Abstract
Liposarcoma can be an aggressive, debilitating and fatal malignancy. In this study, we identifed microRNAs (miRNAs) associated with the differentiation status of liposarcoma to gain insight into the basis for its progression. miRNA expression profiles determined in human tumors and normal fat specimens identified a de-differentiated tumor expression signature consisting of 35 miRNAs. Deregulated miRNA expression was confirmed in a second independent sample cohort. The miR-155 was the most overexpressed miRNA and functional investigations assigned an important role in the growth of de-differentiated liposarcoma cell lines. Transient or stable knockdown of miR-155 retarded tumor cell growth, decreased colony formation and induced G1-S cell cycle arrest in vitro and blocked tumor growth in murine xenografts in vivo. We identified casein kinase 1α (CK1α) as a direct target of miR-155 control which enhanced β-catenin signaling and cyclin D1 expression, promoting tumor cell growth. In summary, our results point to important functions for miR-155 and β-catenin signaling in progression of liposarcoma, revealing mechanistic vulnerabilities that might be exploited for both prognostic and therapeutic purposes.
Keywords: Liposarcoma, miRNA, CK1α, β-catenin, cyclin D1
Introduction
The adipogenic-origin liposarcomas constitute the most common soft tissue sarcoma histologies, comprising ~15–20% of this relatively uncommon malignancy cohort (1). This group is composed of three categories as per the 2002 WHO guidelines (2): 1) well-differentiated and dedifferentiated liposarcoma (WDLPS/DDLPS); 2) myxoid and round cell liposarcoma (MLS and RCL); and 3) pleomorphic liposarcoma (PLS). The focus of the current study is WDLPS/DDLPS, the most common liposarcoma histologic subtype. The molecular hallmark of WD/DDLPS is the presence of one or more supernumerary circular (“ring”) and/or giant rod chromosomes containing highly amplified (more than 10-fold in most cases) DNA sequences localized to the 12q13-q21 region (3). WDLPS arise in the deep soft tissues and lack a described precursor lesion. While WDLPS developing in the periphery (e.g. extremities) commonly exhibit a favorable outcome and rarely de-differentiate, cases arising in the retroperitoneum or deep soft tissues of the pelvis and abdomen exhibit a more dismal natural history. Retroperitoneal/abdominal WDLPS are usually large upon discovery and frequently cannot be excised with microscopically negative surgical margins (4). While lacking metastatic capacity, the avid propensity of retroperitoneal/abdominal WDLPS for local recurrence is associated with considerable morbidity and mortality; adjuvant and neoadjuvant non-surgical approaches are generally not effective (5). Approximately 25–40% of retroperitoneal/abdominal WDLPS will ultimately manifest a dedifferentiated pattern (DDLPS; 6). Dedifferentiation, as originally described by Evans in 1979 (7), is defined as an area within a well differentiated lesion lacking characteristic lipomatous features with the appearance of an intermediate to high-grade sarcoma. About 90% of DDLPS are diagnosed de novo, as a component of a primary presenting lesion, whereas ~10% are identified in the context of a recurrent WDLPS (6). DDLPS are significantly more aggressive than pure WDLPS, exhibiting a local recurrence rate of >80%, a distant metastasis rate of up to 20%, and a five-year disease-specific survival rate of 40–60% despite an aggressive surgical approach combined with systemic chemotherapy (8). There is a pressing need for improved DDLPS management strategies; consequently, enhanced knowledge of molecular deregulations underlying DDLPS biology is critical.
In the past decade, much attention has focused on the tumorigenesis and cancer progression impact of microRNAs (miRNAs). MiRNAs are a class of evolutionally conserved non-coding small RNAs of 18- to 24-nucleotides in length; they participate in post transcriptional gene expression regulation through mRNA degradation, translational inhibition, or chromatin-based silencing mechanisms (9). MiRNAs exert multiple biological functions including development, differentiation, apoptosis, and cell proliferation (9). MiRNA expression is commonly altered in cancer, contributing to tumor initiation, progression and metastasis (10); miRNAs can function either as tumor suppressors or oncogenes (oncomiRs) (10). Accordingly, there is great interest in developing therapeutic strategies to target tumor-driving miRNA deregulations. Insight into the role of altered miRNA expression in WD/DDLPS is currently limited; to date apparently only two studies have investigated specific WD/DDLPS-associated miRNA altered expression and/or function (11,12). The goal of the current study was to further enhance our current knowledge regarding the role of microRNAs in WD/DDLPS. Utilizing a miRNA array platform an expression signature consisting of four over-expressed and 31 downregulated miRNAs was found to differentiate WD/DDLPS from normal fat. One of the over-expressed miRNAs we identified (miR-155) was further evaluated for its role in promoting DDLPS growth in vitro and in vivo and its molecular mechanisms of function were elucidated.
Materials and Methods
Cell lines and Reagents
The human DDLPS cell lines: lipo224, lipo224B, lipo246, lipo573B and lipo863B (all originating from surgically resected human retroperitoneal DDLPS) were established in our laboratory as recently reported (13). The previously described human DDLPS cell line LPS141 was kindly provided by Dr. Jonathan Fletcher (Brigham and Women’s Hospital, Boston, MA; 14). Cells were cultured and passaged in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with glucose and 10% Fetal Bovine Serum (FBS). Authentication of DDLPS cell lines was conducted utilizing Short Tandem Repeat DNA fingerprinting (STR; Table S1) as previously described (13). Human white preadipocytes (PA) primary cultures were purchased from PromoCell (Heidelberg, Germany). PA were differentiated into adipocytes per manufacturer’s instructions using a commercial preadipocyte differentiation media (serum free media containing: insulin, dexamethasone, IBMX, L-thyroxine, ciglitazone, and heparin) and adipocyte nutrition media (3% Fetal Calf Serum (FCS)) supplemented media containing: insulin, dexamethasone, and 3-isobutyl-1-methylxanthine). Adipogenic differentiation was confirmed via Oil red O staining as previously described (13).
Commercially available antibodies were used for Western Blot analyses or immunohistochemical detection of: CK1α (Novus Biologicals, Littleton, CO), β-catenin (BD Transduction Laboratories, Rockville, MD and BD Biosciences; San Jose, CA), phospho beta-catenin (Ser 45) and Foxo3a (Cell Signaling, Danvers, MA), cyclin D1 (Santa Cruz Biotechnology; Santa Cruz, CA) Ki67 (Dako, Carpentaria, CA); Cleaved caspase 3 (BioCare medical, Concord, CA) and β-actin (Santa CruzBiotechnology)
MiRNA expression profiling and miR155 in situ hybridization
For miRNA expression profiling, frozen tissues acquired under an Institution Research Board approved protocol derived from 17 surgically resected WD/DDLPS samples, 5 lipoma samples, and eight frozen normal fat tissues were utilized. In all cases WD/DDLPS histology, as initially clinically diagnosed, was confirmed by a soft tissue and bone pathologist (AJL). Furthermore, in 11 of the cases 12q15/MDM2 fluorescence in situ hybridization (FISH) was done as previously described (13) and demonstrated 12q15/MDM2 amplification as would be expected (Fig S1); in the remaining six cases hybridization was not successful. Tissue was homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and microRNA fraction was purified from total RNA using RT2 qPCR-Grade miRNA isolation kit (SABiosciences, Frederick, MD) per manufacturer’s protocol. De-identified RNA samples were submitted to Exiqon (Vedbaek, Denmark) for quality control and high throughput miRNA profiling; all experiments were conducted as per facility protocols. In brief, RNA quality was assessed by an Agilent 2100 Bioanalyzer profile; only samples with RNA Integrity Numbers (RIN) values>7 were included in the final analysis. One μg total RNA from sample and reference RNA (a mixture of RNA from all samples included) were labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labeling kit. Next, Hy3™-labeled samples and a Hy5™-labeled reference RNA samples were mixed pair-wise and hybridized to the miRCURY™ LNA array version 11.0 containing capture probes targeting all miRBASE version 13 (Sanger Institute, Hinxton, Cambridge, UK) registered human, mouse, and rat miRNAs. Hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station (Tecan, Männedorf, Switzerland). Post hybridization microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) to prevent potential bleaching of the fluorescent dyes. Next, slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA) and image analyses were carried out using the ImaGene 8.0 software (BioDiscovery, El Segundo, CA). Quantified signals were normalized using the global Lowess (Locally Weighted Scatterplot Smoothing) regression algorithm. Raw and normalized miRNA array data were uploaded to GEO (GSE31045). A tissue microarray (TMA) consisting of cores derived from WD/DDLPS FFPE specimens (representing 59 different patients) and eight normal fat control samples was constructed as previously described (15). In situ hybridization was conducted using double-DIG labeled mercury LNA microRNA probe (Exiqon) and a U6 control probe. Tissue slides were hybridized for 2h at 55C on Discovery Ultra (Ventana Medical Systems, Tucson, Arizona). The digoxigenins were then detected with a polyclonal anti-DIG antibody and Alkaline Phosphatase conjugated secondary antibody (Ventana) using NBT-BCIP (Ventana) as the substrate.
Quantitative Real Time PCR (qRT-PCR), western blot analyses, immunohistochemistry, and cellular growth related assays
These assays were all conducted as previously described (13,16). More detailed information is provided in Supplemental Data.
Transfection/transduction procedures
Transient miR-155 knockdown was conducted utilizing peptide nucleotide acid (PNA) based oligomers specifically targeting has-miR-155 (Panagene, Deajeon, Korea); a non-targeting scrambled construct was used as control. Anti-miR-155 PNA sequence is RRRQRRKKR-OO-CTATCACGATTAGCATTA, and scramble control sequence is RRRQRRKKR-OO-ATTATTGTCGGACAA. RRRQRRKKR is the cell penetrating peptide. Transfection procedure followed manufacturers’ instructions; Lipofectamin2000 was utilized as transfection reagent (Invitrogen). A miRNA sponge method was used in order to stably knockdown miR-155 as previously described (17). In Brief, long DNA oligos containing seven repeats of the antisense miR-155 sequence were synthesized and ligated into retroviral pSuperior.retro.neo-GFP vector with HindIII and BglII (New England Biolabs, Ipswich, MA) to create expression plasmids. MiR-155 primer sequences are: Forward (BglII): 5′-acgcagatctacccctatctaattagcattaaacccctatctaattagcattaaacccctatctaattagcattaacgaacacccctatc-3′; Reverse (HindIII): 5′-gcataagcttttaatgctaattagataggggtttaatgctaattagataggggtttaatgctaattagataggggtgttcgttcgttaatg-3′. Retroviruses were generated by co-transfection of the packing plasmids pCGP and pVSV-G (gifted by Dr. Zweidler-McKay, MD Anderson Cancer Center) and the above expression plasmids into 293T cells. DDLPS cell lines were cultured in six-well plates to which 8μg/mL polybrene and virally infected supernatant was added for four hours and after 48 hours selected by fluorescence activated cell sorter (FACS) for green fluorescent protein (GFP) expression. Small interfering RNAs (siRNAs) (20nM pools targeting β-catenin L-003482-00-0005 and control non-targeting constructs D-001810-01-05; Thermo Scientific, Rockford, IL) were introduced into cells using Lipofectamine 2000 (Invitrogen) per manufacturer’s instructions. Briefly, 2×105 cells were plated in each well of a six-well plate and incubated overnight. A mixture of siRNA (20nM) and Lipofectamine 2000 (10μl) diluted in DMEM was added for 24 hr, followed by incubation in regular medium. Knockdown was confirmed by western blot analysis. Forced cyclin D1 expression was conducted using a pCMV-cyclin D1 plasmid (Addgene, Cambridge, MA); empty pCMV plasmid was used as control. Of note, DDLPS cell lines used for transfection/transduction all exhibited relatively high miR155 expression and were selected based on ease of transfection/transduction and high transfection/transduction efficiency. Lipo246 was specifically selected for stable knockdown as this cell lines exhibits 100% tumor take when injected to immunocompromised mice.
Luciferase reporter assays
pMIR-REPORT plasmids containing miR-155 target sequence in the Foxo3a 3′ UTR region and mutant miR-155 target sequence were kindly provided by Dr. Cheng (18). pRL plasmid was used as internal control. To clone the 3′ UTR region of CK1α cDNA, the following primers were synthesized: CK1α Forward: 5′atcgctcgagAGTACTGCAACTGCCAGAAC3′ and CK1α Reverse: 5′gccagcggccgcCAAGGGTGAAAGTCAACTGCT3′. PCR was used to amplify the target 3′ UTR sequence containing the potential binding site of miR-155. The PCR product was inserted into a psiCHECK-2 vector (Promega) using the XhoI and NotI sites. Luciferase reporter assays were conducted as previously described (19). In brief, DDLPS cells were transfected with the relevant constructs and internal controls using Lipofectamine 2000 (Invitrogen). After 48 hours, cells were lysed using passive lysis buffer; firefly and Rennilla luciferase activities in cell lysates were determined using a Dual-luciferase assay kit (Promega) on DTX800 Multimode Detector (Beckman Coulter, Brea, CA). Reporter assay results are depicted as relative luciferase activities; i.e. average ratios of Firefly luciferase to Renilla± standard deviation (SD) for pMIR-REPORT Foxo3a constructs and average ratios of Renilla luciferase to Firefly ± SD for pCHECK-2 CK1α construct.
In vivo DDLPS xenograft experiments
All animal procedures/care was approved by UTMDACC Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals”. Further information is provided in Supplementary Data.
Statistical analysis
The goal of the miRNA gene expression array statistical analysis was to identify the most significantly differentially expressed miRNAs comparing human WD/DDLPS and normal fat specimens. For each miRNA array probe, two-tailed t-test between human WD/DDLPS and normal fat specimens was calculated (using log-transformed data). The difference in LogMedianRatios (ΔLMR) between sample groups was calculated. Java TreeView represented the results as a heatmap diagram.
Cell culture based assays and qRT-PCR analyses were repeated at least twice and mean ± SD was calculated. Cell lines were examined separately. For outcomes that were measured at a single time point, two-sample t tests were used to assess the differences. Differences in xenograft size and weight between xenograft groups at study termination were assessed using a two-tailed Student’s t-test. Significance was set at P ≤0.05.
Results
High throughput miRNA expression profiling reveals potential WD/DDLPS associated deregulated miRNAs
The small RNA fraction isolated from eight frozen human normal fat (NF) samples and 17 WD/DDLPS samples was characterized using miRNA expression arrays. Statistical analysis identified four miRNAs to be significantly (p<0.0001, fold change>1.5) over-expressed in WD/DDLPS as compared to NF. Thirty-one miRNAs were down-regulated in WD/DDLPSs compared to normal tissues. A heatmap based on a two-way unsupervised hierarchical clustering of tissue samples and miRNAs is depicted in Fig 1A. A complete list of individual deregulated miRNAs is provided in Table S2. Two of the four upregulated miRNAs have previously reported to have potential cancer promoting functions (i.e. oncomiRs) while 11 of the downregulated miRNAs (39%) have been shown to harbor tumor suppressor functions (Table S2). In addition, 4 (11%) of the miRNAs identified in our study were recently found to be potentially deregulated in WD/DDLPS (based on massively parallel sequencing analysis), including miR-143, the focus of the previously published study (Table S2; 11).
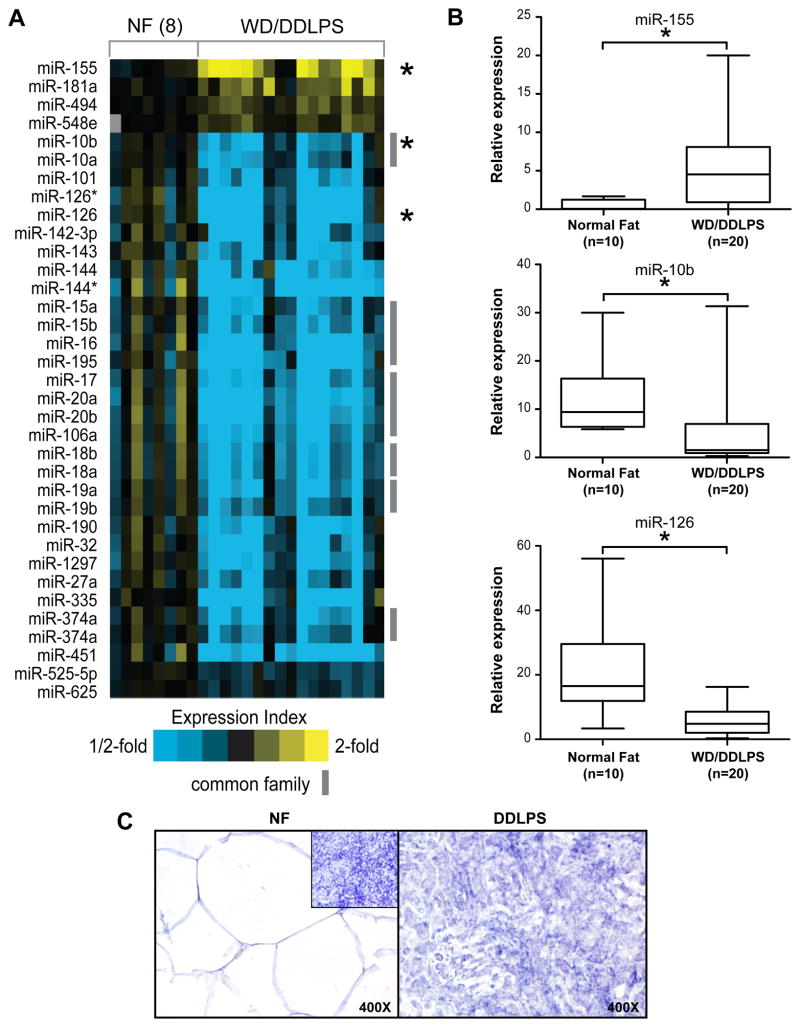
Figure 1. WD/DDLPS miRNA expression signature.
A) MiRNA expression profiles of human liposarcomas (WD/DDLPS) and normal fat tissues (NF) were obtained using high throughput miRNA arrays. A Heat map representation of top overexpressed (yellow) and underexpressed miRNAs (blue) in WD/DDLPS compared to NF. Rows, miRNAs; columns, profiled samples. miRNAs which share a common family (TargetScan) are indicated. (* denotes miRNAs that have been selected for further array validation, see panel B); B) Three of the deregulated miRNAs (miR-155, miR-10b and miR-126) were selected for array validation; qRT-PCR was conducted on an independent set of human samples (WD/DDLPS = 20, NF = 10). The levels of gene expression were normalized using SNORD47 levels based on the comparative threshold cycle method. Results (presented graphically) were in accordance with high throughput miRNA profiling data. (* denotes statistically significant effects, p<0.05); C) Mir-155 in situ hybridization demonstrating high miR-155 expression in DDLPS (right panel) as compared to NF (left panel). Inset represents U6 positive control expression in TMA samples.
Next, expression levels of three of the deregulated miRNAs identified (miR-155, miR-10b, and miR-126) were assessed via qRT-PCR in an independent set of frozen human samples (NF=10, WD/DDLPS=20). As depicted in Fig 1B, miR-155 was found to be, on average, significantly (p<0.05) overexpressed in WD/DDLPS samples as compared to NF (and lipomas; Fig S2) while miR10b and miR126 were both down-regulated in tumor tissues, recapitulating the expression pattern noted in high throughput profiling experiments. Together, these data validate miRNA expression array analyses and justify further investigation of individual identified miRNAs for their role in DDLPS biology.
MiR-155 contributes to enhanced DDLPS cell growth in vitro and in vivo
Of the several WD/DDLPS associated deregulated miRNAs discovered above, miR-155 was chosen for further comprehensive investigation. This selection aligned with our ultimate goal of identifying DDLPS associated molecular deregulations that can be utilized for therapeutic targeting consonant with recent development of compounds targeting oncomiRs as anti-cancer strategies (20). Based on the expression profiling results, miR-155 was the most statistically significantly over-expressed miRNA identified in DDLPS compared to NF as was also confirmed, as depicted above, in an independent set of frozen human samples. Furthermore, we have conducted miR155 in situ hybridization on a liposarcoma focused TMA (Fig 1C). Results demonstrated that miR-155 expression is markedly enhanced in WD/DDLPS samples (50/59 [85%] of samples exhibited high miR-155 expression) as compared to NF samples (high miR155 expression was noted in only one sample [12.5%]).
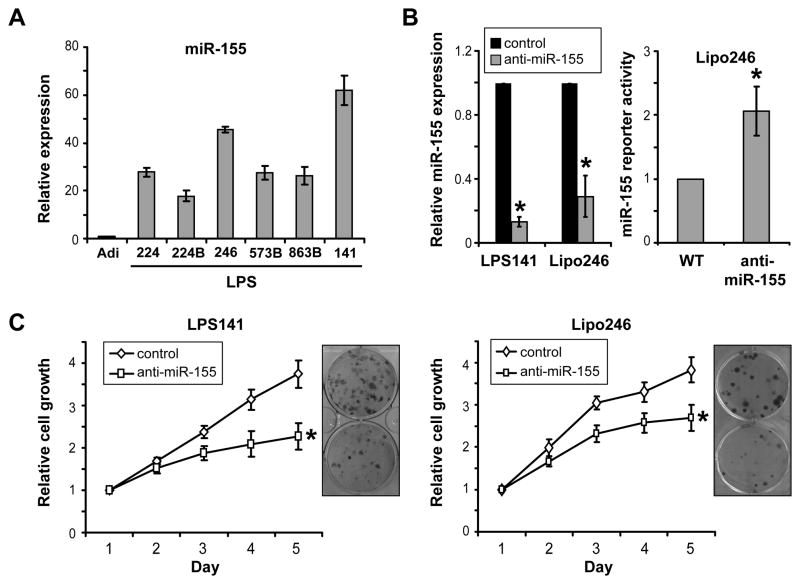
MiR-155 was previously found to act as an oncogene in several hematological and epithelial origin cancers (21–23), however its contribution to DDLPS biology remains to be elucidated. First, we evaluated the expression of miR-155 in a panel of DDLPS cell lines and normal human adipocyte cell cultures. In concordance with the results obtained from the evaluation of human samples, high levels of miR-155 were noted in DDLPS cell lines as compared to adipocytes (Fig 2A). Next, DDLPS cell miR-155 expression was knocked down utilizing an anti-sense construct to determine the contribution of this microRNA to cell growth; qRT-PCR analyses confirmed a significant decreased in miR-155 expression after knockdown when compared to cultures transfected with a non targeting scrambled construct used as control. (Fig 2B). Decrease in miR-155 function was validated utilizing a previously described miR-155 responsive luciferase reporter system consisting of FOXO3a 3′-UTR constructs (18); increase in reporter-driven luciferase expression was noted in miR-155 knocked down cells compared to control (Fig 2B). Most importantly, this functional knockdown abrogated DDLPS cell growth and decreased clonogenic capacity (Fig 2C), suggesting that miR-155 contributes to DDLPS cell growth.
Figure 2. MiR-155 knockdown inhibits DDLPS cell growth.
A) qRT-PCR analyses identified increased miR155 expression in DDLPS cell lines (=lipo224, lipo224B, lipo246, lipo573B, lipo863B and LPS141) as compared to normal human adipocytes (=Adi); B) Transient miR-155 knockdown in DDLPS cell lines utilizing PNA based antagomir was validated using qRT-PCR (left panel) demonstrating marked decrease in miR-155 expression. A miR-155 reporter activity assay (Foxo3a 3′UTR luciferase construct; right panel) demonstrated increased firefly luciferase expression (normalized to Rennilla luciferase) in response to miR155 inhibition, further confirming efficacy of knockdown; C. Transient miR-155 knockdown was found to significantly inhibit DDLPS cell growth (MTS assays) and colony forming capacity. [Graphs represent the average of at least three repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
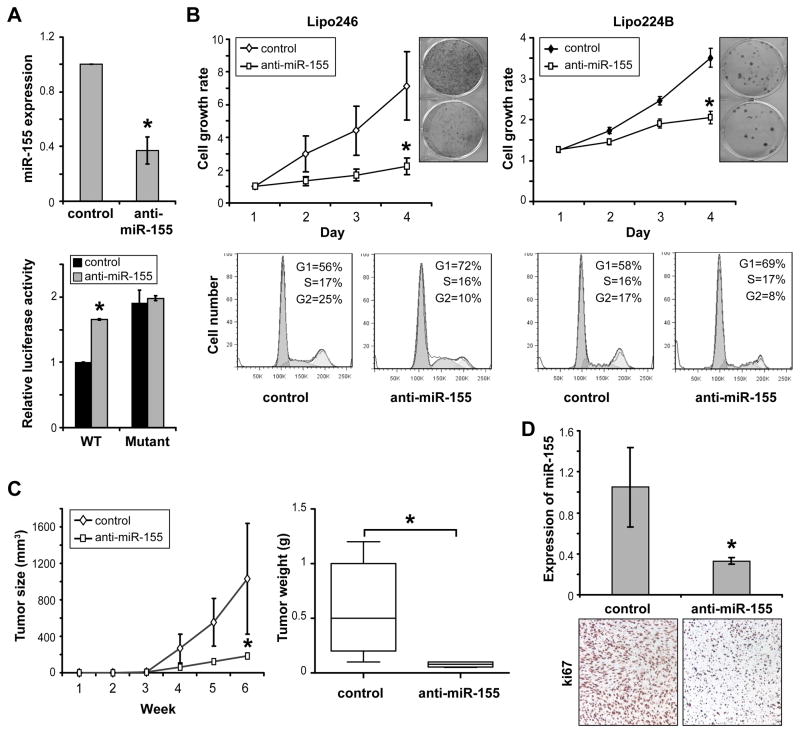
To further determine the potential impact of miR-155 on DDLPS tumorigenesis in vivo, this miRNA was stably knocked down in DDLPS cells utilizing a retroviral pSuperior.retro.neo/GFP miR-155 sponge construct with empty vector as control. Stably transduced cells were isolated using GFP sorting. qRT-PCR and reporter assays were used to confirm functional knockdown (Fig 3A). Similar to the findings above, stable miR-155 knockdown resulted in decreased DDLPS cell growth and colony formation (Fig 3B). Furthermore, miR-155 knockdown resulted in marked G1 cell cycle arrest in stably transduced DDLPS cells (Fig 3B). However, no evidence of miR-155 knockdown-induced apoptosis was noted in annexin V/PI FACS analyses (data not shown). Next, the growth of anti-miR-155 transduced Lipo246 cells was evaluated in vivo. As depicted in Figure 3C, miR-155 knockdown xenografts demonstrated slower growth and a significantly decreased volume and tumor weight at study termination compared to tumors originating from Lipo246 cells stably transduced with control vector (1031mm3 ± 608 vs. 185mm3 ± 40 [p<0.05] and 0.5g ± 0.2 vs. 0.08g ± 0.01 [p<0.05]). qRT-PCR confirmed decreased miR-155 expression in anti-miR-155 transduced DDLPS xenografts (Fig 3D); Ki67 immunohistochemical analysis demonstrated a marked decrease in DDLPS cell proliferation in these tumors compared to controls (Fig 3D). No significant increase in cleaved caspase-3 expression was found in miR-155 knockdown xenografts (data not shown). In sum, these loss-of-function experiments support an oncogenic role for miR-155 in DDLPS contributing to proliferation and cell cycle progression in vitro and in vivo.
Figure 3. MiR155 knockdown inhibits DDLPS xenograft growth in vivo.
A) Stable miR-155 in DDLPS cells utilizing a retroviral sponge construct was confirmed using qRT-PCR (upper panel) demonstrating marked decrease in miR-155 expression. A miR-155 reporter activity assay (WT and mutated Foxo3a 3′UTR luciferase construct; lower panel) demonstrated increased firefly luciferase expression (normalized to Rennilla luciferase) in response to miR155 inhibition after the transfection of the WT construct but no difference when a mutated reporter construct was used (results in lipo246 cells are shown). These data further validated knockdown efficacy; B) similar to the effects of transient miR-155 knockdown, stable blockade was found to decrease the growth and colony forming capacity of DDLPS cells. Moreover, miR-155 knockdown inhibits DDLPS G1 cell cycle progression (PI staining/ FACS analysis; lower panels); C) Anti-miR-155 transduced lipo246 cells were evaluated for local growth in vivo. miR-155 knocked down xenografts exhibited slower growth, decreased size at study termination (left graph; p<0.05), and a significantly decreased weight (right graph; p<0.05) at study termination as compared to control empty vector transduced xenografts; D) qRT-PCR confirmed decreased miR-155 expression in xenografts derived from anti-miR-155 transduced Lipo246 cells (upper panel) and ki67 IHC analysis demonstrated decreased tumor cell proliferation in these xenografts (images were captured at x200 magnification). [Graphs represent the average of at least three repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
Casein kinase 1α (CK1α) is a novel miR-155 target; a potential molecular mechanism driving Wnt/β-catenin pathway deregulation in WD/DDLPS
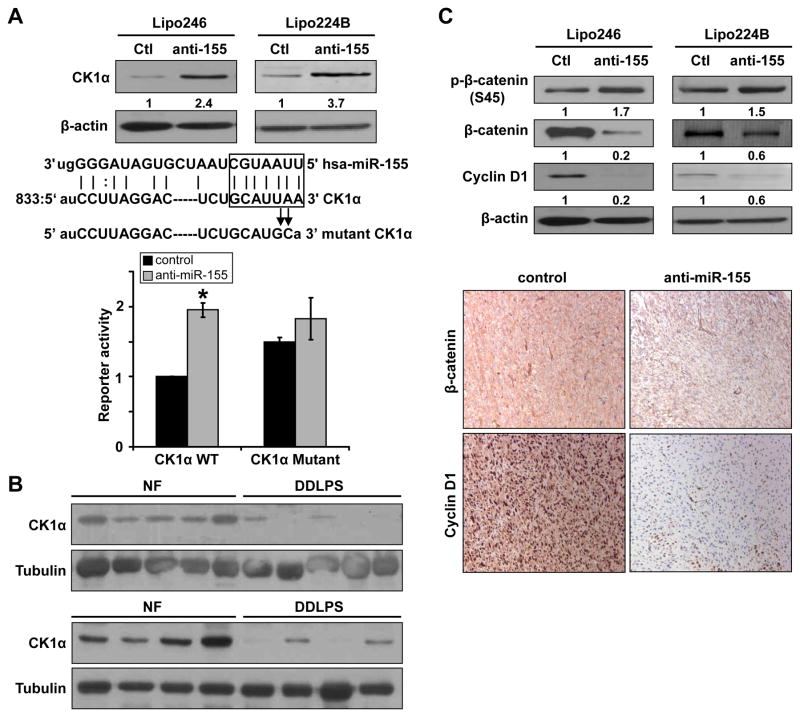
Next, we wanted to identify the molecular mechanisms underlying miR-155 oncogenic function in DDLPS. Previous studies have reported Foxo3a and SCOS1 as miR-155 targets in epithelial origin cancer (18, 24). Therefore, we evaluated the impact of miR-155 knockdown on the expression of these targets in DDLPS. WB analyses failed to demonstrate increased Foxo3a expression in response to miR-155 knockdown (Fig S3). Similarly, no increase, and even a decrease in SOCS1 expression was observed (qRT-PCR; Fig S3). Realizing that other novel miR155 targets might be operative in DDLPS we conducted a search of the miRNA Target Prediction database miRGen (25). Amongst multiple potential targets, this in silico search identified CK1α, a key regulator of the Wnt/β-catenin pathway, to be a predicted miR-155 target, containing a highly conserved motif (833–847 bp, NM_001892.4) matching with the miR155 “seed” sequence (7 nucleotide: gcauuaa) (Figure 4A). Taking into account the well established importance of the CK1α/β-catenin axis in tumorigenesis and tumor progression, we selected to focus on this potential downstream target. Western blot analysis demonstrated increased CK1α protein expression in both lipo246 and lipo224B cells stably transduced with anti-miR-155 (Fig 4A). To confirm an interaction between miR-155 and the 3′UTR of the CK1α mRNA, a luciferase reporter system consisting of a wild type CK1α 3′-UTR miR-155 binding region containing construct and a mutated construct were developed (sequences are depicted in Fig 4A). These were used to determine whether miR-155 directly impacts CK1α expression. DDLPS cells stably transduced with anti-miR-155 exhibited increased WT reporter activity as compared to control vector transduced cells while no significant difference in luciferase readout was found when the mutated construct was evaluated (Fig 4A). These results suggest that CK1α is a direct miR-155 target. To further confirm that miR155 induced modulation of CK1α is of clinical relevance we assessed the protein expression levels of this target in a cohort of snap frozen human DDLPS (i.e. miR155 overexpressing; n=8) and normal fat (n=8) samples via WB analyses. As depicted in Fig 4B DDLPS exhibit markedly lower CK1α expression.
Figure 4. CK1α is a direct miR-155 target.
A) WB analysis demonstrated increased CK1α expression in anti-miR-155 transduced DDLPS cells (relative protein expression levels per densitometry are depicted; upper panel). Human CK1α 3′UTR sequence alignment with miR-155 sequence is depicted; miR155 seed sequence and corresponding matching region in the CK1α 3′UTR is marked. To determine the potential effect of miR-155 on the CK1α 3′UTR, a reporter assay system was developed to include wild-type (WT) and mutated constructs (mutation sites are indicated). An increase in the activity of the WT reporter but not the mutated reporter was found in miR-155 knocked down cells (lower panel); B) WB analyses identified markedly lower CK1α protein expression in human DDLPS samples as compared to normal fat; C) WB analyses demonstrated increased β-catenin phosphorylation and decreased total β-catenin and cyclin D1 expression in anti-miR-155 transduced DDLPS cells reflecting the functional consequences of miR-155 knockdown induced CK1α expression (upper panel; relative protein expression levels per densitometry are depicted). Decreased β-catenin and cyclin D1 expression was noted in anti-miR-155 transduced DDLPS xenografts (lower panel); [Graphs represent the average of at least three repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
In the “off” (inactive) state of Wnt/β-catenin pathway signaling, CK1α binds and phosphorylates β-catenin at serine 45, consequentially leading to the ubiquitination and degradation of this protein (26). To determine whether the above noted impact of miR-155 on CK1α expression functionally impact β-catenin signaling, the expression of phosphorylated β-catenin (S45), β-catenin, and cyclin D1 (a β-catenin downstream target) protein levels in miR-155 knocked-down DDLPS cell lines were examined. As depicted in Fig 4C, an increase in phosphorylated β-catenin expression and a marked decrease in β-catenin and cyclin D1 were found. These cell culture-based observations were further confirmed to occur in vivo: immunohistochemical analysis demonstrated decreased β-catenin and cyclin D1 expression in xenografts derived from miR-155 knocked down DDLPS cells (Fig 4C).
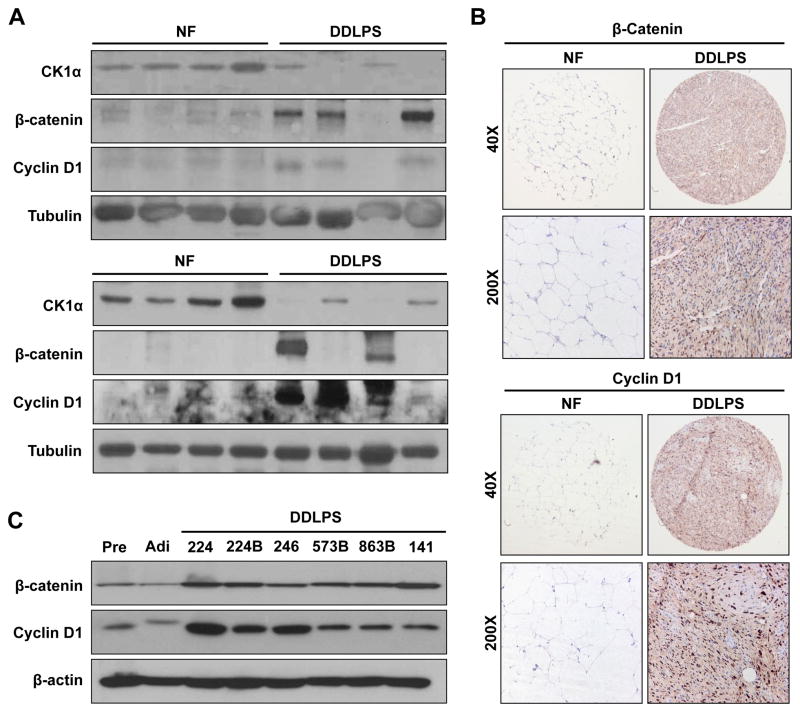
To determine if CK1α expression correlates with the expression of β-catenin and cyclin D1 in human samples we utilized the WB panel depicted in Fig 4B and evaluated the expression of these potential downstream targets. As shown in Fig 5A increased β-catenin/cyclin D1 expression can be identified in DDLPS as compared to NF especially in tumor samples expressing lower CK1α. While β-catenin/cyclin D1 over-expression and enhanced signaling has been identified as a common molecular event in several malignancies (27), little is known about the status of these proteins in WD/DDLPS. To that end, we have assessed the expression of these proteins in our TMA. β-catenin expression was identified in 48 WD/DDLPS samples (81%) to varying levels: low expression was noted in 52% of positively staining tumors and moderate to high expression in 48%. In contrast only one NF samples expressed β-catenin to a low level. Similarly, 95% (n=56) of WD/DDLPS samples exhibited cyclin D1 expression: low levels were identified in 29% of positively staining tumors and 91% expressed moderate to high levels. None of the NF samples expressed cyclin D1 as per our scoring schema (Fig 5B). Next, WB analyses demonstrated increased β-catenin and cyclin D1 expression in a panel of human DDLPS cell lines as compared to preadipocytes and adipocytes (Fig 5C), highlighting the potential relevance of this pathway to DDLPS. Taken together, our data identified CK1α as a novel miR-155 target and suggest that miR-155 overexpression enhances the activation of β-catenin in these malignancies.
Figure 5. β-catenin and cyclin D1 are highly expressed in human liposarcoma.
A) WB analyses conducted on the sample cohort depicted in Fig 4B identified increased β-catenin and cyclin D1 expression in DDLPS as compared to NF, especially in samples expressing low CK1α; B) Representative photographs of β-catenin and cyclin D1 immunostained NF and DDLPS spots from TMA; C) Human DDLPS cell lines exhibit increased β-catenin and cyclin D1 expression as compared to human pre-adipocytes (Pre) and adipocytes (Adi).
MiR-155 pro-WD/DDLPS effects are driven by cyclin D1
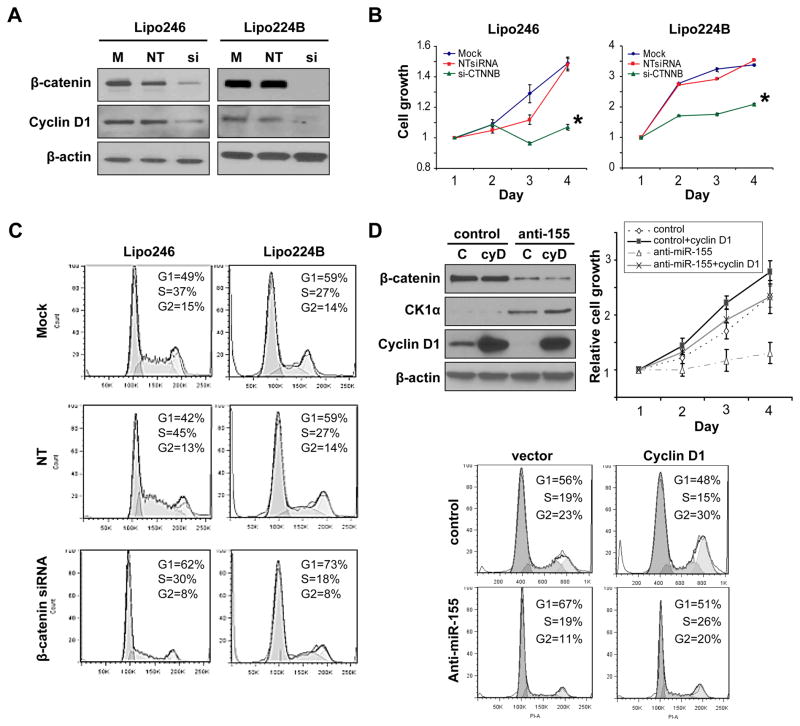
The above findings led to the hypothesis that miR-155 DDLPS associated oncogenic functions are mediated via enhanced β-catenin signaling. To further investigate this possibility, the impact of β-catenin over-expression on DDLPS cell growth was evaluated in loss of function experiments. Knockdown was achieved using a pool of siRNA constructs specifically targeting β-catenin; non targeting constructs were used as controls (Fig 6A). A marked decrease in cyclin D1 was noted after β-catenin knockdown, confirming the effects of this genetic manipulation on β-catenin signaling (Fig 6A). Most importantly, β-catenin knockdown resulted in a significant (p<0.05) decrease in DDLPS cell growth (Fig 6B) and in pronounced G1 cell cycle arrest (Fig 6C). These functional effects align with those identified as a consequence of miR-155 knockdown.
Figure 6. enhanced β-catenin signaling/cyclin D1 expression mediates, at least in part, miR-155 pro-DDLPS effects.
A) Anti-β-catenin siRNA (si; 20nM pool) knockdown resulted in a significant decrease in β-catenin and cyclin D1 expression in DDLPS cells. Non targeting (NT) siRNA construct transfection was used as control; B) β-catenin knockdown abrogates DDLPS cell growth (MTS assays); C) β-catenin knockdown inhibits DDLPS cell G1 cell cycle progression (PI staining/FACS analysis); D) Cyclin D1 forced expression rescues DDLPS cells from the anti-proliferative effects induced by miR-155 knockdown. WB analyses confirmed cyclin D1 (cyD) over-expression and demonstrate no effect on CK1α and β-catenin expression levels. A significant increase in cell growth (upper graph) and G1 cell cycle progression (lower panels) was noted in anti-miR-155 transduced lipo246 after cyclin D1 re-expression. [Graphs represent the average of at least three repeated experiments ±SD; * denotes statistically significant effects (p<0.05)]
To determine whether miR-155 regulates DDLPS cell growth, at least in part, via β-catenin signaling (i.e. through cyclin D1) rescue experiments were performed. Cyclin D1 was forcefully expressed into anti-miR-155 transduced lipo246 cell and empty vector transduced cells (Fig 6D); the effect on cell growth and cell cycle progression was determined. As depicted in Fig 6D, increased cyclin D1 expression was able to overcome the growth and cell cycle progression blockade imposed by miR-155 knockdown. Together, these data support a role for β-catenin signaling/cyclin D1 in the DDLPS pro-tumorigenic, pro-proliferative effects mediated via miR-155 over-expression.
Discussion
The current study highlights several interconnected DDLPS-associated molecular deregulations of potential major translational and clinical relevance. Specifically, we found that miR-155 is highly expressed in human WD/DDLPS and contributing to the tumorigenic phenotype of these unfavorable malignancies via a previously not described regulation of CK1α which results in activation of the β-catenin pathway. Multiple lines of evidence strongly support the significant role that miR-155 plays in tumorigenesis and progression, acting predominantly as an oncogenic miRNA (oncomiR; 18,21–24,28). MiR-155 over-expression has been previously identified in several hematological (e.g. B-cell lymphomas, acute myelocytic leukemia; 22,29) and epithelial-origin carcinomas (e.g. colon, papillary thyroid, pancreatic ductal adenocarcinoma, breast, and lung; (21,30–36). Moreover, miR155 as a molecular prognosticator has been identified in pancreatic carcinoma and lung cancer, where high expression levels are associated with poor patient outcome (36,37). Supporting mir-155’s role as an oncomiR is the finding that mice genetically engineered to express miR-155 expressed from a B-cell-specific promoter spontaneously develop B-cell malignancy (38). Similarly, transduction of miR-155 in hematopoietic stem cells was shown to result in the development of a myeloproliferative disorder (39). Loss of function experiments conducted in several cancer models determined a role for miR-155 in tumor cell proliferation, migration, and invasion (18,24,28). To the best of our knowledge, our study is the first to report a role for miR-155 in mesenchymal-origin solid malignancy.
miRNAs are robust regulators of gene expression functioning by directly inducing mRNA degradation or either repressing the translation targeted mRNAs (9). Translational inhibition may occur via mechanisms such as mRNA uncapping that lead to increased mRNA turnover and decreased target gene expression. OncomiRs such as miR-155 directly target and repress the expression of tumor suppressor or tumor suppressor–like genes (23). To date, several mRNAs coding for transcriptional regulatory proteins, receptors, kinases, and nuclear and DNA binding proteins have been identified as miR-155 direct targets in the cancer context. For example, miR-155 was shown to induce B-cell malignancies by targeting SHIP and C/EBPβ (40); miR-155 regulation of TP53INP1 was found to be a major mediator of pancreatic cancer development (41). Additional cancer-associated targets include: FOXO3, RHOA, MSH2, MSH6, MLH1, SOCS1 and others (18,24,42,43). Interestingly, several targets including Meis1, c-MAF, AID, interleukin-1, IKKε, and Ets-1, have been identified to be regulated by miR-155 in the context of the immune and hematopoietic systems but have not been shown to have a role in miR-155–mediated oncogenesis (23). Together, these insights possibly suggest that miR-155 only negatively regulates a small number of dominant mRNA targets in a particular disease context and in a cancer cell type-specific manner. Identifying the dominant pathways/targets regulated by a miRNA in a given malignancy may shed light on molecular deregulations driving tumor biology. In this study we identified CK1α as a novel miR-155 direct target, possibly implicating β-catenin signaling as a DDLPS molecular driver.
Human β-catenin, a homolog of the Drosophila Armadillo protein, is a multifunctional protein that plays essential roles in development and tissue maintenance (27). β-catenin signaling is tightly regulated in normal cells (44). Generally, cytoplasmic β-catenin levels are kept low via continuous ubiquitin-proteasome-mediated degradation of this protein, a process which is regulated by a multiprotein complex containing CK1α, axin, APC, and GSK-3β (27). CK1α and GSK-3β mediate the degradation of β-catenin molecules by phosphorylating specific amino N terminal residues (Ser45 by CK1α, and Thr41, Ser 37, and Ser33 by GSK-3β in this sequential order), thereby marking the protein and rendering it as recognizable by β-transducin repeat-containing protein (β-TrCP), a component of the E3 ubiquitin ligase, leading to its degradation by the 26S proteasome complex (27). Physiological Wnt pathway activation as well as pathological, aberrant, signaling (for example secondary to CK1α down-regulation as demonstrated in this study) results in β-catenin stabilization, increased expression and enhanced activity leading to the over-expression of down-stream effectors such as cyclin D1 (27). β-catenin pathway deregulation has been frequently noted in multiple cancer types, where aberrant signaling is subverted to provide advantages in growth and survival (44). However, the role of β-catenin in liposarcomagensis and progression has not been widely studied. A recent report identified upregulated canonical WNT signaling in a large cohort of human sarcomas including liposarcoma (45). Sakamoto et al evaluated a small human DDLPS sample cohort (n=12 human specimens) and reported β-catenin protein over-expression in 42% of cases; 17% of samples harbored CTNNB1 (the gene encoding for β-catenin) mutations (46). Our studies further support a role for this pathway in DDLPS: β-catenin and its downstream effector cyclin D1 were found to be over-expressed in all human DDLPS cell lines as compared to pre-adipocytes and adipocytes and were shown to induce DDLPS cell proliferation and cell cycle progression, thereby supporting further investigation of this pathway in DDLPS. While we have focused our study on the impact of decreased CK1α expression on β-catenin signaling, CK1α has also been shown to regulate other cancer contributory targets including MDM2, p53, and E2F1 proteins (47,48). MDM2 amplification is a hallmark of WD/DDLPS contributing to inception of these malignancies (3). Consequently, miR155:CK1α contribution to MDM2 activity may warrant further investigation.
The possibility that miRNA deregulations can be exploited as nodes of tumor vulnerability, i.e. can form the basis for novel anti-cancer therapies, is an exciting potential strategy to modulate gene expression programs in an immediate and reversible manner (20). Several types of antisense-based miRNA inhibitors have been developed to target oncomiRs, seeking to turn off their expression; e.g., ‘antagomirs’, locked nucleic acid oligonucleotides, and various types of 2′-O-modifed oligonucleotides (20). Such approaches have shown initial promise in various pre-clinical cancer models. Our study demonstrates a potential role for miR-155 in DDLPS biology and supports further development and testing of anti-miR-155 therapeutic strategies for this malignancy. However, miR-155 plays important roles in physiological immune surveillance modulating TLR- or IFNb-mediated innate immune responses and affecting T-, B- and dendritic cell functions (23). Thus, systemic miR-155 inhibition could negatively affect immune proficiency. To overcome such concerns, DDLPS-specific delivery agents; e.g., viral vectors or tumor-targeted nanoparticles, may be applicable. Alternatively, strategies targeting miR-155-induced down-stream effects in DDLPS, i.e. β-catenin signaling, might abrogate possible deleterious side effects of miR-155 inhibition. Based on data presented in our study, switching off β-catenin signaling upon which DDLPS cells appear to depend could potentially elicit significant anti-tumor effects. However, agents directly down-regulating β-catenin are not yet available, and such interventions, if developed, may not be clinically useful if they compromise normal β-catenin physiological functioning. For these reasons, approaches that target specific β-catenin pro-tumorigenic downstream effects rather than directly down-regulate β-catenin per se may be more applicable. The current study highlights a potential role for the β-catenin downstream effector cyclin D1 in DDLPS. Cyclin D1 inhibitors are currently under development and at least one agent (ON 013105) has recently reached Phase I clinical testing (49) after demonstrating pre-clinical efficacy in several cancer models in vitro and in vivo (50). Our study supports evaluation of such compounds in DDLPS experimental models.
In conclusion, we found that miR-155 functions as an oncomiR in DDLPS by targeting CK1α, consequently resulting in enhanced β-catenin/cyclin D1 expression and driving DDLPS cell proliferation and cell cycle progression. Together, these data suggest novel DDLPS-associated molecular deregulations that could be exploited for development of more effective therapies on behalf of patients afflicted by these devastating malignancies.
Supplementary Material
Acknowledgments
We thank Jin Q Cheng (H. Lee Moffitt Cancer Center, Tampa, Florida) for providing the pmiR-FOXO3a constructs. Dr Jonathan Fletcher (Brigham and Women’s Hospital, Boston, MA) is thanked for providing the LPS141 cell line and Ms. Kim Vu is thanked for her aid in figure preparation. We would like to thank the Lobo, Margolis, and Jackson families for their philanthropic support of our liposarcoma studied. This manuscript was supported in part by a NIH/NCI RO1CA138345 (to DL) and a Liddy Shriver Foundation Seed Grant (to DL). MDACC cell-line characterization core facility was further supported by an NCI Cancer Center Support Grant (CA#16672)
Footnotes
Conflicts of Interest: None
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher CDMKU, Mertens F. Genetics of Tumors of Soft Tissue and Bone. IARC Press; Lyon, France: 2002. Pathology and Genetics of Soft Tissue and Bone: World Health Organization Classification of Tumors. [Google Scholar]
- 3.Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes and Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 4.Stojadinovic A, Leung DHY, Hoos A, Jaques DP, Lewis JJ, Brennan MF. Analysis of the prognostic significance of microscopic margins in 2,084 localized primary adult soft tissue sarcomas. Ann Surg. 2002;235:424–34. doi: 10.1097/00000658-200203000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zagars GK, Ballo MT, Pisters PWT, Pollock RE, Patel SR, Benjamin RS. Surgical margins and reresection in the management of patients with soft tissue sarcoma using conservative surgery and radiation therapy. Cancer. 2003;97:2544–53. doi: 10.1002/cncr.11367. [DOI] [PubMed] [Google Scholar]
- 6.Singer SM, Cristina R, Antonescu MD, Elyn Riedel MA, Murray F, Brennan MD. Histologic Subtype and Margin of Resection Predict Pattern of Recurrence and Survival for Retroperitoneal Liposarcoma. Ann Surg. 2003;238:358–71. doi: 10.1097/01.sla.0000086542.11899.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans HL, Soule EH, Winkelmann RK. Atypical lipoma, atypical intramuscular lipoma, and well differentiated retroperitoneal liposarcoma. A reappraisal of 30 cases formerly classified as well differentiated liposarcoma. Cancer. 1979;43:574–84. doi: 10.1002/1097-0142(197902)43:2<574::aid-cncr2820430226>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 8.Fabre-Guillevin E, Coindre JM, De Saint Aubain Somerhausen N, Bonichon F, Stoeckle E, Bui NB. Retroperitoneal liposarcomas: Follow-up analysis of dedifferentiation after clinicopathologic reexamination of 86 liposarcomas and malignant fibrous histiocytomas. Cancer. 2006;106:2725–33. doi: 10.1002/cncr.21933. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Ugras S, Brill ER, Jacobsen A, Hafner M, Socci ND, Decarolis PL, et al. Small RNA sequencing and functional characterization reveals microRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011:ePub. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 13.Peng T, Zhang P, Liu J, Nguyen T, Bolshakov S, Belousov R, et al. An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors. Lab Invest. 2011;91:392–403. doi: 10.1038/labinvest.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder EL, Sandstrom DJ, Law K, Fiore C, Sicinska E, Brito J, et al. c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol. 2009;218:292–300. doi: 10.1002/path.2564. [DOI] [PubMed] [Google Scholar]
- 15.Zou C, Smith KD, Liu J, Lahat G, Myers S, Wang WL, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Ann Surg. 2009;249:1014–22. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 16.Lopez G, Liu J, Ren W, Wei W, Wang S, Lahat G, et al. Combining PCI-24781, a Novel Histone Deacetylase Inhibitor, with Chemotherapy for the Treatment of Soft Tissue Sarcoma. Clin Cancer Res. 2009;15:3472–83. doi: 10.1158/1078-0432.CCR-08-2714. [DOI] [PubMed] [Google Scholar]
- 17.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Meth. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–79. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DPM. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–26. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habbe N, Koorstra J-BM, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–6. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: A typical multifunctional microRNA. Biochimi Biophys Acta - Molecular Basis of Disease. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Jiang S, Zhang H-W, Lu M-H, He XH, Li Y, Gu H, et al. MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 25.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Research. 2006;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, et al. Axin-mediated CKI phosphorylation of β-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–76. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald BT, Tamai K, He X. Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang X, Zhuang X, Ju S, Zhang S, Jiang H, Mu J, et al. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30:3340–53. doi: 10.1038/onc.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA Expression Profiling of Thyroid Tumors: Biological Significance and Diagnostic Utility. J Clin Endocrinol Metab. 2008;93:1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 33.Yan L-X, Huang X-F, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Tang S, Le S-Y, Lu R, Rader JS, Meyers C, et al. Aberrant Expression of Oncogenic and Tumor-Suppressive MicroRNAs in Cervical Cancer Is Required for Cancer Cell Growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 36.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 37.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 38.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgantas RW, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein β are targeted by miR-155 in B cells of Eμ-MiR-155 transgenic mice. Blood. 2009;114:1374–82. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gironella M, Seux Mn, Xie M-J, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 Is Regulated by the Transforming Growth Factor β/Smad Pathway and Contributes to Epithelial Cell Plasticity by Targeting RhoA. Mol Cell Biol. 2008;28:6773–84. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–7. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 45.Vijayakumar S, Liu G, Rus Ioana A, Yao S, Chen Y, Akiri G, et al. High-Frequency Canonical Wnt Activation in Multiple Sarcoma Subtypes Drives Proliferation through a TCF/β-Catenin Target Gene, CDC25A. Cancer Cell. 2011;19:601–12. doi: 10.1016/j.ccr.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto A, Oda Y, Adachi T, Saito T, Tamiya S, Iwamoto Y, et al. β-Catenin Accumulation and Gene Mutation in Exon 3 in Dedifferentiated Liposarcoma and Malignant Fibrous Histiocytoma. Arch Pathol Lab Med. 2002;126:1071–8. doi: 10.5858/2002-126-1071-CAAGMI. [DOI] [PubMed] [Google Scholar]
- 47.Huart A-S, MacLaine NJ, Meek DW, Hupp TR. CK1α Plays a Central Role in Mediating MDM2 Control of p53 and E2F-1 Protein Stability. J Biol Chem. 2009;284:32384–94. doi: 10.1074/jbc.M109.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Würl PU, et al. The Role of the Casein Kinase 1 (CK1) Family in Different Signaling Pathways Linked to Cancer Development. Onkologie. 2005;28:508–14. doi: 10.1159/000087137. [DOI] [PubMed] [Google Scholar]
- 49.Clinical trials [Internet] Bethesda (MD): National Library of Medicine (US); 1993. [cited 2012 Jan 23]. Available from: http://clinicaltrials.gov. [Google Scholar]
- 50.Onconova.com [Internet] Newtown (PA): Onconova Therapeutics, Inc; p. c2009. [cited 2012 Jan 23]. Available from: http://www.onconova.com/on013105.shtml. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.