Abstract
We have used a yeast two-hybrid approach to uncover protein interactions involving the D2-like subfamily of dopamine receptors. Using the third intracellular loop of the D2S and D3 dopamine receptors as bait to screen a human brain cDNA library, we identified filamin A (FLN-A) as a protein that interacts with both the D2 and D3 subtypes. The interaction with FLN-A was specific for the D2 and D3 receptors and was independently confirmed in pull-down and coimmunoprecipitation experiments. Deletion mapping localized the dopamine receptor–FLN-A interaction to the N-terminal segment of the D2 and D3 dopamine receptors and to repeat 19 of FLN-A. In cultures of dissociated rat striatum, FLN-A and D2 receptors colocalized throughout neuronal somata and processes as well as in astrocytes. Expression of D2 dopamine receptors in FLN-A-deficient M2 melanoma cells resulted in predominant intracellular localization of the D2 receptors, whereas in FLN-A-reconstituted cells, the D2 receptor was predominantly localized at the plasma membrane. These results suggest that FLN-A may be required for proper cell surface expression of the D2 dopamine receptors. Association of D2 and D3 dopamine receptors with FLN-A provides a mechanism whereby specific dopamine receptor subtypes may be functionally linked to downstream signaling components via the actin cytoskeleton.
Imbalances in dopaminergic signaling are implicated in many neuropsychiatric and motor disorders, including schizophrenia and Parkinson's disease (1). In mammalian brain, dopaminergic signaling is mediated via a cohort of dopamine receptors. Among the cloned dopamine receptor subtypes, the D2-like receptors (D2, D3, and D4) are the major target of antipsychotics, both typical and atypical, as well as anti-Parkinson's drugs (1). These receptors mediate intracellular signaling cascades by coupling to inhibitory subsets of heterotrimeric GTP-binding (G) proteins. In a variety of cell types, D2-like receptor signaling modulates calcium, potassium, and sodium currents through specific regulation of ion channel activities (2, 3). The activation of D2-like receptors also has been implicated in the regulation of cellular morphogenesis (4) and in the maintenance of neuronal structure in adult brain (5–7). Although the D2-like receptors appear to activate discrete signal transduction pathways, the question of whether individual D2-like receptors subserve distinct functional roles is an issue that has not yet been satisfactorily addressed.
To better understand the regulation of dopamine receptor signaling, we are interested in identifying dopamine receptor-interacting proteins. Identification of dopamine receptor-interacting proteins unique to specific receptor subtypes may provide important clues to how functional differences between dopamine receptor subtypes are manifested. We conducted yeast two-hybrid screens of a human brain cDNA library, with the third intracellular loop of the D2S and D3 receptors as bait. Using this approach, we identified filamin A (FLN-A) as a dopamine receptor-interacting protein that interacts specifically with the D2 and D3 dopamine receptor subtypes. Interaction of D2 and D3 with FLN-A may be important for establishing the correct subcellular localization of D2 and D3 receptors and for linking components of dopaminergic signaling complexes with the appropriate receptor subtypes.
Materials and Methods
DNA Constructs and Protein Interaction Assays.
All constructs were generated by subcloning PCR amplification products into appropriate vectors, and each construct was verified by DNA sequence analysis. D2S (amino acids 211–344) and D3 (amino acids 211–329) cDNA fragments encoding the third intracellular (IC3) domains were constructed in the yeast GAL4 DNA-binding domain expression vector pAS2–1 (D2) or pAS-1 (D3). Additional constructs encoding IC3 domains of D1, D2L, D4, and D5 dopamine receptors, as well as M1-muscarinic and β2-adrenergic receptors, were constructed in pAS2–1. PAS/D3nf encodes a polypeptide containing the complete IC3 domain through the C terminus of the D3 splice variant D3nf.
For the D2S library screen, the bait pAS/D2S was transformed into the yeast strain PJ69–2A (MATa). Transformants then were mated against yeast strain Y187 (MATα) expressing adult human brain library cDNAs in the GAL4-activation domain vector pACT2 (CLONTECH). A total of 1.5 × 106 independent clones were screened by selective growth on Leu−/Trp−/His−/Ade− plates. β-Galactosidase activity was determined with the nitrocellulose filter lift assay (8). For the D3 library screen, the pAS/D3 bait plasmid and human brain library cDNAs in pACT2 (CLONTECH) were sequentially introduced into yeast strain MaV103 by the use of standard lithium acetate transformation. Approximately 1 × 106 independent clones were screened, and positive clones were identified by growth on Leu−/Trp−/His−/Ura− selection plates and expression of β-galactosidase activity.
To map sites within D2, D3, and filamin that contribute to dopamine receptor–FLN-A interaction, truncated segments of the D2 and D3 IC3 domain were constructed in pAS2–1, and segments of FLN-A backbone repeats were inserted into pACT2. Bait and prey plasmids were simultaneously transformed into the yeast strain MAV103 and interactions were assayed as described above.
Preparation of Glutathione S-Transferase (GST) Fusion Proteins and in Vitro Binding Assays.
Fusion constructs GST-D2 (amino acids 211–344), GST-D3 (amino acids 211–329), and GST-FLN (amino acids 1788–2121 of FLN-A backbone repeats 16–19) were constructed in the expression vector pGEX-4T-1 (Amersham Pharmacia). Fusion proteins were induced in Escherichia coli strain BL21 (DE3) and purified by using glutathione-Sepharose (Amersham Pharmacia) according to the manufacturer's instructions. FLN-A polypeptide (amino acids 1788–2121) was generated by thrombin cleavage of the GST-FLN fusion protein, and excess thrombin was removed by using benzamidine-Sepharose (Amersham Pharmacia). Binding assays were carried out as described (9).
Eluted proteins were separated by SDS/PAGE and transferred to a nitrocellulose filter (10). The filter was probed with either a 1:1,000 dilution of the anti-FLN-A monoclonal antibody (Chemicon) or a 1:500 dilution of a goat polyclonal anti-GST antibody (Amersham Pharmacia) and developed with horseradish peroxidase-conjugated goat anti-mouse (1:10,000) or rabbit anti-goat (1:2,000) secondary antibodies, respectively (Jackson ImmunoResearch). Immunoreactivity was detected by enhanced chemiluminescence with an ECL Plus kit (Amersham Pharmacia).
Coimmunoprecipitation.
Crude membranes were prepared from HEK293 cells stably expressing FLAG-tagged D2L dopamine receptors as described (10). D2L receptors were immunoprecipitated by using anti-D2 rabbit polyclonal antibody (Santa Cruz Biotechnology) essentially as described by Karpa et al. (10). Western analysis of immunoprecipitated complex was performed by using the anti-FLN-A monoclonal antibody or anti-actin goat polyclonal antibody (1:500, Santa Cruz Biotechnology).
Cell Culture.
A primary culture from rat striatum was prepared essentially as described by Ivkovic and Ehrlich (11). Briefly, striata from late embryonic (embryonic days 17–18) Sprague–Dawley rat pups were dissected into ice-cold Hanks' balanced salt solution (Life Technologies, Rockville, MD). The tissue was dissociated with 1% trypsin (Worthington) and incubated for 10 min at room temperature. Trypsin was deactivated by the addition of 5% horse serum (Sigma). The tissue was resuspended in Neurobasal media (Life Technologies) and mechanically triturated. Dissociated cells were plated on glass coverslips precoated with poly-l-lysine (12.5 μg/ml) and laminin (5 μg/ml). Cells were maintained for 2 weeks in vitro in Neurobasal media supplemented with 2% B27 (Life Technologies) and 5% horse serum.
Immunofluorescence.
For immunostaining of the striatal cell culture, cells were fixed in a solution containing 4% paraformaldehyde, 250 mM sucrose, 25 mM Hepes, 2.5 mM Mg(Ac)2, 2.5 mM KCl and permeabilized in blocking buffer (1× PBS/10% donkey serum/0.3% Triton X-100). Triple labeling was performed by sequentially staining with antibodies against D2 receptor (1:500 dilution of a rabbit polyclonal antibody; Santa Cruz Biotechnology), FLN-A (1:1,000 dilution of a goat polyclonal antibody; Santa Cruz Biotechnology), and the neuronal marker TuJ1 (1:500 dilution of a mouse mAb; Babco, Richmond, CA). D2, filamin, and tubulin staining were visualized with FITC-donkey anti-rabbit (1:200), Red-X-conjugated donkey anti-goat (1:200), or AMCA-conjugated donkey anti-mouse (1:200) secondary antibodies (Jackson ImmunoResearch), respectively. Astrocytes were stained using a 1:200 dilution of antiglial fibrillary acidic protein mouse mAb (ICN) and visualized with a 1:200 dilution of Red-X-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResarch). Fluorescent images were obtained with a Zeiss Axiophot system and captured with qed imaging software.
Results
Interaction of FLN-A with D2 and D3 Dopamine Receptors.
To identify subtype-specific dopamine receptor-interacting proteins, we used the IC3 domain from the D2S and D3 dopamine receptor as bait to screen an adult human brain cDNA library. Several of the D2- and D3-interacting clones contained an identical ≈1.2-kb cDNA insert that encoded a 334-aa-long fragment (amino acids 1788–2121) corresponding to repeats 16–19 of the actin-binding protein FLN-A, also known as nonmuscle filamin/actin-binding protein 280. FLN-A is a ubiquitously expressed, dimeric actin-cross-linking phosphoprotein that promotes orthogonal branching of actin filaments and links membrane glycoproteins to the actin cytoskeleton (14).
To examine the specificity of FLN-A–dopamine receptor interaction, we used the yeast two-hybrid system to test the interaction of FLN-A with additional G-protein-coupled receptors. Bait constructs encoding the IC3 domains of the D1, D2S, D2L, D3, D4, and D5 dopamine receptors, as well as the M1 muscarinic and β2-adrenergic receptors, were tested for interaction with the cDNA clone. FLN-A–dopamine receptor interaction was restricted to the D2 and D3 receptors. FLN-A did not interact with D1, D4, or D5 dopamine receptors or with the M1-muscarinic or β2-adrenergic receptors. D2 splice variants D2S and D2L, as well as the complete carboxyl-terminal domain of the D3 splice variant D3nf, showed comparable interaction with the FLN-A clone, based on the intensity of the β-galactosidase colorimetric assay (data not shown). Together, these results suggest that FLN-A interacts specifically with the D2 and D3 dopamine receptor subtypes.
Verification of Filamin–Dopamine Receptor Interaction.
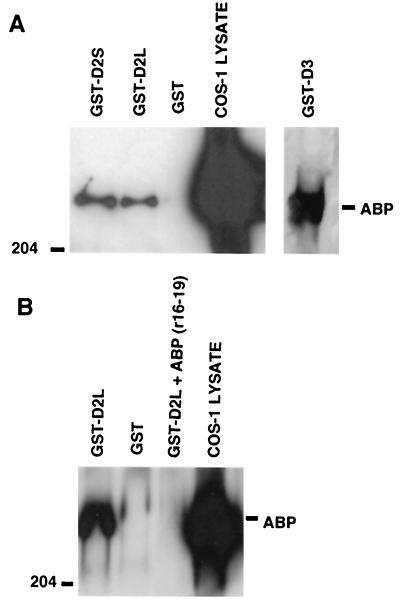
To verify interaction between FLN-A and the D2 and D3 receptors, we tested the ability of naturally expressed full-length FLN-A to associate with GST fusion proteins containing the D2 short, D2 long, or D3 IC3 domain sequences (GST-D2S, GST-D2L, or GST-D3, respectively). As shown in Fig. 1A, a Western blot containing lysate prepared from COS-1 cells produced an ≈280-kDa band immunoreactive with anti-FLN-A antibody. This band represents FLN-A that is endogenously expressed by COS-1 cells. The same band was detected in pull-down assays after the cell lysate was incubated with either the GST-D2S or GST-D2L fusion proteins, but not with GST alone (Fig. 1A). FLN-A was also capable of associating with GST-D3 fusion protein in the pull-down reaction (Fig. 1A). Preincubation of the GST-D2L fusion protein with a fragment of FLN-A (amino acids 1788–2121) prevented association of the endogenously expressed full-length FLN-A with GST-D2L (Fig. 1B).
Figure 1.
Association of D2 and D3 receptors with FLN-A. GST-D2 and GST-D3 (A) fusion proteins were used to pull down FLN-A from COS-1 cell lysates. Endogenously expressed FLN-A (ABP, actin binding protein 280) was pulled down in the presence of GST-D2L, GST-D2S, and GST-D3, but not with GST alone. (B) Preincubation of GST-D2L with FLN-A repeat fragment 16–19 abolished pulldown of full-length FLN-A protein.
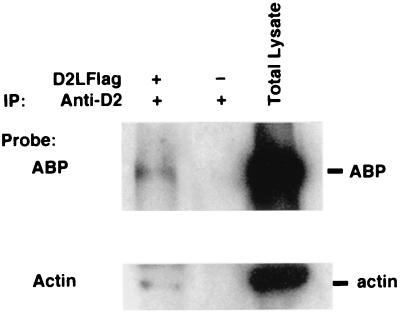
To verify the interaction of full-length D2 dopamine receptor with FLN-A, we tested the ability of anti-D2 antibody to coimmunoprecipitate FLN-A from crude membranes prepared from HEK293 cells stably expressing FLAG-tagged D2L dopamine receptors. These cells endogenously express FLN-A (ABP). As shown in Fig. 2, the anti-D2 antibody was capable of coimmunoprecipitating FLN-A. FLN-A was not coimmunoprecipitated from cells that were not transfected with the D2 dopamine receptors. These results suggest that FLN-A and D2 dopamine receptors are capable of association in intact cells. The anti-D2 dopamine receptor antibody was also capable of coimmunoprecipitating actin from the same membranes, suggesting that FLN-A may physically link D2 dopamine receptors to the submembranous actin skeleton. Together with the pull-down assays, these results provide strong biochemical evidence for FLN-A interaction with the D2 and D3 dopamine receptors.
Figure 2.
Coimmunoprecipitation of D2 dopamine receptors with FLN-A. Anti-D2 rabbit polyclonal antibody was used to immunoprecipitate D2 receptors from crude membranes prepared from HEK293 cells stably expressing D2L-receptor. FLN-A and actin were both detectable in solubilized membranes from D2L-expressing cells, but not from parental HEK293 cells.
Mapping Protein–Protein Interaction Domains.
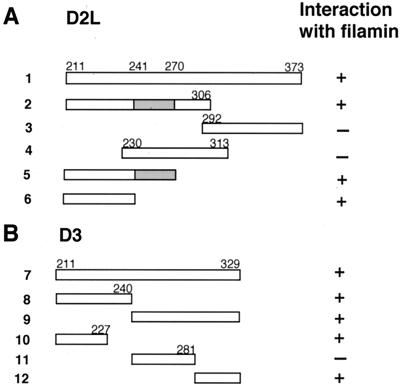
We carried out deletion mapping studies to pinpoint specific binding domains within the D2 and D3 receptors that contribute to dopamine receptor–FLN-A interaction. Truncated fragments of the D2L (Fig. 3A) and D3 (Fig. 3B) IC3 domains were tested for interaction with FLN-A (amino acids 1788–2121) in the yeast two-hybrid assay. Construct 2 (amino acids 211–306) tested positive in the β-galactosidase assay, and constructs 3 (amino acids 292–373) and 4 (amino acids 230–313) failed to interact with FLN-A, thus localizing the binding site for FLN-A within the N-terminal region of the IC3 domain of D2L. Constructs 5 (encoding residues 211–270) and 6 (amino acids 211–241) interacted with FLN-A, indicating that amino acids 211–241 of the D2L receptor comprise a core domain sufficient for interaction with FLN-A.
Figure 3.
Interaction of D2 and D3 deletion mutants with FLN-A. Schematic representation of D2L (A) and D3 (B) IC3 domain constructs tested for interaction with FLN-A (residues 1788–2121) in the two-hybrid assay. Interaction with FLN-A is indicated by the presence (+) or absence (−) of β-galactosidase activity. The alternatively spliced 29-aa exon in D2L is indicated by a shaded box.
Deletion analysis also was used to map the site within the IC3 domain of D3 that contributes to interaction with FLN-A (Fig. 3B). Constructs 7, 8, and 9 were all capable of interacting with FLN-A, suggesting that sequences within both the proximal and distal segments of the D3 IC3 domain contribute to FLN-A–D3 interaction. Further truncations revealed that construct 10 (amino acids 211–227) and construct 12 (amino acids 281–329) interacted with FLN-A, whereas construct 11 (amino acids 240–281) failed to interact. These results are consistent with the idea that within the D3 receptor, both the N- and C-terminal segments of the IC3 domain contribute to the interaction with FLN-A.
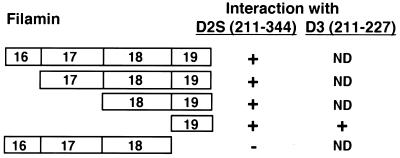
To map the domains within FLN-A that interact with the dopamine receptors, we carried out interaction assays of truncated versions of FLN-A and the IC3 domain of the D2S receptor (Fig. 4). Constructs encoding repeat units 16–19, 17–19, 18 and 19, or 19 alone all showed interaction with D2S in the two-hybrid assay. Repeat unit 19 also showed positive interaction with a construct encoding residues 211–227 of the D3 IC3 domain (Fig. 4). In contrast, a construct encoding FLN-A repeat units 16–18 failed to interact with the D2S IC3 domain. These results indicate that sequences within repeat unit 19 of FLN-A contribute to the interaction of FLN-A with the D2 and D3 receptors.
Figure 4.
Mapping of the D2 and D3 receptor binding site on FLN-A: schematic representation of constructs encoding truncations of FLN-A repeat units 16–19. Constructs were tested for interaction with the entire IC3 domain of the D2S receptor (residues 211–344) or a fragment of the D3 receptor IC3 domain (residues 211–227). Interaction with the D2 or D3 receptor is indicated by the presence (+) or absence (−) of β-galactosidase activity. ND, Not determined. Numbers within boxes indicate repeat units within FLN-A.
FLN-A and Dopamine Receptors Colocalize in Cultures of Rat Striatum.
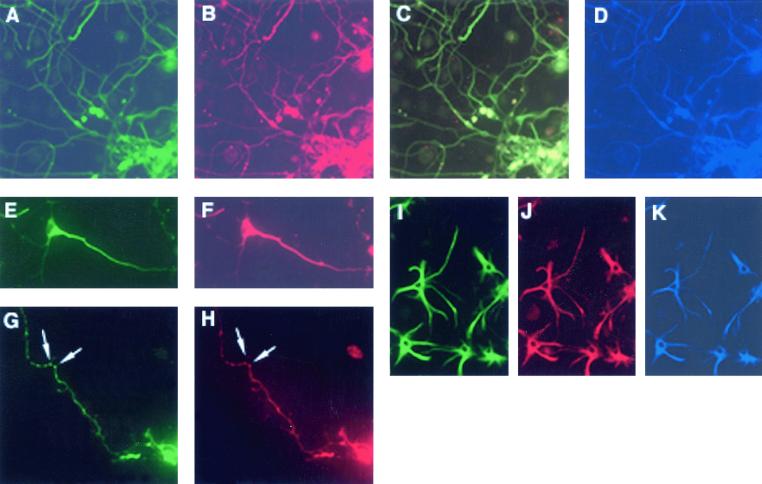
To further characterize FLN-A–dopamine receptor interaction, we examined the expression of FLN-A and D2 dopamine receptors in primary culture derived from late embryonic rat striatum. With the use of an anti-D2 antibody (reactive with both the D2L and D2S isoforms), D2 receptor labeling was particularly abundant in perikarya and neurites (Fig. 5A). We used a triple-labeling protocol to incubate cells with anti-D2, anti-FLN-A, and anti-TuJ1 (the neuron-specific marker, beta tubulin III) antibodies. As shown in Fig. 5 A–F, many striatal neurons were intensely labeled with all three antibodies. Labeling with D2 (Fig. 5 A and E) and FLN-A (Fig. 5 B and F) antibodies was readily visible in cell bodies and proximal neurites, whereas distal processes produced less intense staining (Fig. 5C). Apparent along axons were varicosities that exhibited both FLN-A and D2 receptor immunoreactivity (Fig. 5 G and H, arrows). A small number of FLN-A-D2-positive and TuJ1-negative cells within the culture exhibited immunoreactivity with the astrocyte-specific marker glial fibrillary acidic protein, indicating coexpression of FLN-A and D2 receptors in astrocytes (Fig. 5 I–K). These observations are consistent with the view that FLN-A/D2 receptors are likely to be found to be associated within mammalian brain.
Figure 5.
Expression of D2 receptors and FLN-A in primary striatal cultures from rat brain. Epifluorescent detection of D2 receptor (A), FLN-A (B), and tubulin (D) triple labeling within neurons. The merged image (C) shows D2 receptor and FLN-A coexpression in virtually all neurons. D2 receptor (E and G) and FLN-A (F and H) are coexpressed within neuronal perikarya and neurites, whereas D2 and FLN-A-immunoreactive clusters are detected within axonal varicosities as indicated by arrows (G and H). Detection of D2 receptor (I), FLN-A (J), and glial fibrillary acidic protein (K) triple labeling shows D2 receptor/FLN-A coexpression in astroglia.
Differential Subcellular Localization of D2 Dopamine Receptor in FLN-A-Deficient and FLN-A-Reconstituted Cells.
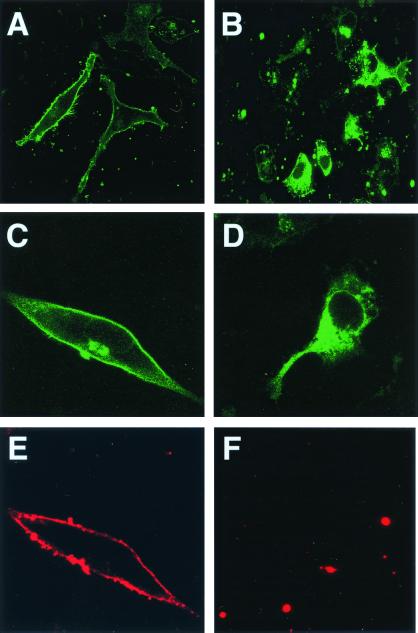
To examine the biological significance of D2–FLN-A interaction, we expressed C-terminal EGFP-tagged D2L dopamine receptors in FLN-A-deficient (M2) and FLN-A-rescued (A7) cells. As shown in Fig. 6A, D2 receptors in A7 cells were found predominantly at the plasma membrane. Cell surface staining of D2 receptors in A7 cells with an antibody directed against the extracellular amino terminus of the D2 receptor produced a staining pattern coincident with EGFP fluorescence (Fig. 6 C and E). In contrast, expression of D2 receptors in FLN-A-deficient M2 cells was predominantly intracellular (Fig. 6B). In these cells, little if any cell surface label was detectable with the anti-D2 antibody (Fig. 6 D and F). These findings suggest that FLN-A may be required for the proper plasma membrane localization of D2 dopamine receptors.
Figure 6.
Expression of D2 dopamine receptors in M2 and A7 melanoma cell lines. Confocal images of D2L-EGFP expression in FLN-A-deficient (M2; B, D, and F) and FLN-A-rescued (A7) cells (A, C, and F). Higher magnification of D2 receptor expression in A7 (C) and M2 (D) cells. Cell surface expression of D2-EGFP was detected in A7 cells (E), but not in M2 cells (F).
Discussion
We have used a yeast two-hybrid approach to identify FLN-A as a protein that interacts with the D2 and D3 dopamine receptor subtypes. The interaction was verified by the ability of GST-D2 and -D3 fusion constructs to pull down endogenous FLN-A from whole cell lysates. Coimmunoprecipitation studies further verified that D2 dopamine receptors interact with FLN-A in intact cells. The presence of a complex containing D2 dopamine receptor, FLN-A, and actin suggests that FLN-A may mediate a physical linkage between membrane-associated D2 dopamine receptors and the submembranous cytoskeletal network. Immunohistochemical studies using a primary rat striatal culture provide evidence that FLN-A and D2 dopamine receptors colocalize within striatal neurons and astrocytes. The colocalization studies, together with the biochemically defined interaction, strongly suggest the likelihood of a physiological interaction between FLN-A and dopamine receptors in mammalian brain.
FLN-A is a ubiquitously expressed actin cross-linking phosphoprotein that is an important structural determinant of the submembranous cytoskeleton of animal cells. It is composed of an N-terminal actin-binding domain, a C-terminal homodimerization domain, and a central rod-like backbone that comprises 23 tandem repeats, each approximately 96 aa in length (14). Dimerization of FLN-A is necessary for cross-linking actin, a feature that is essential for maintaining the integrity of the cortical cytoplasm. Dopamine receptors are the first group of G-protein-coupled receptors for which an interaction with FLN-A has been described. FLN-A has been shown to directly link other membrane-associated receptors, including platelet glycoprotein Ibα (17), β-integrin (18), and FcγRI (19), to the actin cytoskeleton.
At present, the functional significance of dopamine receptor–FLN-A interaction is unknown. Several possible roles for dopamine receptor–FLN-A interaction are illustrated in the model shown in Fig. 7. Linkage of D2 and D3 dopamine receptors via FLN-A to the actin skeleton may help anchor these receptors at or near sites of synaptic activity. Our studies of D2 receptor localization in M2/A7 cell lines suggest that FLN-A is required for proper plasma membrane distribution of D2 dopamine receptors. Future expression studies using D2 or FLN-A interaction mutants will help to clarify whether the correct plasma membrane localization of D2 dopamine receptors depends on or is regulated by specific interaction with FLN-A. It is possible that direct interaction of D2 and D3 dopamine receptors with cytoskeletal proteins such as FLN-A may influence receptor localization as well as the stability of the receptor at the plasma membrane. Association with specific cytoskeletal proteins has been shown to be a mechanism underlying the synaptic localization of several different neurotransmitter receptors. For example, NMDA receptors are clustered and localized to postsynaptic sites by interaction with postsynaptic density protein-95 (20) and α-actinin (21). The anchoring of γ-aminobutyric acid type c receptors at retinal synapses is mediated by microtubule-associated protein 1B (22), whereas the somatostatin receptor subtype 2 has been shown to associate with cortical actin filaments via interaction with cortactin-binding protein 1 (23). It will clearly be of interest to determine whether FLN-A serves to anchor D2 and D3 dopamine receptors at sites of synaptic activity. The ultrastructural localization of FLN-A/dopamine receptor complexes should help resolve this issue.
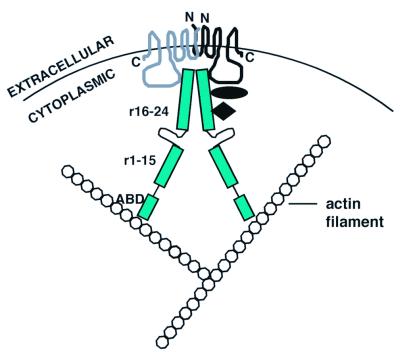
Figure 7.
Schematic representation of dopamine receptor–FLN-A interaction. The model predicts that FLN-A serves as a molecular adapter linking D2 and/or D3 receptors to the actin cytoskeleton. FLN-A may also serve as a platform for assembling additional components (black diamond and oval) of the dopamine signaling complex. The model also predicts that FLN-A may link dopamine receptor monomers (Top, wavy lines). N and C refer to the N and C termini, respectively, of the two dopamine receptor monomers. ABD indicates the actin binding domain, and r1–15 and r16–24 indicate repeat units 1–15 and 16–24 on the FLN-A backbone.
A second possible role for FLN-A is that of a molecular scaffold upon which different components of dopamine signaling complexes may be assembled. Consistent with this notion is the observation that in the Drosophila embryo, FLN-A appears to function as an adaptor, linking components of the dorsoventral signaling pathway with the transmembrane receptor Toll (24). In mammalian systems, FLN-A binds a number of signaling proteins, including ras-related small GTPases (23), the GTPase regulator protein trio (25), the stress-activated protein kinase activator SEK-1 (9), and tumor necrosis factor receptor-associated factor 2 (26). The extensive backbone structure of FLN-A suggests that it could serve as a platform to link D2 and D3 dopamine receptors with receptor-specific regulatory proteins as well as other components of the dopaminergic signaling machinery. The organization of signaling components into such a complex would presumably enhance the specificity of protein–protein interactions and the efficiency of signal transduction by positioning signaling molecules close to their substrates. The observation that D2 dopamine receptor-mediated inhibition of cAMP production is reduced in a FLN-A-deficient melanoma cell line (27) is consistent with this view. In the future, identifying additional FLN-A-interacting proteins within the brain may uncover downstream dopaminergic signaling components that confer D2 or D3 receptor-specific function.
The dimeric structure of FLN-A suggests an additional role in the organization of dopamine receptors. Recent studies have shown that D2 (28, 29) and D3 receptors (10) may exist as homodimers and that dopamine receptors can heterodimerize with other G-protein-coupled receptors (30–32). It is therefore tempting to speculate that FLN-A dimerization could promote either direct association between dopamine receptor monomers or indirect interaction of receptor monomers through a FLN-A bridge. Expression of mutant forms of FLN-A (or D2 or D3 receptors) in which the FLN-A/dopamine receptor binding sites were disrupted should allow us to address this issue.
In animal cells, cytoskeletal components are important determinants of cellular/subcellular morphology, polarity, motility, and secretion, and their functions are subject to regulation by extracellular signals transduced via cell surface receptors (33). Neurotransmitter receptors have been implicated in the regulation of neuronal morphology and connectivity in the adult brain via reorganization of cytoskeletal components. For example, activation of the glutamate receptors results in calpain-mediated breakdown of fodrin (brain spectrin) and has been proposed as a cellular mechanism underlying dendritic remodeling (34). Moreover, long-term antipsychotic drug treatment has been shown to cause alterations in neuronal structure within the adult brain (5–7, 35). Changes in synaptic architecture have been reported in the substantia nigra and the striatum as a consequence of long-term treatment with the typical antipsychotic haloperidol (5–7). Treatment of monkeys with haloperidol has been found to result in a significant decrease in the level of the dendritic D2 receptor-interacting protein spinophilin (35). Because haloperidol is a high-affinity antagonist at D2-like dopamine receptor sites, these findings suggest an important link between the effects of antipsychotic drugs and alterations in dopamine receptor–cytoskeleton interactions. It will thus be of considerable interest to determine whether antipsychotic drugs also affect neuronal morphology and plasticity via effects on FLN-A interaction with D2 and D3 dopamine receptors.
Finally, our observation that FLN-A interacts with both D2 and D3 dopamine receptors is not surprising, in view of the higher degree of sequence similarity within the IC3 domain between D2 and D3 (80%) versus D4 (50% for D2-D4 and 46% for D3-D4). Our mapping analyses confirm that a core sequence of approximately 19 residues within the N-terminal segment of IC3 loop of the D2 and D3 receptors may be required for FLN-A association, although an additional segment within the C-terminal portion of D3 IC3 domain may also contribute to the interaction. A blast search with this 19-aa peptide sequence of the D2 receptor demonstrates a close match with the D3 receptor but not with any other G-protein-coupled receptor, suggesting that the interaction may be specific to the two dopamine receptor subtypes. Expression of D2 or D3 receptors carrying mutations that abrogate interaction with FLN-A should help clarify the physiological significance of this interaction.
Acknowledgments
We are grateful to Dr. Victor Canfield for valuable suggestions and critical comments. We thank Drs. John Hartwig and Mark von Zastrow for generously providing us with the M2/A7 and D2L/HEK293 cells, respectively. This work was funded by Grant P50-MH44866 from the National Institute of Mental Health.
Abbreviations
- FLN-A
filamin A
- IC3
third intracellular
- GST
glutathione S-transferase
References
- 1.Civelli O, Bunzow J R, Grandy D K. Annu Rev Pharmacol Toxicol. 1993;32:281–307. doi: 10.1146/annurev.pa.33.040193.001433. [DOI] [PubMed] [Google Scholar]
- 2.Williams P J, Macvicar B A, Pittman Q J. Neuroscience. 1989;31:673–681. doi: 10.1016/0306-4522(89)90432-6. [DOI] [PubMed] [Google Scholar]
- 3.Surmeier D J, Eberwine J, Wilson C J, Cao Y, Stefani A, Kitai S T. Proc Natl Acad Sci USA. 1992;89:10178–10182. doi: 10.1073/pnas.89.21.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swarzenski B C, Tang L, Oh Y J, O'Malley K H, Todd R D. Proc Natl Acad Sci USA. 1994;91:649–653. doi: 10.1073/pnas.91.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uranova N A, Orlovskaya D D, Apel K, Klintsova A J, Haselhorst U, Schenk H. Synapse. 1991;7:253–259. doi: 10.1002/syn.890070402. [DOI] [PubMed] [Google Scholar]
- 6.Benes F M, Paskevich P A, Domesick V B. Science. 1983;221:969–971. doi: 10.1126/science.6879197. [DOI] [PubMed] [Google Scholar]
- 7.Benes F M, Paskevich P A, Davidson J, Domesick V B. Brain Res. 1985;348:15–20. doi: 10.1016/0006-8993(85)90353-1. [DOI] [PubMed] [Google Scholar]
- 8.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 9.Marti A, Luo Z, Cunningham C, Ohta Y, Hartwig J, Stossel T P, Kyriakis J M, Avruch J. J Biol Chem. 1997;272:2620–2628. doi: 10.1074/jbc.272.5.2620. [DOI] [PubMed] [Google Scholar]
- 10.Karpa K, Lin R, Kabbani N, Levenson R. Mol Pharmacol. 2000;58:677–683. doi: 10.1124/mol.58.4.677. [DOI] [PubMed] [Google Scholar]
- 11.Ivkovic S, Ehrlich M. J Neurosci. 1999;19:5409–5419. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan Z U, Mrzljak L, Gutierrez A, De La Calle A, Goldman-Rakic P S. Proc Natl Acad Sci USA. 1998;95:7731–7736. doi: 10.1073/pnas.95.13.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan Z U, Gutierrez A, Martin R, Penafiel A, Rivera A, De La Calle A. J Comp Neurol. 1998;402:353–371. doi: 10.1002/(sici)1096-9861(19981221)402:3<353::aid-cne5>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Gorlin J, Yamin R, Egan S, Stewart M, Stossel T, Kwiatkowski D, Harwig J. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Hans S, Mckell D, Goate A, Wu J. J Neurosci. 1998;18:914–922. doi: 10.1523/JNEUROSCI.18-03-00914.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox J W, Lamperti E D, Eksioglu Y Z, Hong S E, Feng Y, Graham D A, Scheffer I E, Dobyns W B, Hirsch B A, Radtke R A, et al. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 17.Meyer S C, Zuerbig S, Cunningham C C, Hartiwig J H, Bissell T, Gardner K, Fox J E B. J Biol Chem. 1997;272:2914–2919. doi: 10.1074/jbc.272.5.2914. [DOI] [PubMed] [Google Scholar]
- 18.Sharma C, Ezzell R, Arnaout M. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
- 19.Ohta Y, Stossel T P, Hartwig J H. Cell. 1991;67:275–282. doi: 10.1016/0092-8674(91)90179-3. [DOI] [PubMed] [Google Scholar]
- 20.Wyszynski M, Lin J, Rao A, Nigh E, Beggs A H, Craig A M, Sheng M. Nature (London) 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 21.Hanley J G, Koulen P, Bedford F, Gordon-Weeks P R, Moss S J. Nature (London) 1999;397:66–72. doi: 10.1038/16258. [DOI] [PubMed] [Google Scholar]
- 22.Zitzer H, Richter D, Kreienkamp H. J Biol Chem. 1999;274:18153–18156. doi: 10.1074/jbc.274.26.18153. [DOI] [PubMed] [Google Scholar]
- 23.Ohta Y, Suzuki N, Nakamura S, Hartwig J H, Stossel T P. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards D N, Towb P, Wasserman S. Development (Cambridge, UK) 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 25.Bellanger J, Zugasti O, Lazaro J, Diriong S, Lamb N, Sardet C, Debant A. C R Seances Soc Biol Fil. 1998;192:367–372. [PubMed] [Google Scholar]
- 26.Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U. J Biol Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Bermak J C, Wang W, Zhou Q Y. Mol Pharmacol. 2000;57:446–452. doi: 10.1124/mol.57.3.446. [DOI] [PubMed] [Google Scholar]
- 28.George S R, Lee S P, Varghese G, Zeman P R, Seeman P, Ng G Y K, O'Dowd B F. J Biol Chem. 1998;273:30244–30248. doi: 10.1074/jbc.273.46.30244. [DOI] [PubMed] [Google Scholar]
- 29.Zawarynski P, Tallerico T, Seeman P, Lee S P, O'Dowd B F, George S R. FEBS Lett. 1998;441:383–386. doi: 10.1016/s0014-5793(98)01588-9. [DOI] [PubMed] [Google Scholar]
- 30.Rocheville M, Lange D C, Kumar U, Patel S C, Patel R C, Patel Y C. Science. 2000;288:154–157. doi: 10.1126/science.288.5463.154. [DOI] [PubMed] [Google Scholar]
- 31.Gines S, Hillion J, Torvinen M, Le Crom S, Casado V, Canela E, Rondin S, Lew J Y, Watson S, Zoli M, et al. Proc Natl Acad Sci USA. 2000;97:8606–8611. doi: 10.1073/pnas.150241097. . (First Published July 11, 2000; 10.1073/pnas.150241097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, Wan Q, Pristupa Z B, Yu X, Wang Y T, Niznik H B. Nature (London) 2000;403:274–280. doi: 10.1038/35002014. [DOI] [PubMed] [Google Scholar]
- 33.Carraway K L, Carraway C A C, Carraway K L., III . Signaling and the Cytoskeleton. Berlin: Springer; 1998. [Google Scholar]
- 34.Siman R, Noszek J C. Neuron. 1988;1:279–287. doi: 10.1016/0896-6273(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 35.Lidow M S, Song Z, Castner S A, Allen P B, Greengard P, Goldman-Rakic P S. Biol Psychiatry. 2001;49:1–12. doi: 10.1016/s0006-3223(00)01058-1. [DOI] [PubMed] [Google Scholar]
- 36.Kornau H, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]