Abstract
Osteocalcin (OC) is a major noncollagenous bone matrix protein and an osteoblast marker whose expression is limited to mature osteoblasts during the late differentiation stage. In previous studies we have shown osteosarcomas to lose p53 function with a corresponding loss of osteocalcin gene expression. Introduction of wild type p53 resulted in re expression of the osteocalcin gene. Using gel shift and chromatin immunopreciptation assays, we have identified a putative p53 binding site within the rat OC promoter region and observed an increase in OC promoter activity when p53 accumulates using a CAT assay. The p53 inducible gene Mdm2 is a well-known downstream regulator of p53 levels. Our results showed a synergistic increase in the OC promoter activity when both p53 and MDM2 were transiently overexpressed. We further demonstrate that p53 is not degraded during overexpression of MDM2 protein. Increased OC expression was observed with concomitantly increased p53, VDR, and MDM2 levels in ROS17/2.8 cells during treatment with differentiation promoting (DP) media, but was significantly decreased when co-treated with DP media and the small molecule inhibitor of MDM2-p53 interaction, Nutlin-3. We have also observed a dramatic increase of the OC promoter activity in the presence of p53 and Mdm2 with inclusion of Cbfa-1 and p300 factors. Our results suggest that under some physiological conditions the oncoprotein MDM2 may cooperate with p53 to regulate the osteocalcin gene during osteoblastic differentiation.
Keywords: p53, Mdm2, osteocalcin, osteoblast differentiation, gene regulation
Introduction
Osteocalcin (OC) is a bone gamma-carboxyglutamic acid (GLA)-containing protein which is encoded by the OC gene (also designated as BGLAP in human) [1,2]. Human osteocalcin (hOC) is a small (6 kDa) protein that consists of 100 amino acid residues precursor and 51 in the mature peptide, containing a vitamin K-dependent GLA domain which strongly binds to calcium [1,2]. OC is one of the most abundant noncollagenous bone matrix proteins, constituting 1–2% of total protein in human bone tissue [1]. The OC protein is specifically expressed by osteoblasts and is a marker of osteoblast differentiation during the later stages of bone formation where it regulates mineral deposition and turnover in bone [3]. Of note is that the OC gene expression is generally low or absent in osteosarcomas that originate in osteoblasts [4] and also seen in serum of prostate cancer patients with metastatic lesions in bone [5]. Its expression level has been shown to be related to the differentiation status or metastatic potential in these tumor cells [4,5,6].
In osteoblasts the OC gene is transcriptionally regulated by vitamin D receptor (VDR) through its binding to the vitamin D response element (VDRE) in OC promoter [2]. Multiple VDRE augmenting sequences (VDRE-AS) have been identified close to the VDRE site, which can enhance the transcriptional activation by vitamin D [7]. Many other regulatory elements have also been identified in the proximal OC promoter region, including AP-1 [8], OSE1 - 4 [9,10], Msx2 [11], YY1 [12], and GRE [13]. The OSE2-binding factor Cbfa-1 is the principal regulator of osteocalcin expression [14], and the transcriptional coactivator p300 is recruited to the osteocalcin promoter by Cbfa-1 [15]. Although regulatory elements in the OC gene have been extensively studied, the complex process of the regulation of OC transcription still remains unclear.
The tumor suppressor p53 is a key regulator in growth control, repair and apoptosis [16]. A number of recent studies have linked p53 to differentiation processes in different tissues including osteobalstic differentiation and bone remodeling [17,18,19,20,21] Several bone specific genes like osteopontin, VDR and GDF-15 have also been shown to contain p53 responsive elements (p53REs) and be regulated by p53 [22,23,24,25,26]. We have previously found several bone specific genes to be related to the p53 status of the cell, including OC and bone morphogenetic proteins (BMPs) [27,28,29]. In our previous study, we have identified a potential p53RE site within the hOC promoter and we have observed that wild-type p53 is associated with the hOC promoter activity [30].
The murine double minute (Mdm2) gene is a p53 inducible gene and encodes a type E3 ubiquitin ligase responsible for the degradation of p53 in the 26S proteosome [16]. A well-established role for MDM2 is its feedback regulation of p53 activity: p53 binds to the p53REs in the secondary promoter for Mdm2 and activates MDM2 expression, while the subsequent increase of MDM2 protein results in its binding to p53 at the N-terminal 1–52 residues leading to p53 degradation [31]. However, MDM2 also possesses numerous p53-independent activities, and is also known to interact with a number of other proteins, e.g. Numb, RB, p300, insulin like growth factor receptor, estrogen receptor, and androgen receptor, involved in different cellular activities such as cell fate determination, differentiation, and signaling [32,33,34,35]. In the case of bone, it has been reported that targeted disruption of Mdm2 in this tissue causes skeletal abnormalities, osteopenia and osteoporosis [33].
In this study we conducted experimental analyses in vitro and in vivo to further investigate the effect of p53 and Mdm2 on the OC gene regulation. We observed that p53 exerted a positive effect on OC expression, while inclusion of Mdm2 did not decrease but synergistically increased the OC promoter activity. Our findings suggest that Mdm2 appears to have a physiological role in the regulation of osteoblast gene expression.
Materials and Methods
Cell Lines, Plasmids and Cell Culture
The rat osteosarcoma cell line ROS17/2.8 was kindly provided by Dr. Rodan (Merck Research Laboratory, West Point, PA). This cell line contains a wild-type endogenous p53 and can be induced to express genes associated with advanced stages of differentiation and mineralization in culture [26]. The plasmid pOSCAT3, which contains the −884- to +34-bp region of the hOC gene, was kindly provided by Dr. Morrison [36]. One pOSCAT3-stable ROS17/2.8 cell line was generated with transfection of the pOSCAT3 plasmid in order to monitor the hOC promoter activity during differentiation. Wild-type p53 and Mdm2 expression vectors were kind gifts from Dr. Oren (Weisman Institute, Israel). Cbfa-1 expression vector was a kind gift of Dr. Karsenty [37], and p300 expression vector was provided by Dr. Hottiger [38]. We obtained several p53 and Mdm2 specific short hairpin RNA plasmids (p53shRNA and Mdm2shRNA) from Origene Company (Origene Co, Rockville, MD) and used them in transient transfection in ROS17/2.8 cells. The ROS17/2.8 cell line was grown in DMEM/F-12 media (Gibco, Great Island, NY) with 10% fetal bovine serum in a modified atmosphere of 95% air and 5% CO2 at 37°C.
Differentiation Promoting Media (DP media) and Nutlin Treatment
Differentiation promoting media (DP media) consisted of 10% FBS in basal media plus 10 mM β-glycerophosphate and 50 μg/ml ascorbic acid. The cells were maintained in DP media with periodic changes of every 2 days for one week. For Nutlin treatment, Nutlin-3 (Sigma-Aldrich) was added to cultures at the final concentration of 10 μM simultaneously with DP media.
Transient Transfections and Reporter Assays
The cell lines were transfected with pOSCAT-3 and other expression vectors using Superfect transfection reagent (Qiagen). Fourty-eight hours after transfection the cells were lysed and equal amounts of protein were used to measure chloramphenicol acetyltransferase (CAT) activity using n-Butyryl CoA and 14C Chloramphenicol. The product was extracted with xylene and radioactivity measured using a liquid scintillation counter. Nutlin treatments (10 μM) were done 24 h after transfection. All measurements were carried out on triplicate samples and experiments were repeated at least thrice.
Quantitative RT-PCR
Total RNAs were isolated using TRI reagent (Sigma Chemical Company, St Louis, MI), and reverse-transcribed with the High Capacity cDNA kit (Ambion). Quantitative Real-time PCR was carried out in a ABI Prism 7300 thermocycler with specific primer sets synthesized for rat Mdm2 and p53 (Qiagen, Valencia, CA). Cycling parameters were as follows: 50° C for 2 min, 95° for 10 min, 45 cycles X 95° for 15 sec, and 60° for 1min, followed by the dissociation stage. Relative Quantification of gene expression was determined by using the 2−ΔΔCT method, where fold change in gene expression was relative to Day 0 and normalized against 18sRNA.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was conducted to investigate the binding of p53 to the rOC promoter in ROS17/2.8 cells in vivo by using an EZ-ChIP™ Assay Kit (Upstate, Temecula, CA) as described previously [30]. Mouse antibody 1C12 against p53 (Cell Signaling Tech) was used in ChIP assays. The PCR primers used to detect target sequences were as follows: 5’-TTCATGTGGGGTGTCTCTGA-3’ (forward) and 5’-CTGGGCCTTGGTCTTGAGT-3’ (reverse) for the rOC gene; and 5’-TTCTCTGAGACCAGCA GCAAA-3’ (forward) and 5’-GTCTCTGCCTCC ATTCATGC-3’ (reverse) for the rat p21 gene, which was used as a p53RE positive control.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was conducted to test the if p53 could physically bind to the putative p53 binding sites within the rOC promoter in vitro. A 30-bp double-stranded oligonucleotide probe, 5’- tacTGGCTTGTTCatcaTGGCTTGCTCagc-3’, covering the potential p53 binding site (core nucleotides in uppercase) within the rOC promoter (Fig. 1A) were synthesized and labeled with biotin using Biotin 3’ End DNA Labeling Kit (Pierce, Rockford, IL), and purified by using QIAquick Extraction Kit (Qiagen). The known core p53RE sequence of the rat p21 gene, 5’-cagGAACAAGTCAAGACATGTTCagc-3’, was used as a positive control for p53 binding. The nuclear extract was prepared from ROS17/2.8 cells transfected with p53 expression plasmid by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). The EMSA assay was conducted by using LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL) according to the instructions. For competitive EMSA, the cold unlabeled probe was incubated in the nuclear extract for 15 min at 200-fold concentration of bio-labeled probe before the latter was added.
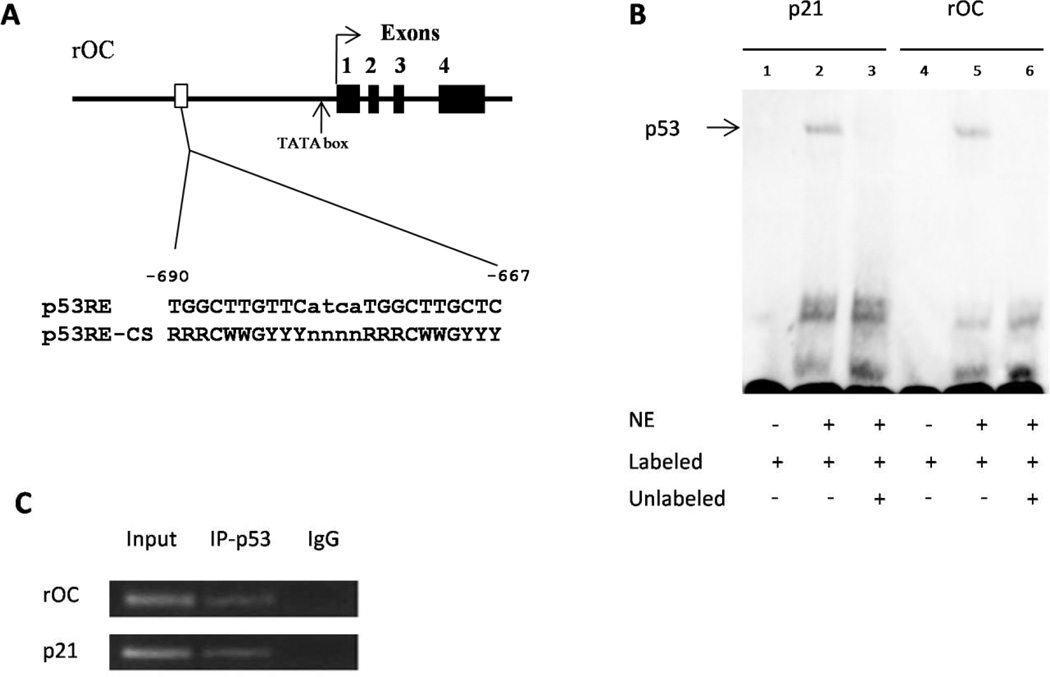
Figure 1. p53 binds to the rOC promoter region.
(A) The rat OC promoter region contains a putative p53 response element at the position of −690 to −667 bp. The TATA box site is also indicated. Black boxes and Numbers, exons. (B) EMSA assay. NE, nuclear extracts from p53 over-expressed ROS17/2.8 cells; Labeled, Biotin-labeled probes; Unlabeled, competitive probes. (C) ChIP assay. Rat ROS17/2.8 cells were cross-linked, sonicated and then immunoprecipitated using an anti-p53 monclonal antibody. The known p53RE sequence in the rat p21 promoter was used as a p53RE positive control. Input, sonicated and precleared supernatant; IP-p53, p53 antiboby-immunoprecipitated samples; IgG, negative controls treated with normal mouse IgG antibody.
Suggestion for this figure: use a TRANSFAC matrix motif in place of the old paper, as it is more current and gives a better representation of the possible nucleotides at each location. Also, for 1B, you cannot be certain that p53 is binding to the EMSA motif as there is no supershift by an antibody specific to p53 and a cell lysate was used.
Western Blot Assays
Protein lysates were prepared using MPER reagent (Pierce, Rockford, IL). A Bradford assay was performed to determine protein concentration. Twenty-five to 50 μg of the protein was run out on an SDS page gel, transferred onto a nitrocellulose membrane, and blocked in 5% milk solution. Primary antibodies used in this study were mouse monoclonal p53 (Pab240) IgG (Santa Cruz Biotech, CA), mouse monoclonal Mdm2 (SMP14) antibody (Santa Cruz Biotech, CA), rabbit VDR (N-20) antibody (Santa Cruz Biotech), and rabbit β-Actin antibody (Imgenex, San Diego, CA). The secondary antibodies used in Western blot were ImmunoPure Antibodoies (Thermo Scientific, Rockford, IL) against rabbit and mouse IgGs, respectively. The blot was developed using SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo Scientific). Relative pixel densitometry was conducted using UNSCAN-IT (Silk Scientific, Orem, UT) for p53, MDM2 and VDR protein after normalization to β-Actin.
Immunoprecipitation (IP) and Immunoblotting (IB) Assays
One-hundred μg of protein lysate was incubated with primary antibody overnight. The complex was then purified on protein A agarose beads for 2 hours at 4°C. Following incubation the complex was washed several times, spun down, and resuspended for Western Blot analyses. Antibodies against p53 and MDM2 (Santa Cruz Biotech, CA) as described above were used in IP and IB assays.
Statistical analysis
Statistical analysis was performed using software GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA). The values given are mean ± S.E.M. Statistical analysis between two samples was performed using Student's t-test. In all cases, P < 0.05 was considered as significant.
Results
p53 binds to the rat osteocalcin promoter region
We have previously showed that p53 is able to bind to the hOC promoter region by using ChIP and DNA affinity immunoblotting assays [30]. Even though the 20-bp palindromic p53RE sequence on hOC promoter is conserved in multiple primate species, it is not present in rat. To investigate whether there are some other p53 binding sites in the rOC gene, we searched for a consensus p53 binding sequence in the proximal promoter region of the rOC gene. We noticed a potential p53 binding site at the −690 to −667-bp position of the rOC gene (Fig. 1A). This 24-bp potential p53 binding site, TGGCTTGTTCatcaTGGCTTGCTC, contains two, 10-nt p53-binding consensus RRRCWWGYYY motifs separated by a 4-nt spacer (lowercase letters) with only two mismatched nucleotides (underlined) in the noncritical position [39].
To further investigate whether the p53 protein can bind to the predicted p53 binding site within the rOC promoter, we carried out an EMSA study using a double strand probe spanning the predicted p53 binding site with a probe containing the known p53RE site on the rat p21 promoter as a positive control. As shown in Fig. 1B, a band shift was seen when either the probe for rat p21 or for rOC was incubated with the nuclear extracts isolated from p53 over-expressed ROS17/2.8 cells (Lane 2 and 5). Competition experiments demonstrated that the binding of nuclear lysate and the rOC or p21 probes can be abolished by unlabeled probes of an extremely high concentration (Lane 3 and 6). No band shift was observed without the incubation of nuclear lysate (Lane 1 and 4). This result indicated that the predicted p53 binding site within the rOC promoter can be bound by ROS17/2.8 cell lysate in vitro.
We have previously showed that rOC is expressed during differentiation in rat ROS17/2.8 cells which contain an endogenous wild-type p53 [26]. In order to determine whether wild-type p53 protein can bind to the putative 24-nt p53 binding site within the rOC promoter in vivo, we then conducted a ChIP assay to investigate the occupancy of the rOC promoter by endogenous p53 in ROS17/2.8 cells. ROS17/2.8 cells were cross-linked with 1% formaldehyde and sheared, and DNA-protein complexes were precipitated with anti-p53 antibodies. PCR was run using primers designed to span the putative p53 binding site within the rOC gene. A rat p21 promoter sequence containing a known p53RE site was also included in ChIP assay to show the presence of p53 binding as a positive control. As shown in Fig. 1C, the rOC promoter sequence (−769 to −630 bp) containing the putative 24-bp p53 binding site was selectively amplified by PCR from the chromatin-protein complex immunoprecipitated by anti-p53 antibody, but not from the negative control precipitated by normal mouse anti-IgG antibody. This result further supported that endogenous p53 occupies the rOC promoter region spanning the putative p53 binding site in ROS17/2.8 cells.
Stabilization and accumulation of p53 increase hOC promoter activity
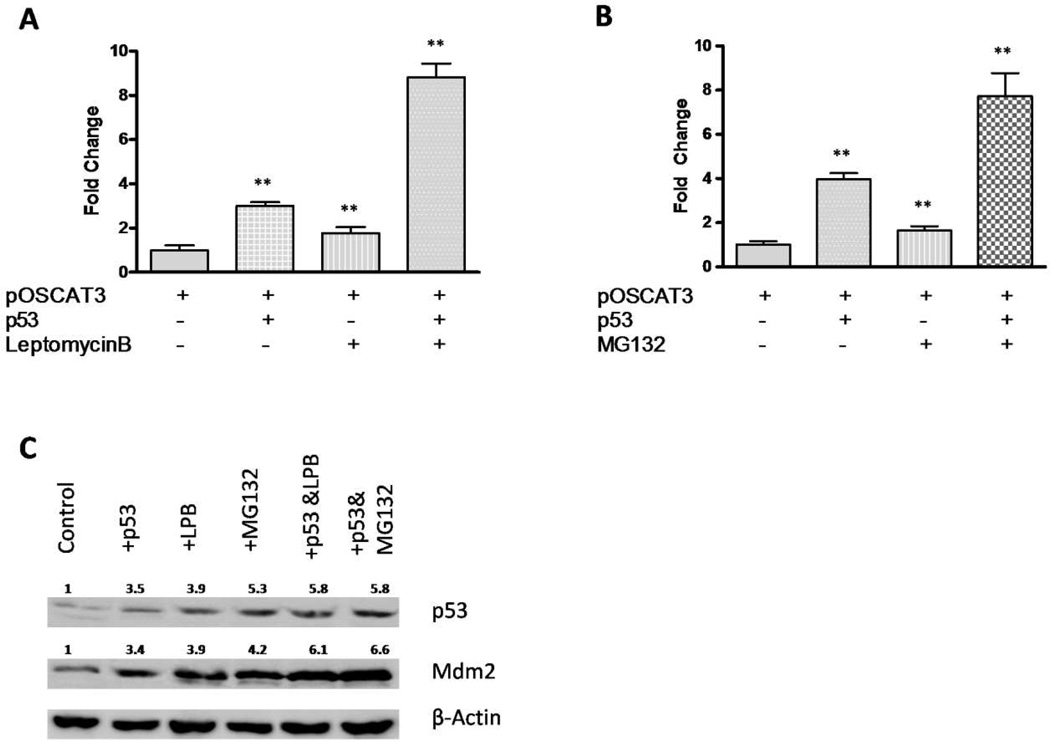
In our previous studies we have showed that wild-type p53 positively regulates the hOC gene expression by using the pOSCAT3 reporter transfected in ROS17/2.8 cells [30]. To further understand the role of p53 on the OC gene expression, we carried out transfections which included either an inhibitor of nuclear transport, leptomycin B, or a proteasome inhibitor, MG132, into ROS17/2.8 cells co-transfected with pOSCAT3 and p53 expression plasmid. Leptomycin B prevents the export of p53 protein from the nucleus to the cytoplasm where MDM2-mediated degradation occurs, and thus promotes the p53 regulatory function on its target genes [40]. MG132, a membrane-permeable peptide aldehyde, inhibits the chymotrypsin-like activity of the proteasome, which stabilizes p53 and induces apoptosis in a number of tumor cells in a time- and dose-dependent manner [41]. In the present study we observed that both leptomycin B and MG132 significantly enhanced the human osteocalcin promoter activity when p53 was exogenously expressed, with 8.8 and 7.6-fold increase when compared to the control (p < 0.05 for both), respectively (Fig. 2A and 2B). Western blot assays confirmed the increased expression of p53 protein in ROS17/2.8 cells treated with leptomycin B or MG132 (Fig. 2C). These observations further demonstrated that the presence of p53 facilitated the osteocalcin gene expression in a dose-dependent manner.
Figure 2. Leptomycin B and MG132 both increase the hOC promoter activity.
The 884-bp pOSCAT3 construct was transiently co-transfected into ROS17/2.8 cells with p53 expression vector, with or without 5 nM leptomycin B (A) or 20 μM MG132 (B). The CAT assays were done 48 hr after transfection, and the data are representative of 6–8 independent experiments. P values were determined by Student’s t-test. **, P < 0.05. (C) p53 and Mdm2 proteins were detected by Western blotting with β-Actin as a control. Relative expression levels of p53 and Mdm2 are indicated after normalizing to β-Actin. LPB, leptomycin B.
MDM2 has a positive effect on the osteocalcin promoter activity
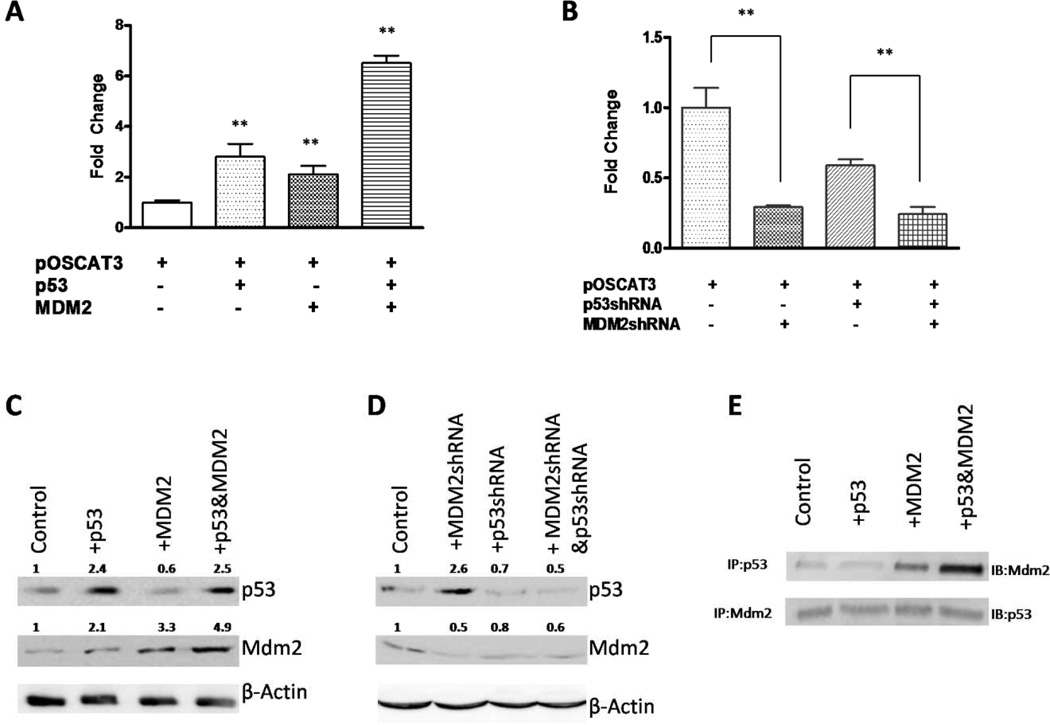
MDM2 regulates p53 turnover by acting as an ubiquitin ligase and normally causes the degradation of p53 in cytoplasm. In order to determine if MDM2 down regulates p53-mediated up-regulation of the osteocalcin promoter activity, we introduced Mdm2 expression vector alone or with p53 into pOSCAT3-transfected ROS17/2.8 cells. CAT activity was measured 48 hrs after transfection. As shown in Fig. 3, our results were contrary to what we had expected in hOC promoter activity when Mdm2 and p53 were co-transfected and over-expressed. By itself, p53 increased the hOC promoter activity by 2.8-fold compared to the control (p < 0.05), but Mdm2 alone had a non-significant, 2.1-fold increase (p = 0.16) in its effect on the hOC promoter activity. However, inclusion of p53 with Mdm2 produced a synergistic up-regulation in hOC promoter activity with 6.5-fold increase (p < 0.05) when compared to the non-transfected control (Fig. 3A). We further noticed that the pOSCAT3 activity was significantly decreased when endogenous MDM2 was knocked down using MDM2 specific shRNAs (Fig. 3B). As shown in Fig. 3B, transient knockdown of p53 produced a 50% drop in activity of the osteocalcin promoter (p), whereas downregulation of Mdm2 expression caused a 70% drop in activity (p < 0.05) compared to the non-transfected control. Using a double knockdown of p53 and Mdm2, a greater drop in activity, to about 30% of the non-transfected control (p < 0.05) was seen. This suggestes the loss of optimal levels of these proteins decreases osteocalcin promoter activity. Western blot assays further revealed that p53 protein did not degrade when over-expressed with MDM2 (Fig. 3C), whereas levels of p53 and MDM2 were both reduced after transfection of shRNAs (Fig. 3D), with an exception of the increase of p53 when MDM2shRNA was transfected alone. These results were surprising as MDM2 is known to inhibit p53 function and reduce its level by transporting p53 out of the nucleus and aiding in p53 degradation through the proteosomal pathway [42].
Figure 3. Mdm2 synergizes with p53 in the regulation of the hOC promoter activity.
(A) CAT assay for the human OC promoter activity was conducted in ROS17/2.8 cells with transient transfections of pOSCAT3, p53 and/or Mdm2 expression vectors. All the data is representative of 6–8 independent experiments. (B) Reduction of endogenous p53 and Mdm2 levels reduces the osteocalcin promoter activity. p53 and Mdm2 expression was reduced using respective shRNA in transient transfections with the pOSCAT3 and the CAT activity was measured 48 hr later. The results represent average +SE of triplicates of an experiment that was repeated thrice. P values were determined by Student’s t-test. **, P < 0.05. (C) and (D), p53 and Mdm2 levels were detected by Western blotting in ROS17/2.8 cells co-transfected with p53 and Mdm2 expression vectors or shRNAs. Relative expression levels of p53 and Mdm2 are indicated after normalizing to β-Actin. (E) Immunoprecipitation (IP) and immunoblotting (IB) assays for detection of p53-Mdm2 binding complex. Equal amounts of lysates from the above experiments were immunoprecipitated with p53 (the upper panel) or Mdm2 (the bottom panel) and followed by IB with Mdm2 or p53 antibodies, respectively.
To further investigate the interaction of p53 and MDM2, we immunoprecipitated p53 protein from whole ROS17/2.8 cell lysate after transfections followed by immunoblotting for MDM2. As shown in Fig. 3E (upper panel), exogenous expression of MDM2 produced higher levels of p53-MDM2 complexes as detected by IB with anti-MDM2 antibody. We also carried out immunoprecipitation with MDM2 specific antibody followed by immunoblotting with p53 specific antibody (Figure 3E bottom panel). In this case, we noticed the presence of p53 in all the samples irrespective of the amount of Mdm2 expressed. Overexpression of Mdm2 did not actually produce any dramatic reduction in p53 levels as would be expected if MDM2 caused degradation of p53. These results suggest that there is no substantial loss of p53 with over-expression of Mdm2, at least under conditions used in this present study.
Disruption of p53-MDM2 binding by Nutlin-3 reduces osteocalcin promoter activity
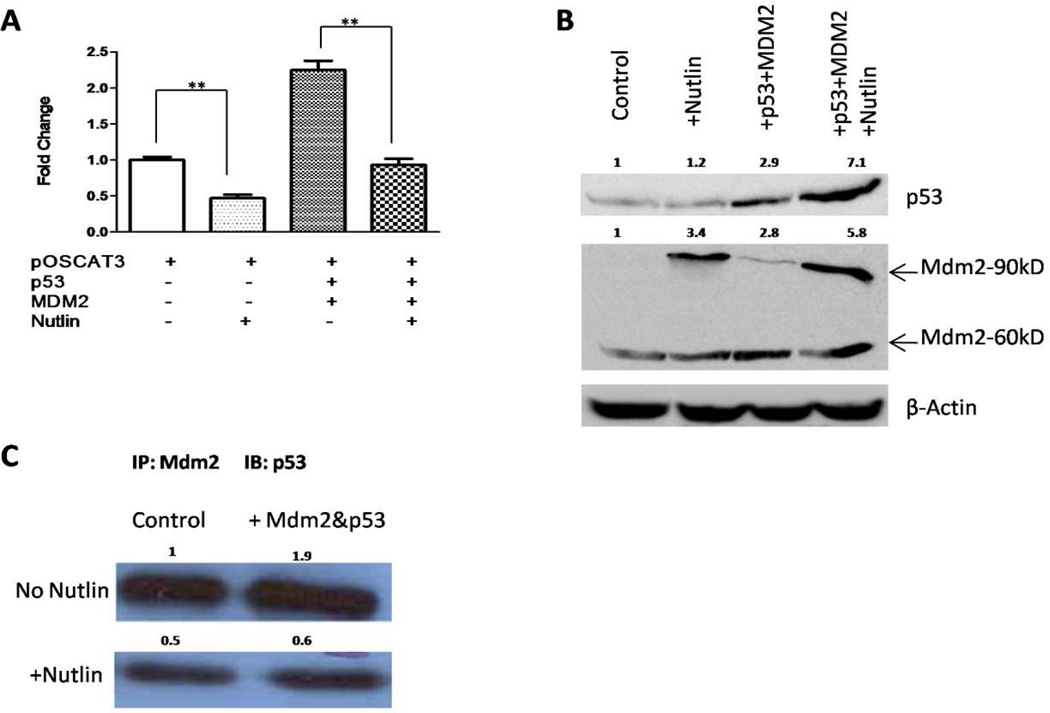
Since p53-MDM2 complex appeares to positively regulate OC gene expression, we attempted to determine whether and how disruption of the complex by the inclusion of a small molecule inhibitor Nutlin-3 affected hOC promoter activity. As shown in Fig. 4, the treatment of pOSCAT3-transfected cells with Nutlin-3 caused about a 50% reduction in basal activity of the hOC promoter when compared to the control cells (p < 0.05), while the increase produced by co-transfection of p53 and Mdm2 was also significantly (p < 0.05) reduced by Nutlin-3 (Fig. 4A). Western blot assays further revealed that levels of p53 and MDM2 proteins were both individually increased after transfection. Higher levels of both proteins were observed when Nutlin-3 treatment was also included (Fig. 4B). In the case of p53 this may result from stabilization of the protein while in the case of MDM2, an increased amount of the 90kDa band is a result of increased p53 inducing its expression, also see Realtime PCR results below. These observations suggest that the p53-MDM2 complex is required for the upregulation of osteocalcin expression while disruption of the p53-MDM2 complex by Nutlin-3 was not conducive to osteocalcin activity.
Figure 4. Disruption of p53-Mdm2 interaction with Nutlin-3 reduces the hOC promoter activity.
(A)Transient transfections were carried out with pOSCAT3, p53 and/or Mdm2 in the presence and absence of Nutlin-3. The experiment was repeated thrice in triplicates and a representative result is shown. P values were determined by Student’s t-test. **, P < 0.05. (B) p53 and Mdm2 proteins were detected by Western blotting with β-Actin as a control. Relative expression levels of p53 and Mdm2 are indicated after normalizing to β-Actin. (C) Treatment with Nutlin-3 reduces p53 association with Mdm2. Equal amounts of lysates from the above experiment was immunoprecipitated with Mdm2 followed by Western blotting with p53 antibodies to determine the amount of p53 bound to Mdm2 under the conditions of the experiment.
To further investigate the effect of Nutlin-3 on p53-Mdm2 interaction, we immunoprecipitated whole cell lysates from the co-transfection of p53 and Mdm2 with MDM2 antibody and then probed for the presence of p53 associated with MDM2. As shown in Fig. 4C, treatment of Nutlin-3 produced a substantial decrease in the amount of p53 bound to MDM2 in both control and co-transfected cell lysates, suggesting that Nutlin-3 was indeed able to disrupt the binding of p53 and MDM2, and consequently the loss of p53 in complex with MDM2 caused a reduction in the hOC promoter activity.
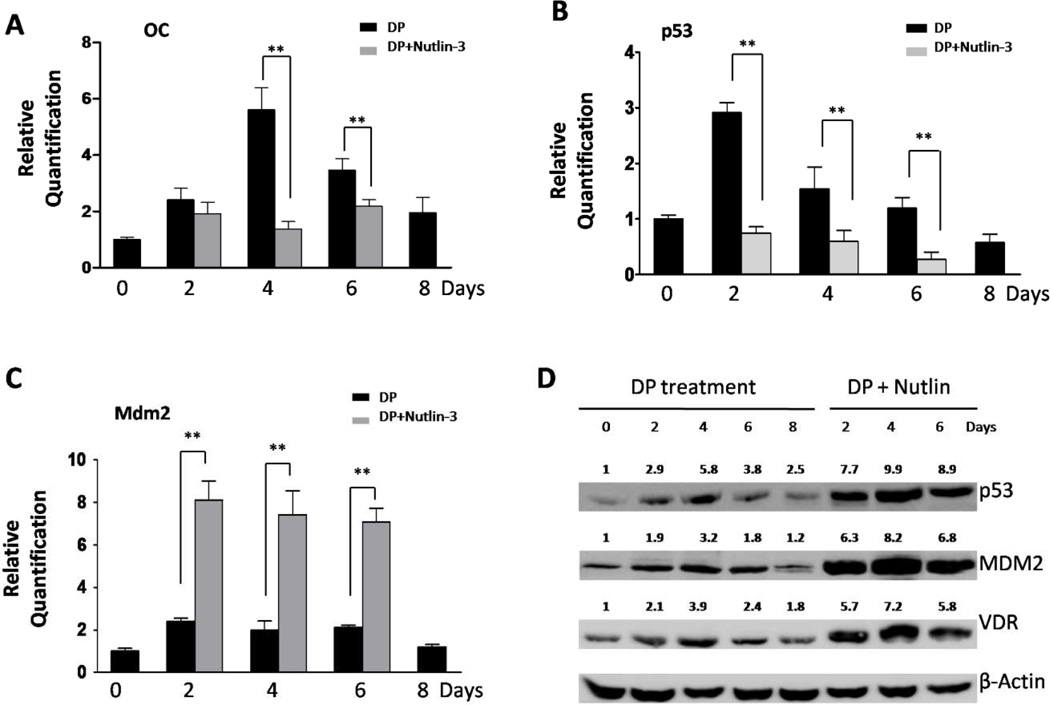
Differentiation promoting (DP) media increase osteocalcin, p53 and Mdm2 gene expression
We have previously showed that human osteocalcin promoter activity concomitantly increases with p53 levels in a time-dependent manner when the cells are exposed to a DP media [30]. To further investigate changes in the rat osteocalcin expression, we measured rOC mRNA expression using real-time quantitative RT-PCR. Relative quantification of gene expression is shown as fold change relative to Day 0 and normalized against 18sRNA. We also measured expression of p53 and Mdm2 in the rat ROS17/2.8 cells after periodic exposure to DP media (0, 2, 4, 6, and 8 days), at both the level of transcript by quantitative RT-PCR and protein level by using Western Blot assays. As shown in Fig. 5, increased rOC mRNA levels were observed after the second day exposed to DP media, with the maximal level on Day 4 of about 5.6-fold increase when compared to Day 0 (p < 0.05; Fig. 5A). Elevated rOC mRNA expression was also seen on Day 2 and Day 6 with 2.4- and 3.5-fold increase (p < 0.05) respectively when compared to the Day 0. Increased levels of both p53 mRNA and protein were also observed during DP treatment (Fig. 5B and 5D). Quantitative RT-PCR showed the highest p53 mRNA level on Day 2 with 2.9-fold increase (p < 0.05) when compared to Day 0 (Fig. 5B), while Western blot showed the highest protein level was observed on Day 4 (Fig. 5D). At the transcriptional level, the expression of Mdm2 was observed with periodic increase on Day 2, 4 and 6 after DP treatment by using quantitative RT-PCR with 2.4, 2, and 2.1-fold increase (p < 0.05), respectively (Fig. 5C). Western blot assay also showed increases of MDM2 protein expression with the highest level on Day 4 (Fig. 5D). These observations show the parallel increase of p53 and rOC expression in a time-dependent manner in ROS17/2.8 cells during osteoblastic differentiation, with simultaneous changes of Mdm2 expression in ROS17/2.8 cells during DP treatment.
Figure 5. DP media and Nutlin-3 treatments cause to changes in the expression levels of rOC, p53, and MDM2.
(A) to (C), Rat ROS17/2.8 cells were exposed to DP media with or without 10 μM Nutlins, and the mRNA levels of p53, Mdm2 and osteocalcin were determined using a quantitative RT-PCR method. Relative quantification of gene expression was determined by using fold change relative to Day 0 and normalized against 18sRNA. DP experiment with Nutlins could not be extended longer than 6 days due to massive apoptosis of cells. P values were determined by Student’s t-test. **, P < 0.05. (D) The expression of endogenous p53, VDR and Mdm2 proteins were detected using Western blotting in ROS17/2.8 cells subjected to the treatment of DP media with or without Nutlins. Relative expression levels of p53, Mdm2 and VDR proteins are indicated after normalizing to β-Actin.
Nutlin-3 decreases osteocalcin and p53 but increases Mdm2 expression
To test the impact of Nutlin-3 on the expression of rat OC gene, we then treated ROS17/2.8 cells with DP media combined with Nutlin-3 at the final concentration of 10 μM, and detected mRNA transcripts for OC, p53 and Mmd2 on day 2, 4 and 6 after Nutlin-3 treatment by using real-time PCR. Protein expression was also detected by Western blot for p53 and Mdm2. We were unable to obtain sufficient amount of good quality RNA from Nutlin-treated cells on Day 8 due to extensive cell death as result of high p53 levels. As shown in Fig. 5A, compared to Nutlin-3 untreated cells, a significant decrease of OC mRNA expression was observed in Nutlin-3 treated ROS17/2.8 cells on Day 4 (5.6±1.3 vs. 1.4±0.4, p < 0.05) and Day 6 (3.5±0.7 vs. 2.2±0.4, p<0.05) while a slight decrease on Day 2 (2.4±0.7 vs. 1.9±0.7, p>0.05). We also found that p53 mRNA levels were significantly decreased on Day 2 (0.7±0.2 vs. 2.9±0.3, p < 0.05), Day 4 (0.6±0.2 vs. 1.6±0.5, p < 0.05) and Day 6 (0.3±0.3 vs. 1.2±0.3, p < 0.05) after Nutlin-3 treatment (Fig. 5B). However, p53 was stabilized by Nutlin-3 treatment, due to inhibition of MDM2 by the drug, allowing it to avoid degradation (Fig. 5D). Enhanced levels of p53 were probably responsible for inducing higher levels of both Mdm2 mRNA and protein expression in Nutlin-3 treated cells (Fig. 5C and 5D). These results show that the basal levels of OC expression correlated well to the amount of p53-MDM2 complex rather than that of individual p53 or MDM2 levels. This suggests that the two proteins need to be physically bound in the regulation of OC expression.
Activation of OC expression through VDR depends on the presence of p53-MDM2 complex
VDR is known as an important activator of OC gene, while it itself is a target of p53 [23]. We have previously showed an additive effect of VDR and p53 on the OC transcription. To further understand the role of p53 and MDM2 on the activation of VDR for OC expression, we investigated the VDR protein expression in ROS17/2.8 cells treated with DP media with or without Nutlin-3. As shown in Fig. 5D, we observed a similar pattern of VDR protein expression to those of p53 and MDM2 in ROS17/2.8 cells treated with DP media. The highest level of VDR protein was seen on Day 4 with the highest expression level of OC mRNA, p53 protein and MDM2 protein. On the other hand, in ROS17/2.8 cells treated with both DP media and Nutlin-3, VDR protein levels were much higher, as well as, a significant decrease of OC expression when compared to DP treatment alone (Fig. 5A and 5D). These results suggest that high levels of VDR by itself were incapable of up regulating OC gene transcription even though the OC gene contains a Vitamin D response element and is known to be an important regulator of its expression [7].
P53 and MDM2 may function as part of a multiprotein complex in the regulation of the OC gene
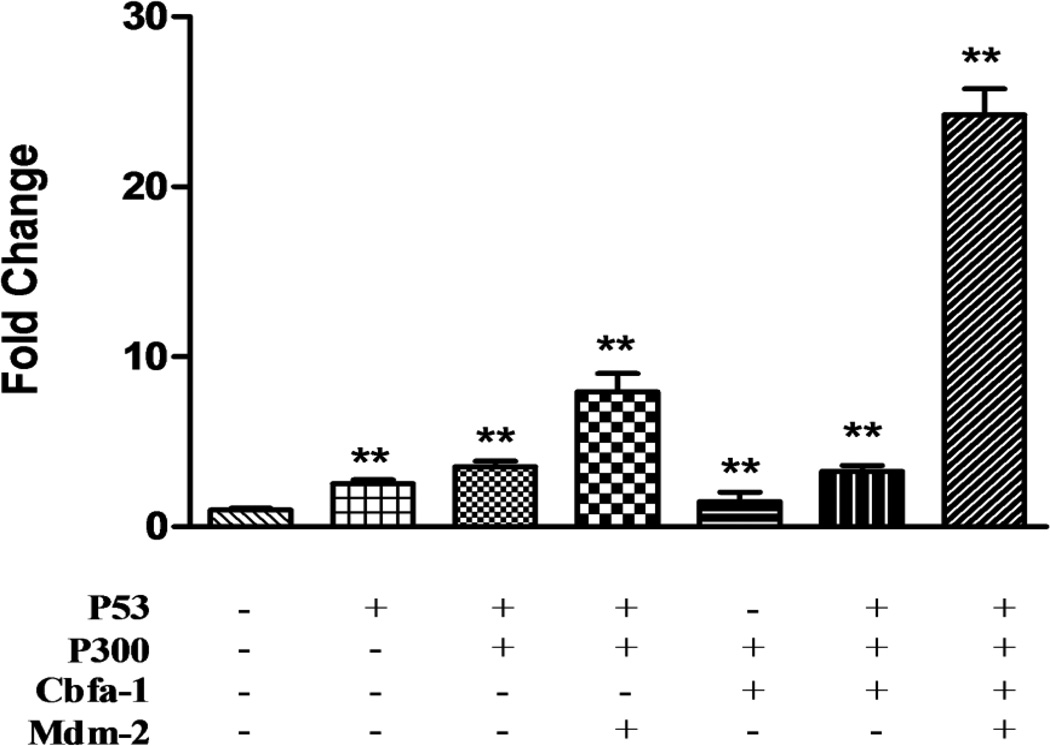
The OC gene regulation is well studied and several different proteins are involved in binding to the 884bp promoter region and affecting gene expression. Cbfa-1 is believed as a principal regulator to bind to its recognition sites on the OC promoter [43]. Additionally the transcriptional coactivator p300 is recruited to the OC promoter by Cbfa-1 [14]. These interactions have been known to stabilize the interactions between multiple protein factors that bind around the promoter [14]. We therefore performed our transfections by including expression constructs for these two factors along with p53 and Mdm2. As expected of a coactivator, the presence of p300 substantially increased osteocalcin basal activity several fold. Individually all constructs had a positive effect on osteocalcin promoter activity (not shown for all but just p53). As shown in Fig. 6 while all of them had an effect on osteocalcin promoter activity, neither one of them individually showed an increase. Cbfa1 is inhibited by p53 [33] and this may partly explain some of these results. The individual gene products also compete for p300 and this might also be responsible for some of the results seen. However, inclusion of the expression constructs simultaneously produced a synergistic increase in osteocalcin promoter activity with about 24-fold when compared to the control, suggesting that optimal activity of the osteocalcin promoter may require a number of different proteins of which p53 and perhaps Mdm2 may represent physiologically important factors.
Figure 6. Inclusion of known Cbfa-1 and p300 optimizes the hOC promoter activity in the presence of p53 and Mdm2.
CAT assay for the human OC promoter activity was conducted ROS17/2.8 cells with transient transfections of pOSCAT3, p53, Mdm2, Cbfa-1 and p300 expression vectors. All the data is representative of 3–4 independent experiments.
Discussion
OC is a small protein expressed and secreted solely by osteoblasts and thought to play a role in the body's metabolic regulation, bone-building, bone mineralization and calcium ion homeostasis [44]. Recent studies have suggested OC may behave as a hormone in influencing insulin secretion from pancreas beta cells [44,45]. More recently, studies have suggested a probable role in male fertility [46]. OC is also expressed in androgen-independent prostate cancer cells. These observations suggest regulatory mechanisms governing OC gene expression may be more global than previously understood.
We have in earlier studies established that ostseosarcomas show p53 gene rearrangements resulting in loss of p53 expression. OC expression is absent with functional loss of p53 in osteosarcoma cells, while introducing wild-type p53 allowed cells to regain OC expression and advanced differentiation both in vivo and in vitro [27,28]. The present study shows that the positive effect of p53 on osteocalcin gene expression may be mediated through its direct binding to the OC promoter. This observation is not surprising, given the fact that several bone specific genes, like VDR, osteopontin and GDF-15, appear to harbor one or more p53 response elements and are also regulated by p53 [22,23,24,25,26]. While there are several studies showing a role for p53 in tissue differentiation [47,48], the exact mechanism of action has not been elucidated. Direct transcriptional activation of key tissue specific genes may be one mechanism. There is still a debate in the literature on the question of whether p53 is an inhibitor or activator of differentiation [33,49]. It is very likely that p53 behaves as an inhibitor during the early phase of osteoblast differentiation when cell proliferation and matrix deposition occurs. Transcription factors Cbfa-1 and osterix are required for the different phases of differentiation and p53 likely acts as an inhibitor during these stages [33,49]. But since bone remodeling is a life-long process and proceeds through cycles or bone growth and resorption [50], it is also easy to see how p53 function may be important to protect the process from deregulation. Osteocalcin is produced during late mineralization stage and its presence is thought to trigger bone resorption [3]. However, newer roles that go beyond bone have been attributed to this gene. The fact that the osteocalcin gene requires changes to its nucleosome structure before it is made transcribable [51] along with its tight regulation suggests that its functional significance may be currently underestimated.
Our results showing an effect with Mdm2 on OC expression were unexpected, given the well-established role for this protein in the down-regulation of p53. Recent studies have highlighted an important role for the p53-Mdm2 network in the maintenance of the progenitor cell population [52]. It is therefore becoming clear that even under unstressed conditions, interactions between p53 and Mdm2 play an important role in normal development. While the exact role of Mdm2 in the regulation of the osteocalcin gene in not completely evident, it appears to facilitate rather than inhibit the effect produced by p53. There is no appreciable loss of p53 when both proteins are expressed together and therefore Mdm2-induced loss of p53 does not happen under unstressed conditions at least in osteoblasts. Rather the presence and balance of both proteins appear to be necessary for this process. In this respect our results with Nutlin-3 treatment are revealing and show that stabilization of free p53 is inhibitory while its presence with Mdm2 promotes osteocalcin gene expression. There are several observations in the literature regarding p53 independent roles for Mdm2 [32,34,53], and this protein has been found as part of a complex with p53 and other proteins on DNA especially in instances steroid hormones are involved [54,55,56]. Mdm2 has also been found to be present on DNA targets of p53 such as p21 along with p53, and it has been suggested that Mdm2 may be important to recruit p53 to chromatin [57].
Recent studies have suggested caspase 2 mediated cleavage of Mdm2’s RING finger domain a region directly responsible for p53 ubiquitination, is important for maintaining p53’s stability and transcriptional activity [58]. In an earlier study investigators report caspase mediated Mdm2 cleavage to be a common feature of non-apoptotic cells [59] suggesting that under physiological conditions Mdm2 may be cleaved to stabilize p53. This would support some of our results reported here. Therefore, under some physiological conditions it is possible that Mdm2 has a role in aiding in the maintenance of tissue homeostasis along with p53.
Acknowledgements
This work is supported by grants from the National Institutes of Health grant R15 AR055362 to NC and funds from Midwestern University. We thank all individuals cited in the text for their kind gifts of reagents. We thank Oliver Couture for his critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Celeste AJ, Rosen V, Buecker JL, Kriz R, Wang EA, Wozney JM. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986;5:1885–1890. doi: 10.1002/j.1460-2075.1986.tb04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price PA, Otsuka AA, Poser JW, Kristaponis J, Raman N. Characterization of a gamma-carboxyglutamic acid-containing protein from bone. Proc Natl Acad Sci U S A. 1976;73:1447–1451. doi: 10.1073/pnas.73.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 4.Hopyan S, Gokgoz N, Bell RS, Andrulis IL, Alman BA, Wunder JS. Expression of osteocalcin and its transcriptional regulators core-binding factor alpha 1 and MSX2 in osteoid-forming tumours. J Orthop Res. 1999;17:633–638. doi: 10.1002/jor.1100170503. [DOI] [PubMed] [Google Scholar]
- 5.Arai Y, Takeuchi H, Oishi K, Yoshida O. Osteocalcin: is it a useful marker of bone metastasis and response to treatment in advanced prostate cancer? Prostate. 1992;20:169–177. doi: 10.1002/pros.2990200302. [DOI] [PubMed] [Google Scholar]
- 6.Levedakou EN, Strohmeyer TG, Effert PJ, Liu ET. Expression of the matrix Gla protein in urogenital malignancies. Int J Cancer. 1992;52:534–537. doi: 10.1002/ijc.2910520406. [DOI] [PubMed] [Google Scholar]
- 7.Sneddon WB, Bogado CE, Kiernan MS, Demay MB. DNA sequences downstream from the vitamin D response element of the rat osteocalcin gene are required for ligand-dependent transactivation. Mol Endocrinol. 1997;11:210–217. doi: 10.1210/mend.11.2.9877. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg D, Polly P, Eisman JA, Morrison NA. Identification of an osteocalcin gene promoter sequence that binds AP1. J Cell Biochem. 1996;60:447–457. doi: 10.1002/(SICI)1097-4644(19960315)60:4%3C447::AID-JCB2%3E3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Tamura M, Noda M. Identification of a DNA sequence involved in osteoblast-specific gene expression via interaction with helix-loop-helix (HLH)-type transcription factors. J Cell Biol. 1994;126:773–782. doi: 10.1083/jcb.126.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo B, Aslam F, van Wijnen AJ, Roberts SG, Frenkel B, Green MR, DeLuca H, Lian JB, Stein GS, Stein JL. YY1 regulates vitamin D receptor/retinoid X receptor mediated transactivation of the vitamin D responsive osteocalcin gene. Proc Natl Acad Sci U S A. 1997;94:121–126. doi: 10.1073/pnas.94.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer T, Gustafsson JA, Carlstedt-Duke J. Glucocorticoid-dependent transcriptional repression of the osteocalcin gene by competitive binding at the TATA box. DNA Cell Biol. 1997;16:919–927. doi: 10.1089/dna.1997.16.919. [DOI] [PubMed] [Google Scholar]
- 14.Paredes R, Arriagada G, Cruzat F, Olate J, Van Wijnen A, Lian J, Stein G, Stein J, Montecino M. The Runx2 transcription factor plays a key role in the 1alpha,25-dihydroxy Vitamin D3-dependent upregulation of the rat osteocalcin (OC) gene expression in osteoblastic cells. J Steroid Biochem Mol Biol. 2004;89–90:269–271. doi: 10.1016/j.jsbmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 15.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, Van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 2007;35:7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzaro G, Bossi G, Coen S, Sacchi A, Soddu S. The role of wild-type p53 in the differentiation of primary hemopoietic and muscle cells. Oncogene. 1999;18:5831–5835. doi: 10.1038/sj.onc.1202962. [DOI] [PubMed] [Google Scholar]
- 18.El-Dahr SS, Aboudehen K, Dipp S. Bradykinin B2 receptor null mice harboring a Ser23-to-Ala substitution in the p53 gene are protected from renal dysgenesis. Am J Physiol Renal Physiol. 2008;295:F1404–F1413. doi: 10.1152/ajprenal.90378.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerone MA, Marchetti A, Bossi G, Blandino G, Sacchi A, Soddu S. p53 is involved in the differentiation but not in the differentiation-associated apoptosis of myoblasts. Cell Death Differ. 2000;7:506–508. doi: 10.1038/sj.cdd.4400676. [DOI] [PubMed] [Google Scholar]
- 20.Aloni-Grinstein R, Zan-Bar I, Alboum I, Goldfinger N, Rotter V. Wild type p53 functions as a control protein in the differentiation pathway of the B-cell lineage. Oncogene. 1993;8:3297–3305. [PubMed] [Google Scholar]
- 21.Brynczka C, Merrick BA. The p53 transcriptional target gene wnt7b contributes to NGF-inducible neurite outgrowth in neuronal PC12 cells. Differentiation. 2008;76:795–808. doi: 10.1111/j.1432-0436.2007.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto I, Sasaki Y, Ishida S, Imai K, Tokino T. Identification of the osteopontin gene as a direct target of TP53. Genes Chromosomes Cancer. 2002;33:270–278. doi: 10.1002/gcc.10020. [DOI] [PubMed] [Google Scholar]
- 23.Maruyama R, Aoki F, Toyota M, Sasaki Y, Akashi H, Mita H, Suzuki H, Akino K, Ohe-Toyota M, Maruyama Y, Tatsumi H, Imai K, Shinomura Y, Tokino T. Comparative genome analysis identifies the vitamin D receptor gene as a direct target of p53-mediated transcriptional activation. Cancer Res. 2006;66:4574–4583. doi: 10.1158/0008-5472.CAN-05-2562. [DOI] [PubMed] [Google Scholar]
- 24.Li PX, Wong J, Ayed A, Ngo D, Brade AM, Arrowsmith C, Austin RC, Klamut HJ. Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J Biol Chem. 2000;275:20127–20135. doi: 10.1074/jbc.M909580199. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Filipovic Z, Brown D, Breit SN, Vassilev LT. Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol Cancer Ther. 2003;2:1023–1029. [PubMed] [Google Scholar]
- 26.Schwartz KA, Lanciloti NJ, Moore MK, Campione AL, Chandar N. p53 transactivity during in vitro osteoblast differentiation in a rat osteosarcoma cell line. Mol Carcinog. 1999;25:132–138. doi: 10.1002/(sici)1098-2744(199906)25:2<132::aid-mc8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Chandar N, Campbell P, Novak J, Smith M. Dependence of induction of osteocalcin gene expression on the presence of wild-type p53 in a murine osteosarcoma cell line. Mol Carcinog. 1993;8:299–305. doi: 10.1002/mc.2940080413. [DOI] [PubMed] [Google Scholar]
- 28.Chandar N, Swindle J, Szajkovics A, Kolman K. Relationship of bone morphogenetic protein expression during osteoblast differentiation to wild type p53. J Orthop Res. 2005;23:1345–1353. doi: 10.1016/j.orthres.2005.04.010.1100230616. [DOI] [PubMed] [Google Scholar]
- 29.Chandar N, Saluja R, Lamar PC, Kolman K, Prozialeck WC. P53 and beta-catenin activity during estrogen treatment of osteoblasts. Cancer Cell Int. 2005;5:24. doi: 10.1186/1475-2867-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Liu W, Roberts W, Hooker S, Fedor H, DeMarzo A, Isaacs W, Kittles RA. 8q24 allelic imbalance and MYC gene copy number in primary prostate cancer. Prostate Cancer Prostatic Dis. 2010;13:238–243. doi: 10.1038/pcan.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Zhang R. p53-independent activities of MDM2 and their relevance to cancer therapy. Curr Cancer Drug Targets. 2005;5:9–20. doi: 10.2174/1568009053332618. [DOI] [PubMed] [Google Scholar]
- 33.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1:1027–1035. [PubMed] [Google Scholar]
- 35.Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, Jones SN. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem. 2004;279:4877–4886. doi: 10.1074/jbc.M305966200. [DOI] [PubMed] [Google Scholar]
- 36.Morrison NA, Shine J, Fragonas JC, Verkest V, McMenemy ML, Eisman JA. 1,25-dihydroxyvitamin D-responsive element and glucocorticoid repression in the osteocalcin gene. Science. 1989;246:1158–1161. doi: 10.1126/science.2588000. [DOI] [PubMed] [Google Scholar]
- 37.Karsenty G. Transcriptional regulation of osteoblast differentiation during development. Front Biosci. 1998;3:834–837. doi: 10.2741/a326. [DOI] [PubMed] [Google Scholar]
- 38.Hasan S, Hassa PO, Imhof R, Hottiger MO. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410:387–391. doi: 10.1038/35066610. [DOI] [PubMed] [Google Scholar]
- 39.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 40.Lain S, Xirodimas D, Lane DP. Accumulating active p53 in the nucleus by inhibition of nuclear export: a novel strategy to promote the p53 tumor suppressor function. Exp Cell Res. 1999;253:315–324. doi: 10.1006/excr.1999.4672. [DOI] [PubMed] [Google Scholar]
- 41.Lauricella M, D'Anneo A, Giuliano M, Calvaruso G, Emanuele S, Vento R, Tesoriere G. Induction of apoptosis in human osteosarcoma Saos-2 cells by the proteasome inhibitor MG132 and the protective effect of pRb. Cell Death Differ. 2003;10:930–932. doi: 10.1038/sj.cdd.4401251. [DOI] [PubMed] [Google Scholar]
- 42.Coutts AS, La Thangue NB. Mdm2 widens its repertoire. Cell Cycle. 2007;6:827–829. doi: 10.4161/cc.6.7.4086. [DOI] [PubMed] [Google Scholar]
- 43.Paredes R, Arriagada G, Cruzat F, Villagra A, Olate J, Zaidi K, van Wijnen A, Lian JB, Stein GS, Stein JL, Montecino M. Bone-specific transcription factor Runx2 interacts with the 1alpha,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol Cell Biol. 2004;24:8847–8861. doi: 10.1128/MCB.24.20.8847-8861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kassi E, Papavassiliou AG. A possible role of osteocalcin in the regulation of insulin secretion: human in vivo evidence? J Endocrinol. 2008;199:151–153. doi: 10.1677/JOE-08-0294. [DOI] [PubMed] [Google Scholar]
- 46.Oury F, Sumara G, Sumara O, Ferron M, Chang H, Smith CE, Hermo L, Suarez S, Roth BL, Ducy P, Karsenty G. Endocrine regulation of male fertility by the skeleton. Cell. 2011;144:796–809. doi: 10.1016/j.cell.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 48.Molchadsky A, Shats I, Goldfinger N, Pevsner-Fischer M, Olson M, Rinon A, Tzahor E, Lozano G, Zipori D, Sarig R, Rotter V. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One. 2008;3:e3707. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, Li B. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dworetzky SI, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Progressive changes in the protein composition of the nuclear matrix during rat osteoblast differentiation. Proc Natl Acad Sci U S A. 1990;87:4605–4609. doi: 10.1073/pnas.87.12.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montecino M, Stein JL, Stein GS, Lian JB, van Wijnen AJ, Cruzat F, Gutierrez S, Olate J, Marcellini S, Gutierrez JL. Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence. Biochem Cell Biol. 2007;85:419–425. doi: 10.1139/O07-070. [DOI] [PubMed] [Google Scholar]
- 52.Liu G, Terzian T, Xiong S, Van Pelt CS, Audiffred A, Box NF, Lozano G. The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. J Pathol. 2007;213:360–368. doi: 10.1002/path.2238. [DOI] [PubMed] [Google Scholar]
- 53.Wawrzynow B, Zylicz A, Wallace M, Hupp T, Zylicz M. MDM2 chaperones the p53 tumor suppressor. J Biol Chem. 2007;282:32603–32612. doi: 10.1074/jbc.M702767200. [DOI] [PubMed] [Google Scholar]
- 54.Saji S, Okumura N, Eguchi H, Nakashima S, Suzuki A, Toi M, Nozawa Y, Hayashi S. MDM2 enhances the function of estrogen receptor alpha in human breast cancer cells. Biochem Biophys Res Commun. 2001;281:259–265. doi: 10.1006/bbrc.2001.4339. [DOI] [PubMed] [Google Scholar]
- 55.Liu G, Schwartz JA, Brooks SC. Estrogen receptor protects p53 from deactivation by human double minute-2. Cancer Res. 2000;60:1810–1814. [PubMed] [Google Scholar]
- 56.Duong V, Boulle N, Daujat S, Chauvet J, Bonnet S, Neel H, Cavailles V. Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress-inducing agents. Cancer Res. 2007;67:5513–5521. doi: 10.1158/0008-5472.CAN-07-0967. [DOI] [PubMed] [Google Scholar]
- 57.White DE, Talbott KE, Arva NC, Bargonetti J. Mouse double minute 2 associates with chromatin in the presence of p53 and is released to facilitate activation of transcription. Cancer Res. 2006;66:3463–3470. doi: 10.1158/0008-5472.CAN-05-1381. [DOI] [PubMed] [Google Scholar]
- 58.Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, Hubbard D, Bhutkar A, Jacks T. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pochampally R, Fodera B, Chen L, Shao W, Levine EA, Chen J. A 60 kd MDM2 isoform is produced by caspase cleavage in non-apoptotic tumor cells. Oncogene. 1998;17:2629–2636. doi: 10.1038/sj.onc.1202206. [DOI] [PubMed] [Google Scholar]