Abstract
Over the past decade, a growing community of researchers has emerged around the use of COnstraint-Based Reconstruction and Analysis (COBRA) methods to simulate, analyze and predict a variety of metabolic phenotypes using genome-scale models. The COBRA Toolbox, a MATLAB package for implementing COBRA methods, was presented earlier. Here we present a significant update of this in silico ToolBox. Version 2.0 of the COBRA Toolbox expands the scope of computations by including in silico analysis methods developed since its original release. New functions include: (1) network gap filling, (2) 13C analysis, (3) metabolic engineering, (4) omics-guided analysis, and (5) visualization. As with the first version, the COBRA Toolbox reads and writes Systems Biology Markup Language formatted models. In version 2.0, we improved performance, usability, and the level of documentation. A suite of test scripts can now be used to learn the core functionality of the Toolbox and validate results. This Toolbox lowers the barrier of entry to use powerful COBRA methods.
Keywords: Systems Biology, Computational Biology, MATLAB, Flux Balance Analysis, Fluxomics, Visualization, Gap Filling, Metabolic Engineering
INTRODUCTION
COnstraint-Based Reconstruction and Analysis (COBRA) methods have been successfully employed in the field of microbial metabolic engineering1–3 and are being extended to modeling transcriptional4–8 and signaling9–11 networks and the field of public health12. Specifically, COBRA methods have been used to guide metabolic pathway engineering, to model pathogens13 and host-pathogen interactions14 and to assess the impact of disease states on human metabolism15. A wide variety of COBRA methods have been developed over the years16, 17. COBRA methods have been employed in hundreds of research articles over the past decade that characterize genome-scale properties of metabolic networks and their phenotypic states18–20.
The COBRA approach focuses on employing physicochemical, data-driven, and biological constraints to enumerate the set of feasible phenotypic states of a reconstructed biological network in a given condition (Figure 1a). These constraints include compartmentalization, mass conservation, molecular crowding21, and thermodynamic directionality22–24. More recently, transcriptome data have been used to reduce the size of the set of computed feasible states14, 25, 26. Although COBRA methods may not provide a unique solution, they provide a reduced set of solutions that may be used to guide biological hypothesis development27. The COBRA Toolbox provides researchers with a high-level interface to a variety of COBRA methods. Detailed descriptions of COBRA methods can be found in a variety of reviews3, 16, 28, 29
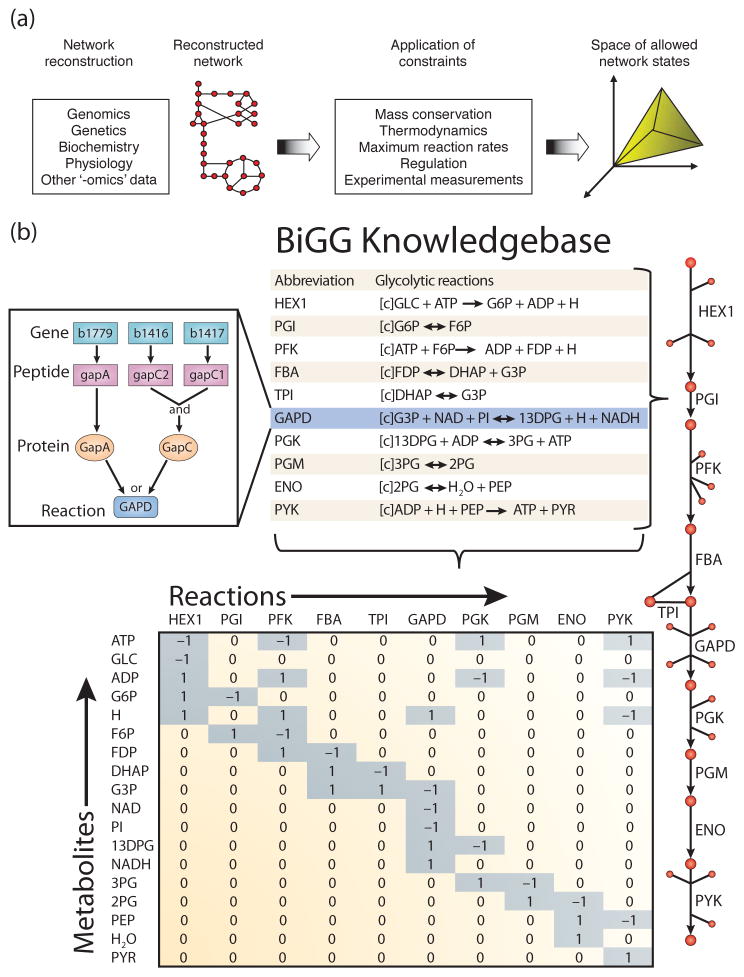
Figure 1. The philosophy of COnstraints-Based Reconstruction and Analysis.
(a) COnstraints-Based Reconstruction and Analysis (COBRA) of biological networks involves the creation of network models from a variety of biological data sources. The capabilities of the model are then assessed in the context of physical, chemical, regulatory, and omics constraints (Reproduced from Becker et al.17 with permission). (b) COBRA models are often derived from BiGG knowledgebases which are essentially 2-D annotations of the genome that relate metabolic activity to genomic loci. (left inset) In Escherichia coli, the glyceraldehyde-3-phosphate dehydrogenase (GAPD) activity can be provided by two isozymes (GapA or GapC); GapC is a heteromeric protein that requires genes from two genomic loci. The contents of a BiGG knowledgebase can be converted to a map (right) to facilitate visual interpretation. Or a mathematical modeling formalism to develop and explore hypotheses, such as a stoichiometric matrix (bottom) that can be used to explore mass flow through the network. (Modified reproduction from Reed et al.33 with permission).
The biological network models that are analyzed with COBRA methods are constructed in a bottom-up fashion from bibliomic and experimental data and thus represent Biochemically, Genetically, and Genomically (BiGG) consistent knowledgebases30, 31. BiGG knowledgebases are manually-curated 2-D genome annotations32 that relate biological functions, such as metabolic reactions, to the genome through the use of the gene-protein-reaction formalism33 (Figure 1b). Application of the BiGG formalism to metabolism has been particularly successful, and metabolic reconstructions are available for many organisms34–38, 39, 40–42. A detailed protocol describing the construction of high-quality BiGG knowledgebases for metabolism, and their transformation into mathematical models has been recently published43.
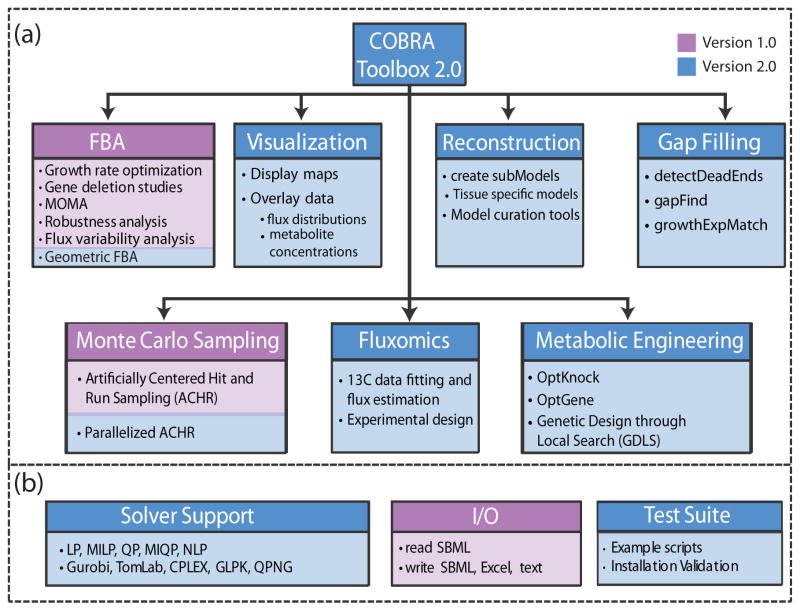
The first release of the COBRA Toolbox in 2007 provided access to a variety of methods, including flux balance analysis, gene essentiality analysis, and minimization of metabolic adjustment analysis (Table 1). Since the release of the first version of the COBRA Toolbox, many additional COBRA-related methods have been published44–48. In version 2.0 of the COBRA Toolbox, we have extended the capabilities to include: geometric FBA44, Loop law49, creation of context-specific subnetwork models using omics data14, 25 Monte Carlo sampling15, 50–52,13C fluxomics, gap filling45, 53, metabolic engineering46–48, and visualization of computational models of metabolism (Table 1 / Figure 2).
Table 1.
Features of the COBRA Toolbox 1.0 and 2.0
| COBRA 1.0 | New in COBRA 2.0 | |
|---|---|---|
| FBA |
|
|
| Fluxomics |
|
|
| Gap Filling |
|
|
| Input / Output |
|
|
| Metabolic Engineering |
|
|
| Reconstruction |
|
|
| Sampling |
|
|
| Test Suite |
|
|
| Visualization |
|
|
Figure 2. Overview of the COBRA toolbox.
(a) Seven categories of COBRA methods contained within version 2.0 of the COBRA Toolbox. (b) The COBRA Toolbox contains solver interface functions for linear, quadratic, mixed integer linear and quadratic, and nonlinear programming problems. Functions to read and write models in several formats are available. A test suite is included to validate installation as well as provide example implementation of many methods.
Additionally, methods5, 24, 54 and resources55 have been developed by community members that can serve as add-ons to the core COBRA Toolbox or provide models or other input. Specifically, Chandrasekaran and Price54 have developed a method—probabilistic regulation of metabolism (PROM)—that incorporates regulatory information from transcriptome, ChIP-chip, or literature data into a metabolic network model. Fleming and Thiele24 developed an extension to thermodynamically constrain reaction directionality And, Henry et al. 55 have developed a web-based resource (http://www.theseed.org/models/) that provides access to draft metabolic network reconstructions for a variety of organisms—these models may be imported into the COBRA Toolbox for further refinement and analysis.
This protocol aims to provide researchers with the ability to use the in silico methods included in the Toolbox with only high-level knowledge of the algorithms. Because of the wide range of creative uses for COBRA methods, not all of the Toolbox’s capabilities are described in this protocol; additional functionalities are described in the Documentation.
The COBRA Toolbox supports models in the Systems Biology Markup Language (SBML) format56. Importation of the models into MATLAB is dependent on libSBML57 and the SBMLToolbox58. Because SBML does not yet provide complete support for a few key COBRA parameters, we provide an explicit description of the COBRA extensions to SBML below and in Supplementary Information. The COBRA Toolbox is available for download from http://www.cobratoolbox.org. Detailed documentation in html format is available in the ‘docs’ folder of the COBRA Toolbox.
Tool Box Installation
There are two options for installing the COBRA Toolbox ‘à la carte’ or bundled. The à la carte version only contains the COBRA Toolbox. The bundled version includes the COBRA Toolbox, libSBML, the SBMLToolbox, GLPK, and glpkmex. The bundled version has been tested on Mac OS X 10.6 Snow Leopard (64-bit) Ubuntu GNU/Linux Lucid (64-bit), Windows XP (32-bit), and Windows 7 (64-bit). Separate installation instructions are provided in Equipment Setup.
COBRA-compliant SBML file
Documentation on the SBML standard is available on the SBML website (http://sbml.org) and a description of a COBRA-compliant SBML file is provided in the Supplementary Material. Sample models in COBRA-compliant SBML may be downloaded from the BiGG knowledgebase (http://bigg.ucsd.edu)31 or draft models may be downloaded from the Model SEED (http://www.theseed.org/models)55. The model files must include the following information for all calculations: stoichiometry of each reaction, upper and lower bounds of each reaction and objective function coefficients for each reaction.
Several functions within the Toolbox47, 48 require information that is not, yet, in the SBML standard or scheduled for removal in SBML 3 and beyond. The gene-reaction associations are essential for relating the metabolic reactions to the genome and the subsystem is useful for ontological classification. Metabolite formulas and charges are necessary to make sure the model is physically consistent (no generation of mass or energy). Additional annotation parameters, such as KEGG or CAS IDs, should be specified in the notes field.
<reaction> … <notes> <html xmlns=” http://www.w3.org/1999/xhtml ”> <p>GENE_ASSOCIATION: ((gene1) and (gene2)) or (gene3)</p> <p>SUBSYSTEM: Transport Inner Membrane</p> <p>KEGGID: …</p> … </html> </notes> </reaction> <metabolite> … <notes> <html xmlns=” http://www.w3.org/1999/xhtml ”> <p>FORMULA: C6H12O6</p> <p>CHARGE: 0</p> <p>CAS: …</p> … </html> </notes> </metabolite>
Metabolic map files
The visualization tools require text files of the coordinates for placing metabolites and reactions on a map. Map coordinate files for many metabolic pathways are available from the BiGG knowledgebase. The COBRA Toolbox relates COBRA SBML models to the map coordinate files via the reaction and metabolite ids. A map file for glycolysis may be used with various SBML models as long as the identifiers match. The format for a map file is described in Supplementary Material.
MATERIALS
Equipment
The COBRA Toolbox version 2.0 or above (http://www.cobratoolbox.org)
A computer capable of running MATLAB
Version 7.0 or above of MATLAB (Mathworks Inc.) numerical computation and visualization software (http://www.mathworks.com)
libSBML programming library 4.0.1 or above (http://www.sbml.org)
SBMLToolbox version 3.1.1 or above for MATLAB to allow reading and writing models in SBML format (http://www.sbml.org)
-
A linear programming (LP) solver. Currently the COBRA Toolbox supports:
Gurobi (Gurobi Optimization. http://www.gurobi.com) through Gurobi Mex (http://www.convexoptimization.com/wikimization/index.php/Gurobi_mex)
CPLEX (ILOG Inc.) through Tomlab (Tomlab Optimization Inc. http://tomopt.com)
GLPK (http://www.gnu.org/software/glpk) through glpkmex (http://glpkmex.sourceforge.net) – Note, GLPK does not provide accurate solutions for OptKnock or GDLS calculations as implemented in the Toolbox.
Caution! Other solvers (such as Mosek, http://www.mosek.com; LINDO, http://www.lindo.com; and PDCO, http://www.stanford.edu/group/SOL/software/pdco.html), may work with the COBRA Toolbox but they have not been validated.
Critical! For best performance it may be necessary to adjust parameters of the installed solver.
-
A quadratic programming (QP) solver. (optional) Currently the COBRA toolbox supports:
CPLEX (ILOG Inc.) through Tomlab
QPNG (part of GLPK) – Note, QPNG does not provide accurate solutions for MOMA as implemented in the Toolbox.
Caution! Other solvers (such as Mosek and PDCO), may work with the COBRA Toolbox but they have not been validated.
Critical! For best performance it may be necessary to adjust parameters of the installed solver.
-
A nonlinear programming (NLP) solver. (optional) Currently the COBRA toolbox supports:
SNOPT through Tomlab
Critical! For best performance it may be necessary to adjust parameters of the installed solver.
Equipment Setup
à la carte installation of Toolbox
Install MATLAB
Install libSBML, the SBML Toolbox, and selected solvers according to their specific instructions.
Unpack the COBRA 2.0 archive
Bundled installation of Toolbox
Install MATLAB
-
Unpack the COBRA 2.0 archive
Cobra_Install_Path is the path to the top level directory for the COBRA Toolbox
-
Update Shared Library Path — Mac OS X and GNU/Linux Only
Mac OS: DYLD_LIBRARY_PATH=Cobra_Install_Path/external/toolboxes/SBMLToolbox_3.1.2/toolbox/
GNU/Linux: LD_LIBRARY_PATH=Cobra_Install_Path/external/toolboxes/SBMLToolbox_3.1.2/toolbox
PROCEDURE
Notes on nomenclature: italics denotes a parameter that is supplied to a function. A bracketed [parameter] is optional. ≫ denotes the Matlab command line; anything following ≫ is meant to be entered on the command line. All time estimates for the functions are predicate on a model of about 1200 genes, 2300 reactions, 1800, metabolites, and a 2.4 GHz Intel Core 2 Duo processor. When substantial preprocessing efforts are required, we provide time estimates based on personal experience.
Initializing the Toolbox
-
1|
Navigate to the directory where you installed the Toolbox:
≫ initCobraToolbox()
-
2|
Save the paths added if desired:
≫ savepath()
Changing COBRA solvers
-
3|
Set the solvers used by the COBRA Toolbox using the following function:
≫ changeCobraSolver(solverName, [solverType]);
Variables are defined as follows: solverName specifies the solver package to use; the COBRA Toolbox currently supports ‘gurobi’, ‘tomlab_cplex’, ‘glpk’, and ‘qpng’. solverType (default ‘LP’) specifies the type of problems (‘LP’, ‘MILP’, ‘QP’, ‘MIQP’, ‘NLP’) to solve with the solver specified by solverName. When changeCobraSolver is called without any arguments, it will return the names of the current solvers settings.
Run COBRA Toolbox test suite [~103 s]
-
4|
The test suite contains scripts that test the functionality of scripts within the Toolbox. The scripts in the the testing directory provide useful examples of many of the Toolbox’s functions.
≫ testAll()
testAll sequentially navigates the test suite directory (testing) and runs each test. Upon completion, it displays which tests were completed successfully and which failed.
Caution! For solver suites other than Gurobi or Tomlab, the user may encounter failures that require tuning of solver parameters.
Read COBRA-compliant SBML models into MATLAB [~102 s]
-
5 |
Load a COBRA-compliant model into MATLAB. To load a model, navigate within MATLAB to the directory containing the model and call the following function from the command window:
≫ model = readCbModel([filename]);
When called with no arguments, readCbModel will prompt the user to select a file using a dialog box. readCbModel supports SBML-formatted (Level 2 versions 1 or 4) files. SBML files for a variety of organisms are available from the BiGG knowledgebase (http://bigg.ucsd.edu)31. The function returns a COBRA Toolbox model structure containing the necessary fields to describe the model for use with subsequent steps. See Supplementary Material for a description of the fields in a COBRA Toolbox model structure; hereafter, model denotes a COBRA Toolbox model structure.
CRITICAL STEP! If the model is not properly loaded into MATLAB, none of the following functions will work. Ensure that libSBML and SBML Toolbox are properly installed and accessible by MATLAB and that the SBML file is formatted correctly.
Saving the model
-
6 |
COBRA Toolbox model structures may be saved as text or SBML files. On Microsoft Windows, the structures may also be written to an Excel (xls) file.
≫ writeCbModel(model, format, [fileName], [compSymbolList], [compNameList], [SBMLLevel], [SBMLVersion]);
For format use ‘sbml’ for SBML file format or ‘xls’ for Excel format (only available on MS Windows). For filename use the name of the file. If not provided, a dialog box will prompt the user to specify name and location of the output file. This feature is dependent on the SBML Toolbox to generate the XML file. The toolbox is able to output SBML level 2 versions 1 or 4.
Modify COBRA Toolbox models
-
7 |
Once the model is loaded into MATLAB by readCbModel, the model can be modified to simulate different conditions such as altering reaction bonds (A), adding (B) or removing reactions (C) or changing the model objective (D).
-
to alter reaction bounds:
≫ model = changeRxnBounds(model, rxnNameList, value, boundType);
rxnNameList is a cell array of reaction ids corresponding to reaction ids in model.rxns; value is a floating point number; boundType specifies which bounds to change for the reactions and can take values of ‘l’, ‘u’, or ‘b’ for lower, upper, or both, respectively. This function is useful for defining the in silico media composition by changing the lower bounds of exchange reactions.
-
New reactions can be added to a COBRA Toolbox model using the following function:
≫ [model] = addReaction(model, rxnName, metaboliteList, stoichCoeffList, [revFlag], [lowerBound], [upperBound], [objCoeff], [subsystem], [grRule], [geneNameList], [systNameList], [checkDuplicate]);
metaboliteList is a list of metabolites involved in the reaction (if a metabolite does not exist in model.mets then this function will add it); stoichCoeffList is the stoichiometric coefficients for the corresponding elements in metaboliteList. This function checks for reactions with the same name or stoichiometic coefficients, however this can be disabled by setting checkDuplicate to false.
-
To remove a reaction, call the following function:
≫ [model] = removeRxns(model, rxnRemoveList)
rxnRemoveList a cell array of reaction ids corresponding to elements in model.rxns. Metabolites that are no longer involved in any reactions are removed from the model. The model may no longer function after reactions have been removed.
-
COBRA modeling often entails performing calculations that focus on a specified objective, such as growth59. To change the objective function, use the following function:
≫ model = changeObjective(model, rxnNameList, [objectiveCoeff]);
rxnNameList is either a string or a cell array of strings containing reaction ids corresponding to elements in model.rxns that should be included in the objective function; objectiveCoeff specifies the weight given to the respective reaction in rxnNameList. If left empty, objectiveCoeff is assumed to be 1.
-
Omics-Guided Creation of Context-Specific Models. Timing ~102 s + >1 hr to format data
-
8 |
An emerging application of genome-scale reconstructions is analyzing omics data in a systems context14, 25, 26. In particular, this procedure is useful for building cell-, tissue-, or condition-specific models. createTissueSpecificModel is designed to map transcriptomic or proteomic data onto a reconstruction using two established algorithms (GIMME25 or Shlomi26). The GIMME algorithm is an LP procedure that best matches high-throughput data to an original flux distribution derived from the full model; thus the algorithm requires a predefined objective function. The Shlomi algorithm is an MILP procedure that best matches high-throughput data to pathway length, thus avoiding the need for a predefined objective function. Novice users can utilize the GIMME algorithm with two inputs: the COBRA model and expression data; while more experienced users can tweak additional parameters.
≫ [tissueModel,Rxns] = createTissueSpecificModel(model,expressionData, [proceedExp],[orphan],[exRxnRemove],[solver],[options], [funcModel]);
Required Inputs: model is a reconstruction with gene-protein-reaction associations; expressionData is a structure that contains two inputs: .Locus (a vector of GeneIDs matching gene ids in model.genes), and .Data (a vector of presence/absence calls). Optional Inputs: proceedExp (default value of 1, to process multiple data sets, set proceedExp to 0); orphan (default value of 1) controls whether or not reactions with no known gene-protein-reaction associated are included when peforming Shlomi-based network trimming (orphan reactions are always included when the GIMME reaction is employed, regardless of the orphan setting); exRxnRemove is a list of select exchange reactions that are excluded (that is if a specific cell or tissue is known not to have a particular metabolite transporter); solver is either ‘GIMME’ or ‘Shlomi’ and defaults to ‘GIMME’; options is only used with the GIMME algorithm, and it specifies which reactions comprise the objective function (by default, the objective function is chosen from model.c with a 90% (0.9) threshold); funcModel controls whether the output tissueModel is fully functional (every reaction can carry a flux) or not when using the GIMME algorithm. Output: tissueModel is the final cell-, tissue-, or condition-specific model generated from the function; Rxns is a structure containing statistics about what reactions were or were not expressed based on the omics data and what reactions were added or removed from the model (see Anticipated Results).
Visualization Timing ~101 s
-
9 |
Visual representation of a metabolic network can aid in understanding the model. Maps for a variety of metabolic pathways are available for many of the models hosted in the BiGG knowledgebase (http://bigg.ucsd.edu). See Supplementary Material for a description of the map file format. These maps may be used for other organisms that have similar metabolic pathways, given that the user uses the same metabolite and reaction ids as the BiGG model that was used to create the map. To load a map the following command is used:
≫ map = readCbMap([filename])
If readCbMap is called with no arguments, a dialog box will prompt the user to select a map file. After the map has been read into MATLAB, it can be viewed as a MATLAB figure or a scalable vector graphic (svg).
-
10|
To view a map as a MATLAB figure the following commands are used
≫ changeCbMapOutput(‘matlab’) ≫ drawCbMap(map,[options],[varargin])
where, options is a map options structure created by setMapOptions. See software documentation for description of optional parameters and ANTICIPATED RESULTS for an example.
-
11|
To save a map as an SVG file the following commands are used.
≫ changeCbMapOutput(‘svg’); ≫ drawCbMap(map,[options],[varargin])
By default, drawCbMap will create a file named ‘target.svg’ in the current working directory. The filename can be set by inputting additional parameters:
≫ drawCbMap(map,’FileName’,filename)
Simulate optimal growth using flux-balance analysis (FBA) Timing <102 s
-
12 |
Simulating optimal growth using FBA is one of the fundamental COBRA phenotypic calculations for metabolic network models. FBA is a method that calculates the flow of metabolites through a metabolic network28. Growth is simulated by optimizing the model for flux through the model’s biomass function; however, it is also possible to perform simulations that focus on optimizing other biological characteristics, such as ATP production. The reaction to optimize is set using the model.c vector (see step 7D).
In addition to specifying an objective, it is also necessary to define the in silico growth medium; this is accomplished by modifying the bounds of exchange reactions. Exchange reactions for metabolites comprising the in silico growth medium should have a lower bound less than 0; all other exchange reactions should have a lower bound of 0. All exchange reactions should have an upper bound greater than 0 to prevent metabolite build up. The solution returned will have units based on the units used in the model (typically mmol · gDW−1 · h−1). FBA can be performed either in (A) standard (B) geometric mode:
-
Standard FBA is performed with:
≫ [solution] = optimizeCbModel(model, [osenseStr], [minNorm], [allowLoops])
where: osenseStr is either ‘max’ or ‘min’ to maximize or minimize the value of the objective; minNorm (default 0, if nonzero, attempt to find a solution that minimizes the presence of loops; allowLoops (default true, if set to false, use the loop law algorithm49 to remove loops—this proceedure can be time consuming).
optimizeCbModel will return a solution structure containing: the objective value ‘f’, the primal solution ‘x’, the dual solution ‘y’, the reduced cost ‘w’, a universal status flag ‘stat’, a solver specific status flag ‘origStat’, and the time to compute the solution ‘time’. The primal solution, ‘x’ represents the flux carried by each reaction within the model. The dual solution, ‘y’ represents the shadow prices for each metabolite and indicates how much the addition of the corresponding metabolite will increase or decrease the objective value28, 60. The reduced cost, ‘w’, indicates how much each reaction affects the objective. A solver status of 1 indicates that an optimal solution was found.
-
Geometric FBA44 is an alternative to standard FBA. Geometric FBA attempts to return the minimal flux distribution central to the bounds of the solution space while still maintaining optimal growth rate. The flux distribution returned should then be reproducible regardless of the solver used.
≫ flux = geometricFBA(model,[varargin])
The function returns the vector ‘flux’ which contains the centered optimal flux distribution.
-
-
13|
Visualizing an Optimal Flux Distribution
The optimal flux distribution obtained using optimizeCbModel or geometricFBA can be overlaid onto an existing map of the model using:
≫ drawFlux(map, model, flux, [options], [varargin]
where: map is a map object created with readCbMap (see Visualization Step 9); model is the COBRA model structure that was used for performing FBA or Geometric FBA; options is a drawCbMap options structure.
-
14|
Classification of Model Genes Based on Optimal FBA Solution
Parsimonious FBA (pFBA) is an FBA approach that incorporates flux parsimony as a constraint to categorize the solution space61. The concept of flux parsimony, in the context of a metabolic network, means to minimize the total material flow required to achieve an objective.
In this method, genes are classified into six categories: (1) essential genes, metabolic genes necessary for in silico growth in the given media; (2) pFBA optima, non-essential genes contributing to the optimal growth rate and minimum gene-associated flux; (3) enzymatically less efficient (ELE), genes requiring more flux through enzymatic steps than alternative pathways that meet the same predicted growth rate; (4) metabolically less efficient (MLE), genes requiring a growth rate reduction if used; (5) pFBA no-flux, genes that are unable to carry flux in the experimental conditions; and (6) Blocked, genes that are only associated with the reactions that cannot carry a flux under any condition (“blocked” reactions).
To categorize the genes and reactions within a model and return a model with flux minimization constraints, execute the following:
≫ [GeneClasses, RxnClasses, modelIrrevFM] = pFBA(model, [varargin])
Where, GeneClasses contains a list of all genes that are within the categories above; RxnClasses contains a list of all reactions that are within the categories above; and modelIrrevFM is a model that contains the flux minimization constraints. If a map is available for the model, the results from this function can be visualized by using the ‘map’ and ‘mapoutname’ flags in the varargin input. A test case may be found in the ANTICIPATED RESULTS section. Additional options are described in the software documentation directory.
CRITICAL STEP
The subsequent steps in this protocol rely on the functionality of optimizeCbModel. If optimizeCbModel fails to return a feasible flux distribution for the examples within this protocol, the problem may be due to the installation of the LP solver. It is not necessary that geometricFBA return a solution for the subsequent steps.
Solving COBRA problem structures (Advanced User) Timing >100 s
-
15 |
The COBRA toolbox has five function calls used for solving different optimization problems. Basic users will not need to call these low level functions directly as higher level functions encapsulate these calls. These functions act as a common interface for different LP, MILP, QP, MIQP, and NLP solvers ensuring that labs can share code even when using different installed solvers.
The five solver functions use a similar input argument structure: problem structure followed by optional argument/value pairs. The required fields in the problem structure vary for each function to supply the required information to solve the type of problem. For example, the mixed integer problem structures require a field which specifies variable type (continuous, integer, binary). A description on the format of COBRA problem structures can be found in Supplementary Material. The COBRA solution structure also provides a common output format regardless of the solver used.
≫ [solution] = solveCobraLP(LPproblem, [varargin]) ≫ [solution] = solveCobraMILP(MILPproblem, [varargin]) ≫ [solution] = solveCobraQP(QPproblem, [varargin]) ≫ [solution] = solveCobraMIQP(MIQPproblem, [varargin]) ≫ [solution] = solveCobraNLP(NLPproblem, [varargin])
Simulating deletion studies Timing ~102–104 s
-
16 |
Deletion studies can be easily simulated with in silico models. Gene deletion methods within the Toolbox are dependent on the proper setup of the gene-reaction matrix as well as the rules defining the Boolean relationship between genes and reactions. Reactions that are affected by a gene deletion have their upper and lower flux bounds set to zero and are therefore not functional. The set of reactions on which a gene deletion has an effect is calculated using the gene reaction association and rules.
It is possible to study either (A) single essential gene deletions or (B) pairs of synthetic lethal genes. The possible results from deletion studies are: 1) unchanged maximal growth, 2) reduced maximal growth, or 3) no growth (lethal). Deletion studies can be used to predict gene/reaction essentiality.
(A) Essential Gene Study
≫ [grRatio, grRateKO, grRateWT, hasEffect, delRxns, fluxSolution] = singleGeneDeletion(model, method, [geneList])
where: method can be either ‘FBA’ (default) ‘MOMA’62 or linear MOMA (‘lMOMA’); geneList, is a cell array of genes corresponding to model.genes (if not provided deletion simulations are performed for all genes in the model); grRatio is the growth rate of the knockout / growth rate of WT; grRateKO is the growth rate of the knockouts; grRateWT is the wild-type growth rate; hasEffect is a Boolean list that contains true for each gene whose deletion alters the growth rate; delRxns contains a list of the reactions, the bounds of which are set to 0 for each gene deletion; and fluxSolution is the flux solution for each deletion.
(B) Synthetic Lethal Study
≫ [grRatioDble, grRateKO, grRateWT] = doubleGeneDeletion(model, method, [geneList1], [geneList2])
where: method can be either ‘FBA’ (default) ‘MOMA’62 or linear MOMA (‘lMOMA’); geneList1 is a cell array of genes corresponding to model.genes (if not provided, the function assumes all genes in model.genes are to be interrogated); geneList2 is a cell array of genes that correspond to the second set of genes in the synthetic lethal pair (if not provided, the function assumes that all genes in model.genes are to be interrogated); grRatioDble is the growth rate of the knockout / growth rate of WT; grRateKO is the growth rate of the knockouts; and grRateWT is the wild-type growth rate.
Flux Variability Analysis (FVA) Timing ~102 s
-
17 |
FBA only returns a single flux distribution that corresponds to maximal growth under given growth conditions. However, alternate optimal solutions may exist which correspond to maximal growth. FVA calculates the full range of numerical values for each reaction flux within the network63.
To determine the minimum and maximum flux values that the reactions within the model can carry while obtaining a specific percentage of optimal growth rate:
≫ [minFlux maxFlux] = fluxVariability(model, optPercentage, [rxnNameList], [verbFlag], [allowLoops])
where optPercentage (default 100) specifies the percentage of optimal that an alternate flux distribution must realize to be considered an acceptable alternative flux distribution.
Visualization of Flux Variability Analysis Results
-
18|
To visualize the results from this function, a flux variability map can be generated from an existing reaction map, color coding reactions based on flux directionality.
≫ drawFluxVariability(map, model, minFlux, maxFlux, [options])
where: map is the map structure corresponding to the model read in using readCbMap; model is the COBRA model structure used in the fluxVariability function; minFlux and maxFlux are vectors generated by the fluxVariability function described above; and options is a structure containing optional parameters such as edge and node color and size: bi-directional reversible reactions are colored green, unidirectional reversible reactions that carry flux in the forward direction are colored magenta, unidirectional reversible reactions that carry flux only in the reverse direction are colored cyan, and irreversible fluxes are colored blue.
Sampling the solution space [>102 s] (Advanced User)
-
19|
FBA only returns a single optimal point and thus gives little information about the entire solution space. An alternative approach is to characterize the solution space using sampling27. The generalized parallel sampler samples any arbitrary linearly-constrained space by moving a fixed number of points in parallel.
≫ [sampleStructOut, mixedFrac] = gpSampler(sampleStruct, [nPoints], [bias], [maxTime], [maxSteps])
where: sampleStruct is the COBRA Toolbox problem structure for linear programming problems (see Supplementary Information); nPoints is the number of sample points is set through; maxTime is the maximum sampling time; bias is a structure that imposes marginal distributions on reactions; sampleStructOut is sampleStruct with the addition of the ‘points’ field containing the solutions; and mixedFrac gives an estimate of how mixed the sampling solution is relative to the warmup points—a mixedFrac value of 0.5 indicates complete mixing.
Fluxomics (Advanced User) Timing >102 s
-
20 |
Carbon 13 tracing experiments provide the ability to measure internal flux rates in a metabolic network64. To use this data, additional information about carbon tracking must be added to the COBRA model. This is stored in the .isotopomer field as described in Supplementary Information Section S.3. In order to use the C13 solver, the functions must be generated:
≫ [experiment] = generateIsotopomerSolver(model, inputMet, [experiment], [FVAflag])
where: model is the COBRA model with an .isotopomer field; inputMet is a string corresponding to the C13 labeled input; experiment is a list of metabolites that must be measured; and FVAflag removes reactions that cannot carry a flux.
-
21|
Two solvers are generated, one based on the cumomer method65 and one on the faster EMU method66. The solvers are called internally during the scoreC13Fit function below. A given flux distribution can be scored against a set of C13 data:
≫ output = scoreC13Fit(v0,expdata,model)
where: vo is the initial guess for fitting; and expdata is one or more sets of experimental data described in Supplementary Innformation Section S.3.
-
22|
Next, the most optimal flux distribution can be found with a non-linear optimization as such:
≫ [vout] = fitC13Data(v0,expdata,model, [majorIterationLimit])
This function will return the flux with the lowest experimental score found by the NLP solver. Very often it is useful to compute the confidence intervals of reactions which are consistent with C13 data.
≫ [vs, output, v0] = C13ConfidenceInterval(v0, expdata, model, max_score, [directions], [majorIterationLimit]) (~102 s)
where: v0 is the initial guess; expdata is the experimental data that must be fit; max_score is the highest acceptable score; and directions is the list of reactions and reaction ratios which will be maximized and minimized (by default all reactions).
Gap Filling [~103 s]
-
23 |
Due to incomplete knowledge, a metabolic model may possess gaps. A gap is defined as missing biochemical information which can explain discrepancies between model predictions and experimental data. Gaps are typically reactions that facilitate the conversion of an available metabolite in the model to one necessary to achieve an objective. Identifying gaps in metabolic models can be attempted using either (A) detectDeadEnds or (B) gapFind.
(A) Detect Dead Ends in a Model
≫ outputMets = detectDeadEnds(model, [removeExternalMets])
The detectDeadEnds function searches the model.S matrix for metabolites that participate in only one reaction (can only be produced or only be consumed) and returns the corresponding indices for the metabolites in the model.mets field. Setting removeExternalMets to true removes external metabolites from the results. Not all gaps can be identified by simply inspecting the model.S matrix.
(B) Find All Gaps in a Model
The GapFind algorithm45 allows one to find all gaps in a model and all metabolites that are downstream from a model gap.
≫ [allGaps, rootGaps, downstreamGaps] = gapFind(model, findNCgaps, verbFlag)
where: allGaps is a list of the metabolite indices for a metabolite at a gap; rootGaps is a list of metabolites that cannot be produced; and downstreamGaps is a list of metabolites that are produced in a reaction that requires a metabolite that cannot be produced.
This function is run in an interactive and iterative fashion to guarantee that all gaps are identified. Set the lower bound of all exchange reactions within model to −1, the upper bound on all reactions to a relatively large positive number (for example 105), and the lower bound of all reversible reactions to a relatively large negative number (for example -105) within model. The appropriate bound magnitude required varies from model to model. If the bound magnitudes are too small, the algorithm will incorrectly identify many metabolites as gaps; if this occurs, increase the bound magnitudes by 10-fold. Repeat this process as necessary until the algorithm does not identify all metabolites as gaps.
-
24|
In addition to these two gap identification functions, the Toolbox includes an optimization-based algorithm (growthExpMatch) that identifies candidate reactions to fill gaps in the model53. growthExpMatch identifies the minimum number of reactions from a universal reaction database that are required for a metabolic model to grow on a specified substrate.
≫ [solution] = growthExpMatch(model, KEGGFilename, compartment, iterations, dictionary, logFile, threshold)
where: KEGGFilename is the name of the reaction .lst file downloaded from KEGG (http://www.genome.jp/kegg)67, 68; compartment is a string denoting which compartment to generate exchange reactions for; iterations controls the number iterations to run the function; dictionary is an n by 2 cell array that maps metabolites to KEGG IDs; logFile is the name of the .mat file to save the solution to; and threshold is the minimum value that the biomass function can take for the model to be considered growing.
-
25|
Display the growthExpMatch solution by printing the log file using the following function:
≫ printSolutionGEM(dir, matFile)
where: dir is the directory containing the growthExpMatch solution .mat file; and matFile is the name of the growthExpMatch solution .mat file.
Metabolic Engineering Timing ~102–103 s
-
25 |
The COBRA Toolbox version 2.0 provides three methods for in silico metabolic engineering: (A) OptKnock46, (B) OptGene47, and (C) GDLS48.
(A) OptKnock
OptKnock runs the OptKnock algorithm46 to determine reaction sets to knock out for the overproduction of a specific product when the model is optimized for internal cellular objectives.
≫ [OptKnockSol, biLevelMILPproblem] = OptKnock(model, selectedRxnList, [options], [constrOpt], [prevSolutions], [verbFlag])
where: OptKnockSol contains the best knockout set; and biLevelMILPproblem is the MILP problem generated by the algorithm and subsequently solved. See ANTICIPATED RESULTS for an example setup of options and constrOpt structures.
There are several things to take note of when calling the OptKnock function. First the function does not use the upper and lower bounds set within the model that is passed in. The model is first converted into irreversible format, splitting reactions with a lower bound < 0 and upper bound > 0. The resulting set of reactions has its lower bounds set to 0 and upper bounds set to options.vMax. Use the constrOpt structure to apply constraints on reactions, such as a minimal flux through the biomass function or ATP maintenance. Failure to set the proper constraints may lead to incorrect predictions generated by the function.
(B) OptGene
OptGene is an evolutionary programming-based method to determine gene knockout strategies for overproduction of a specific product47. It can handle non-linear objective functions such as product flux multiplied by biomass.
≫ [x, population, scores, optGeneSol] = OptGene(model, targetRxn, substrateRxn, generxnList, maxKOs, [population])
where: targetRxn specifies the reaction to optimize; substrateRxn specifies the exchange reaction for the growth; generxnList is a cell array of strings that specifies which genes or reactions are allowed to be deleted; and maxKOs sets the maximum number of knockouts; x is the best scoring set as determined by the functions optGeneFitness or optGeneFitnessTilt; population is the binary matrix representing the knockout sets; and optGeneSol is the structure summarizing the results. If resuming a previous simulation, the binary matrix (population) can be specified.
(C) Genetic Design Local Search
The Genetic Design Local Search (GDLS) algorithm48 may be used to identify what to knock out to increase in silico production of desired metabolites
≫ [gdlsSolution, biLevelMILPproblem, gdlsSolutionStructs] = GDLS(model, targetRxns, [vargin])
where: targetRxns is a specific list of genes, gene sets, or reactions to knock; gdlsSolution is the knockout solution; biLevelMILPproblem is the bi-level MILP problem for the solution; and gdlsSolutionStructs containes the intermediate solutions. This approach typically runs faster than the global search performed by OptKnock, however, it is not guaranteed to identify the global optima.
TROUBLESHOOTING
Troubleshooting for several steps in the protocol is available (Table 2). If your problem is not addressed here, see The openCOBRA Project website(http://opencobra.sourceforge.net/) for discussion forums and support. See also: testOptKnock().
Table 2.
Troubleshooting Table
| Issue | Solution |
|---|---|
| Tests failed in testing suite | Check that all necessary requirements are met for the function to run:
|
| SBML file not read in correctly | Check that libSBML 4.0.1+ and SBMLToolbox 3.1.1+ are installed and accessible in MATLAB. Is the file a COBRA compliant SBML file? (see Supplementary Material) If compartment abbreviations are not used as compartment ids within the file, specify all compartment abbreviations using compSymbolList input in readCbModel. |
| Unknown compartment error using writeCbModel | Specify all compartment abbreviations and names using compSymbolList and compNameList respectively when calling writeCbModel. |
| drawCbMap requires significant time to produce output | Try changing output to SVG. Text rendering in MATLAB figures is consuming. |
| optimizeCbModel returns an infeasible or infinite solution | For an infeasible solution, check that the constraints on the model allow for experimental growth. For an infinite solution, check that at least one constraint is limiting. |
| SolveCobraLP/MILP/QP/MIQP fails because csense or vartype is not a character array | Be sure that csense and/or vartype is initialized as a character array. MATLAB may cast the variable as a double instead. Run verifyCobraProblem to verify the format of the problem structure. |
Additional troubleshooting solutions can be found at the COBRA toolbox Google group (http://groups.google.com/group/cobra-toolbox).
ANTICIPATED RESULTS
Here, we provide examples of Toolbox functionality that is commonly used for our research or by external collaborators. For the most part, the default settings for the functions will suffice. Advanced users interested in additional features of the COBRA Toolbox should explore the Documentation in the COBRA Toolbox as well as the forums and bug-trackers available at: http://opencobra.sourceforge.net
Displaying and saving metabolic maps
The format for a metabolic map coordinate file is described in Supplementary information. Maps are available for download from the BiGG database (http://bigg.ucsd.edu/).
Load a map coordinate file
Navigate to the directory (testing/testMaps) containing the map file ‘ecoli_core_map.txt’ and then execute the following commands:
≫ map = readCbMap(‘ecoli_core_map.txt’);
Display a metabolic map
≫ changeCbMapOutput(‘matlab’); ≫ drawCbMap(map);
The following example illustrates one of the many ways to change the appearance of a map. Colors are based on the RGB style. To change all of the nodes to black and the edges to red: First, create an n × 3 matrix where n is the number of nodes in the map and each row is the RGB for black ([0,0,0]).
≫ node_colors = repmat([0,0,0],size(map.molName,1),1);
Next, create an n × 3 matrix where n is the number of edges in the map and each row is the RGB for red ([1,0,0]).
≫ edge_colors = repmat([1,0,0],size(map.connectionName,1),1);
The create a map options structure. The first argument is either an empty matrix or a previously created map options structure.
≫ options = setMapOptions([], map, ‘nodeColor’, node_colors, ‘edgeColor’, edge_colors); ≫ drawCbMap(map, options);
Save a metabolic map
≫ changeCbMapOutput(‘svg’) ≫ drawCbMap(map);
The file ‘target.svg’ will be saved in the working directory.
Optimal flux distributions and growth rates for E. coli core model
To read in the E. coli core model and predict a flux distribution for optimal growth, navigate to the directory (testing/testMaps) containing the E. coli core model and map file and execute the following functions:
≫ model = readCbModel(‘ecoli_core_model.xml’); ≫ map = readCbMap(‘ecoli_core_map.txt’); ≫ changeCbMapOutput(‘svg’); ≫ solution = optimizeCbModel(model);
The expected optimal biomass flux (solution.f) is ~0.87.
≫ drawFlux(map, model, solution.x, [], ‘FileName’, ‘EcoreOptFlux1.svg’);
The drawFlux function call generates an svg file named EcoreOptFlux1.svg in the working directory. The reactions are color coded using a linear scale from cyan (corresponding to a flux of −29.17) to magenta (corresponding to a flux of 45.51).
To more easily extract data from the map, change the width of reactions arrows corresponding to reactions carrying zero flux to 1 point. In addition, set the lower and upper bounds to −15 and 15 respectively.
≫ drawFlux(map, model, solution.x, [], ‘ZeroFluxWidth’, 1, ‘lb’, −15, ‘ub’, 15, ‘FileName’, ‘EcoreOptFlux2.svg’);
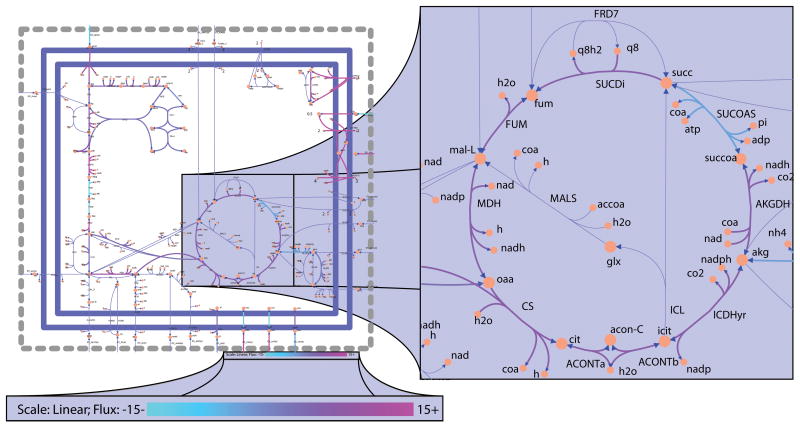
An svg file named EcoreOptFlux2.svg should be saved in the working directory with reactions color coded from cyan (flux of −15 or less) to magenta (flux of 15 or greater) and reactions carrying zero flux have their corresponding arrows narrowed (Figure 3).
Figure 3. Flux balance analysis of E. coli core model.
(left) Full E. coli core map. (right) Zoom in on the optimal flux distribution map of the citric acid cycle. (bottom) Zoom in on the flux color scale. Reactions are colored according to a scale of cyan (flux of 15 mmol · gDW−1 · h−1 or greater in the reverse direction) to magenta (flux of 15 mmol · gDW−1 · h−1 or greater in the forward direction). Reactions carrying zero flux have their corresponding arrows narrowed.
Parsimonious FBA (pFBA) Categorization of Genes for E. coli growth on acetate
To perform pFBA on the E. coli core model growing in an acetate minimal medium and plot the results, navigate to the directory (testing/testMaps) containing the E. coli core model and execute the following commands.
≫ map = readCbMap(‘ecoli_core_map.txt’); ≫ model = readCbModel(‘ecoli_core_model.xml’);
Remove glucose from the minimal medium.
≫ model = changeRxnBounds(model,’EX_glc(e)’,0,’l’);
Add acetate to the minimal medium.
≫ model = changeRxnBounds(model,’EX_ac(e)’,-10,’l’); ≫ [pFBAGeneClasses pFBARxn] = pFBA(model, ‘map’, map, ‘mapOutName’, ‘Ecore_pFBA_ac.svg’);
A reaction map with reactions color coded according to the reaction class will be saved as ‘Ecore_pFBA_ac.svg’ in the working directory. Inspecting the map will show that pyruvate kinase (PYK) is classified by pFBA as enzymatically less efficient (ELE). This is because its use does not reduce the computed growth rate, but it increases the amount a flux through the network. Interestingly, it has been reported that this enzyme is down-regulated in wild-type E. coli when grown on acetate minimal media69. Simulation time was ~20 s with the Gurobi LP solver.
Flux Variability Analysis (FVA) of E. coli core model
To perform FVA for the E. coli core model under glucose limited aerobic growth conditions with a minimum cellular growth of 90% of optimal, navigate to the directory (testing/testMaps) containing the E. coli core model and execute the following commands.
≫ model = readCbModel(‘ecoli_core_model.xml’); ≫ [minFlux maxFlux] = fluxVariability(model,90);
Simulation time is approximately 2 s with the Gurobi LP solver.
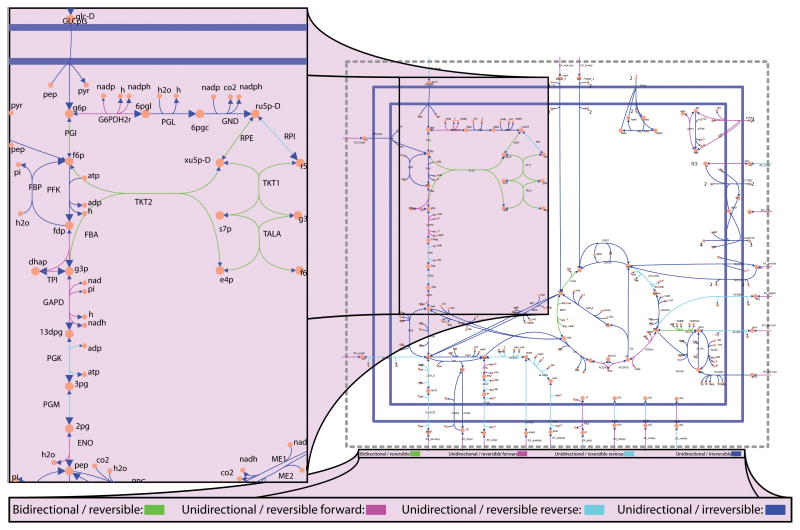
≫ map = readCbMap(‘ecoli_core_map.txt’); ≫ changeCbMapOutput(‘svg’); ≫ drawFluxVariability(map, model, minFlux, maxFlux, [], ‘fileName’, ‘EcoreFluxVariability.svg’);
A reaction map with reactions color coded according to the flux directionality it can carry will be saved to the file ‘EcoreFluxVariability.svg’ in the current working directory (Figure 4). Bi-directional reversible reactions are colored green. Unidirectional reversible reactions which carry flux in the forward direction are colored magenta. Unidirectional reversible reactions which carry flux only in the reverse direction are colored cyan. Irreversible fluxes are colored blue. Unidirectional fluxes have enlarged arrowheads in the direction of the flux.
Figure 4. Flux variability analysis of E. coli.
(right) Reaction map of E. coli core model. (left) Flux variability analysis of part of glycolysis and pentose phosphate pathway in the E. coli core model when growth rate is constrained to 90% of optimal. Bi-directional reversible reactions are colored green. Unidirectional reversible reactions which carry flux in the forward direction are colored magenta. Unidirectional reversible reactions which carry flux only in the reverse direction are colored cyan. Irreversible fluxes are colored blue. Unidirectional fluxes have enlarged arrowheads in the direction of the flux.
Sampling of the solution space of E. coli core model for growth in aerobic versus anaerobic conditions
To read in the E. coli core model and sample its solution space under glucose minimal media and aerobic conditions with 200 points for 2 minutes, navigate to the directory (testing/testMaps) containing the E. coli core model and execute the following commands:
≫ model_aerobic = readCbModel(‘ecoli_core_model.xml’); ≫ sampleStruct_aerobic = gpSampler(model_aerobic,200,[],120);
Simulation time of ~120 seconds with the Gurobi LP solver.
≫ model_anaerobic = changeRxnBounds(model_aerobic,’EX_o2(e)’,0,’l’); ≫ sampleStruct_anaerobic = gpSampler(model_anaerobic,200,[],120);
Simulation time of ~120 seconds with the Gurobi LP solver.
Sampling results will be returned in the two structures sampleStruct_aerobic and sampleStruct_anaerobic within the field points.
Visualize Sampling Results for a Set of Reactions
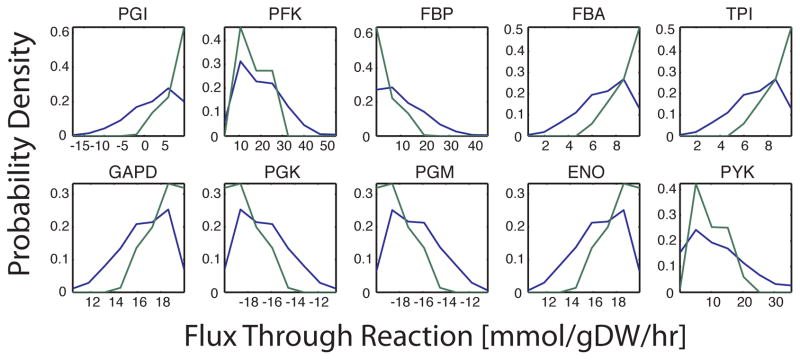
≫ rxnList = {‘PGI’, ‘PFK’, ‘FBP’, ‘FBA’, ‘TPI’, ‘GAPD’, ‘PGK’, ‘PGM’, ‘ENO’, ‘PYK’};
≫ plotSampleHist(rxnList, {sampleStruct_aerobic.points, sampleStruct_anaerobic.points }, {model_aerobic, model_anaerobic},[],[2,5]);
A MATLAB figure will also be generated showing the histograms for glycolysis with aerobic in blue and anaerobic in green (Figure 5).
Figure 5. Sampling histogram of glycolysis using the E. coli core model under aerobic and anaerobic glucose minimal media conditions.
For growth in aerobic (–) vs. anaerobic (–) medium, there is a large shift in the probable flux through many of the reactionsIn general, the range of flux probabilities for each reaction became more constrained. Phosphoglucose Isomerase (PGI) switched from being able to carry flux in either direction with aerobic conditions to only carrying flux in the forward direction with anaerobic conditions.
Identifying gaps in the metabolic network
To find gaps in the Ec_iJR904 model using the gapFind function, navigate to the directory (testing/testReadWrite) containing the Ec_iJR904.xml and execute the following commands:
≫ model = readCbModel(‘Ec_iJR904.xml’); ≫ exchangeRxns = model.rxns(findExcRxns(model));
Set the lower bound for all reversible reactions to −1 × 106.
≫ model = changeRxnBounds(model, model.rxns(logical(model.rev)), -1e6, ‘l’);
Set the upper bound for all reactions to 1 × 106.
≫ model = changeRxnBounds(model, model.rxns, 1e6, ‘u’);
Set lower bound for all exchange reactions to −1 to allow for uptake.
≫ model = changeRxnBounds(model, exchangeRxns,−-1, ‘l’); ≫ [allGaps, rootGaps, downstreamGaps] = gapFind(model);
Using the Gurobi MILP solver (simulation time 1.5 s), there are 64 metabolites identified as gaps: 28 root gaps and 36 downstream gaps; using the GLPK solver, only 20 of the 36 downstream gaps are identified.
Filling gaps using growthExpMatch
Remove the PGK reaction from the E. coli core model and use growthExpMatch to propose candidate reactions required to allow growth on glucose. Navigate to the directory (testing/testGrowthExpMatch) containing the E. coli core model and the univesal reaction database and execute the following commands:
≫ model = readCbModel(‘ecoli_core_model.xml’);
≫ modelKO = removeRxns(model,{‘PGK’});
≫ KEGGFilename = ‘2010_07_30_KEGG_reaction.lst’;
≫ load(‘Dictionary.mat’);
≫ growthExpMatch(modelKO, KEGGFilename, ‘[c]’, 5, dictionary,’GEMLog.txt’);
The PGK reaction is removed from the E. coli core model, removing the ability of the model to produce biomass from glucose. Updated versions of the KEGG reaction list should be downloaded from the KEGG website (http://www.genome.jp/kegg)67, 68. The resulting GEMLog file should contain 5 solutions (Table 3)—note, that if no solutions are found then the log file will not be generated. The first solution R01512 corresponds to the PGK reaction which was removed previously. The remaining four solutions are alternate reaction sets that when added allow the model to grow on glucose. With the Gurobi MILP solver, simulation time was ~2×103 seconds.
Table 3.
growthExpMatch gap filling solutions
| # | KEGG ID | Reaction Name | Reaction Formula |
|---|---|---|---|
| 1 | ‘R01512_b’ | phosphoglycerate kinase | ATP + 3-Phospho-D-glycerate <=> ADP + 3-Phospho-D-glyceroyl phosphate |
| 2 | ‘R02188_f’ | 3-Phospho-D-glyceroyl-phosphate:polyphosphate phosphotransferase | 3-Phospho-D-glyceroyl phosphate + (Phosphate)n <=> 3-Phospho-D-glycerate + (Phosphate)n |
| 3 | ‘R01515_f’ | acylphosphatase | 3-Phospho-D-glyceroyl phosphate + H2O <=> 3-Phospho-D-glycerate + Orthophosphate |
| 4 | ‘R00761_f’ | fructose-6-phosphate phosphoketolase | D-Fructose 6-phosphate + Orthophosphate <=> Acetyl phosphate + D-Erythrose 4-phosphate + H2O |
| 5 | ‘R00024_f’ | ribulose-bisphosphate carboxylase | D-Ribulose 1,5-bisphosphate + CO2 + H2O <=> 2 3-Phospho-D-glycerate |
| ‘R01523_f’ | phosphoribulokinase | ATP + D-Ribulose 5-phosphate <=> ADP + D-Ribulose 1,5-bisphosphate |
Solutions from five iterations of growthExpMatch on a PGK knockout growing on glucose using the E. coli core model. The first solution returned is the knocked out reaction. Solutions from iterations 2, 3, and 5 are reactions to utilize 3-Phospho-D-glycerol phosphate while the solution from iteration 4 bypasses the gap by converting D-Fructose 6-phosphate to D-Erythrose 4-phosphate and Acetyl phosphate.
Optimize product secretion using the E. coli core model
Set Up Model for Strain Design
Navigate to the directory (testing/testMaps) containing the E. coli core model and execute the following commands.
≫ model = readCbModel(‘ecoli_core_model.xml’);
Adjust the minimal medium composition to be anaerobic and contain a supply of glucose (20 mmol · gDW−1 ·· h−1).
≫ model = changeRxnBounds(model, {‘EX_o2(e)’, ‘EX_glc(e)’}, [0–20], ‘l’);
Build a list of candidate reactions for deletion to optimize product formation. It is wise to exclude exchange and transport reactions, and biomass and ATP maintenance requirements.
≫ selectedRxns = {model.rxns{ [1, 3:5, 7:8, 10, 12, 15:16, 18, 40:41, 44, 46, 48:49, 51, 53:55, 57, 59:62, 64:68, 71:77, 79:83, 85:86, 89:95]}};
Optknock Analysis of Model
To optimize for lactate secreation with 5 deletions or less using the optKnock method:
≫ options.targetRxn = ‘EX_lac-D(e)’;
≫ options.vMax = 1000;
≫ options.numDel = 5;
≫ options.numDelSense = ‘L’;
≫ constrOpt.rxnList = {‘Biomass_Ecoli_core_N(w/GAM)-Nmet2’, ‘ATPM’}
≫ constrOpt.values = [0.05, 8.39];
≫ constrOpt.sense = ‘GE’;
≫ optKnockSol = OptKnock(model, selectedRxns, options, constrOpt);
The resulting knockout list (optKnockSol.rxnList) is alcohol dehydrogenase, fumarase, glutamate dehydrogenase, malic enzyme (NADP), pyruvate kinase (Table 4). The resulting knockout predicted growth rate of ~0.142 and product excretion rate of ~37.7. The computational time required for this simulation was ~4 seconds with the Gurobi MILP solver.
Table 4.
Expected results from OptKnock and GDLS optimizations for lactate, succinate and pyruvate production growing on glucose
| OptKnock | GDLS | |||||
|---|---|---|---|---|---|---|
| Target | Lactate | Succinate | Pyruvate | Lactate | Succinate | Pyruvate |
| Knockout List | ALCD2x, FUM, GLUDY, ME2, PYK | ALCD2x, GLUDy, LDH_D, PFL, THD2 | ACALD, LDH_D, PTAr | ACALD, FRD7, GLUDy, ME2, PYK | ALCD2x, GLUDy, LDH_D, PFL, THD2 | ACALD, LDH_D, PTAr |
| Product (mmol · gDW−1 · h−1) | 37.7 | 20.2–26.1 | 18.9 | 37.7 | 20.2–26.1 | 18.9 |
| Biomass (mmol · gDW−1 · h−1) | 0.142 | 0.120 | 0.151 | 0.142 | 0.120 | 0.151 |
| Computational Time (s) | 28.1 | 24.5 | 5.6 | 4.9 | 2.9 | 1.8 |
The solutions for succinate and pyruvate are the same for both methods. The lactate solutions vary by two reactions; however, both resulting models have the same production and growth rates.
Strain Design using GDLS
Execute the following commands to optimize for the lactate secretion using the GDLS algorithm.
≫ [gdlsSolution, bilevelMILPproblem, gdlsSolutionStructs] = GDLS(model, ‘EX_lac-D(e)’, ‘minGrowth’, 0.05, ‘selectedRxns’, selectedRxns, ‘maxKO’, 5, ‘nbhdsz’, 3);
The resulting knockout list (gdlsSolution.KOs) is acetaldehyde dehydrogenase, fumarate reductase, glutamate dehydrogenase, phosphotransacetylase, and NAD(P) transhydrogenase (Table 4). The resulting knockout predicted growth rate of ~0.14 and product excretion rate of ~37.7. The computational time required for this simulation was ~2 seconds with the Gurobi LP solver.
Both methods were also used to optimize for succinate product with a maximum of 5 knockouts and pyruvate with a maximum of 3 knockouts. For succinate and pyruvate, the list of reactions to knock out was the same for both OptKnock and GDLS, however, two reactions were different when the target product was lactate (Table 4). For lactate, OptKnock chose alcohol dehydrogenase and fumarase, while GDLS chose acetaldehyde dehydrogenase, fumarate reductase. However, both result in the same optimal flux distribution.
Build a Draft Tissue-Specific Human Macrophage Model from the Global Human Metabolic Network and Omics Data
Navigate to testing/testTissueModel. To save time, we have provided a MAT file (testTissueModel.mat) that contains the global human metabolic network model and a formatted expressionData structure. The model is the version of the human metabolic network reconstruction70 that was used to create an alveolar macrophage model14 using expression data from Kazeros et al.71
≫ load(‘testTissueModel.mat’)
Build a Draft Model with GIMME
The GIMME algorithm retains reactions from Recon 1 that are orphans or are present in the high-throughput data. The reactions with no detected expression are minimized and those not required to retain flux through the objective reaction are removed.
≫ [tissueModel,Rxns] = createTissueSpecificModel(model,expressionData);
Where: tissueModel is the GIMME algorithm-derived draft model; and Rxns is a structure with lists of all the reactions. The reactions fall into the following categories: Expressed - 1769 potentially active reactions based on transcriptome data; UnExpressed - 497 reactions without requisite gene products based on transcriptome data; unknown -41 reactions unable to be predicted by transcriptome data; Upregulated - 52 UnExpressed reactions added back into model; Downregulated - 0 Expressed reactions removed from model; and UnknownIncluded - 1476 orphan reactions included. The calculations should take ~50 seconds with Gurobi LP solver for GIMME.
Supplementary Material
Acknowledgments
We would like to thank individuals who have contributed to or tested COBRA 2.0 –Steinn Gudmundsson, Tom Conrad, Neema Jamshidi, Richard Notebaart, and Jake Feala.
Funding was provided in part by the National Institute of Allergy and Infectious Diseases NIH/DHHS through interagency agreement Y1-AI-8401-01, NIH grants : GM68837-05A1, DE-PS02-08ER08-01, GM057089-12, and GM057089-11S1, and the CalIT2 Summer scholars program. RF and IT were funded by U.S. Department of Energy, Offices of Advanced Scientific Computing Research and the Biological and Environmental Research as part of the Scientific Discovery Through Advanced Computing program, grant DE-SC0002009.
Footnotes
Author Contributions
JS, RQ, RMTF, IT, JDO, AMF, DCZ, AB, NEL, SR, JK, and DRH contributed modules to the COBRA Toolbox v2.0. DRH, JS, RQ, AB, JDO, NEL, and BØP wrote the manuscript
Competing Interests
Bernhard Ø. Palsson serves on the scientific advisory board of Genomatica, Inc.
Contributor Information
Jan Schellenberger, Email: jschelle@ucsd.edu, Bioinformatics Program, University of California San Diego, La Jolla, CA, USA.
Richard Que, Email: rque@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA.
Ronan M. T. Fleming, Email: ronan.mt.fleming@gmail.com, Science Institute & Center for Systems Biology, University of Iceland, Reykjavik, Iceland
Ines Thiele, Email: ithiele@hi.is, Faculty of Industrial Engineering, Mechanical Engineering & Computer Science & Center for Systems Biology, University of Iceland, Reykjavik, Iceland.
Jeffrey D. Orth, Email: jorth@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
Adam M. Feist, Email: afeist@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
Daniel C. Zielinski, Email: dczielin@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
Aarash Bordbar, Email: aabordba@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA.
Nathan E. Lewis, Email: n1lewis@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
Sorena Rahmanian, Email: srahmani@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA.
Joseph Kang, Email: j9kang@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA.
Daniel R. Hyduke, Email: hyduke@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
Bernhard Ø. Palsson, Email: palsson@ucsd.edu, Bioengineering Department, University of California San Diego, La Jolla, CA, USA
References
- 1.Feist AM, et al. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng. 2010;12:173–86. doi: 10.1016/j.ymben.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26:659–67. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 5.Gianchandani EP, Joyce AR, Palsson BO, Papin JA. Functional states of the genome-scale Escherichia coli transcriptional regulatory system. PLoS Comput Biol. 2009;5:e1000403. doi: 10.1371/journal.pcbi.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brynildsen MP, Wong WW, Liao JC. Transcriptional regulation and metabolism. Biochem Soc Trans. 2005;33:1423–1426. doi: 10.1042/BST0331423. [DOI] [PubMed] [Google Scholar]
- 7.Thiele I, Fleming RM, Bordbar A, Schellenberger J, Palsson BO. Functional characterization of alternate optimal solutions of Escherichia coli’s transcriptional and translational machinery. Biophys J. 2010;98:2072–81. doi: 10.1016/j.bpj.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiele I, Jamshidi N, Fleming RM, Palsson BO. Genome-scale reconstruction of Escherichia coli’s transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5:e1000312. doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Thiele I, Jamshidi N, Palsson BO. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput Biol. 2009;5:e1000292. doi: 10.1371/journal.pcbi.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyduke DR, Palsson BØ. Towards genome-scale signalling-network reconstructions. Nat Rev Genet. 2010;11:297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- 12.Raman K, Vashisht R, Chandra N. Strategies for efficient disruption of metabolism in Mycobacterium tuberculosis from network analysis. Mol Biosyst. 2009;5:1740–51. doi: 10.1039/B905817F. [DOI] [PubMed] [Google Scholar]
- 13.Becker SA, Palsson BO. Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation. BMC Microbiol. 2005;5:8. doi: 10.1186/1471-2180-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bordbar A, Lewis NE, Schellenberger J, Palsson BO, Jamshidi N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol Syst Biol. 2010;6:422. doi: 10.1038/msb.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiele I, Price ND, Vo TD, Palsson BO. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280:11683–95. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 16.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Micro. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 17.Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protocols. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 18.Notebaart RA, Teusink B, Siezen RJ, Papp B. Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput Biol. 2008;4:e26. doi: 10.1371/journal.pcbi.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev. 2009;33:164–90. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raman K, Yeturu K, Chandra N. targetTB: A target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. 2008;2:109. doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez A, et al. Impact of the solvent capacity constraint on E. coli metabolism. BMC Syst Biol. 2008;2:7. doi: 10.1186/1752-0509-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90:1453–61. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming RM, Thiele I, Nasheuer HP. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophys Chem. 2009;145:47–56. doi: 10.1016/j.bpc.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming RM, Thiele I. von Bertalanffy 1.0: a COBRA toolbox extension to thermodynamically constrain metabolic models. Bioinfomatics. 2010 doi: 10.1093/bioinformatics/btq607. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker SA, Palsson BO. Context-Specific Metabolic Networks Are Consistent with Experiments. PLoS Comput Biol. 2008;4:e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotech. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 27.Schellenberger J, Palsson BO. Use of randomized sampling for analysis of metabolic networks. J Biol Chem. 2009;284:5457–61. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- 28.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotech. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orth JD, Palsson BØ. Systematizing the generation of missing metabolic knowledge. Biotechnology and Bioengineering. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palsson B. Metabolic systems biology. FEBS Lett. 2009;583:3900–4. doi: 10.1016/j.febslet.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schellenberger J, Park J, Conrad T, Palsson B. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palsson BO. Two-dimensional annotation of genomes. Nat Biotechnol. 2004;22:1218–9. doi: 10.1038/nbt1004-1218. [DOI] [PubMed] [Google Scholar]
- 33.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 34.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong SH, et al. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotech. 2004;22:1275–1281. doi: 10.1038/nbt1010. [DOI] [PubMed] [Google Scholar]
- 36.Mo M, Palsson B, Herrgard M. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Systems Biology. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogales J, Palsson B, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Systems Biology. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nookaew I, et al. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol. 2008;2:71. doi: 10.1186/1752-0509-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuepfer L, Sauer U, Blank LM. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005;15:1421–30. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez O, et al. Reconstruction, modeling & analysis of Halobacterium salinarum R-1 metabolism. Mol Biosyst. 2008;4:148–59. doi: 10.1039/b715203e. [DOI] [PubMed] [Google Scholar]
- 42.Heinemann M, Kummel A, Ruinatscha R, Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92:850–64. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- 43.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smallbone K, Simeonidis E. Flux balance analysis: A geometric perspective. Journal of Theoretical Biology. 2009;258:311–315. doi: 10.1016/j.jtbi.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Satish Kumar V, Dasika M, Maranas C. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgard AP, Pharkya P, Maranas CD. Optknock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnology and Bioengineering. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 47.Patil K, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lun DS, et al. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schellenberger J, Lewis NE, Palsson BØ. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophysical J. 2011;200:544–53. doi: 10.1016/j.bpj.2010.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price ND, Schellenberger J, Palsson BO. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys J. 2004;87:2172–86. doi: 10.1529/biophysj.104.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiback SJ, Famili I, Greenberg HJ, Palsson BO. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. J Theor Biol. 2004;228:437–47. doi: 10.1016/j.jtbi.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–43. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 53.Reed JL, et al. Systems approach to refining genome annotation. Proceedings of the National Academy of Sciences. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:17845–50. doi: 10.1073/pnas.1005139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–82. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 56.Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 57.Bornstein BJ, Keating SM, Jouraku A, Hucka M. LibSBML: an API Library for SBML. Bioinformatics. 2008;24:880–881. doi: 10.1093/bioinformatics/btn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keating SM, Bornstein BJ, Finney A, Hucka M. SBMLToolbox: an SBML toolbox for MATLAB users. Bioinformatics. 2006;22:1275–1277. doi: 10.1093/bioinformatics/btl111. [DOI] [PubMed] [Google Scholar]
- 59.Feist AM, Palsson BO. The biomass objective function. Current Opinion in Microbiology. 2010;13:344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varma A, Palsson BO. Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. Journal of Theoretical Biology. 1993;165:477–502. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 61.Lewis NE, et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metabolic Engineering. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Fischer E, Zamboni N, Sauer U. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Analytical Biochemistry. 2004;325:308–316. doi: 10.1016/j.ab.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 65.Wiechert W, Möllney M, Isermann N, Wurzel M, de Graaf AA. Bidirectional reaction steps in metabolic networks: III. Explicit solution and analysis of isotopomer labeling systems. Biotechnology and Bioengineering. 1999;66:69–85. [PubMed] [Google Scholar]
- 66.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): A novel framework for modeling isotopic distributions. Metabolic Engineering. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waygood EB, Sanwal BD. The control of pyruvate kinases of Escherichia coli. I. Physicochemical and regulatory properties of the enzyme activated by fructose 1,6-diphosphate. J Biol Chem. 1974;249:265–74. [PubMed] [Google Scholar]
- 70.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104:1777–82. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazeros A, et al. Overexpression of apoptotic cell removal receptor MERTK in alveolar macrophages of cigarette smokers. Am J Respir Cell Mol Biol. 2008;39:747–57. doi: 10.1165/rcmb.2007-0306OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.