI alone samples, leading to the conclusion that they likely represent the formin domains bound to actin filaments.
TgPRF Inhibits Formin-mediated TgACTI Polymerization
To determine what effect TgPRF plays on formin-mediated enhancement of TgACTI polymerization, we used fluorescence microscopy to observe TgACTI filament formation in the presence of TgPRF. Addition of TgPRF to TgACTI alone resulted in complete loss of filaments observed by microscopy (Figure 3B). Addition of TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 to the TgACTI - TgPRF reaction restored filaments slightly, but polymerization was not as robust as TgACTI alone or that seen in the presence of either formin without profilin (Figure 3B). This decrease in polymerization was surprising since yeast profilin Bni1 has been shown to interact with formin to enhance actin polymerization (23), and similar activity has also been described for a variety of other formins (24). We therefore added profilin and formin domains to TgACTI and examined polymerization by light scattering. Addition of TgPRF inhibited the marked increase in light scattering seen with addition of either TgFRM1-FH1-FH2 or TgFRM2-FH1-FH2 to TgACTI (Figure 4A). Addition of both TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 simultaneously with TgACTI resulted in an even larger increase in light scattering than with either formin alone (Figure 4B). However, addition of TgPRF prevented this increase (Figure 4B). Therefore, at equimolar concentration to TgACTI, TgPRF does not contribute to but rather inhibits formin-induced polymerization of TgACTI. To determine if this inhibition was concentration-dependent, TgPRF was added at a 1:10 ratio to TgACTI in the presence of either TgFRM1-FH1-FH2 or TgFRM2-FH1-FH2. At this lower ratio of TgPRF, there was a slight enhancement in light scattering in the presence of TgFRM2-FH1-FH2 and minimal change in the presence of TgFRM1-FH1-FH2 (Figure 4C), results seen in repeated experiments (data not shown). Overall, these results suggest that TgPRF serves largely to inhibit the actin polymerizing activities of formin FH1-FH2 domains in T. gondii.
DISCUSSION
Previous studies have shown that while apicomplexan motility relies on polymerized actin, filaments are only transiently detected in vivo and instead the majority of actin remains unpolymerized at steady state (6, 11). Apicomplexan parasites contain few actin-binding proteins, but do express proteins such as formins, profilin, ADF, and capping protein (16). Here, we have analyzed the effects of two T. gondii formins and profilin on polymerization of TgACTI in vitro using highly purified recombinant proteins. TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 enhanced polymerization by inducing filaments that were otherwise inefficiently assembled by TgACTI alone. In contrast, TgPRF was shown to function primarily in sequestration of TgACTI, and surprisingly TgPRF did not substantially enhance formin-mediated actin assembly. In combination with the previously observed instability and rapid turnover of parasite actin filaments (13), the sequestering actions of TgPRF shown here, and TgADF reported previously (18), are expected to maintain a high G-actin pool in the parasite. In contrast, formin proteins likely play the major role in inducing actin filament formation in vivo, hence supporting gliding motility and cell invasion. Since disruption of either of these functions (i.e. PRF and ADF sequestering TgACTI monomers or FRMs inducing filament polymerization) impairs gliding motility, it is evident that a balance of regulatory activities is key to maintaining actin dynamics in vivo.
Previous studies have used in vitro biochemical assays based on assembly of pyrene-labeled actin to demonstrate that TgPRF inhibits pointed end polymerization but contributes to barbed end assembly of rabbit actin in vitro (25). In contrast, we observed a dose-dependent inhibition of TgACTI polymerization by TgPRF as monitored by light scattering, sedimentation, and fluorescence microscopy. Importantly, in the assays conducted here, TgACTI filaments were uncapped and the resulting inhibition reflects net assembly-disassembly at both ends of the filament. We have not examined gelsolin-capped filaments here since this protein is absent from the T. gondii assemblage of actin-binding proteins (17), and heterologous gelsolin has not been shown to interact with this divergent actin. Similarly, we have not compared elongation rates using spectrin-actin seeds since this protein is also not found in T. gondii and mammalian spectrin-actin seeds do not support TgACTI polymerization (7). Nonetheless, the dramatically different effects of TgPRF on parasite actin (inhibition via sequestration) vs. mammalian (supports barbed end growth) indicate that studying interactions between homologous protein partners is paramount for correctly determining the functional attributes of actin-binding proteins.
In contrast to the conventional function of profilins in enhancing ATP exchange on actin, TgPRF has been reported to inhibit nucleotide exchange on rabbit actin (26). We observed a similar inhibition of ATP exchange for both rabbit actin and TgACTI, although this effect was somewhat modest. Although the inhibition of nucleotide exchange diverges from the function of yeast and mammalian systems, the profilins from Arabidopsis (37) and Chlamydomonas (38) also either inhibit or have no effect on actin nucleotide exchange. The Kobs for inhibition of ATP-TgACTI exchange by TgPRF was much higher (~ 6 μM) than that previously reported for TgADF (0.8 μM), indicating that of the two, ADF likely plays a more prominent role in inhibiting nucleotide exchange by TgACTI.
Our data based on in vitro studies suggest that TgPRF acts primarily to sequester G-actin rather than promote filament formation. Previous studies have shown that TgPRF is essential for motility, host cell invasion, and egress in vivo (25). Although TgPRF is likely required for actin regulation, its precise role in controlling motility in vivo has not been defined. One interpretation of our in vitro data is that without TgPRF to aid in keeping the actin monomer pool high, filaments may form more readily than normal, hence disrupting motility. Similarly, TgADF functions primarily to sequester TgACTI (18). Depletion of TgADF in a conditional knockout leads to formation of long actin filaments that extend throughout the parasite, resulting in aberrant motility and impaired host cell invasion and egress (19). The effect of depleting TgADF would appear to be more significant in promoting aberrant actin filament formation since the conditional knockout of TgPRF was not reported to develop stable actin filaments (25). It was previously estimated that the intracellular concentration of TgACTI is around 40 μM (19). Similar concentrations have been calculated for TgADF (35 μM) (18) and TgPRF (38 μM) (this study). Consequently, sequestration by both TgPRF and TgADF is expected to complex nearly all of TgACTI in a globular form, ensuring there is only a small free monomer pool for polymerization. Other recent studies have highlighted that key structural differences in actin influence filament stability and contribute to the lack of stable actin filaments in the parasite (13).
In contrast to other systems where profilin enhances formin-mediated actin polymerization (20), TgPRF decreased polymerization caused by FH1-FH2 domains of FRM1 and FRM2 in T. gondii. This effect was most pronounced at equimolar concentrations between TgPRF and TgACTI, a ratio chosen because it is similar to in vivo concentrations. However, even at 1:10 ratio of profilin to actin, only modest enhancement of formin-activated polymerization was seen. This was an unexpected finding as conventional profilins have been reported to interact synergistically with formins to enhance actin polymerization (23). It is possible that the lack of polymerization enhancement by the formin domain constructs used here could be due to the degenerate sequences of the putative FH1 domains in T. gondii, which contain fewer prolines than most conserved FH1 domains (30). Consistent with this, biochemical analysis has previously revealed that TgPRF has a very low binding affinity for peptides of the TgFRM2 putative FH1 domain (26), and TgPRF failed to co-precipitate with T. gondii formins (30). Importantly, our inability to observe a role for PRF in enhancing FRM-mediated actin polymerization in vitro does not preclude such a role in vivo, since the constructs used here contain only part of the full-length proteins, and additional regulatory proteins may modulate activity in vivo. Additionally, since apicomplexan parasites have multiple developmental stages, it is possible that differences in expression or regulation of formins, and other actin-binding proteins, result in differences in actin dynamics between motile and non-motile stages.
Formins typically aid in nucleation of actin filaments through dimerization and interaction with the actin filament via the FH2 domain and interaction with profilin-actin dimers via the FH1 domain (28). Previous work has shown that the T. gondii formins TgFRM1 and TgFRM2 increase polymerization of heterologous actin (30). In the present study, we demonstrate that FH1-FH2 domain constructs from these formins act to enhance polymerization of TgACTI in vitro. The increase in polymerization observed upon addition of TgFRM1-FH1-FH2 or TgFRM2-FH1-FH2 is the first observation of robust TgACTI polymerization without a requirement for chemical stabilizing agents (13). Apicomplexan parasites contain a limited subset of actin binding proteins and for example lack Arp2/3 (17). Hence, formins are likely to be the key mediators of actin polymerization in vivo. Consistent with this, disruption of TgFRM1 results in impaired gliding, cell invasion, and egress, and dominant negative expression of mutants of TgFRM1 and TgFRM2 also disrupt similar processes (30).
When the effects of TgFRM1-FH1-FH2 and TgFRM2-FH1-FH2 on actin were studied using rabbit actin, TgFRM1 proved to be a more potent nucleator than TgFRM2 (30). Similar results were also observed when the formins from P. falciparum were tested for their impact on chicken actin polymerization (32). Conversely, in the current studies, TgFRM2-FH1-FH2 appears about 10 times more potent than TgFRM1-FH1-FH2 in increasing light scattering of TgACTI. Addition of forming domains to TgACTI induced greater polymerization as seen by fluorescence microscopy, sedimentation and light scattering, although the greatly enhanced signals in the latter assay suggested a contribution of cross-linking. Consistent with this, TgFRM1 contributes to actin filament cross-linking as observed in the fluorescence actin filament assay and by quick-freeze EM. In particular, EM images revealed that FRM1 domains likely bind both to the end of the filament and along the sides. Cryo-EM images of TgACTI polymerized in the presence of TgFRM2 domains also revealed filaments that were bundled and heavily decorated along their sides. Bundling has been previously reported to occur with mammalian formins FRL1 and mDia2 (39) and with Formin1 from Arabidopsis thaliana (AFH1) (40, 41). AFH1 interacts with the barbed end of the actin filament in a non-processive manner and binds to the side of the filament following nucleation, facilitating formation of filament bundles (40). The Formin2 family of apicomplexan formins is most related to AFH1 (32) consistent with the possibility that apicomplexan formins also contribute to filament cross-linking. The similar functions of apicomplexan formins and profilin to those in plants is not surprising considering other unconventional actin dynamics shared between this phylum of parasites and plants. Like apicomplexans, plant cells maintain a highly unpolymerized population of G-actin (42, 43). Plant profilins are also maintained at a high concentration within the cell and occur at ~ 1:1 ratio with actin (44). Hence, the function and regulation of actin assembly in apicomplexans may be more closely related to that of plants, rather than yeast or mammals. Whether this is due to the ancient shared ancestry of protozoans and plants (45), or an example of convergent evolution is uncertain. These differences in actin dynamics might be exploited to specifically interfere with parasite motility and hence prevent infection of mammalian cells.
CONCLUSION
The transition between unpolymerized actin and filament formation is a highly regulated process in T. gondii, despite a minimal set of actin-binding proteins. Our studies reveal that two classes of actin-binding proteins within the parasite have opposing impacts on TgACTI polymerization. TgPRF primarily sequesters TgACTI monomers while TgFRM1 and TgFRM2 enhance polymerization. The presence of regulatory proteins with these opposite functions likely aids in maintaining the precise balance between monomeric actin and polymerized filaments within the parasite.
Acknowledgments
Funding
Partially supported by a predoctoral fellowship from the American Heart Association to KMS, grants from the NIH (AI073155) to LDS, and the Swiss National Foundation (FN3100A0-116722, DS and WD).
We thank John Cooper, David Sept, and John Heuser for helpful suggestions, Keliang Tang for technical assistance. Deep-etch electron microscopy was performed by Robyn Roth, Laboratory of Electron Microscopy Sciences, Department of Cell Biology, Washington University School of Medicine.
ABBREVIATIONS
- TgADF
T. gondii actin depolymerizing factor
- TgACTI
T. gondii actin
- FH1
formin homology 1
- FH2
formin homology 2
- TgFRM1
T. gondii formin 1
- TgFRM2
T. gondii formin 2
- TgPRF
T. gondii profilin
References
- 1.Sibley LD. How apicomplexan parasites move in and out of cells. Curr Opin Biotechnol. 2010;21:592–598. doi: 10.1016/j.copbio.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Starnes GL, Coincon M, Sygusch J, Sibley LD. Aldolase is essential for energy production and bridging adhesin-actin cytoskeletal interactions during parasite invasion of host cells. Cell Host Microbe. 2009;5:353–364. doi: 10.1016/j.chom.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 5.Dobrowolski JM, Niesman IR, Sibley LD. Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell Motil Cytoskel. 1997;37:253–262. doi: 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Wetzel DM, Håkansson S, Hu K, Roos DS, Sibley LD. Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol Biol Cell. 2003;14:396–406. doi: 10.1091/mbc.E02-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahoo N, Beatty WL, Heuser JE, Sept D, Sibley LD. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol Biol Cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller LH, Aikawa M, Johnson JG, Shiroishi T. Interaction between cytochalasin B-treated malarial parasites and erythrocytes. Journal of Experimental Medicine. 1979;149:172–184. doi: 10.1084/jem.149.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wetzel DM, Schmidt J, Kuhlenschmidt M, Dubey JP, Sibley LD. Gliding motility leads to active cellular invasion by Cryptosporidium parvum sporozoites. Infect Immun. 2005;73:5379–5387. doi: 10.1128/IAI.73.9.5379-5387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell DG, Sinden RE. The role of the cytoskeleton in the motility of coccidian sporozoites. J Cell Sci. 1981;50:345–359. doi: 10.1242/jcs.50.1.345. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz S, Grainger M, Howell SA, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C. Malaria parasite actin filaments are very short. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- 12.Schüler H, Mueller AK, Matuschewski K. Unusual properties of Plasmodium falciparum actin: new insights into microfilament dynamics of apicomplexan parasites. FEBS Letters. 2005;579:655–660. doi: 10.1016/j.febslet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 13.Skillman KM, Diraviyam K, Khan A, Tang K, Sept D, Sibley LD. Evolutionarily divergent, unstable filamentous actin is essential for gliding motility of apicomplexan parasites. PLoS Pathogens. 2011;7:e1002280. doi: 10.1371/journal.ppat.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 15.Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat Rev Microbiol. 2006;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- 16.Schüler H, Matuschewski K. Regulation of apicomplexan microfilament dynamics by minimal set of actin-binding proteins. Traffic. 2006;7:1433–1439. doi: 10.1111/j.1600-0854.2006.00484.x. [DOI] [PubMed] [Google Scholar]
- 17.Gordon JL, Sibley LD. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genomics. 2005;6:e179. doi: 10.1186/1471-2164-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta S, Sibley LD. Toxoplasma gondii actin depolymerizing factor acts primarily to sequester G-actin. J Biol Chem. 2010;285:6835–6847. doi: 10.1074/jbc.M109.068155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta S, Sibley LD. Actin depolymerizing factor controls actin turnover and gliding motility in Toxoplasma gondii. Molec Biol Cell. 2011;22:1290–1299. doi: 10.1091/mbc.E10-12-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 22.Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 23.Sagot I, Rodal AA, Mosely J, Goode BL, Pellman D. An actin nucleation mechanism mediated by bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 24.Goode BL, Eck MJ. Mechansim and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 25.Plattner F, Yarovinsky F, Romero S, Didry D, Carlier MF, Sher A, Soldati-Favre D. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe. 2008;3:77–87. doi: 10.1016/j.chom.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Kucera K, Koblansky AA, Saunders LP, Frederick KB, De La Cruz EM, Ghosh S, Modis Y. Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11. J Mol Biol. 2010;403:616–629. doi: 10.1016/j.jmb.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kursula I, Kursula P, Ganter M, Panjikar S, Matuschewski K, Schuler H. Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 2008;16:1638–1648. doi: 10.1016/j.str.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Daher W, Plattner F, Carlier MF, Soldati-Favre D. Concerted action of two formins in gliding motility and host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010;6:e1001132. doi: 10.1371/journal.ppat.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daher W, Klages N, Carlier MF, Soldati-Favre D. Molecular characterization of Toxoplasma gondii formin 3, an actin nucleator dispensable for tachyzoite growth and motility. Eukaryot Cell. 2011 doi: 10.1128/EC.05192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baum J, Tonkin CJ, Paul AS, Rug M, Smith BJ, Gould SB, Richard D, Pollard TD, Cowman AF. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe. 2008;3:188–198. doi: 10.1016/j.chom.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Lopez MF, Berggren K, Chernokalskaya E, Lazarev A, Robinson M, Patton WF. A comparison of silver stain and SYPRO Ruby Protein Gel Stain with respect to protein detection in two-dimensional gels and identification by peptide mass profiling. Electrophoresis. 2000;21:3673–3683. doi: 10.1002/1522-2683(200011)21:17<3673::AID-ELPS3673>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues CO, Ruiz FA, Rohloff P, Scott DA, Moreno SNJ. Characterization of isolated acidocalcisomes from Toxoplasma gondii tachyzoites reveals a novel pool of hydrolyzable polyphosphate. J Biol Chem. 2002;277:48650–48656. doi: 10.1074/jbc.M208990200. [DOI] [PubMed] [Google Scholar]
- 35.Gershman LC, Newman J, Selden LA, Estes JE. Bound-cation exchange affects the lag phase in actin polymerization. Biochemistry. 1984;23:2199–2203. doi: 10.1021/bi00305a015. [DOI] [PubMed] [Google Scholar]
- 36.Heuser JE. Development of the quick-freeze, deep-etch, rotary replication technique of sample preparation for 3-D electron microscopy. Prog Clin Biol Res. 1989;295:71–83. [PubMed] [Google Scholar]
- 37.Perelroizen I, Didry D, Christensen H, Chua NH, Carlier MF. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J Bio Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- 38.Kovar DR, Yang P, Sale WS, Drobak BK, Staiger CJ. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. J Cell Sci. 2001;114:4293–4305. doi: 10.1242/jcs.114.23.4293. [DOI] [PubMed] [Google Scholar]
- 39.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 40.Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006;16:1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 41.Michelot A, Guerin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger CJ, Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005;17:2296–2313. doi: 10.1105/tpc.105.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbon BC, Kovar DR, Staiger CJ. Latrunculin B has different effects on pollen germination and tube growth. Plant Cell. 1999;11:2349–2363. doi: 10.1105/tpc.11.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ. Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell. 2002;14:2613–2626. doi: 10.1105/tpc.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staiger CJ, Blanchoin L. Actin dynamics: old friends with new stories. Curr Opin Plant Biol. 2006;9:554–562. doi: 10.1016/j.pbi.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]