Abstract
EIF2AK3 is a type I transmembrane protein that functions as an endoplasmic reticulum (ER) stress sensor to regulate global protein synthesis. Rare mutations in EIF2AK3 cause Wolcott-Rallison syndrome (OMIM 226980), an autosomal recessive disorder characterized by diabetes, epiphyseal dysplasia, osteoporosis, and growth retardation. To investigate the role of common genetic variation in EIF2AK3 as a determinant of bone mineral density (BMD) and osteoporosis, we sequenced all exons and flanking regions and then genotyped 6 potentially functional single nucleotide polymorphisms (SNPs) in this gene in 997 Amish subjects for association analysis, with attempted replication in 887 Mexican Americans. We found that the minor allele of a nonsynonymous SNP rs13045 had borderline associations with decreased forearm BMD in both discovery and replication cohorts (unadjusted P = 0.036 and β = −0.007 for the Amish; unadjusted P = 0.031 and β = −0.008 for Mexican Americans). A meta-analysis indicated this association achieved statistical significance in the combined sample (unadjusted P = 0.003; Bonferroni corrected P = 0.009). Rs13045 and three other potentially functional SNPs, a promoter SNP (rs6547787) and two nonsynonymous SNPs (rs867529 and rs1805165), formed two haplotypes (a low-BMD associated haplotype, denoted haplotype B (minor allele frequency (MAF) = 0.311) and a common haplotype A (MAF = 0.676)). There were no differences in mRNA expression from lymphoblastoid cell lines between the two haplotypes. However, after treating lymphoblastoid cell lines with thapsigargin to induce ER stress, cell lines with haplotype B showed increased sensitivity to ER stress (P = 0.014) compared to cell lines with haplotype A. Taken together, our results suggest that common nonsynonymous sequence variants in EIF2AK3 have a modest effect on ER stress response and may contribute to the risk for low BMD through this mechanism.
Keywords: EIF2AK3, polymorphisms, haplotype, ER stress, bone mineral density, osteoporosis
Introduction
Osteoporosis, which increases significantly with aging, is a major public health burden. It is characterized by reduced bone strength and susceptibility to low trauma fractures [1]. Osteoporosis aggregates in families, and the heritability of BMD has been estimated to range from 0.5 to 0.8 [2–5]. Genetic association studies have identified a number of susceptibility loci and variants; however, these largely have very small effects and the mechanisms whereby they act are poorly understood. Delineating the genetic influence in osteoporosis and BMD is important to improve our understanding of the pathophysiology and to provide better prevention and treatment for the disease.
Endoplasmic reticulum (ER) stress is the consequence of the deleterious accumulation of unfolded proteins in the lumen of the ER, which exceeds the capacity of ER-associated protein degradation [6]. There are three known ER stress-sensor proteins, IRE1, ATF6, and EIF2AK3. While the Ire1 knockout is an embryonic lethal [7] and Atf6 knockout mice have no overt phenotype [8, 9], EIF2AK3 mutations in man and mice lead to severe developmental defects. In humans, mutations in EIF2AK3 cause Wolcott-Rallison syndrome (WRS, OMIM 226980), a rare autosomal recessive disorder characterized by infancy-onset insulin dependent diabetes, multiple epiphyseal dysplasia, and osteoporosis [10]. Complete loss-of-function mutations or deletions/insertions in EIF2AK3 segregated with WRS in several families [11, 12]. Eif2ak3 knockout mice also exhibit insufficient proliferation of pancreatic islets, which results in diabetes mellitus, and skeletal defects, including deficient mineralization, osteoporosis, and abnormal compact bone development [13, 14]. Therefore, EIF2AK3 is essential in bone and pancreas, two organs that have a high demand for protein synthesis and processing and thus susceptibility to dysfunction by high and/or prolonged ER stress with aging.
EIF2AK3 is a type I transmembrane protein and activated by ER stress [15–17]. Activation of EIF2AK3 promptly leads to phosphorylation of its single known substrate, the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α), on serine residue 51, which then results in attenuation of the rate of translation and consequent repression of global protein synthesis [18]. EIF2AK3 expression is ubiquitous but especially abundant in osteoblasts and pancreatic islet cells [13, 14]. The ability of osteoblasts to cope with the high endogenous levels of ER stress is critical for type I collagen synthesis to maintain bone mass. Therefore, we hypothesized that common functional sequence variants in EIF2AK3 might alter response to ER stress and thus influence BMD and susceptibility to osteoporosis. Here we report a consistent association between common nonsynonymous EIF2AK3 variants and BMD in two independent populations and further show that the low BMD haplotype marked by the associated SNPs exhibits increased phosphorylation of eIF2α during ER stress compared to the alternate haplotype.
Materials and Methods
Old Order Amish Subjects
The Amish Family Osteoporosis Study (AFOS) was initiated in 1997 with the goal of identifying the genetic determinants of osteoporosis in the Old Order Amish. Detailed information about recruitment, phenotyping and clinical characteristics of participants has been previously described [19, 20]. Briefly, any Amish individuals found to have a T score of −2.5 or less in either the hip or spine were recruited and designated as probands. The probands' spouses and all first-degree relatives aged 20 years and over were invited to participate in the study. Thirteen individuals diagnosed with osteogenesis imperfecta on the basis of having a COL1A2 mutation were excluded from the analysis [21]. This report contains data from 972 AFOS subjects (617 women and 385 men), including 57 osteoporotic probands. All AFOS participants can be connected into a single 14-generation pedigree by including additional ancestors. BMD was measured by dual energy X-ray absorptiometry (DXA), using a Hologic Model 4500 W (Hologic Inc., Bedford, MA). The region of BMD measurements included the lumbar spine (L1–L4), hip (trochanter, intertrochanter, femoral neck and total) and forearm (the radius and ulna combined ultradistal, 1/3, midpoint, and total). The protocol was approved by the Institutional Review Board of the University of Maryland.
Mexican American Subjects
The San Antonio Family Osteoporosis Study (SAFOS) was initiated in 1997, with the goal of identifying susceptibility genes for osteoporosis in Mexican Americans. Details of sampling and recruitment procedures have been reported previously [5, 22]. A total of 887 individuals from 34 families were recruited, with ages ranging from 18 to 96 years. Prior to participation, all subjects provided written informed consent under protocols approved by the Institutional Review Board at the University of Texas Health Science Center at San Antonio. Phenotypes including height, weight, BMD, physical activity and medical history (such as diabetes status) were collected at the General Clinical Research Center located at the Audie Murphy Veteran's Administration Hospital on the campus of the Health Science Center in San Antonio. BMD was measured using a Hologic Model 1500W DXA (Hologic, Inc., Bedford, MA) at the same sites as for the AFOS. Physical activity of each study participant was scored into weekly metabolic equivalents (METs) using a modified version of the Stanford 7-Day Physical Activity Recall Instrument [23, 24].
Sequencing and Genotyping
EIF2AK3 is approximately 71 kilobases (kb) in length, containing 16 exons. To capture potentially functional variants, we sequenced approximately 7 kb of the 5' flanking region, all exons, 5' and 3' UTR, splice junctions and 2 kb of the 3' flanking region. Genomic DNA samples were obtained from 46 Amish subjects (23 with spine BMD Z-score > 2 and 23 with spine BMD Z-score < −2.5). Nineteen sets of primers were designed using Primer 3.0 software (Whitehead Institute for Biomedical Research, Boston, MA). PCR products were amplified and then sequenced as previously described [25]. All SNP genotyping was performed using TaqMan SNP genotyping assays, according to the manufacturer's instructions, and analyzed using SDS 2.1 software (Applied Biosystems, Foster City, CA). The genotype concordance rate was greater than 98% in 180 duplicate samples for both the Amish and Mexican American samples.
Cell Culture
Epstein-Barr (EB) virus-transformed lymphoblastoid cell lines derived from haplotype-selected Amish individuals were cultured in RPMI-1640 (Sigma) and grown at 37° C with 5% CO2. The media was supplemented with 2 mM L-glutamine, 10% heat-inactivated fetal bovine serum (Invitrogen), and antibiotics (penicillin/streptomycin, 100 units/ml, Invitrogen).
Allele-Specific Expression
Allele-specific expression of the promoter SNP rs6547787 was examined in transformed lymphoblasts from 7 individuals heterozygous for the coding SNP rs13045 (G/A) located within the mRNA sequence and in linkage disequilibrium (LD) with rs6547787 (r2 = 0.99). Total RNA was obtained and was used for cDNA synthesis using the gene-specific primer (GAACATCGATGACAAGCTTAGGTATCGATAATTTTCCTCCAACCAAAACAAC) and Superscript III reverse transcriptase (Invitrogen). The 194 bp PCR products containing rs13045 were amplified from both cDNA and genomic DNA (Forward: TTGGGAATAAGATGATCATTCCTT; Reverse: TTTCCACTATATGCACTGAGTCC). Allelic expression of rs13045 in cDNA or genomic DNA was measured using Pyrosequencing (sequencing primer: CATGCTTTCACGGTCT) and quantified using Allele Quantification software (Pyrosequencing, Inc., Uppsala, Sweden) according to the manufacturer's methods. The allelic expression ratio (G/A) in cDNA was obtained for each sample, and correction was performed by dividing the mRNA ratio by the ratio obtained from amplification of genomic DNA from the same heterozygous subject. Each sample was assayed 3 times and the mean values calculated.
ER Stress and Immunoblot Analyses
To determine if three nonsynonymous variants affect EIF2AK3 function, we first measured changes in phospho-eIF2α levels in lymphoblasts before and after induction of ER stress by thapsigargin (TG), an ER Ca2+-ATPase inhibitor [16, 17, 26, 27]. eIF-2α is the only known substrate of EIF2AK3, and its phosphorylation levels have been widely used to measure EIF2AK3 activity [16, 17, 26, 27]. Initially we carried out assay optimization studies in which lymphoblastoid cells were cultured and treated with TG at different concentrations and phospho-eIF2A was measured at varying time intervals (see Supplementary Materials).
A total of 14 lymphoblastoid cell lines were selected from Amish participants with the two major haplotypes based on both nonsynonymous rs13045 and rs6547787 genotypes, including 7 homozygotes for the major alleles (GG+AA; Haplotype A) and 7 homozygotes for the minor alleles (AA+CC; Haplotype B). The 14 cell lines were randomly grouped into 7 pairs of haplotype A vs. haplotype B. Each pair was treated in parallel, and each cell line was treated in duplicate. Cells were seeded onto 10-cm dishes at a density of ~2×105 /ml to maintain low cell density after about 15 hours of growth [28]. To induce ER stress, cells were treated with thapsigargin (TG, Calbiochem) dissolved in dimethysulfoxide (Sigma). The TG concentrations and the treatment intervals are indicated in Figure 4. After treatment, cells were pelleted by centrifugation and lysed in lysis buffer (100 μl/1×106 cells) (50 mM Tris-HCl pH 6.8, 2% SDS with freshly added phosphatase inhibitors: 5 mM sodium orthovanadate and 20 mM β-glycerophosphate). Lysates were boiled for 3 min, vortexed for 30 sec and frozen at −80 °C; the boiling-freezing process was then repeated. Lysates were cleared by centrifugation at 14,000×g for 15 min at 4°C. The supernatant (20 μl of each sample) were resolved by SDS-PAGE electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were incubated with anti-phospho-eIF2α antibody (1:1000 dilution; Cell Signaling) or anti-EIF2AK3 (1:200, Santa Cruz Biotechnology) overnight at 4°C and were detected using enhanced chemiluminescence reagent (Thermo Fisher) and fluorography. Membranes were then stripped (Western Blot Stripping Buffer, Thermo Fisher), reprobed anti-eIF2α antibody (1:1000 dilution; Cell signaling) for 2 h at room temperature, and detected as described. Densitometric measurements were conducted using Quantity One software (Bio-Rad). Phospho-eIF2α levels were quantitated from the immunoblot analysis and were corrected for the level of total eIF2α. One of the cell lines treated with 2 μM TG was used as a control for each immunoblot analysis to normalize for interexperimental variability in signal strength.
Figure 4.
Activation of eIF2α signaling in lymphoblastoid cell lines with differing haplotypes.
(A) Lymphoblastoid cells homozygous for the common haplotype A (HapA) or for the low-BMD haplotype B (HapB) were untreated as control (Con.) or treated with indicated concentrations of TG for 15 minutes. Shown are the typical results of phosphorylated eIF2α, total eIF2α, and EIF2AK3, detected by Western blots with indicated antibodies. (B) Densitometric analysis of phosphorylated eIF2α (P-eIF2α) and total eIF2α (T-eIF2α) (n = 7 for each genotype). AU, arbitrary units. The basal levels were used as the correction for after treatment levels. (* P = 0.014)
Statistical Analyses
Summary statistics of population characteristics and phenotypes were generated using SAS version 9.1 (SAS Institute Inc, Cary, NC). Association analysis between SNPs and BMD were performed under a variance component model that evaluates the effect of each genotype as an additive effect on BMD, while simultaneously estimating the effects of measured environmental covariates and a polygenic component to account for phenotypic correlation due to relatedness [29, 30]. When analyzing the Amish data set, age, age2, sex, and BMI were included as covariates. The polygenic component was modeled using the relationship matrix derived from the complete Amish pedigree structure available through published genealogical records [31]. For analyses of the SAFOS data set, the relationship matrix included 887 individuals from 34 families, and genotype effects were additionally adjusted for the effects of physical activity (expressed as total METs) and diabetes status (present/absent) because these variables were previously found to be significantly associated with BMD in this population [5]. We used a Bonferroni approach to set a threshold for statistical significance in which we divided the test-wide threshold of 0.05 by the number of independent SNPs tested (r2<0.8) in the discovery sample. SNPs having P-values less than 0.05 in both discovery and replication populations and also having the same direction of effect sizes were considered to be replicated. Statistical power, estimated using QUANTO software [32], indicated that each of our family-based cohorts has 80% power to detect common SNPs (MAF>0.05) accounting for approximately 2% of the quantitative phenotypic variation at α = 0.017 level.
A meta-analysis of the combined Amish and Mexican American association results, was performed using the METAL software, applying inverse-variance methodology to quantify the heterogeneity between studies [33]. Population heterogeneity tests were conducted using Cochran's Q-test as implemented in METAL (Supplementary Table 1) [34]. The random effects model was used when heterogeneity test was significant; otherwise the meta-analysis was conducted using fixed effects methods [35]. Pairwise LD correlation statistics (r2 and |D'|) and haplotype frequencies were assessed using Haploview software (http://www.broad.mit.edu).
In allele-specific expression, we tested for distortion of allelic ratios in cDNA by comparing the observed sample mean to a hypothesized population mean (μ0 = 1). The Z statistic was used to compute the P value. For immunoblot analyses, the results are presented as mean ± s.e.m. Statistical significance was estimated by a 2-tailed Student's t-test using GraphPad PRISM version 5.00 for Windows (GraphPad Software, San Diego, CA).
Results
BMD Association Studies
Characteristics of the discovery set (972 Amish) and the replication set (887 Mexican Americans) are shown in Table 1. On average, the Amish subjects were about 8 years older and had lower BMI than the Mexican-American subjects. The hip and spine BMDs of the Amish were lower than those of Mexican Americans, whereas the forearm BMD was higher in the Amish.
Table 1.
Participant characteristics in the Amish and Mexican Americans (presented as mean and standard deviation).
| Old Order Amish | Mexican Americans | |
|---|---|---|
| Sample Size | 972 | 887 |
| Female (%) | 596 (61.3%) | 528 (59.5%) |
| Age (year) | 50.4 ± 16.1 | 42.8± 16.0 |
| BMI (kg/m2) | 27.4 ± 5.4 | 30.9± 7.2 |
| Spine BMD (g/cm2) | 0.921 ± 0.149 | 1.028 ± 0.147 |
| Femoral Neck BMD (g/cm2) | 0.831 ± 0.148 | 0.860 ± 0.139 |
| Forearm BMD (g/cm2) | 0.603 ± 0.087 | 0.581± 0.079 |
| Spine Z-score | −0.57 (1.26) | 0.08 (1.17) |
| Femoral neck Z-score | −0.17 (1.14) | 0.19 (1.15) |
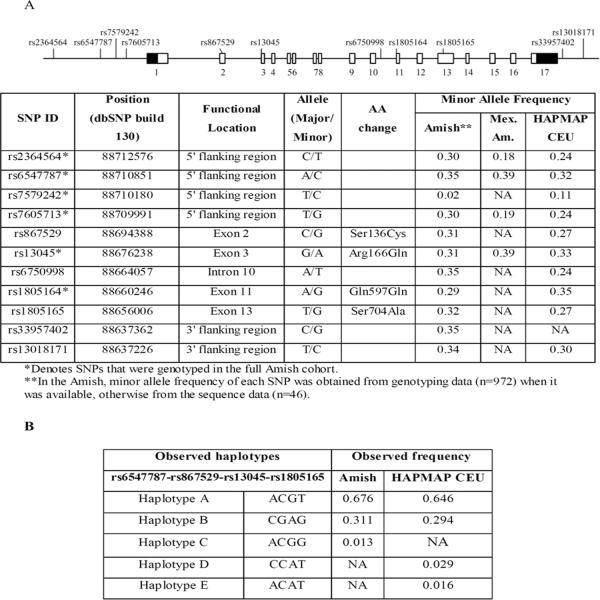
A total of 11 SNPs in EIF2AK3 were identified by sequencing, including four SNPs located in the promoter region (rs2364564, rs6547787, rs7579242, and rs7605713), three nonsynonymous polymorphisms (rs867529 (S136C), rs13045 (R166Q), and rs1805165 (S704A)), and one synonymous SNP (rs1805164 (Q579Q)) (Figure 1A). We genotyped six potentially functional SNPs (four SNPs in the promoter, a nonsynonymous SNP rs13045, and a synonymous SNP rs1805164) in 972 subjects from the AFOS and attempted replication by genotyping these SNPs in 887 Mexican Americans. The other nonsynonymous variants (rs867529 and rs1805165) were not genotyped because they were in high LD (r2 ≥ 0.94) with two genotyped SNPs (rs6547787 and rs13045). The genotype frequencies of all SNPs were in Hardy-Weinberg equilibrium and were not significantly different between Amish and the HAPMAP CEU data (Figure 1A). As shown in Figure 2, the 6 SNPs genotyped included 3 sets of highly correlated SNPs: set 1 (rs2364564 and rs7605713), set 2 (rs6547787 and rs13045), and set 3 (rs1805164). SNP rs7579242 was not highly correlated with either set, but had a MAF of only 0.02 in the Amish. Because of its low allele frequency and subsequently low power to detect associations with this SNP in our data, we adjusted for only 3 sets of `independent' comparisons in our analyses, leading to a multiple adjusted significance threshold of 0.017 (0.05/3).
Figure 1.
EIF2AK3 SNPs and haplotypes identified by DNA sequencing in the Amish.
(A) Diagram of the EIF2AK3 gene and location and frequency of SNPs. (B) Haplotypes observed in the Amish and HAPMAP CEU dataset.
Figure 2.
Linkage disequilibrium plots of 11 SNPs in EIF2AK3 in the Amish presented as |D'| (A) and r2 (B).
Approximate location of each SNP is shown at the top. Value in each box indicates |D'| or r2 value between two SNPs (intersection) and ranges from 0 to 0.99. No value in the box indicates complete LD (r2 = 1).
One of the six SNPs was significantly associated with BMD at the total hip (P < 0.017) in the Amish, and three additional SNPs were nominally associated with variation in BMD at one or more skeletal sites (P = 0.017–0.037) (Table 2 and Supplementary Table 1). Among these, the association of a nonsynonymous SNP rs13045 with forearm BMD was nominally replicated in Mexican-Americans (P = 0.031, Table 2). Although this association just missed the Bonferroni-corrected threshold for statistical significance in both populations, the direction of this association was the same and the effect sizes were similar in the two populations (β = −0.007, P = 0.036 for the Amish, and β = −0.008, P = 0.031 for Mexican Americans). A meta-analysis combining results from both populations revealed the minor alleles of rs13045 (A allele) and rs6547787 (C allele) to be associated with decreased forearm BMD (P = 0.003 for both) (Table 2 and Supplementary Table 1). These two SNPs were in high LD and formed a haplotype that also included nonsynonymous SNPs rs867529 and rs1805165 (all r2 ≥ 0.94; see Figure 2B). These 4 SNPs comprised two common haplotypes (denoted haplotype A (ACGT, frequency = 0.676) and haplotype B (CGAG, frequency = 0.311)) that accounted for over 98% of all haplotypes comprising these four SNPs, consistent with the HAPMAP CEU data. We thus hypothesized that the associations of rs13045 and rs6547787 with BMD could arise from functional haplotype B (CGAG) leading to lower BMD.
Table 2.
Associations between EIF2AK3 SNPs (rs6547787, rs7579242, and rs13045) with BMD in the Amish and Mexican Americans.
| Marker ID | Skeletal Site BMD | Discovery set: Old Order Amish | Replication set: Mexican American | Meta Analysis | ||||
|---|---|---|---|---|---|---|---|---|
| N | Beta* | Pval** | N | Beta* | Pval** | P value | ||
| rs6547787 | Lumbar spine | 879 | −0.017 | 0.017 | 752 | 0.000 | 0.919 | 0.068 |
| Total hip | 879 | −0.004 | 0.467 | 751 | 0.000 | 0.919 | 0.642 | |
| Femoral neck | 879 | −0.008 | 0.203 | 751 | 0.001 | 0.919 | 0.387 | |
| Total forearm | 811 | −0.007 | 0.018 | 766 | −0.006 | 0.068 | 0.003 | |
| Forearm midpoint | 810 | −0.007 | 0.037 | 766 | −0.004 | 0.168 | 0.014 | |
| rs7579242 | Lumbar spine | 933 | 0.026 | 0.215 | 764 | −0.013 | 0.919 | 0.340 |
| Total hip | 934 | 0.045 | 0.013 | 763 | −0.020 | 0.321 | 0.24 | |
| Femoral neck | 934 | 0.031 | 0.089 | 764 | −0.030 | 0.098 | 0.881 | |
| Total forearm | 867 | 0.015 | 0.078 | 776 | 0.006 | 0.919 | 0.140 | |
| Forearm midpoint | 866 | 0.016 | 0.081 | 776 | 0.010 | 0.232 | 0.035 | |
| rs13045 | Lumbar spine | 917 | −0.017 | 0.019 | 748 | −0.002 | 0.919 | 0.069 |
| Total hip | 919 | −0.008 | 0.202 | 746 | −0.001 | 0.919 | 0.310 | |
| Femoral neck | 919 | −0.011 | 0.082 | 746 | 0.000 | 0.919 | 0.174 | |
| Total forearm | 844 | −0.007 | 0.036 | 762 | −0.008 | 0.031 | 0.003 | |
| Forearm midpoint | 843 | −0.006 | 0.058 | 762 | −0.006 | 0.052 | 0.007 | |
Bonferroni-adjusted threshold for statistical significance = 0.017 for three independent comparisons (see text).
Beta represents the effect of the minor allele of each SNP.
Functional Analysis of EIF2AK3 Haplotypes
We performed functional studies to examine if expression or activity of the EIF2AK3 differed between the two common haplotypes defined by the promoter SNP and three non-synonymous SNPs.
When a promoter variant and a coding SNP are in strong LD, the coding SNP may be used to determine if there is differential expression resulting from the promoter variant by examining the relative amounts of RNA transcripts produced by each allele in heterozygous individuals (allelic distortion) [36, 37]. To test whether the promoter sequence variant (rs6547787) is associated with altered gene expression, we compared the mRNA ratio of two alleles of rs13045 (r2 = 0.99 with rs6547787) in lymphoblastoid cell lines derived from Amish individuals heterozygous for both SNPs. The allelic ratio from genomic DNA from the same individual was tested in parallel to cDNA and used for correction of possible unequal amplification. In seven samples that we tested, the mean standardized ratio of G/A (cDNA/DNA) for rs13045 was 0.93 ± 0.03, which was not significantly different from 1 (P = 0.14) (Table 3). Therefore, we did not observe a distortion in allele-specific expression due to the presence of rs6547787.
Table 3.
Allele-specific expression of two alleles (G/A) of rs13045 in lymphoblastoid cell lines from heterozygous individuals.
| Sample ID | Ratio of G:A In DNA template | Ratio of G:A In cDNA template | Standardized ratio of G/A (cDNA:DNA) |
|---|---|---|---|
| 1 | 0.85 | 0.74 | 0.87 |
| 2 | 0.89 | 0.72 | 0.82 |
| 3 | 0.74 | 0.66 | 0.89 |
| 4 | 0.76 | 0.81 | 1.07 |
| 5 | 0.71 | 0.70 | 1.00 |
| 6 | 0.76 | 0.85 | 1.11 |
| 7 | 0.85 | 0.68 | 0.80 |
| Mean ± s.e.m | 0.93 ± 0.03 |
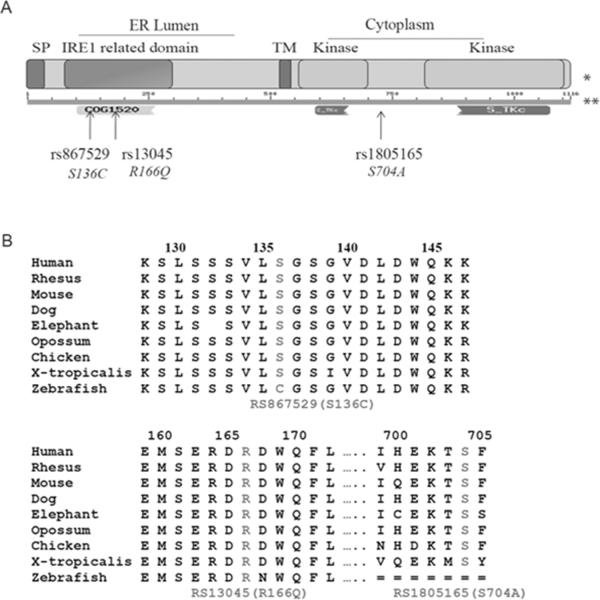
EIF2AK3 is a serine/threonine protein kinase that spans the ER membrane. As shown in Figure 3A, while rs1805165 is in an unknown functional region of the protein, nonsynonymous variants rs867529 and rs13045 are located in a highly conserved region encoding a WD40-like repeat (COG1520). Based on multispecies alignment, the amino acids coded by common alleles of the two SNPs (serine for rs867529 and arginine for rs13045) are highly conserved from zebrafish to human (Figure 3B). Furthermore, this domain (amino acids 102–257) is responsible for dimerization of EIF2AK3 [16], functions as an ER stress sensing domain, and shares homology with IRE1 [15, 17], another ER stress transducer. Therefore, we hypothesized that the amino acid changes in this domain might affect kinase activity of EIF2AK3 by influencing its sensitivity to ER stress.
Figure 3.
Effect of three amino acid changing variants in EIF2AK3.
(A) The location of three nonsynonymous SNPs on the protein. SP, signal peptide; TM, transmembrane domain. The diagram of the protein domains (*) was adapted from [16]. The conserved domains (**) were predicted by multiple sequence alignments in the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?seqinput=NP_004827.4).
(B) Multispecies alignment of amino-acid sequences around three nonsynonymous SNPs, obtained from the UCSC genome browser.
We obtained transformed lymphoblastoid cell lines from Amish individuals who are homozygous for haplotype A or haplotype B and examined the changes in phospho-eIF2α after ER stress in those cell lines (n = 7 for each haplotype). There were no differences in the levels of the EIF2AK3 protein between cell lines of the two haplotypes, either before or after TG treatment (Figure 4A). We observed similar low basal levels of phosphorylated eIF2α in cells with the different haplotypes before TG treatment. After treatment with TG, the relative amount of phosphorylated eIF2α in cells with haplotype B increased about 3-fold, while those with haplotype A increased less than 2-fold, compared to basal levels (P = 0.014) (Figure 4B). We observed a similar response pattern with 2 μM TG treatment, although it did not reach statistical significance (P = 0.076). These data suggest that EIF2AK3 activity is increased in response to ER stress in lymphoblast cells with haplotype B in humans (i.e., cells harboring haplotype B are more sensitive to ER stress).
Discussion
Motivated by a large body of prior data suggesting that alteration of EIF2AK3 function leads to abnormal bone phenotypes in both human patients and knockout mouse models, we tested associations between common SNPs in EIF2AK3 and BMD. We observed several associations in this region, including a nominal association between rs13045, which alters a conserved amino acid in the ER sensing domain of the protein, and lower forearm BMD in two independent populations, with an estimated effect size of ~0.07 g/cm2. This effect size is well within the range (0.06–0.17) of the strongest associations of other loci reported from recent BMD GWA studies in which over 10,000 study subjects were included [38–43].
To date, EIF2AK3 has not been identified by any published GWAS as a bona fide locus associated with BMD even though the BMD-associated variants we identified in EIF2AK3 are common and well represented in the HAPMAP CEU data. However, forearm BMD has not been included in many of the current BMD GWAS studies. The forearm is not affected by weight bearing as are the spine and hip, and forearm BMD is less correlated with lumbar spine and femoral neck BMD, both of which were studied by most BMD-GWAS. It is possible that the SNPs we identified are more strongly correlated with BMD at the forearm than they are with BMD at these other sites. Indeed, although we found rs13045 to be associated with BMD at spine and total forearm in the Amish, only the association with forearm BMD was replicated in Mexican Americans. Possibly, EIF2AK3 may have differential effects on unique skeletal sites or different effects among different populations with different genetic backgrounds or environmental exposures, although this is speculative. BMD, particularly of the forearm, is regulated by parathyroid hormone (PTH), whose concentrations are influenced by vitamin D. However, adjusting for serum intact PTH levels did not alter the observed association of rs13045 and BMD (data not shown). We also performed analyses without adjusting for BMI, and the association results remained the same for forearm BMD and were more significant for femoral neck and spine BMD as we expected (data not shown).
Of note, a GWAS meta-analysis recently identified a SNP rs11684404 in EIF2AK3 to be associated with height (meta-analysis P = 9.9×10−14) [44]. Rs11684404 is in perfect LD with our BMD-associated rs13045 (r2 = 1 in HAPMAP CEU data). Since skeletal structure is an important determinant of height, these data are consistent with our BMD data. However, we did not replicate the association of rs13045 and height in our cohort, likely due to limited study power. A recent progressive supranuclear palsy (PSP) study also identified a genome-wide significant signal (rs7571971) in high LD with rs13045 (r2 > 0.8) [45]. This same study found that EIF2AK3 gene expression in brain tissue was not influenced by this DNA variant [45], which is consistent with our finding that it is ER stress sensitivity, but not protein expression, affected by SNPs in this LD block.
Bone cells such as osteoblasts and osteoclasts produce and secrete large amounts of proteins and experience persistent ER stress to maintain proper protein synthesis and folding. Thus defects in the ability to deal with ER stress may lead to dysfunction in global protein synthesis [18], impaired cell viability and apoptosis [26, 27], and thus low BMD. During ER stress challenge experiments in lymphoblastoid cell lines from AFOS participants, we observed that cell lines homozygous for the low BMD-associated haplotype B had more elevated levels of phosphorylated eIF2α than those cells homozygous for haplotype A, suggesting that they were more sensitive to ER stress. Since this phenomenon was only examined in lymphoblasts, we can only infer cell types such as osteoblasts and osteoclasts would respond similarly. Therefore, further investigation will be necessary to clarify the influence of haplotype B in the function of EIF2AK3 and bone homeostasis.
The amino acid change caused by rs867529 (S136C) is predicted by the Polyphen software to be possibly damaging, while rs13045 (R166Q) and rs1805165 (S704A) are predicted to be benign (http://genetics.bwh.harvard.edu/pph/index.html). However, these three nonsynonymous variants are highly correlated with each other (r2 ≥ 0.94) and constitute a single functional haplotype. Therefore, we could not specify which particular SNP led to an alteration in phosphorylation of eIF2α in response to ER stress.
We did not observe a difference in expression between the two alleles of rs6547787, located in the promoter region of EIF2AK3 and also in linkage disequilibrium with the aforementioned nonsynonymous variants. Interestingly, as predicted by the Patch program (http://www.gene-regulation.com/pub/programs.html#patch), the A-to-C substitution at rs6547787 alters a polypurine tract (from GGAAG to GGCAG) that is a consensus DNA binding site for nuclear factor of activated T-cells (NFAT). All NFAT family members bind to similar DNA sequence sites [46], and NFATc1 has been recognized as a master transcription factor to regulate osteoclast differentiation [47]. Therefore, it is possible that rs6547787 or other unstudied SNPs in LD with rs6547787 may impact gene expression in osteoclasts, but not in lymphoblastoid cells.
EIF2AK3 is highly expressed in both osteoblasts and islet cells, but it is not clear if it functions in the same way in the two cell types. Recent studies showed that EIF2AK3 in β cells regulates insulin secretion by affecting proinsulin's ER-to-Golgi trafficking and degradation, rather than by inhibiting protein synthesis. The Akita-Ins2 mutant mice with decreased EIF2AK3 gene dosage (EIF2AK3+/−) exhibited a delayed diabetic progression compared with the same mutant mice with EIF2AK3+/+[48]. It has been suggested that EIF2AK3 may function differently in response to demand for insulin under normal physiological and stress conditions [49]. Unfortunately, we did not measure glucose and insulin concentrations in this study to enable us to examine possible pleiotropic effects of EIF2AK3 on both BMD and insulin secretion.
In summary, our study explored associations between BMD at various skeletal sites and potentially functional variants identified in EIF2AK3. We found that the minor allele of a nonsynonymous SNP rs13045 was nominally associated with low forearm BMD in two independent populations (and significantly associated in a combined analysis), and that this SNP tags a larger haplotype containing other potentially functional SNPs. In follow-up functional studies, we observed that lymphoblastoid cell lines homozygous for the low BMD-associated haplotype displayed increased sensitivity to TG-induced ER stress. Our results suggest that sequence variants in EIF2AK3 may have a modest effect on variation of BMD and may contribute to risk of osteoporosis through this mechanism.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the families participating in the AFOS and SAFOS. This work was supported by National Institutes of Health, R01-AR46838 and R01 AG18728 for The Amish Family Osteoporosis Study and R01-AR43351 for The San Antonio Family Osteoporosis Study. Additional support was provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488).
This work was supported by National Institutes of Health, R01-AR46838 and R01 AG18728 for The Amish Family Osteoporosis Study and R01-AR43351 for The San Antonio Family Osteoporosis Study. Additional support was provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488).
Footnotes
Authors' roles: Study design: ARS, BDM, JCM and JL. Study conduct: JL and NH. Data collection: JL, BDM, ARS, and EAS. Data analysis: JL, HW, BDM, and JRO. Drafting of manuscript: JL, NH, BDM, and ARS. Revising of manuscript: JL, BDM, ARS, EAS, JRO, HW, JCM, and NH. Obtained funding: ARS and BMD.
There are supplemental materials included with the submission.
Disclosures: The authors state that they have no conflicts of interest.
References
- 1.Consensus development conference: diagnosis, prophylaxis and treatment of osteoporosis. American Journal of Medicine. 1991;90:107–110. [Google Scholar]
- 2.Smith DM, Nance WE, Kang KW, Christian JC, Johnston CC., Jr Genetic factors in determining bone mass. J Clin Invest. 1973;52(11):2800–2808. doi: 10.1172/JCI107476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 4.Brown LB, Streeten EA, Shuldiner AR, Almasy LA, Peyser PA, Mitchell BD. Assessment of sex-specific genetic and environmental effects on bone mineral density. Genet Epidemiol. 2004;27(2):153–161. doi: 10.1002/gepi.20009. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell BD, Kammerer CM, Schneider JL, Perez R, Bauer RL. Genetic and environmental determinants of bone mineral density in Mexican Americans: results from the San Antonio Family Osteoporosis Study. Bone. 2003;33(5):839–846. doi: 10.1016/s8756-3282(03)00246-1. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13(10):1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 7.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106(39):16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Boot-Handford RP, Briggs MD. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res. 339(1):197–211. doi: 10.1007/s00441-009-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickwood S, Bonthron DT, Al-Gazali LI, Piper K, Hearn T, Wilson DI, Hanley NA. Wolcott-Rallison syndrome: pathogenic insights into neonatal diabetes from new mutation and expression studies of EIF2AK3. J Med Genet. 2003;40(9):685–689. doi: 10.1136/jmg.40.9.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25(4):406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 13.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 15.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275(32):24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 16.Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277(21):18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 17.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 18.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 19.Streeten EA, McBride DJ, Lodge AL, Pollin TI, Stinchcomb DG, Agarwala R, Schaffer AA, Shapiro JR, Shuldiner AR, Mitchell BD. Reduced incidence of hip fracture in the Old Order Amish. J Bone Miner Res. 2004;19(2):308–313. doi: 10.1359/JBMR.0301223. [DOI] [PubMed] [Google Scholar]
- 20.Streeten EA, McBride DJ, Pollin TI, Ryan K, Shapiro J, Ott S, Mitchell BD, Shuldiner AR, O'Connell JR. Quantitative trait loci for BMD identified by autosome-wide linkage scan to chromosomes 7q and 21q in men from the Amish Family Osteoporosis Study. J Bone Miner Res. 2006;21(9):1433–1442. doi: 10.1359/jbmr.060602. [DOI] [PubMed] [Google Scholar]
- 21.Daley E, Streeten EA, Sorkin JD, Kuznetsova N, Shapses SA, Carleton SM, Shuldiner AR, Marini JC, Phillips CL, Goldstein SA, et al. Variable Bone Fragility Associated with an Amish COL1A2 Variant and a Knock-in Mouse Model. J Bone Miner Res. 2009 doi: 10.1359/jbmr.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O'Connell JR, Perez R, Dyer TD, Almasy L, Blangero J, et al. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18(12):2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 23.Blair SN, Haskell WL, Ho P, Paffenbarger RS, Jr, Vranizan KM, Farquhar JW, Wood PD. Assessment of habitual physical activity by a seven-day recall in a community survey and controlled experiments. Am J Epidemiol. 1985;122(5):794–804. doi: 10.1093/oxfordjournals.aje.a114163. [DOI] [PubMed] [Google Scholar]
- 24.Sallis JF, Haskell WL, Wood PD, Fortmann SP, Rogers T, Blair SN, Paffenbarger RS., Jr Physical activity assessment methodology in the Five-City Project. Am J Epidemiol. 1985;121(1):91–106. doi: 10.1093/oxfordjournals.aje.a113987. [DOI] [PubMed] [Google Scholar]
- 25.Hoppman N, McLenithan JC, McBride DJ, Shen H, Bruder J, Bauer RL, Shaffer JR, Liu J, Streeten EA, Shuldiner AR, et al. A common variant in fibroblast growth factor binding protein 1 (FGFBP1) is associated with bone mineral density and influences gene expression in vitro. Bone. 2010;47(2):272–280. doi: 10.1016/j.bone.2010.04.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4(1):e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mak BC, Wang Q, Laschinger C, Lee W, Ron D, Harding HP, Kaufman RJ, Scheuner D, Austin RC, McCulloch CA. Novel function of PERK as a mediator of force-induced apoptosis. J Biol Chem. 2008;283(34):23462–23472. doi: 10.1074/jbc.M803194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam N, Sandberg ML, Sugden B. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J Virol. 2004;78(4):1657–1664. doi: 10.1128/JVI.78.4.1657-1664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. Jama. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, O'Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, et al. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106(1):226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwala R, Biesecker LG, Hopkins KA, Francomano CA, Schaffer AA. Software for constructing and verifying pedigrees within large genealogies and an application to the Old Order Amish of Lancaster County. Genome Res. 1998;8(3):211–221. doi: 10.1101/gr.8.3.211. [DOI] [PubMed] [Google Scholar]
- 32.Gauderman WJMJ. QUANTO 1.2: A computer program for power and sample size calculations for genetic-epidemiology studies. 2007 http://hydra.usc.edu/gxe.
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10(1):101–129. [Google Scholar]
- 35.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2(9):e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Zhang Z, Chu W, Hale T, Cooper JJ, Elbein SC. Molecular screening and association analyses of the interleukin 6 receptor gene variants with type 2 diabetes, diabetic nephropathy, and insulin sensitivity. J Clin Endocrinol Metab. 2005;90(2):1123–1129. doi: 10.1210/jc.2004-1606. [DOI] [PubMed] [Google Scholar]
- 37.Shuldiner AR, Tanner K, Moore CA, Roth J. RNA template-specific PCR: an improved method that dramatically reduces false positives in RT-PCR. Biotechniques. 1991;11(6):760–763. [PubMed] [Google Scholar]
- 38.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371(9623):1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, Tang ZH, Pan F, Yang TL, Chen XD, et al. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84(3):388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358(22):2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 43.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Snorradottir S, Center JR, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41(1):15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 44.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43(7):699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20(19):2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 47.Takayanagi H. The role of NFAT in osteoclast formation. Ann N Y Acad Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 48.Gupta S, McGrath B, Cavener DR. PERK (EIF2AK3) regulates proinsulin trafficking and quality control in the secretory pathway. Diabetes. 2010;59(8):1937–1947. doi: 10.2337/db09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavener DR, Gupta S, McGrath BC. PERK in beta cell biology and insulin biogenesis. Trends Endocrinol Metab. 2010;21(12):714–721. doi: 10.1016/j.tem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.