Abstract
We report on the highly unusual case of a 75-year-old woman who developed a biphasic right axillary mass of apparent melanoma and adenocarcinoma 13 years after a diagnosis of primary melanoma on her right upper back. The differential diagnosis included a collision tumor and metastatic melanoma with adenocarcinomatous transdifferentiation. We utilized immunohistochemical staining, DNA sequencing, and comparative genomic hybridization (CGH) to characterize this unusual tumor. By immunohistochemistry, the melanomatous component was positive for S100 and Melan-A, and had patchy positivity for cytokeratin. The adenocarcinomatous component was negative for melanoma markers, but was strongly positive for cytokeratin. In addition, the glandular component was positive for CDX-2 and Ber-EP4, giving the distinct histologic and immunohistochemical impression of a gastrointestinal metastasis nested within a deposit of metastatic melanoma. Clinical and radiologic work-up failed to reveal a primary gastrointestinal malignancy. Molecular genetic analysis, including DNA sequencing and CGH, revealed that both areas contained an identical NRAS Q61K mutation and had highly similar CGH profiles, including gains of chromosome 1q, and losses of 1p, 4, 9, and 10, which are archetypical of melanoma. The NRAS mutation was also identified in a deposit of metastatic melanoma resected twelve years earlier, but was not seen in the patient’s non-tumorous tissue, indicating that it was somatically acquired. Genetic analyses demonstrate that two morphologically distinct tumors arose from a common ancestor melanoma cell that harbored an NRAS mutation and subsequently divergently evolved by the acquisition of additional genomic alterations. Our findings illustrate the ability of molecular analyses to resolve lineage in complex neoplasms and illustrate the phenotypic plasticity of cancer cells.
Keywords: Melanoma, Adenocarcinoma, Transdifferentiation, Comparative Genomic Hybridization
INTRODUCTION
The juxtaposition or intermingling of two histomorphologically distinct tumors is relatively rare, and can raise diagnostic problems in distinguishing between a collision tumor and transdifferentiation of a neoplastic clone into a different phenotype. The term collision tumor is used to reflect the finding of two or more apparently distinct neoplastic cell populations that are thought to have different origins. Collision tumors can manifest as synchronous primary malignancies at the same site, as metastasis within a primary tumor of another type, or two contiguous metastases from different primary neoplasms. Examples of primary collision tumors include reports of a coincident gastric adenocarcinoma and gastrointestinal stromal tumor,8 invasive ductal carcinoma of the breast and small lymphocytic lymphoma,11 and an anaplastic oligodendroglioma with a gangliocytoma,31 whereas examples of colliding metastases include the intracranial collision of prostate and esophageal carcinomas,16 breast and ovarian adenocarcinomas metastasizing to the same axillary lymph node,30 and a breast carcinoma and melanoma admixed in the same axillary lymph node.9 Collision tumors between melanoma and various epithelial malignancies have recently been reviewed.29
While divergent differentiation is commonly seen in cancers originating from pluripotent cells such as teratomas, blastomas, and sex-cord stromal tumors, it is also occasionally seen in urothelial carcinomas, neuroendocrine tumors, histiocytic sarcomas,18 and melanoma. Banerjee and Eyden have defined this phenomenon in melanomas as the development of morphologically, immunohistochemically, and/or ultrastructurally recognizable non-melanocytic cell or tissue components within an existing melanoma.4 The wide-ranging types of divergent differentiation reported in melanomas include fibroblastic/myofibroblastic, schwannian, smooth muscle, rhabdomyosarcomatous, osteocartilaginous, ganglionic and ganglioneuroblastic, and neuroendocrine,1–4,6,10,17,19,20,26 as well as rare reports of epithelial differentiation.4,14,25,33
There are general limitations to the utility of histomorphology and immunohistochemistry in establishing the clonal relationship between two morphologically distinct tumor cell populations and in distinguishing transdifferentiation from collision. In this paper, we illustrate the utility of molecular genetic analyses to circumvent these limitations and demonstrate that a metastatic tumor deposit with histologic and immunohistochemical features of adenocarcinoma arose by transdifferentiation of metastatic melanoma. Such analyses provide powerful ancillary information to resolve the lineage of origin in complex tumors and to establish the clonal origin of histopathologically and immunohistochemically distinct tumors.
CASE REPORT
We present a case of a 75-year-old woman who initially sought medical attention in 1993 for a 1.2-cm pigmented skin lesion of the upper back, just right of the midline, which was diagnosed on biopsy as melanoma, 1.9 mm in thickness, with an in situ component that extended to the lateral specimen margins. The patient underwent surgical re-excision of the biopsy site, without lymph node dissection, approximately one month later; this re-excision specimen was free of residual melanoma. The patient then presented 15 months later with left axillary lymphadenopathy and a slightly enlarged right axillary lymph node, prompting bilateral axillary lymph node dissections. One of thirteen left axillary lymph nodes contained metastatic melanoma comprised of sheets of dyshesive polygonal cells with abundant amphophilic cytoplasm and enlarged, irregular nuclei; these cells stained positively for melanoma markers by immunohistochemistry (S100 +, melanoma cocktail (HMB-45/tyrosinase) +; Table 1), and were negative for cytokeratin (Table 1). Nodal metastases were not identified on the right side. The patient then underwent an autologous vaccine protocol for one year.
Table 1.
Immunohistochemical Results for Left Axillary Lymph Node Metastasis and Right Axillary Lymph Node Biphasic Metastatic Deposit with Features of Melanoma and Adenocarcinoma
| Component | Immunohistochemical Staina |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S100 | Melan-A | HMB-45 | MiTF | NSE | Keratin cocktail | CK7 | CK19 | CK20 | HMWK-903 | CDX-2 | Ber-EP4 | mCEA | TTF-1 | HepPar-1 | Glypican-3 | |
| Left Axillary Lymph Node | +b | +/−c | +d | + | + | −e | − | − | − | − | − | − | − | − | − | − |
| Right Axilla: Melanoma component | +/− | + | − | − | + | −/+ | − | − | − | − | − | − | − | − | − | −/+f |
| Right Axilla: Adenocarcinoma component | − | − | − | − | − | + | − | + | + | − | + | + | − | − | − | + |
Vendors and dilutions used: Ber-EP4 (Dako, Carpinteria, CA; 1:400 dilution), monoclonal carcinoembryonic antigen (mCEA, Dako, Carpinteria, CA; 1:2 dilution), cytokeratin cocktail [AE1/AE3 (Dako, Carpinteria, CA; 1:100 dilution) and Cam5.2 (Becton Dickinson, Franklin Lakes, NJ; 1:50 dilution)], cytokeratin 19 (CL19. Dako, Carpinteria, CA; 1:60 dilution), cytokeratin 20 (CK20, Dako, Carpinteria, CA; 1:100 dilution), cytokeratin 7 (CK7, Dako, Carpinteria, CA; 1:500 dilution), glypican-3 (Biomosaics, Burlington, VT; undiluted), HepPar-1 (Dako, Carpinteria, CA; 1:100 dilution), HMB-45 (Enzo Life Sciences, Farmingdale, NY; 1:2 dilution), high molecular weight keratin-903 (HMWK-903, Enzo Life Sciences, Farmingdale, NY; 1:2 dilution), Melan-A (Dako, Carpinteria, CA; undiluted), microphthalmia transcription factor-1 (MiTF-1, Dako, Carpinteria, CA; 1:200 dilution), neuron-specific enolase (NSE, Dako, Carpinteria, CA; 1:2 dilution), S100 (Dako, Carpinteria, CA; 1:1000dilution), thyroid transcription factor-1 [TTF-1 (Signet, Dedham, MA; 1:8 dilution)].
A stain is considered positive (+) if diffusely and strongly positive staining is present in greater than 90% of cells.
A stain is considered patchy positive (+/−) if staining is present in 50%–90%of cells.
HMB-45 present in a cocktail with tyrosinase in the original left axillary lymph node metastasis.
A stain is considered negative (−) if fewer than 1% of cells stain positively.
A stain is considered weakly positive (−/+) if staining is present in 1–50% of cells.
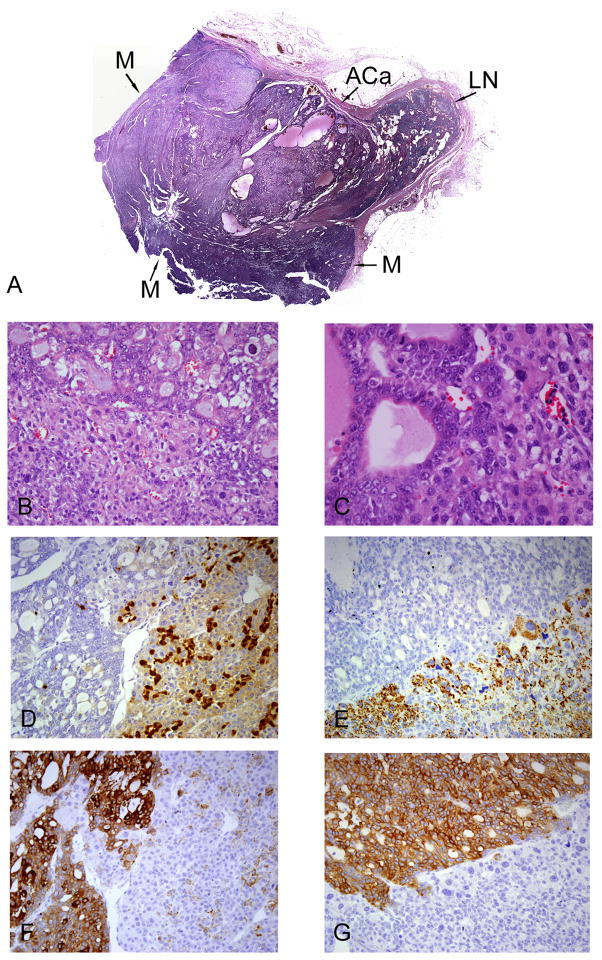
The patient was followed closely and had an uneventful medical history until 12 years later, when she presented with another axillary mass, now on the right side, that was enlarging over a two-week interval. Upon right axillary lymph node dissection, a single 4.9-cm lymph node contained metastatic tumor, and the remaining 14 lymph nodes were cancer-free. Histologic examination of the positive lymph node revealed a biphasic metastasis with histologic and immunohistochemical characteristics of both melanoma and adenocarcinoma (Figure 1A). Interestingly, the adenocarcinomatous component was nested within the melanomatous component, with no clear intervening stroma or fibrous capsule. The outer rim of tumor contained sheets and nests of dyshesive polygonal cells with abundant amphophilic cytoplasm, enlarged pleomorphic nuclei with densely hyperchromatic chromatin and prominent nucleoli, and without significant pigment deposition (Figures 1B and 1C). This aspect of the tumor exhibited patchy, but strong cytoplasmic and nuclear immunohistochemical staining for S100 (Figure 1D), as well as staining for Melan-A (Figure 1E), NSE, and weak, patchy staining with a cytokeratin cocktail (AE1/AE3 and Cam 5.2, Figure 1F), but was negative for the melanoma markers HMB-45andMiTF-1, as well asCK7, CK19, CK20, HMWK-903, CDX-2, Ber-EP4, mCEA, glypican-3, HepPar-1, and TTF-1 (Table 1). By contrast, the central portion of the tumor consisted of somewhat smaller cells arranged in gland-like and microcystic structures with apparent mucin, colloid, or proteinaceous debris within the lumina (Figures 1B and 1C). The nuclei of these cells were smaller and had a more vesicular chromatin pattern than the nuclei in the adjacent melanomatous component. Quite intriguingly, the cells in the adenocarcinomatous area stained strongly with the keratin cocktail (Figure 1F), Ber-EP4 (Figure 1G), CK19, CK20, CDX-2, and glypican-3 (Table 1), but were negative for S100 (Figure 1D), Melan-A (Figure 1E), HMB-45, MiTF-1, NSE, mCEA, HepPar-1, HMWK-903, CK7, and TTF-1 (Table 1). The differential diagnosis of this area thus included metastasis of an adenocarcinoma, possibly of gastrointestinal, hepatic, or pancreatobiliary origin, verses true transdifferentiation of the adjacent melanoma into adenocarcinoma. Importantly, neither the patient’s history nor a careful work-up revealed any evidence of a second malignancy beside the prior melanoma, which arose in a drainage area of this right axillary lymph node metastasis. No evidence of adenocarcinomatous differentiation was noted in the patient’s primary melanoma or the left axillary metastasis from 12 years earlier, and immunohistochemical analysis of this material showed no cytokeratin expression (Table 1). The patient was next seen six months later, at which time a follow-up PET-CT study revealed widespread metastases in the pelvis, vertebrae, and lungs. No colonic, hepatic, gastric, or pancreatobiliary lesions were seen. At this point, the patient was lost to follow-up.
Figure 1. Biphasic metastatic tumor deposit in a right axillary lymph node.
H&E-stained section (Panel A, 1x); ACa = Adenocarcinoma component, M = Melanomatous component, LN = Normal residual lymph node. H&E-stained section of the interface of adenocarcinoma (Panel B, top) and melanoma components (Panel B, bottom, 200x). H&E-stained section of the interface of adenocarcinoma (Panel C, left) and melanoma components (Panel C, right, 400x). Immunohistochemical staining for S100 demonstrating patchy staining in the melanoma component (Panel D, right), and negative staining in the adenocarcinoma component (Panel D, left, 200x). Immunohistochemical staining for Melan-A demonstrating positive staining in the melanoma component (Panel E, bottom), and negative staining in the adenocarcinoma component (Panel E, top, 200x). Immunohistochemical staining for keratin cocktail (AE1/AE3 and Cam5.2) demonstrating strong cytoplasmic staining in the adenocarcinoma component (Panel F, left) and weak, patchy staining in the melanoma component (Panel F, right, 200x). Immunohistochemical staining for Ber-EP4 demonstrating strong staining in the adenocarcinoma component (Panel G, top), and negative staining in the melanoma component (Panel G, bottom, 200x).
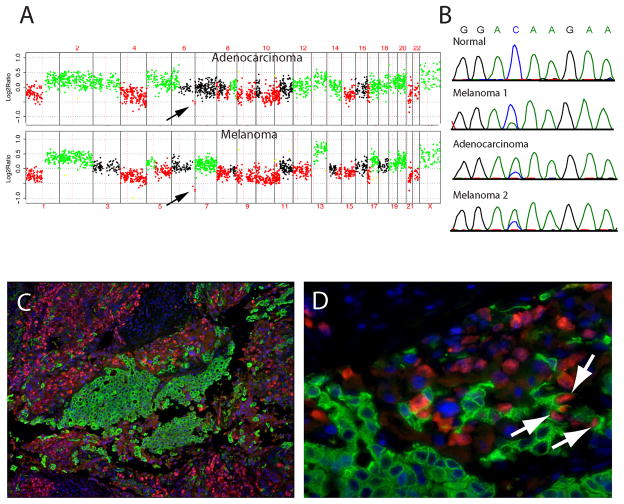
To further evaluate these two tumors at a molecular level, DNA was extracted from both tumor areas and from the original melanoma metastasis from the left axilla, resected 12 years earlier, and subjected to comparative genomic hybridization (CGH)13 and DNA sequence analysis for mutations in BRAF and NRAS,12 oncogenes commonly mutated in the majority of melanomas originating from the trunk.13 Multiple chromosomal aberrations in both tumor areas were identified by CGH, the majority of which were shared between the two areas indicating a clonal relationship: loss of chromosomes 1p, 4, 9, 10q, 11p, 15, 17p, 21, and 22, as well as gains of chromosomes1q, 2, 5p, 13, 17q, 19, and 20 (Figure 2A); this pattern of chromosomal aberrations is typical for melanoma.7 A common clonal origin of the two tumor areas was further suggested by the presence of a narrow deletion of the distal part of chromosome 6q, which was present in both histologically distinct tumors (see arrows in Figure 2A); analysis of the original metastasis by CGH was unsuccessful due to the small size of the tumor and normal tissue contamination. Finally, the notion of a clonal relationship between the different tumor populations was confirmed by detecting an NRAS Q61K mutation caused by an identical nucleotide substitution in both tumor areas (Figure 2B, third and fourth panels), as well as in the metastasis resected 12 years earlier (Figure 2B, second panel). This mutation was somatically acquired as the patient’s normal tissue showed no abnormalities (Figure 2B, top panel). Both tumors were wild type for BRAF. We note that cross-contamination of our microdissected sample can be ruled out as an explanation for finding the mutation in both areas from the biphasic right axillary lymph node metastasis. A careful manual microdissection was performed on keratin-stained slides to help distinguish the two regions. Also, the peak heights of the sequencing traces indicate that the mutant allele is the most abundant allele in both samples (Figure 2B, third and fourth panels). The opposite would be expected if a minority of mutant cells from one area had been accidently included in the microdissected tissue of the other area. NRAS maps to chromosome 1p, which showed loss of one copy in both tumor areas by CGH. The increased abundance of the mutant allele in both samples thus indicates that the chromosomal arm with the wild type copy was deleted. Interestingly, the chromosomal alterations in both tumor areas are not entirely identical by CGH. Some aberrations, such as a loss of chromosome 5q, gain of chromosomes 7 and 18, and loss of distal chromosome 14, were only found in the area of overt melanoma, whereas the adenocarcinomatous portion demonstrated gains of 6p and distal 8q, which are absent in the melanoma (Figure 2A). Together, the genetic findings demonstrate that the two morphologically distinct tumor cell populations arose from a common ancestor cell, but represent genetically distinct subclones. Dual-color immunofluorescence using S100 (red) and keratin cocktail (green), performed as previously described,15 showed strong keratin positivity in the adenocarcinomatous area and S100 positivity in the melanoma region as expected (Figures 2C and 2D). Interestingly, a few cells along the transition zone between the two areas were positive for both S100and keratin (arrows in Figure 2D).
Figure 2. Molecular genetic studies of metastatic melanoma with striking adenocarcinomatous differentiation.
Array CGH profile (Panel A) from adenocarcinomatous component (top panel) and melanomatous component (botton panel). DNA extracted from tumor was compared to normal male reference DNA. The y-axis represents the mean log2 ratio for each clone normalized to the genome median log2 ratio. The x-axis represents individual clones ordered by genomic position from chromosome 1 to 22, X, Y with the use of data obtained from the University of California at Santa Cruz Genome Browser. Gains are shown in green, losses in red, and, clones which are outliers in a region are in yellow. Sequence traces for NRAS around codon 61 for DNA extracted from normal tissue, left axillary metastasis from 1994 (Melanoma 1), right axillary adenocarcinomatous component from 2006 (Adenocarcinoma), and right axillary melanomatous component from 2006 (Melanoma 2) (Panel B). Dual-color immunofluorescence of right axillary metastasis with antibodies to S100 and keratin (Panels C and D). The tissue section was stained using labeled antibodies against S100 (red) and keratin (green). Arrows indicate the cells that are positive for both S100 and keratin (Panel D).
These molecular results also verify that this metastasis is from the patient’s primary melanoma on the back, as the metastatic melanoma resected from the contralateral axilla 12 years prior harbors the same NRAS mutation (Figure 2B). Approximately 20% of melanomas on the trunk harbor NRAS mutations, most of which occur at codon 61, as in our case.21 By contrast, NRAS mutations are infrequent in adenocarcinoma, with the exception of follicular thyroid carcinoma, in which codons 12 and 13 are frequently involved.27
METHODS
Immunohistochemical stains were performed on paraffin-embedded, 4-μm sections cut from the patient’s initial left axillary lymph node metastasis, as well as the biphasic neoplasm from the right axilla. Antibody vendor and dilution information is included in footnotes to Table 1, along with the criteria employed to evaluate the immunohistochemical stains. Manual micro-dissection of the tumor-bearing region from the right axilla was performed on five unstained 10-μm paraffin sections using a stereo-microscope (Leica MZ12) to avoid significant normal cell contamination and to avoid cross-contamination. DNA was extracted from both of these tumor areas using chloroform-phenol extraction. Mutational analysis was performed by direct sequencing of polymerase chain reaction (PCR) – amplified products generated with primers specific for NRAS exon 3 and BRAF exon 15 as previously described.12 Array CGH was performed using 600 to 2000 ng of genomic DNA, labeled by random priming and hybridized on a whole genome human BAC array (HumArray 3.1), and analyzed as previously described.13 Immunofluorescence of paraffin-embedded sections was also performed as previously described15 using antibodies to S100 (Dako, Carpinteria, CA) and cytokeratins AE1/AE3 (BioCare Medical, Concord, CA).
DISCUSSION
This unusual case report illustrates the remarkable phenotypic plasticity of cancer cells. During development, lineage specific transcription factors and epigenetic modifications lead to lineage commitment with stable patterns of gene expression and phenotypes in cells. Recent work has demonstrated that the combinatorial expression of no more than three transcription factors can convert, for example, fibroblasts to neurons.32 It is conceivable that in cancer cells, transdifferentiation, as seen in our case, may in part be due to somatic activation of lineage-foreign transcription factors. For example, transcription factors of the ETS family play a critical role in lineage determination of epithelial cells24 and have recently been demonstrated to be somatically activated in melanoma.23 Mechanisms driving such de- or transdifferentiation likely involve epigenetic alterations caused by malfunction of genes involved in maintaining chromatin marks an/or DNA methylation, as recently shown in several cancer types including melanoma.22
Finding lineage-atypical gene expression and morphologic signs of transdifferentiation in melanoma is not unprecedented. Cytokeratin expression in melanoma has been reported previously,5,10,28 but usually involves simple keratins; the expression of CK20, as was found in this case, is distinctly unusual in melanoma. In a series of melanoma cell lines and tissues analyzed for mRNA and protein expression of a panel of cytokeratins, none was found to express CK20.10 Wen et al. recently reported a 27-year-old woman with a primary, non-pigmented right scalp melanoma containing intimately admixed glandular elements.33 The melanoma component of that tumor stained for classic melanocyte markers (S100, HMB-45, MiTF-1, and PNL2), whereas the carcinomatous component was positive for cytokeratins AE1/AE3 and Cam5.2, as well as MiTF-1, suggesting true transdifferentiation of melanoma into carcinoma, however, no genetic analysis was performed to prove a clonal relationship.33 In our case, the finding of a shared point mutation in NRAS coupled with the shared presence of multiple chromosomal aberrations in both areas clearly establishes that the two morphologically distinct tumors arose from the same precursor that already had a substantial number of chromosomal abnormalities. While the different phenotypes of the two areas may in part be due to the occurrence of additional genetic alterations, the observation of cells that strongly express both lineage markers (S100and keratin; Figure 2D), raises the interesting possibility of epigenetic reprogramming of the cellular phenotype, as the position of cells at the interface between the two areas indicate that the transdifferentiation process may be actively ongoing. The remarkable phenotypic plasticity of melanoma is also illustrated by recent reports showing true rhabdomyosarcomatous differentiation in both primary and metastatic melanomas.19,20
The utility of molecular genetic methods to assist in the classification of neoplasms with ambiguous histomorphology is illustrated in this complicated case. A diagnosis of metastatic adenocarcinoma would have been rendered using routine histopathologic and immunohistochemical analysis of solely the adenocarcinomatous component of this highly unusual metastatic melanoma.
Acknowledgments
Supported by grants from the National Cancer Institute (R01 CA1315241 to BCB) and the American Skin Association (Abby S. and Howard P. Milstein Innovation Award to BCB)
References
- 1.Abbott JJ, Amirkhan RH, Hoang MP. Malignant melanoma with a rhabdoid phenotype: histologic, immunohistochemical, and ultrastructural study of a case and review of the literature. Arch Pathol Lab Med. 2004;128:686–8. doi: 10.5858/2004-128-686-MMWARP. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee SS, Bishop PW, Nicholson CM, et al. Malignant melanoma showing smooth muscle differentiation. J Clin Pathol. 1996;49:950–1. doi: 10.1136/jcp.49.11.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee SS, Coyne JD, Menasce LP, et al. Diagnostic lessons of mucosal melanoma with osteocartilaginous differentiation. Histopathology. 1998;33:255–60. doi: 10.1046/j.1365-2559.1998.00494.x. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee SS, Eyden B. Divergent differentiation in malignant melanomas: a review. Histopathology. 2008;52:119–29. doi: 10.1111/j.1365-2559.2007.02823.x. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee SS, Harris M. Morphological and immunophenotypic variations in malignant melanoma. Histopathology. 2000;36:387–402. doi: 10.1046/j.1365-2559.2000.00894.x. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee SS, Menasce LP, Eyden BP, et al. Malignant melanoma showing ganglioneuroblastic differentiation: report of a unique case. Am J Surg Pathol. 1999;23:582–8. doi: 10.1097/00000478-199905000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–5. [PubMed] [Google Scholar]
- 8.Bi R, Sheng W, Wang J. Collision tumor of the stomach: gastric adenocarcinoma intermixed with gastrointestinal stromal tumor. Pathol Int. 2009;59:880–3. doi: 10.1111/j.1440-1827.2009.02460.x. [DOI] [PubMed] [Google Scholar]
- 9.Carswell KA, Behranwala KA, Nerurkar AA, et al. Breast carcinoma and malignant melanoma metastasis within a single axillary lymph node. Int Semin Surg Oncol. 2006;3:32. doi: 10.1186/1477-7800-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N, Gong J, Chen X, et al. Cytokeratin expression in malignant melanoma: potential application of in-situ hybridization analysis of mRNA. Melanoma Res. 2009;19:87–93. doi: 10.1097/CMR.0b013e3283252feb. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KJ, Tam W, Chuang E, et al. Concurrent invasive ductal carcinoma and chronic lymphocytic leukemia manifesting as a collision tumor in breast. Breast J. 2007;13:413–7. doi: 10.1111/j.1524-4741.2007.00451.x. [DOI] [PubMed] [Google Scholar]
- 12.Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 13.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 14.Davis JP, Maclennan KA, Schofield JB, et al. Synchronous primary mucosal melanoma and mucoepidermoid carcinoma of the maxillary antrum. J Laryngol Otol. 1991;105:370–2. doi: 10.1017/s0022215100116032. [DOI] [PubMed] [Google Scholar]
- 15.Denoyelle C, Abou-Rjaily G, Bezrookove V, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–63. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 16.Dewan S, Alvarez VE, Donahue JE, et al. Intracranial collision metastases of prostate and esophageal carcinoma. J Neurooncol. 2009;95:147–50. doi: 10.1007/s11060-009-9909-1. [DOI] [PubMed] [Google Scholar]
- 17.Eyden B, Pandit D, Banerjee SS. Malignant melanoma with neuroendocrine differentiation: clinical, histological, immunohistochemical and ultrastructural features of three cases. Histopathology. 2005;47:402–9. doi: 10.1111/j.1365-2559.2005.02240.x. [DOI] [PubMed] [Google Scholar]
- 18.Feldman AL, Arber DA, Pittaluga S, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111:5433–9. doi: 10.1182/blood-2007-11-124792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gattenlohner S, Brocker EB, Muller-Hermelink HK. Malignant melanoma with metastatic rhabdomyosarcomatoid transdifferentiation. N Engl J Med. 2008;358:649–50. doi: 10.1056/NEJMc0707079. [DOI] [PubMed] [Google Scholar]
- 20.Gharpuray-Pandit D, Coyne J, Eyden B, et al. Rhabdomyoblastic differentiation in malignant melanoma in adults: report of 2 cases. Int J Surg Pathol. 2007;15:20–5. doi: 10.1177/1066896906295775. [DOI] [PubMed] [Google Scholar]
- 21.Haluska FG, Tsao H, Wu H, et al. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12:2301s–7s. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- 22.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jane-Valbuena J, Widlund HR, Perner S, et al. An Oncogenic Role for ETV1 in Melanoma. Cancer Res. 2010;70:2075–84. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jedlicka P, Sui X, Gutierrez-Hartmann A. The Ets dominant repressor En/Erm enhances intestinal epithelial tumorigenesis in ApcMin mice. BMC Cancer. 2009;9:197. doi: 10.1186/1471-2407-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajo K, Zubor P, Spacek J, et al. Carcinosarcoma of the uterus with melanocytic differentiation. Pathol Res Pract. 2007;203:753–8. doi: 10.1016/j.prp.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Magro CM, Crowson AN, Mihm MC. Unusual variants of malignant melanoma. Mod Pathol. 2006;19 (Suppl 2):S41–S70. doi: 10.1038/modpathol.3800516. [DOI] [PubMed] [Google Scholar]
- 27.Nikiforova MN, Nikiforov YE. Molecular diagnostics and predictors in thyroid cancer. Thyroid. 2009;19:1351–61. doi: 10.1089/thy.2009.0240. [DOI] [PubMed] [Google Scholar]
- 28.Ohsie SJ, Sarantopoulos GP, Cochran AJ, et al. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008;35:433–44. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 29.Satter EK, Metcalf J, Lountzis N, et al. Tumors composed of malignant epithelial and melanocytic populations: a case series and review of the literature. J Cutan Pathol. 2009;36:211–9. doi: 10.1111/j.1600-0560.2008.01000.x. [DOI] [PubMed] [Google Scholar]
- 30.Sughayer MA, Zakarneh L, Abu-Shakra R. Collision metastasis of breast and ovarian adenocarcinoma in axillary lymph nodes: a case report and review of the literature. Pathol Oncol Res. 2009;15:423–7. doi: 10.1007/s12253-008-9141-x. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi Y, Kanamori M, Kumabe T, et al. Collision tumor of anaplastic oligodendroglioma and gangliocytoma: a case report. Brain Tumor Pathol. 2009;26:89–93. doi: 10.1007/s10014-009-0255-y. [DOI] [PubMed] [Google Scholar]
- 32.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen YH, Giashuddin S, Shapiro RL, et al. Unusual occurrence of a melanoma with intermixed epithelial component: a true melanocarcinoma? : case report and review of epithelial differentiation in melanoma by light microscopy and immunohistochemistry. Am J Dermatopathol. 2007;29:395–9. doi: 10.1097/DAD.0b013e31812f5235. [DOI] [PubMed] [Google Scholar]