Abstract
We report the computational and rational design of new generations of several tryptophan-rich peptides from the compstatin family. The binding efficacy of the peptides has been tested using extensive molecular dynamics-based structural and physicochemical analysis, using 32 atomic-detail trajectories in explicit water for 22 peptides bound to human, rat, or mouse target protein C3, to a total of 257 nanoseconds. The criteria for the new designs are: (i) optimization for high binding affinity and for the balance between hydrophobicity and polarity to improve solubility compared to known compstatin analogs; and (ii) development of dual specificity anti–human-rat/mouse C3 analogs, which is important for use in animal models for disease, given the species specificity of known compstatin analogs. Three of the new analogs have been analyzed in more detail as they possess strong and novel binding characteristics and are promising candidates for further optimization. This work paves the way for the development of an improved therapeutic for age-related macular degeneration, and other complement system-mediated diseases, compared to known compstatin variants.
1. Introduction
The complement system provides the first line of defense against the invasion of foreign pathogens [1]. Nevertheless, its inappropriate or excessive activation may cause or aggravate several pathological conditions, such as age-related macular degeneration, asthma, adult respiratory distress syndrome, hemolytic anemia, rheumatoid arthritis, rejection of xeno-transplantation, stroke and heart attack [2,3,4,5]. Therefore, the development of drugs for the control of complement activation is of considerable importance.
Complement activation proceeds via three biochemical pathways (classical, alternative and lectin), which converge to a common point, the cleavage of protein C3 to fragments C3b and C3a. The large fragment, C3b, tags pathogen surfaces for recognition by phagocytic cells (opsonization), and the small fragment, C3a, aids in immune cell recruitment (chemotaxis) and inflammation. The C3b fragment also participates in complexes, called convertases, which are responsible for cleavage of C3 to C3a and C3b, as well as cleavage of complement protein C5 to C5a and C5b. C5 is the starting protein of the common activation pathway, which ends with the formation of the membrane attack complex (MAC), a multicomponent protein assembly involved in lysis of pathogen membranes. Protein C3 is essential in all pathways and represents a good target for complement inhibition [6,7,8]. For example, regulation of C3 cleavage would control the effects of C3a and C3b, and the progression of complement activation to C5, and, eventually, to MAC. Altogether, regulation of C3 would affect the opsonization, chemotactic, inflammatory, and lytic capabilities of the complement system.
The peptide compstatin binds to human and primate C3 and prevents its cleavage to C3a and C3b, a key step in complement activation. Compstatin also binds to the C3b fragment as well as the inactive C3c fragment, both of which contain the C3 β-chain. Compstatin was first discovered by a phage-displayed random peptide library for binding against C3b through truncation of an initial 32-residue peptide, named “Clone 9” in ref. [9]. Compstatin has the sequence Ile1-Cys2-Val3-Val4-Gln5-Asp6-Trp7-Gly8-His9-His10-Arg11-Cys12-Thr13-NH2 and is maintained in a cyclic conformation via the disulfide bridge Cys2-Cys12. Compstatin is a promising candidate for the treatment of unregulated complement activation [4, 10, 11]. Importantly, it is active against C3 from primate mammals, but inactive against C3 of non-primate mammals [12]. This species specificity precludes the development of related disease models in non-primate animals. Thus, the development of active compstatin analogs against non-primate targets, such as rat C3 (rC3) or mouse C3 (mC3), is an important, unaccomplished to-date goal.
The structure of the complex between the proteolytic fragment C3c of human C3 (hC3) and compstatin analog W4A9 (the N-terminal acetylated, double mutant Ac-Val4Trp/His9Ala) has been determined at 2.4-Å resolution [4]. C3c is convenient for co-crystallization studies as it maintains the structural characteristics of C3 and C3b at the β-chain level, which contains the compstatin binding site, and offers the advantage of a smaller size fragment than C3 and C3b. In addition, C3c is free of the C3d domain (site of the opsonization thioester bond), which is the activation domain of intact C3. We recently employed de novo design methods to identify novel, compstatin-based inhibitors of human C3 [13]. Using activity measurements, we demonstrated that selected analogs with aromatic (Trp) substitutions at one or both terminal ends had near-W4A9 activity [14]. Furthermore, using atomistic molecular dynamics (MD) simulations of W4A9 complexes with hC3 or rC3, we developed a mechanistic interpretation for the species-specificity of compstatin at molecular level [15] and we designed a “transgenic” mC3 with human-like binding site characteristics [16]. A main goal of our present work is to identify compstatin-based compounds with promising inhibitory activity against non-primate proteins (rC3 or mC3). The most promising inhibitors are also studied in complex with hC3 and compared with W4A9.
Our work combines de novo design [13] and atomistic MD simulations [15] and draws on a large body of accumulated knowledge on compstatin-based inhibitors, from NMR experiments [17, 18, 19, 20, 21], MD simulations [22, 23], mutational studies [20, 21, 24, 25, 26, 27, 28], computational studies [29, 30], rational, experimental-combinatorial and computational-combinatorial design methods [10, 13, 14, 20, 21, 24, 26, 27, 31, 32, 33, 34, 35, 36, 37]. Previous studies showed that the two cysteines 2 and 12, and native residues 5–8 were critical for activity. Here we focus mainly on substitutions that introduce new interaction capabilities at the N-terminal end (positions 1, 3 and 4); in some cases, we study mutations at other positions (9–11 and 13).
Insertion of a charged (Arg) or aromatic (Trp) amino acid at position 1 improves the computed affinity for both hC3 and rC3. The Arg1 insertion also improves the peptide solubility, an important factor in drug-development. Interestingly, an extension at the N-terminal end of the ligand enables the formation of interactions in the simulations, which compensate for interactions eliminated or lost in non-primate complexes and improve dramatically the computed ligand affinities for rC3 and mC3.
2. Methodology
De novo design
The de novo design focused on the identification of inhibitors against rC3, and followed the methodology outlined in ref. [13]. In its general implementation setup, this protocol has a sequence selection stage and a sequence validation stage. The selection stage employs integer linear optimization to identify amino acid sequences, which correspond to global energy minima of a given (rigid or flexible) template fold. In the present application we use several structures from a simulation of the rC3:W4A9 complex [4, 15] as a structural template (see below). The validation stage computes the approximate binding affinities of these sequences to the target protein, without restricting the conformational space or the binding mode of the complex. For the purposes of the present design (the identification of compstatin analogs with a binding mode similar to the one in the hC3c:W4A9 complex), sequences identified by the second de novo stage underwent detailed atomistic MD simulations in explicit water, which assess the structural behavior and stability of complexes involving the sequences from the selection stage.
I. Selection Stage
Definition of structural template
Our sequence-selection template consisted of a truncated rC3 protein model with residues 329–534 (the compstatin binding site in the hC3:W4A9 complex). This template was based on the simulation system of the rC3:W4A9 complex, employed in ref. [15]. To model flexibility, we placed the template into eight distinct conformations, taken at 1-ns intervals from a 7-ns molecular dynamics (MD) simulation of the rC3:W4A9 complex [15]. All conformations were aligned along the structure of the hC3c:W4A9 complex (PDB code: 2QKI [3]); after this alignment, the coordinates of the compstatin variant were extracted from the hC3c:W4A9 complex and combined with each of the rC3 conformations, to create eight distinct rC3:compstatin complexes.
Mutation Sets
The next step in the selection stage is to specify a list of possible amino acid types for each position in the designed sequence. This mutation set can be general (e.g., all twenty natural amino acids at every position), or restricted by knowledge-based considerations or other criteria, such as the solvent-accessible-surface-area (SASA). Two mutation sets were used in the present design. The first set allowed mutations at positions 1, 3 and 4 of compstatin. The amino acid types at each position were based upon the observed SASA of the corresponding W4A9 residue in the rC3c complex simulations. If a residue of the bound W4A9 was more than 50% exposed to solvent, only hydrophilic amino acids were allowed. If a position was less than 20% exposed, only hydrophobic amino acids were allowed; otherwise, all amino acids were allowed. Based on this criterion, position 1 was allowed to select from a set of hydrophilic amino acids (G, A, P, R, K, D, E, N, Q, H, S, T) and the native amino acid Ile, position 3 was allowed to select from a set of hydrophobic amino acids (A, C, G, V, I, L, M, F, Y, W, T), and position 4 was allowed to select from all amino acids. The small amino acids A, T and G were allowed in all positions. This led to a total of 2860 possible sequences. The second mutation set was based upon results from the first. Of the three positions allowed to mutate in the first set, position 3 showed the least variability, with W being the dominant amino acid (26% probability); other mutations in this position included F (21.4%), M (14.9%), Y (13.5%), I (7.2%), T (5.3%), C (4.1%), V (3.7%), L (3%), A (0.9%). Thus, the second mutation set fixed W at position 3 and allowed the other two positions (1 and 4) to mutate as before. This led to a total of 260 sequences.
Forcefield
The energy calculations of the selection stage employed the 6-bin Centroid-Centroid force field [38, 39].
Determination of low-energy sequences
Because the design template was flexible, the distance bin sequence selection model was used to find low-energy sequences (see [40, 41] for details on the model). The model is described in detail in ref. [40]
| (1) |
The model minimizes the pair-wise energy between amino acid j in position i and amino acid l in position k. The binary variable is an interaction variable, and equals the product of the binary variables and . equals one only if both and equal one, indicating that amino acid j is in position i and amino acid l is in position k. The final binary variable bikd equals one if the distance between position i and position k falls into distance bin d and zero otherwise. In this way, the model is allowed to select one distance bin from among the multiple distance bins that residues i and k may span in the eight MD templates.
Validation Stage
Selected sequences from the previous stage were subjected to approximate binding affinity calculations [13, 42]. This allowed us to re-rank the sequences from stage one according to approximate binding affinities and identify candidate binders.
The approximate binding affinity of a protein P and peptide L is defined as
| (2) |
where qPL, qP and qL are, respectively, the partition functions of the protein-peptide complex PL, the free protein and the free peptide. These partition functions are defined in Eq. (3), where the sets B, F, and L contain rotameric conformation ensembles of the complex, free protein, and free peptide, respectively:
| (3) |
The temperature T entering in the Boltzmann factors of Eq. (3) was set to 298 K. In order to calculate the approximate binding affinity for a mutant sequence, the three-dimensional (3D) structure of each sequence was predicted by the Rosetta ab initio method [43, 44, 45]. A total of 2,000 3D peptide structures were generated. Cluster analysis of their main chainφ and ψ torsional angles with OREO [46, 47] determined 11 representative structures of the whole ensemble. Each of these structures was docked to the target protein (the last snapshot of the 7-ns rC3:W4A9 run) using Rosetta Dock [48,49,50]. For each docking run, 2,000 docked conformers were generated. The ten lowest-energy conformers were selected as seeds for the complex-ensemble generation using Rosetta Design [48]. Likewise, the ten lowest-energy structures from each of the peptide clusters were selected as seeds for the peptide ensemble generation. For the free protein ensemble, only the crystal structure of the last MD snapshot was selected as the seed. Rosetta Design used these seeds to generate rotamerically-based conformation ensembles (22,000 for the peptide, 22,000 for the complex, and 2000 for the protein). The energies of each of the members of the ensemble were used to calculate the corresponding partition functions [Eq. (3)] and the approximate binding affinity [Eq. (2)].
Choice of complexes
We simulated twenty-two compstatin-based analogs in complex with human (Homo sapiens), rat (Rattus norvegicus) and mouse (Mus musculus) C3. The analogs were classified into four groups (“generations”). The first generation contained selected promising analogs from the above described de novo design, whereas the other three explored modifications (substitutions or extensions), introduced predominantly at the N-terminal end. The choice of analogs and their classification into generations is detailed in the Results section. The most promising complexes were subjected to more than one run, based on their ability to retain the crystallographic structure and interactions of the hC3c:W4A9 complex [4], and the magnitude of their association free energies, as quantified by a MM-GB/SA analysis. A comprehensive list of all simulations is included in Table 1.
Table 1.
Summary of simulations conducted in the present work.
| Generationa | Runb | Analog sequence | Protein species | Run duration (ns) |
|---|---|---|---|---|
| 1 | H1W3Y4:R | Ac-HCWYQDWGAHRCT-NH2 | Rat | 7 |
| Q1W3Y4:R1 | Ac-QCWYQDWGAHRCT-NH2 | Rat | 7 | |
| Q1W3Y4:R2 | Ac-QCWYQDWGAHRCT-NH2 | Rat | 7 | |
| W3P4:R | Ac-ICWPQDWGAHRCT-NH2 | Rat | 7 | |
| T1W3F4:R | Ac-TCWFQDWGAHRCT-NH2 | Rat | 7 | |
| S1W3:R | Ac-SCWWQDWGAHRCT-NH2 | Rat | 7 | |
| T1W3:R | Ac-TCWWQDWGAHRCT-NH2 | Rat | 7 | |
|
| ||||
| 2 | R1W3:R1 | Ac-RCWWQDWGAHRCT-NH2 | Rat | 7 |
| R1W3:R2 | Ac-RCWWQDWGAHRCT-NH2 | Rat | 7 | |
| R1:H1 | Ac-RCVWQDWGAHRCT-NH2 | Human | 10 | |
| R1:H2 | Ac-RCVWQDWGAHRCT-NH2 | Human | 7 | |
| R1:R | Ac-RCVWQDWGAHRCT-NH2 | Rat | 10 | |
| R1H11:R1 | Ac-RCVWQDWGAHHCT-NH2 | Rat | 7 | |
| R1H11:R2 | Ac-RCVWQDWGAHHCT-NH2 | Rat | 7 | |
| R1K9H11:R1 | Ac-RCVWQDWGKHHCT-NH2 | Rat | 7 | |
| R1K9H11:R2 | Ac-RCVWQDWGKHHCT-NH2 | Rat | 7 | |
| R1K10H11:R1 | Ac-RCVWQDWGKKHCT-NH2 | Rat | 7 | |
| R1K10H11:R2 | Ac-RCVWQDWGKKHCT-NH2 | Rat | 7 | |
|
| ||||
| 3 | W1:H1 | Ac-WCVWQDWGAHRCT-NH2 | Human | 10 |
| W1:H2 | Ac-WCVWQDWGAHRCT-NH2 | Human | 7 | |
| W1:R | Ac-WCVWQDWGAHRCT-NH2 | Rat | 10 | |
| W1:M | Ac-WCVWQDWGAHRCT-NH2 | Mouse | 7 | |
| W13:H | Ac-ICVWQDWGAHRCW-NH2 | Human | 7 | |
| W13:M | Ac-ICVWQDWGAHRCW-NH2 | Mouse | 10 | |
| W1W13:H | Ac-WCVWQDWGAHRCW-NH2 | Human | 7 | |
| W1W13:M | Ac-WCVWQDWGAHRCW-NH2 | Mouse | 7 | |
|
| ||||
| 4 | S-1S0: H1 | Ac-SSICVWQDWGAHRTC-NH2 | Human | 10 |
| S-1S0: H2 | Ac-SSICVWQDWGAHRTC-NH2 | Human | 10 | |
| S-1S0: R1 | Ac-SSICVWQDWGAHRTC-NH2 | Rat | 10 | |
| S-1S0: R2 | Ac-SSICVWQDWGAHRTC-NH2 | Rat | 10 | |
| S-1S0: M1 | Ac-SSICVWQDWGAHRTC-NH2 | Mouse | 10 | |
| S-1S0: M2 | Ac-SSICVWQDWGAHRTC-NH2 | Mouse | 10 | |
|
| ||||
| Native compstatin | Ac-ICVVQDWGHHRCT-NH2 | |||
| W4A9 analog: | Ac-ICVWQDWGAHRCT-NH2 | |||
The classification into generations is explained in the Methodology section.
Nomenclature: Each substitution, with respect to the parent compound W4A9, is denoted by its one-letter amino acid code and its position; the letter (H/R/M) after the colon “:” denotes the C3 species (human/rat/mouse). A number following this last letter denotes the run number, in case of multiple runs. The diserine extension at the N-terminal end of analogs in generation 4 is denoted as “S-1S0”.
Simulation Systems
All compstatin derivatives were maintained in a cyclic conformation via a Cys2-Cys12 disulfide patch of the CHARMM topology file. Human, rat and mC3 were modeled by the same truncated C3c system of ref. [15]. The various complexes were immersed in a box of water molecules in the shape of a 89-Å truncated octahedron; overlapping water molecules were omitted. Titratable residues were assigned their most common ionization state at physiological pH. The total charge of the simulation systems was set to zero, by addition of appropriate numbers of chloride anions.
Force Field Specifications
The peptide atomic charges, van der Waals and stereochemical parameters were taken from the CHARMM22 all-atom force field [51], including a CMAP backbone φ/ψ energy correction [52] and indole parameters from ref. [53]. Force field specifications and simulation protocols were the same as in ref. [15]. All simulations were conducted with the molecular mechanics program CHARMM, versions c35a2, c35b5 [54].
Initial Coordinates
With the exception of the diserine N-terminal extension in one analog (see below), and protein loop 369–378, the initial positions of all other backbone heavy atoms were taken from the crystallographic structure of the hC3:W4A9 complex (PDB code 2QKI) [4]. As explained in ref. [15], with this choice, we avoid introducing any a priori structural differences from the human complex. Loop 369–378 contains a deletion at position 372 of the rat and mouse protein (Fig. S1). In non-primate complexes, the initial conformation of this loop was constructed with the program Modeller [55]; its root-mean-square difference (RMSD) from the corresponding conformation in hC3c was 1.39 Å. Heavy atoms of all invariant side chains outside of the 369–378 loop were initially placed at the corresponding coordinates of the human complex; mutated side chains were modeled with the SCWRL4 program [56]. Initial hydrogen positions were determined by the HBUILD algorithm of the CHARMM program.
Simulation Protocols
To avoid structural deformations at the protein boundary due to the truncation, the main chain heavy atoms of an external protein shell, with atoms at least 20 Å away from any atom of compstatin, were harmonically restrained to their initial crystallographic positions. Segments 373–377 of the reconstructed loop (373–376 in rat and mouse) were also harmonically restrained in all simulations, with the exception of the complexes of generation 4, in which the ligand N-terminal extension (particularly Ser-1) can interact with the loop. The structures were initially optimized by 150 energy minimization steps with the steepest-descent and adopted-basis Newton-Raphson (ABNR) algorithms. This was followed by an equilibration run, consisting of: (i) 30 ps of dynamics, with all protein and ligand heavy atoms harmonically restrained by a force constant of 10 kcal/mol·Å2; (ii) five 50-ps segments, in which the harmonic force constants were gradually lowered to 1.5 kcal/mol·Å2 in the external shell, and to 0 kcal/mol/Å2 elsewhere. The systems were then simulated further, retaining the harmonic-restraint setup from the end of equilibration. The length of this “production run” (7–10 ns) was chosen to ensure that the affinities of the most promising complexes (Table 1) converged to stable values.
Initial conformation of the diserine N-terminal end extension
One of the simulated analogs (S-1S0, corresponding to the fourth generation) contained a two-serine N-terminal extension, combined with the W4A9 sequence. The initial conformation of the extension was optimized as follows. At first, we extended the analog by two alanine residues. To construct initial conformations of the dialanine, we varied its four main chain torsional angles in the range −180° to 180, using a grid of 30°. The initial conformations of atoms outside the extension were prepared as described above, for human and rat complexes; for the mouse complex, they were positioned to the coordinates of a low-affinity structure from the W4A9:rC3 simulations [56]. For each of the resulting dialanine conformations, we minimized the entire complex by 100 steepest-descent minimization steps; during this minimization, atoms outside the dialanine extension were harmonically restrained by 100 kcal/mol/Å2. The lowest-energy conformation was finally selected as the optimum structure of the extension. The alanine side chains were replaced by serines, and their optimum orientations were determined by the program SCWRL4 [56].
Analysis of side-chain contacts
Probability-density maps of intermolecular side-chain contacts were computed with the WORDOM package [57]. Two side-chains were considered in contact if the distance of their geometric centers was smaller than 6.5 Å.
Computation of association free energies
The association free energies (second column in Table 3) were computed by the relation
Table 3.
Association free energies of selected complexes (in kcal/mol). A complete list of association free energies for the remaining complexes of Table 1 is given in Table S3 of the Supplementary Information.
| Run | Binding Free Energy | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Total | Polar Componenta | Non-polar Componenta | Polar Interactionb | |||||
| Std Dev | Std Dev | Std Dev | Std Dev | |||||
| R1:H1 | −58.3 | 0.6 | 0.4 | 0.6 | −58.8 | 1.2 | −65.2 | 0.7 |
| R1:H2 | −60.2 | 2.3 | −1.1 | 1.9 | −59.1 | 0.4 | −71.0 | 0.1 |
| Average | −59.3 | 1.9 | −0.3 | 1.6 | −58.9 | 0.9 | −68.1 | 2.9 |
| W1:H1 | −56.8 | 1.1 | 3.4 | 0.1 | −60.2 | 1.2 | −51.0 | 1.5 |
| W1:H2 | −54.6 | 0.1 | 6.0 | 0.6 | −60.6 | 0.8 | −46.8 | 0.9 |
| Average | −55.7 | 1.4 | 4.7 | 1.4 | −60.4 | 1.0 | −48.9 | 2.4 |
| S-1S0:H1 | −58.8 | 0.0 | 1.1 | 0.3 | −59.9 | 0.2 | −47.8 | 4.1 |
| S-1S0:H2 | −55.7 | 0.1 | 6.4 | 0.3 | −62.2 | 0.4 | −32.0 | 0.5 |
| Average | −57.3 | 1.6 | 3.8 | 2.7 | −61.0 | 1.2 | −39.9 | 8.4 |
| S-1S0:R1 | −55.7 | 0.8 | 3.4 | 0.5 | −59.0 | 0.3 | −47.1 | 0.5 |
| S-1S0:R2 | −53.9 | 1.7 | 3.4 | 0.6 | −57.3 | 1.1 | −45.4 | 1.4 |
| Average | −54.8 | 1.6 | 3.4 | 0.5 | −58.2 | 1.2 | −46.2 | 1.4 |
| S-1S0:M1 | −54.4 | 0.0 | 4.8 | 0.2 | −59.2 | 0.2 | −45.3 | 0.9 |
| S-1S0:M2 | −59.0 | 0.0 | 4.8 | 0.2 | −63.8 | 0.2 | −46.5 | 0.8 |
| Average | −56.7 | 2.3 | 4.8 | 0.2 | −61.5 | 2.3 | −45.9 | 1.1 |
Values are averaged over 700 snapshots (last 7-ns of each run).
Computed with Eq. (1), assuming identical protein and ligand conformations in the complex and dissociated states (see Methods). bDefined in Eq. (2).
The polar-interaction components [Eq. (3)] measure the strength of intermolecular polar (Coulomb and GB) interactions in the complexes.
| (4) |
where PL, P and L denote, respectively, the complex, protein, and ligand. The individual free energies were computed in the Molecular Mechanics-Generalized Born/Surface Area (MM-GB/SA) approximation [58]. In this approximation, representative coordinates of each state X (X=PL, P, or L) are extracted from the corresponding simulation trajectories. The corresponding free energies are computed from the relation.
| (5) |
The first term on the right-hand side of Eq. (5) describes the dependence of the internal energy on the molecular geometry (bond lengths, bond angles, torsional angles); the second term describes Coulombic interaction energies between the atomic charges of the molecule; the third term represents the electrostatic contribution to the solvation free energy, and is modeled by the GBSW generalized-Born approximation [59,60]. The next term describes van der Waals interactions; the final term describes non-polar contributions to the solvation free energy, assumed proportional to the solvent-accessible surface area, Sx, of the molecule. The proportionality coefficient σ was set to 0.005 kcal/mol/Å2, as in the GBSW parameterization. Note that for the complexes studied here, the contribution from this last term to the obtained affinities was approximately constant among complexes; thus, the value used for σ affects the individual but not the relative association free energies.
In the application of Eq. (4) we make the “one-trajectory” approximation [61, 62], which assumes that the protein and ligand have identical structures in the complex and free (dissociated) states. This assumption ignores structural relaxation, which might contribute a few kcal/mol to relative affinities. On the other hand, it also eliminates contributions from intramolecular (bonded, intramolecular van der Waals and intramolecular Coulomb energies, which contribute thousands of kcal/mol to the energies of the complex and free protein [Eq. (5)], and may introduce large uncertainties in the relative affinities [61,62]; in the “one-trajectory approximation” these contributions cancel out in the association free energies [Eq. (4)].
The MM-GB/SA approximation and the related Molecular Mechanics/Poisson Boltzmann Surface Area (MM-PBSA) approximation [58] have been extensively used in affinity estimates ([63, 64] and references therein). Their performance is fragile [65], as they are based on numerous assumptions: they combine a molecular mechanics energy function with an implicit treatment of solvation effects, and include solute conformational entropy effects in an approximate manner. Here (see Results), we obtain for W4A9 a +9 kcal/mol relative affinity disfavoring rC3 over hC3 [15]. This estimate has the correct sign, since W4A9 is experimentally inactive against rC3; in fact, it is probably a lower bound to the relative affinity, since a “three-trajectory” approximation increases the value to ~ +19 kcal/mol [15]. Thus, relative affinities of this magnitude (~ +10 kcal/mol) are indicative of ligands specific for human (vs non-primate) C3.
The interaction energies between two groups of atoms (R and R′) (were computed by the relation.
| (6) |
The first and second group of terms on the right-hand side of Eq. (6) describe, respectively, polar and nonpolar interactions between R and R′; in our calculations, R corresponded to a ligand residue and R′ to the entire protein model; alternatively, R was a protein residue and R′ was the entire ligand. To compute the GB term in Eq. (6), we included all protein and ligand atoms and set the charges of atoms outside the two groups R and R′ to zero. The last term contains the difference in solvent accessible surface areas of groups R and R′ in the complex and unbound states.
The generalized-Born energies and the atomic accessible-surface areas (ΔSi) entering in Eq. (6) depend on the location of R and R′ in the complex. The polar component contains a Coulombic term and a GB contribution, modeling the interaction between group R and the solvent polarization potential induced by R′. Similarly, the nonpolar component contains a van der Waals interaction between R, R′ and a surface term, expressing cavity contributions and nonpolar interactions with the surrounding solvent. The sum of the two components reflects the total direct interaction between R and R′ in the solvated complex.
Free-energy values were averaged over the last 7 ns of each trajectory; to estimate the free-energy uncertainties, we partitioned these 7 ns into two blocks of 3.5 ns, and computed the standard deviation of averages over the 3.5-ns segments.
3. Results
I. De novo design
The sequence-selection stage employed two mutation sets at positions 1, 3 and 4. The first mutation set generated 1,000 sequences; 26% of all the sequences and 48% of the top 500 sequences had a W substitution at position 3. This prompted our use of a second set, in which position 3 was fixed to W. All 260 sequences of the second set were generated, and the top quarter with respect to energy was analyzed. Position 4 had a dominance of F or W (19% each), while position 1 was more variable, favoring H, N, F, Q, R, S, or T (10% each). We selected 17 sequences from the second mutation set for the validation step. These included: (i) the best 8 sequences with respect to their energy (E-) rank, the seven top sequences contained a W substitution at position 4, and a variety of substitutions at position 1; and (ii) 9 more sequences, combining a non-W substitution at position 4 together with residues at position 1 encountered in the best 8 sequences (Table 2). The approximate binding affinities of all sequences were computed as described in the Methodology section, together with the W4A9 compound (V4W/H9A). The results are summarized in Table 2. For each sequence, the E-rank and an approximate binding affinity (K*-) rank are given. Sequences with small values of E-ranks had low-energies in stage 1; sequences with small values of K*-ranks were predicted to have high-affinities in the second stage.
Table 2.
Sequence selection and approximate binding affinity results for inhibitors of rC3 from the second mutation set. Rankings are given for sequence selection (lowest energy-rank = 1, E) and approximate binding affinity (highest affinity-rank = 1, K*). Mutations (with respect to W4A9) are indicated in bold face.
| Name | Rank | Sequence | |
|---|---|---|---|
| E | K* | 1 13 | |
| H1W3Y4 | 39 | 1 | HCWYQDWGAHRCT |
| Q1W3Y4 | 25 | 2 | QCWYQDWGAHRCT |
| W3P4 | 64 | 3 | ICWPQDWGAHRCT |
| T1W3F4 | 53 | 4 | TCWFQDWGAHRCT |
| SQ081 | 81 | 5 | PCWMQDWGAHRCT |
| S1W3 | 4 | 6 | SCWWQDWGAHRCT |
| T1W3 | 7 | 7 | TCWWQDWGAHRCT |
| R1W3 | 3 | 8 | RCWWQDWGAHRCT |
| SQ001 | 1 | 9 | PCWWQDWGAHRCT |
| SQ005 | 5 | 10 | HCWWQDWGAHRCT |
| SQ006 | 6 | 11 | NCWWQDWGAHRCT |
| SQ002 | 2 | 12 | QCWWQDWGAHRCT |
| SQ042 | 42 | 13 | HCWFQDWGAHRCT |
| W4A9 | - | 14 | ICVWQDWGAHRCT |
| SQ014 | 14 | 15 | PCWYQDWGAHRCT |
| SQ019 | 19 | 16 | ICWWQDWGAHRCT |
| SQ080 | 80 | 17 | PCWHQDWGAHRCT |
| SQ008 | 8 | 18 | PCWPQDWGAHRCT |
Several sequences have higher approximate binding affinity than W4A9, the best inhibitor of hC3 comprising entirely of natural amino acids. The top two sequences (in terms of affinity-rank) have an aromatic residue at position 4 and differ only at position 1. The top five sequences also have high energy-ranks. In general, there is no strict correlation between energy- and affinity-rank throughout the set. This is partly due to the fact that the energies are computed from the selection stage, which assumes that the ligand binding mode (location/orientation) is similar to the one in the hC3c:W4A9 complex; on the other hand, affinities are computed in the validation stage by a more general docking procedure, in which the free protein, free ligand and the protein-ligand complex are allowed to explore a larger number of conformations and binding modes. This expanded search uncovers a more diverse set of binding modes, which change the predicted relative stability of the various complexes summarized in Table 2.
II. Molecular Dynamics Simulations
The structure and interactions of the complex between W4A9 and hC3 were analyzed by X-ray crystallography in ref. [4] and our MD simulations in ref. [15]. We first present them briefly; later, we use them to justify our choice of analogs and to assess the behavior of the simulated complexes.
A. The human complex with W4A9
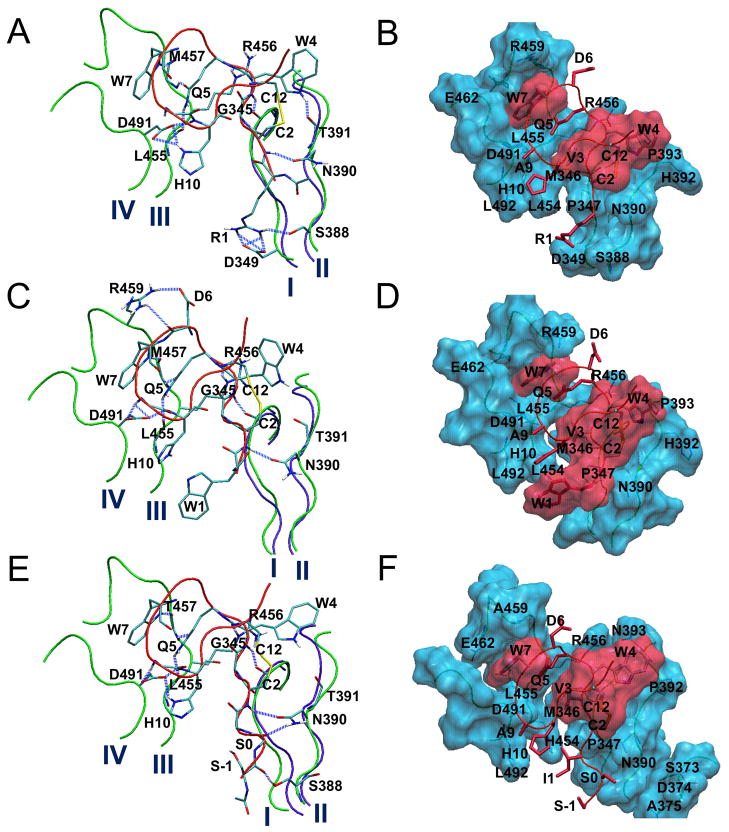
In the crystal structure (PDB code 2QKI [4]) and the MD simulations [15], the ligand segment 1–10 forms extensive interactions with the four protein sectors 344–349, 388–393, 454–462, and 488–492. These interactions are preserved in the R1 and W1 complexes investigated in the current work (runs: R1:H1, W1:H1) and are shown in Figs. 1(A–D). The main chain moieties of N-terminal residues Ile1 and Cys2 make competing hydrogen bonds with the Asn390 side chain. The Val3 side chain is buried in a very stable hydrophobic cluster formed by residues Met346, Pro347 and Leu454. The Trp4 main chain participates in stable hydrogen bonds with Gly345 CO and the side chain of Arg456; the Trp4 side chain packs against the Cys2–Cys12 disulfide-bridge and residue Pro393. The Gln5 side chain makes two intermolecular hydrogen bonds with main chain groups of Leu455 and Met457. The Trp7 side chain intercalates between segments 455–458 and 488–491, making a stable hydrogen bond with Met457 CO and non-polar contacts with Gln5, Met457, Arg459, and Glu462. Finally, the main chain NH groups of Ala9 and His10 form very stable hydrogen bonds with the Asp491 side chain; an additional hydrogen-bond is often observed among the side chain of His10 and Asp491; also, the His10 side chain makes frequent non-polar contacts with Leu454 and Leu492.
Figure 1.
Simulation structures of the compstatin binding site for the complexes R1:H (A, B), W1:H (C, D) and S-1S0:R (E, F) at the end of runs R1:H1, W1:H1 and S-1S0R1, respectively. Important hydrogen bonds and nonpolar contacts are shown, respectively, in the left and right panel. The labels I–IV (in A) indicate four protein sectors with atoms at least within 7 Å from the ligand (344–349, 388–393, 454–462 and 488–492). Compstatin is shown in red tubes and sticks. The violet tubes show the initial conformations of sectors I and II. The blue lines in plots A, C, E denote important hydrogen bonds. In plots B, D and F, protein residues are indicated by a cyan surface, and ligand residues Cys2, Val3, Trp4, Trp7 and Cys12 are indicated by a red surface.
B. Choice of analogs
The de novo design identified a number of sequences capable of forming good interactions with the structural template. We assessed selected promising sequences by atomistic MD simulations. In particular, we investigated the ability of the designed analogs to bind non-primate (rat or mouse) C3, with a similar “binding mode” (location/orientation) to the one in the W4A9:hC3 complex [4, 15]; such analogs are likely to inhibit C3 by interfering with the formation of the C3:C3-convertase complex in a manner analogous to W4A9 [4], and thus constitute promising rC3 inhibitors. The possibility that successful inhibitors might bind preferentially at different positions on non-primate (and/or human) C3 cannot be excluded; the systematic and accurate comparison of such diverse binding modes by atomistic MD simulations is very difficult, given the size of C3, and is beyond the scope of this article.
We studied four “generations” of compstatin-based compounds, in complex with human or non-primate C3. All simulations are summarized in Table 1. Throughout the paper, an analog is denoted by the set of substitutions along each sequence, relative to W4A9 (see Table 1).
The first generation included six compounds (H1W3Y4, Q1W3Y4, W3P4, T1W3F4, S1W3, T1W3). These analogs had high-affinity ranks in the de novo sequence selection stage (Table 2); the first four had also large energy ranks. All analogs had significant variability at position 1. Earlier simulations showed that key interactions between compstatin N-terminal residues 1 or 2 and the Asn390 side chain were lost in the rat complex, due to the displacement of sector 388–393 away from the ligand [15]. It was thus deemed probable that the insertion of bulky or hydrogen-bonding side chains at position 1 might restore interactions with Asn390 in the rat complex. This was not fully accomplished in the simulations of this generation (see below), but was pursued further in the analogs of the next three generations, which contained, respectively, charged or aromatic substitutions at position 1, or a diserine extension before residue 1. Sequences with a Pro substitution at position 1 were also suggested by the design. They were not studied further, as it was judged that this change might interfere with the hydrogen-bonding capacity of the ligand N-terminal end.
The second generation contained analogs with an Arg substitution (in place of Ile) at position 1. The parent analog (R1W3) resulted from the de novo design (Table 1). In the MD simulations the Arg1 side-chain formed a salt-bridge with proximal residue Asp349, which improved the computed affinity for rC3. In an attempt to improve affinity even further, additional polar or charged substitutions at positions 9–11 were also explored (analogs R1H11, R1K9H11 and R1K10H11). The analogs were simulated in complex with rat and/or hC3, to assess the impact of these substitutions across species.
The third generation included analogs with Trp substitutions at positions 1 and/or 13, suggested by earlier de novo design [13] of inhibitors against hC3. Subsequent activity measurements showed that analog W1 had near-W4A9 inhibitory activity against hC3, whereas analogs W13 and W1W13 had slightly reduced activities and solubilities, which may be connected [14]. In the experimental structure [4] and the MD simulations of the W4A9:hC3 complex [15], the (native) Ile1 side chain forms weak nonpolar contacts with the protein and the Thr13 side chain is exposed to solvent. Thus, the Trp1/Trp13 substitutions introduce a capability for new interactions (aromatic, π-cation, π-stacking or hydrogen bonding) at the N- and C-terminal positions. Such interactions may improve hC3 inhibition, and/or introduce inhibitory activity against non-primate C3. To check this possibility, and compare ligand affinities across species, we study here complexes of such analogs with human, rat and mC3.
The analogs considered so far contain 13 residues. The last generation included one analog, with a diserine extension at the N-terminal end (positions “−1”, “0”), and the W4A9 sequence at positions 1–13. These analogs were inspired by our earlier simulations [15] and the present results with human and rat/mouse complexes, which showed that sector 388–393 of non-primate proteins has a persistent tendency to deviate from the position seen in the human crystallographic complex [4]. This structural reorganization disrupts hydrogen-bonding interactions of Asn390 and the main chain of the first two N-terminal residues (Ile1 and Cys2 in W4A9), and weakens the affinity of the rat and mouse – compstatin complexes. Based on this knowledge, we reasoned that an elongation of the compstatin main chain at the N-terminal end might help retain hydrogen-bonding interactions with the displaced protein sector, and yield compstatin analogs with human-like affinity for non-primate C3. As a first guess, we included serine side chains in the extension, due to their hydrogen-bonding capacity. Note that a diserine extension was also part of the initial phage-display peptide (Clone 9 in ref [9]), before its truncation to the 13-residue compstatin. Ongoing work investigates other side chains at positions “0” and “−1”.
C. Structural behavior of the simulated complexes
Root-mean-square differences (RMSD) between the simulation and crystallographic [4] coordinates of protein and ligand main chain atoms N Cα, C are listed in Table S1 of the Supplementary Information (SI). The first two rows report the RMSD values for the two runs of the human complex W4A9:hC3, conducted in ref. [15].
The protein main chain (second column) remains near the crystallographic conformation in all complexes, as indicated by the small RMSD values (below or near 1 Å); the original secondary structure is also well retained (not shown). Sector 388–393 has the largest deviation in all rC3 and mC3 complexes studied here, due to its tendency to move away from the conformation seen in the human complex and towards the solvent. An example is shown in Fig. 1(E), where the 388–393 conformation at the end of run S-1S0:R1 (green) is juxtaposed to the initial conformation (red). This behavior was first observed in the simulations of the W4A9:rC3 complex [15], and is consistently reproducible. The fragment 388–396 contains four substitutions in rC3 and mC3 (Pro392His, Asn393Pro, Arg395Gln and Gln396Lys; an alignment of human, mouse and rat sequences is shown in SI Fig. S1). Unpublished simulations show that the insertion of human substitutions in all four positions of mC3 reduces the structural deviation of 388–393 at the level of the human complex.
In some rat or mouse complexes, sector 345–349 also experiences larger structural deviations relative to the human complexes. This is partly related to the presence of an Ala residue (instead of Gly) at position 345, which alters somewhat the main chain conformation and affects the formation of a hydrogen-bond between the main chain amide group of ligand residue 4 and the carbonyl group of Gly/Ala345 [16]. Deviations in the other two sectors (354–359, 488–493) are similar to the ones in the human complexes.
The displacement of 388–393 in non-primate complexes facilitates the deviation of the ligand away from the original binding mode (position/orientation). This is reflected in the larger ligand RMSD values (next-to-last column of Table S1), especially in the non-primate complexes of generation 1. The diserine extension also affects the binding mode, presumably due to the formation of interactions with 388–393 and the flexible loop 371–376, which is also near the extension. The larger ligand RMSD values are mainly due to shifts/rotations with respect to the original bound location, rather than due to changes in the ligand shape; upon removing the ligand net rotation/translation, they become comparable to the RMSD values of the human runs (last column of Table S1). Furthermore, the ligand maintains its secondary structure in all complexes, with the exception of an intramolecular β-bridge 3–11, which is not well preserved in the non-primate complexes (Fig. S2).
D. Protein - Ligand Interactions
The statistics of important intermolecular hydrogen bonds for all complexes are listed in SI Tables S2A-D. Statistics of residue intermolecular energies are plotted in Fig. S3; nonpolar interactions (side chain contacts) are shown in Fig. S4. We first discuss the results of simulations with non-primate (mouse and rat) C3; human complexes are analyzed next.
Non-primate complexes
First Generation
The analogs of this generation were studied in complex with the rat protein and showed a similar dynamical behavior with the W4A9:rC3 complex of ref. [15]. The ligand lost intermolecular interactions with Sectors 388–393 and 488–492, as documented in Tables S2–S3 and Figs. S3–S4. The only exception was run Q1W3Y4:R1, in which sector 388–383 remained near the starting conformation; this behavior was not reproduced in the second run (Q1W3Y4:R2). The computed affinities (Table S3) were in the range −37–−41 kcal/mol, significantly worse than the corresponding affinities of W4A9 for rC3 (−46 kcal/mol) and hC3 (−55 kcal/mol) [15].
Second Generation
All analogs of this group were studied in complex with the rat protein. With the parent analog R1W3 we conducted two runs (R1W3:R1-2), in which Arg1 formed a frequent salt bridge with Asp349; an example of this interaction is shown in Fig. 1A, for the R1 complex with hC3 (below). The Arg1-Asp349 salt bridge improved R1W3 affinity for rC3 (−47.8 kcal/mol), relative to W4A9 and all other first-generation analogs (Table S3). Still, it was by ~8 kcal/mol weaker than the corresponding affinity of W4A9 for hC3 (−55 kcal/mol [15]). For this reason, we introduced additional substitutions at positions 9–11 (R1, R1H11, R1K9H11, R1K10H11). We also restored (native) Val at position 3, based on the observation that the Trp3 side chain formed extensive non-polar contacts with Met346, Pro347, His454 in the R1W3 runs, but the ligand main chain moiety 1–4 was somewhat shifted with respect to its position in the crystallographic complex; this displacement hindered the formation of hydrogen bonds between the main chain of residues 2 or 4 and the protein (not shown) [4,15].
The interactions of the first analog (R1) with rC3 were similar to the ones of the W4A9:rC3 complex [15]; the analog moiety 1–3 formed somewhat improved non polar contacts with protein residues Met346, Pro347, Ser388 and Asn390. The computed affinity for rC3 was −50.9 kcal/mol (Table S3), improved by ~ −5 kcal/mol compared to W4A9, and by ~ −2 kcal/mol compared to R1W3. The additional substitutions at positions 9–11 (complexes R1K9H11:R, R1K10H11:R, R1H11:R in Table S3) had a worsening effect of affinity, presumably, the original W4A9 residues (A9, H10) were already optimal choices at these positions.
To summarize, the Arg1 substitution improved the computed rC3 affinity (especially for R1), but not to the level of W4A9 affinity for hC3, even when accompanied by polar/charged residue substitutions at positions 9–11; the lost or diminished interactions due to the structural changes in rC3 were not entirely compensated by the Arg1-Asp349 salt bridge and other novel interactions, at least for the complexes studied here.
Third Generation
We simulated various rat and mouse complexes (W1:R, W1:M, W13:M, W1W13:M). We observed improved interactions with respect to the first two generations, and computed an overall enhancement of affinity by ~ −2–−5 kcal/mol, relative to the W4A9 affinity for rC3. This overall enhancement with respect to the first two generations is mainly attributed to improved non-polar interactions between Trp1 and residues Pro347, Ser388, Asn390, His454 and partly Leu492 (an example of these interactions is shown in Fig. 1D, for the W1 complex with hC3 (below). An additional intramolecular (ligand) π-cation interaction between Arg11 and Trp13 was observed in the W13:M and W1W13:M simulations; in both complexes, this interaction seemed to stabilize the bound conformation (especially in the region 8–13) of the ligand despite the displacement of sector 388–393, assisting thus the formation of intermolecular hydrogen bonds, between Ala9 or His10 and Asp491. Nevertheless, a complete recovery of the W4A9:hC3 hydrogen bond (Table S2C) and non-polar interactions (Fig. S3) was not achieved.
Fourth Generation
The results of the first three generations suggest that the introduction of polar, ionic or aromatic interactions at position 1 does not increase the computed affinity for rC3/mC3 to the level of W4A9 affinity for hC3. In the fourth generation, we extended the ligand outside the Cys2-Cys12 ring, by adding two residues at the N-terminal end. This extension could augment ligand affinity for r/mC3 in several ways: first, it could assist sector 388–393 to retain the conformation seen in the crystallographic complex W4A9:hC3 [4]; even if sector 388–393 were displaced, the extension might form novel interactions, replacing the disrupted hydrogen-bonds between residues 1 and 2 and Asn390. Here we employed a diserine extension, to exploit the hydrogen-bonding capacity of the serine side chains. Starting from the N-terminal end of the original compound (position 1), the inserted serines are denoted “Ser0” and “Ser-1” in what follows. Note that the initial phage-display peptide also included a diserine extension at its N-terminal end [9]. A more comprehensive, ongoing study will optimize the extension residue types.
The displacement of sector 388–393 was somewhat smaller relative to other non-primate complexes (runs S-1S0:R and S-1S0:M in Table S1); additional simulations would be needed to check the reproducibility of this result. Notably, the hydrogen bond Cys2 NH - Asn390 OD was maintained and a novel, stable hydrogen bond was formed between the main chain of Ser0 and Asn390 (Fig. 1E, Fig. S5C and Table S2D). The first hydrogen bond is observed for the first time in simulations of a non-primate complex, and is reproduced in all four non-primate runs of this generation (Table S2D); the second hydrogen bond is new, due to the extension.
Some interactions were reduced relative to the W4A9:hC3 complex, particularly non-polar contacts of ligand residues Trp4 and Cys12 with Asn390 and His393 and of Asp6, Trp7 with Pro459 (Figs. S3–S4, S5B); on the other hand, residue Ser-1 formed novel contacts with sector 371–376 (Figs. 1(E–F), S5(C–D)). Nevertheless, the similarity between the diserine extension rC3 and mC3 complexes and the W4A9:hC3 complex is high, and is additionally verified by residue interaction free-energy difference values which are close to zero [Figs. 2(A, B)].
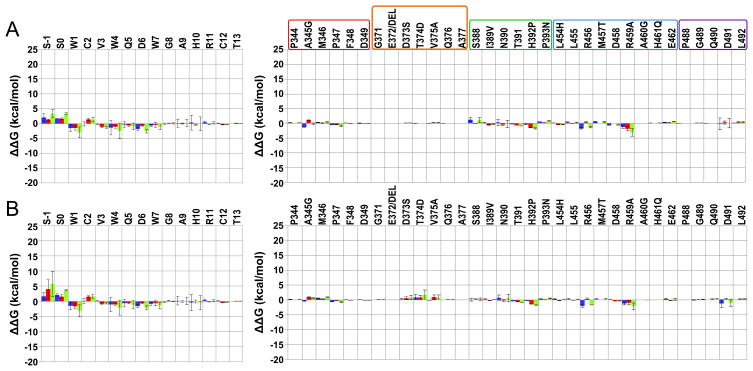
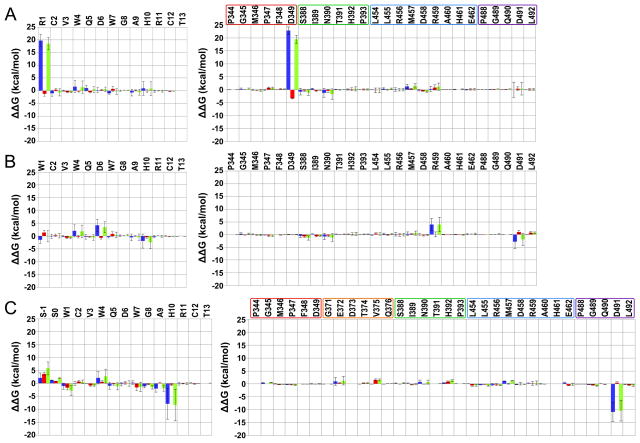
Figure 2.
Residue intermolecular interaction energy differences between the complex W4A9:hC3 [15] and selected complexes of this work. Compstatin and C3 results are in the left and right panel. Data are averaged over all respective runs. The colored code used is: blue – polar; red – non-polar; and green – total. The uncertainties (error bars) are computed as described in methods. (A) W4A9:hC3 – S-1S0:R difference; (B) W4A9:hC3 – S-1S0:M difference. Positive/negative values indicate, respectively, gained/lost interactions in the present complexes, relative to the W4A9:hC3 complex [15]. C3 regions interacting with compstatin analogs are enclosed in boxes in (A), colored as follows: red – sector 344–349; orange – sector 371–376; green – sector 388–393; blue – sector 454–462; and purple – sector 488–492. An additional residue, A377, is shown in the orange box to account for possible compensatory effects due to the E372 deletion with respect to human C3 (not present).
The computed affinities of the diserine analog for non-primate C3 were −54.8–−56.7 kcal/mol, near the affinity of W4A9 for hC3 and by ~ 9–11 kcal/mol stronger than its affinity for rC3.
Human complexes
Second generation
We performed simulations with R1, the strongest non-primate binding analog in this group (runs R1H1/2 in Table S2B). All interactions of the human complex W4A9:hC3 [15] were reproduced [Figs. 1(A, B) and 3A]. Furthermore, the Arg1-Asp349 salt bridge (Fig. 1A) was always present, and intermolecular side chain interactions of Arg1 with Pro347, Asp349 and Ser388, or Val3, Gln5 with Asp491, were slightly improved.
Figure 3.
Residue intermolecular interaction energy differences between the complex W4A9:hC3 [15] and selected compstatin simulations of the present work. Compstatin and C3 results are shown in the left and right panel. Data are averaged over all respective runs. The colored code used is: blue – polar; red – nonpolar; and green – total. The uncertainties (error bars) are computed as described in methods. (A) W4A9:hC3 – R1:H difference; (B) W4A9:hC3 – W1:H difference; (C) W4A9:hC3 – S-1S0:H difference. Positive/negative values indicate, respectively, gained/lost interactions in the new complexes, relative to the W4A9:hC3 complex [15]. C3 regions interacting with compstatin analogs are enclosed in boxes in (A) and (C), colored as follows: red – sector 344–349; green – sector 388–393; blue – sector 454–462; purple – sector 488–492; and orange – sector 371–376.
The computed affinity for hC3 is −59.3 kcal/mol, improved by ~ −3 kcal/mol relative to W4A9 (Table 3). Notably, this is mainly due to the polar component, reflecting the contribution to stability due to the Arg1-Asp349 salt bridge.
Third generation
We studied analog W1, which exhibited the strongest affinity and best solubility in the experiments of ref. [14]. The Trp1 side chain interacted frequently with His10 and Leu492 (Fig. 1D) in both runs (W1:H1 and W1:H2); furthermore, Asp6 formed an electrostatic interaction with Arg459. Even though this interaction was not observed in other simulated complexes (where Asp6 was mainly solvent-exposed), it could be related to an early mutation study, which showed that the Asp6Ala decreased activity (Table 3 of ref. [17]). Despite these additional interactions, all interactions of the human complex W4A9:hC3 [15] were reproduced [Figs. 1(C, D) and 3B] and the computed hC3 affinity was −55.7 kcal/mol, by ~ 4 kcal/mol weaker than the affinity of R1 (second generation) and comparable to the W4A9 affinity (−56 kcal/mol [15]). These results suggest that the Trp1 side chain did not fully exploit its interaction capabilities, presumably due to the lack of a suitable binding pocket in its proximity (unlike Trp4 and Trp7, which intercalate between specific protein sectors [4]).
In both complexes, we observed a π-cation Arg11-Trp13 interaction (not shown); In complex W13:H, this interaction seemed to interfere with the Cys2-Asn390 hydrogen bond, resulting in a somewhat reduced affinity (−51.1 kcal/mol; Table S3), relative to the W4A9 complex (−56 kcal/mol). In complex W1W13:H, the same interaction seemed to stabilize the ligand conformation, facilitating non-polar interactions of Trp1 with residues Pro347, Ser388, Asn390, Leu454 and Leu492 (Figs. S3–S4); the computed W1W13 affinity was considerably increased (−61.4 kcal/mol). Apart from the intramolecular π-cation interaction, Trp13 was solvent-exposed on its other side, without making interactions with the protein.
Fourth generation
The key Cys2-Asn390 interaction was retained in both runs (S-1S0:H1/2 in Table S2D). Furthermore, the main-chain of Ser0 formed an additional hydrogen bond with Asn390; this bond was somewhat less stable than the corresponding interaction in the non-primate complexes (Table S2D), presumably due to the increased proximity of Asn390 to the ligand (Fig. S5(A–B)). In the second run, the intermolecular hydrogen bonds of His10 were not well reproduced (Table S2D, Fig. S5A), resulting in less favorable polar interactions compared to the W4A9:hC3 complex (Fig. 3C). The predicted affinity (−57.3 kcal/mol) is somewhat stronger relative to W4A9, albeit not as good as for the R1 analog (Table 3).
To summarize, the simulations suggested that the diserine analog may be a promising “dual-specificity” inhibitor, estimated to bind with similar strength both human and non-primate C3. In the simulations, the analog forms intermolecular interactions with C3 via its main-chain moiety, whereas the serine side chains remain mostly solvent-exposed. Thus, possibly avenues to improve this compound could be to optimize the side-chains of the extension, and/or increase further the extension length.
4. Discussion
Since the discovery of compstatin [9], its inhibitory activity against hC3 has been examined in-depth by experimental [20, 21, 24, 25, 26, 27] and computational studies [15, 23, 29, 30, 37]. Rational, experimental-combinatorial and computational-combinatorial design methods have suggested numerous analogs with natural or artificial amino acids [13, 14, 20, 21, 24, 26, 27, 31, 32, 33, 34, 35, 36, 37].
In the present work we have employed a combination of de novo design/atomistic MD simulations, to identify compstatin-based analogs with promising affinity for non-primate C3. Furthermore, we have studied complexes of the most promising inhibitors with hC3, and compared with W4A9, the best to-date natural-amino acid inhibitor of the human protein.
We focused on substitutions which inserted additional interaction capabilities at the ligand terminal ends and, sometimes, positions 9–11; the x-ray structure of the W4A9:hC3 complex [4], and our earlier MD simulations of the human and rC3 complexes [15] suggested that the corresponding W4A9 residues did not make optimum intermolecular interactions, leaving room for improvement; this was especially true in the simulations of the rat complex, due to localized, reproducible structural changes of protein sectors near the ligand [15].
The insertion of an Arg side-chain at position 1 was estimated to improve the ligand affinity for both rC3 (Table S3) and hC3 (Table 1), prompting the study of the second-generation analogs. This was mainly due to the formation of a stable salt bridge Arg1-Asp349, together with the reproduction of most other intermolecular interactions seen in the W4A9:hC3 complex [4]. Note that the polar free-energy component for the R1:H complex is negative (Table 3), whereas it is positive for all other complexes (Table 3 and S3); this implies that the salt bridge compensates for the free-energy increase, due to the transfer of the Arg1 charge from water into the complex. Apart from its contribution to affinity, the N-terminal charge improves the solubility of the ligand, an important consideration in drug development.
Our previous experimental studies assumed that the insertion of tryptophan amino acids in compstatin might create avenues for mechanistic binding studies to C3, which would exploit the diverse physicochemical properties of tryptophan, i.e. its hydrophobicity (benzene ring), hydrogen-bond donor capability (indole amide) and its capability for π-stacking or π-cation interactions [14]. Our experiments showed that the Trp1 ligand was soluble and had a near-W4A9 inhibitory activity for hC3. In the present MD simulations of the W1:hC3 complex, the Trp1 side chain formed nonpolar interactions with His10 and Leu492 and W1 had a near-W4A9 affinity (Table 3). Overall, the simulations suggest that the Trp1 substitution improves affinity for both human and non-primate C3, but to a smaller extent than Arg1. The Trp13 substitution reduced the experimental solubility of compstatin analogs, restricting its potential use in compstatin-based drugs [14]. Despite this result, the combination of a Trp13 substitution with solubility-increasing mutations, by introducing polar amino acids, might yield promising inhibitors. To obtain insights on the potential interactions of Trp13 with C3, we included this mutation in two of the simulated analogs. One analog (W13) had somewhat smaller affinity for hC3, relative to W4A9; the second analog (W1W13) had significantly increased affinity (Table S3). In both complexes, most new intermolecular interactions (relative to W4A9) were formed by Trp1. Trp13 was solvent-exposed, or made intramolecular π-cation interactions with Arg11.
The introduction of the diserine extension at the N-terminal end (fourth-generation) was prompted by the displacement of sector 388–393 away from the ligand in the non-primate simulations, which eliminated or weakened interactions with Asn390 and caused the overall destabilization of the non-primate C3 – W4A9 complex [15]. In the simulated non-primate and human complexes, the interaction of the Cys2 main chain with Asn390 was retained and a novel interaction was formed, involving the main-chain of Ser0. The non-primate sector 388–393 was displaced away from the ligand to a smaller extent, relative to other non-primate complexes (Table S1), and the computed affinity for r/mC3 was improved by ~10 kcal/mol. In addition, the polar diserine extension adds much needed solubility at the N-terminus.
As discussed in the Methods, the performance of MM-GB/SA, that is employed here to estimate the complex stabilities, is fragile [65], as it is based on numerous assumptions. In particular, the “one-trajectory” approximation [61,62] eliminates contributions from intramolecular energies, which contribute thousands of kcal/mol to the energies of the complex and free protein [Eq. (2)] and may introduce large uncertainties in the relative affinities [61,62]]; on the other hand, the protein and/or ligand structural relaxation, which are ignored in this approximation, may contribute a few kcal/mol to relative affinities. In the case of W4A9, the “one-trajectory” approximation yields a +9 kcal/mol relative affinity, disfavoring rC3 over hC3 [15]. This estimate has the correct sign, since W4A9 is experimentally inactive against rC3; in fact, it is probably a lower bound to the relative affinity, since a “three-trajectory” approximation increases the value to ~ +19 kcal/mol [15]. Thus, relative affinities of this magnitude (~ +10 kcal/mol) or larger may be indicative of ligands specific for human (vs non-primate) C3.
The first-generation ligands have rC3 affinities in the range −37–−41 kcal/mol (Table S3); these are weaker than the affinity of W4A9 for rC3 and hC3, respectively, by 5–9 kcal/mol and 15–19 kcal/mol. At the same time, the rC3 complexes of these ligands experience localized structural changes (Table S1), which disrupt or eliminate intermolecular interactions seen in the human complex (Table S2A). Based on the above, we conclude that they are not promising inhibitors of rC3.
The second- and third-generation ligands have rC3 affinities in the range −46–−51 kcal/mol and hC3 affinities in the range −56–−59 kcal/mol. Both estimates are similar, or slightly better than the corresponding affinities of W4A9 (−46 and −55 kcal/mol [15]). Thus, the computed affinities of these ligands for hC3 are stronger by at least 5 kcal/mol; the associated free-energy uncertainties (in the “one-trajectory” approximation) are smaller (between 0.2–3.0 kcal/mol). Overall, these results suggest that ligands of these two groups bind human and rC3 with a near-W4A9; in analogy with W4A9, they are not promising inhibitors against non-primate C3, but they are worth exploring further as hC3 inhibitors.
Ligand S-1S0 (fourth generation) has near-W4A9 affinity for hC3 (~−57 kcal/mol) and in the range −55–−57 kcal/mol for non-primate C3. The two-residue extension contains four more main chain torsional angles, compared to 13-residue analogs. Assuming an upper-bound of ~0.6 kcal/mol loss in conformational entropy (−TΔS) per rotatable bond upon binding [66], the S-1S0 affinities could be overestimated with respect to the corresponding W4A9 affinities, possibly by ~ 2.5 kcal/mol. Thus, we do not conclude that S-1S0 inhibits hC3 as strongly as W4A9, despite the similar affinity of these two ligands for this protein. On the other hand, the computed relative (human – rat) affinity of S-1S0 is near ~0 kcal/mol. Conformational-entropy corrections should affect to a similar extent the S-1S0 affinity for human and rC3, leaving the relative affinity estimate unmodified. In comparison, the relative W4A9 affinity is +9 kcal/mol. The dramatic improvement of S-1S0 affinity for rC3 is due to improved interactions, and suggests that S-1S0, or other compounds carrying an N-terminal extension, are worth exploring further as promising inhibitors of non-primate C3.
For some of the peptides studied here, W1, W13, R1, and S-1S0, there is experimental evidence for inhibitory activity against hC3. W1 has been discussed above in view of our recent study, which showed that it had near-W4A9 inhibitory activity and good solubility, whereas W13 suffered from solubility problems in aqueous environment [14]. The peptide R1 was found previously to be active in a study that aimed to delineate the role of acetylation in producing a 3-fold increase in inhibitory activity compared to non-acetylated compstatin [10, 24]. The structural rationale was that the positively charged backbone amino group was disrupting the hydrophobic cluster at the termini (spanning residues I1-C2-V3-V4/C12/T13 of native compstatin), which was deemed necessary for binding and activity before the availability of the structure of the C3c-W4A9 complex, and that upon removal of the positive charge by acetylation the hydrophobic cluster was fortified and contributed to the 3-fold increase in activity. To prove this hypothesis, a positive charge was re-introduced in the vicinity at the side chain level by replacing Ile1 by Arg1 in native compstatin (or V4H9 peptide), which resulted to 2-fold decrease in inhibitory activity, essentially reverting the activity to nearly that of non-acetylated compstatin. The peptide with the diserine extension is expected to be active, as the diserine was present at the N-terminus of the original 32-amino acid peptide that was identified in the phage-displayed and a subsequent study [9, 18]. We have recently initiated experimental studies using ELISA-based complement system inhibition assays with human and rat serum for two of the analogs designed in this study, the R1 and S-1S0 peptides (Table 1). Preliminary data indicate that in human serum the R1 and S-1S0 analogs have comparable inhibitory activities, which are higher than those of native compstatin and slightly lower than those of the W4A9 analog, thus validating the computational design (López de Victoria A, Kieslich CA, Tamamis P, Gorham RD Jr, Bellows-Peterson M, Lo D, Floudas CA, Archontis G, Morikis D, unpublished). However, in rat serum the R1 and S-1S0 analogs were found inactive. This observation suggests that the two amino acid N-terminal extension may not be sufficient to mediate all intermolecular interactions needed for binding to rC3 and that longer extension may be necessary for activity in rat serum. Both R1 and S-1S0 analogs were soluble in aqueous buffer in the in vitro studies compared to reduced solubility of the W4A9 analog at the tested experimental concentrations.
5. Conclusions and Future Directions
The computational predictions of this work suggest that the introduction of novel interaction capabilities at the N-terminus of compstatin analogs augments affinity for non-primate C3, but not to the level of W4A9 affinity for hC3. The introduction of an extension at the N-terminal end seems to be more promising. With respect to hC3 inhibitors, the insertion of a charge at the N-terminal end is promising, as it increases affinity and solubility.
Ongoing and future experimental studies of extension analogs in complex with human and non-primate C3, will aim to further enhance our understanding of compstatin inhibition, a required and vital step towards testing human disease models in animals.
The diserine extension in compstatin aimed to exploit potential hydrogen-bonding interactions with C3. In ongoing and future work, an optimization of the backbone length and amino acid types of the extension, including combinations with Arg1 and Trp1 and incorporation of non-natural amino acids, will be systematically studied.
Supplementary Material
Acknowledgments
Most of the MD simulations were performed on Linux clusters of the Biophysics group at the University of Cyprus. Some MD simulations were performed at an IBM cluster at the Cyprus Institute and a Linux cluster of the BioMoDeL group at the University of California at Riverside. The peptide design calculations were performed in a Linux cluster of the CASL group at Princeton University. PT acknowledges support from a senior Fulbright Scholarship by the Cyprus Fulbright Commission. GA and PT acknowledge support from the Cyprus Research Promotion Foundation grant INFRASTRUCTURE/STRATEGIC/0308/31, “Cy-Tera: A Multi- Teraflop/s computing facility for Science and Technology in Cyprus”, that is co-funded by the European Regional Development fund. GA and PT also acknowledge a research grant from the University of Cyprus. MLBP acknowledges Government support under and awarded by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a. CAF acknowledges financial support from the National Institutes of Health (grant 5R01GM052032). DM acknowledges financial support from the Beckman Initiative for Macular Degeneration (grant 1112). GA and PT would like to thank Prof. Roland Dunbrack for the SCWRL4 program. The authors acknowledge useful discussions with Chris A. Kieslich.
References
- 1.Muller-Eberhard HJ. Complement-chemistry and pathways. In: Gallin JI, Goldstein IM, Synderman R, editors. Inflammation: Basic Principles and Clinical Correlates. Raven Press; New York, USA: 2002. pp. 21–54. [Google Scholar]
- 2.Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, Hauser MA, Rickman CB, Bok D, Hageman GS, Johnson LV. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahu A, Lambris JD. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology. 2000;49:133–148. doi: 10.1016/s0162-3109(00)80299-4. [DOI] [PubMed] [Google Scholar]
- 4.Janssen BJC, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 5.Ricklin D, Lambris JD. Compstatin: a complement inhibitor on its way to clinical application. Adv Exp Med Biol. 2008;632:273–292. doi: 10.1007/978-0-387-78952-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walport MJ. Complement. Second of two parts. N Engl J Med. 2001;344(15):1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 7.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. PNAS. 2006;103(52):19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gros P, Milder FJ, Janssen BJC. Complement driven by conformational changes. Nature Reviews Immunology. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 9.Sahu A, Ray BK, Lambris JD. Inhibition of human complement by a C3- binding peptide isolated from a phage-displayed random peptide library. J Immunol. 1996;157:884–891. [PubMed] [Google Scholar]
- 10.Morikis D, Lambris JD. Structure, dynamics, activity and function of compstatin and design of more potent analogs. In: Morikis D, Lambris JD, editors. Structural Biology of the Complement System. CRC Press/Taylor & Francis Group; Boca Raton, FL, USA: 2005. pp. 317–340. [Google Scholar]
- 11.Soulika AM, Holl MCH, Sfyroera G, Sahu A, Lambris JD. Compstatin inhibits complement activation by binding to the β-chain of complement factor 3. Mol Immunol. 2006;43:2023–2029. doi: 10.1016/j.molimm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Sahu A, Morikis D, Lambris JD. Compstatin, a peptide inhibitor of complement, exhibits species-specific binding to complement component C3. Mol Immunol. 2003;39:557–566. doi: 10.1016/s0161-5890(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 13.Bellows ML, Fung HK, Taylor MS, Floudas CA, López De Victoria A, Morikis D. New compstatin variants through two de novo protein design frameworks. Biophys J. 2010;98:2337–2346. doi: 10.1016/j.bpj.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López de Victoria A, Gorham RD, Jr, Bellows ML, Ling J, Lo DD, Floudas CA, Morikis D. A new generation of potent complement inhibitors of the compstatin family. Chem Biol Drug Des. 2011;77:31–440. doi: 10.1111/j.1747-0285.2011.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamamis P, Morikis D, Floudas CA, Archontis G. Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins: Structure, Function, and Bioinformatics. 2010;78:2655–2667. doi: 10.1002/prot.22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamamis P, Pierou P, Mytidou C, Morikis D, Floudas CA, Archontis G. Design of a Modified Mouse Protein with Ligand Binding Properties of its Human Analog by Molecular Dynamics Simulations: The Case of C3 Inhibition by Compstatin. Proteins: Structure, Function, and Bioinformatics. 2011;79:3166–3179. doi: 10.1002/prot.23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikis D, Assa-Munt N, Sahu A, Lambris JD. Solution structure of compstatin, a potent complement inhibitor. Protein Sci. 1998;7:619–627. doi: 10.1002/pro.5560070311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klepeis JL, Floudas CA, Morikis D, Lambris DJ. Predicting peptide structures using NMR data and deterministic global optimization. J Comput Chem. 1999;20:1354–1370. [Google Scholar]
- 19.Sahu A, Soulika A, Morikis D, Spruce L, Moore WT, Lambris JD. Binding kinetics, structure-activity relationship, and biotransformation of the complement inhibitor compstatin. J Immunol. 2000;165:2491–2499. doi: 10.4049/jimmunol.165.5.2491. [DOI] [PubMed] [Google Scholar]
- 20.Morikis D, Roy M, Sahu A, Troganis A, Jennings PA, Tsokos GC, Lambris JD. The structural basis of compstatin activity examined by structure-function-based design of peptide analogs and NMR. J Biol Chem. 2002;277:14942–14953. doi: 10.1074/jbc.M200021200. [DOI] [PubMed] [Google Scholar]
- 21.Morikis D, Lambris JD. Structural aspects and design of low-molecular-mass complement inhibitors. Biochem Soc Trans. 2002;30:1026–1036. doi: 10.1042/bst0301026. [DOI] [PubMed] [Google Scholar]
- 22.Mallik B, Lambris JD, Morikis D. Conformational interconversion in compstatin probed with molecular dynamics simulations. Proteins: Structure, Function, and Bioinformatics. 2003;52:130–141. doi: 10.1002/prot.10491. [DOI] [PubMed] [Google Scholar]
- 23.Tamamis P, Skourtis S, Morikis D, Lambris JD, Archontis G. Conformational analysis of compstatin analogues with molecular dynamics simulations in explicit water. J Mol Graph Model. 2007;26:571–580. doi: 10.1016/j.jmgm.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Klepeis J, Floudas CA, Morikis D, Tsokos GC, Argyropoulos E, Spruce L, Lambris JD. Integrated comp utational and experimental approach for lead optimization and design of compstatin variants with improved activity. J Am Chem Soc. 2003;125:8422–8423. doi: 10.1021/ja034846p. [DOI] [PubMed] [Google Scholar]
- 25.Soulika AM, Morikis D, Sarrias MR, Roy M, Spruce LA, Sahu A, Lambris JD. Studies of structure-activity relations of complement inhibitor compstatin. J Immunol. 2003;170:1881–1890. doi: 10.4049/jimmunol.171.4.1881. [DOI] [PubMed] [Google Scholar]
- 26.Morikis D, Soulika AM, Mallik B, Klepeis JL, Floudas CA, Lambris JD. Improvement of the anti-C3 activity of compstatin using rational and combinatorial approaches. Biochem Soc Trans. 2004;32:28–32. doi: 10.1042/bst0320028. [DOI] [PubMed] [Google Scholar]
- 27.Mallik B, Katragadda M, Spruce LA, Carafides C, Tsokos CG, Morikis D, Lambris JD. Design and NMR characterization of active analogues of compstatin containing non-natural amino acids. J Med Chem. 2005;48:274–286. doi: 10.1021/jm0495531. [DOI] [PubMed] [Google Scholar]
- 28.Furlong ST, Dutta AS, Coath MM, Gormley JJ, Hubbs SJ, Lloyd D, Mauger RC, Strimpler AM, Sylvester MA, Scott CW, Edwards PD. C3 activation is inhibited by analogs of compstatin but not by serine protease inhibitors or peptidyl alpha-ketoheterocycles. Immunopharmacology. 2000;48(2):199–212. doi: 10.1016/s0162-3109(00)00205-8. [DOI] [PubMed] [Google Scholar]
- 29.Mallik B, Morikis D. Development of a quasi-dynamic pharmacophore model for anti-complement peptide analogues. J Am Chem Soc. 2005;127:10967–10976. doi: 10.1021/ja051004c. [DOI] [PubMed] [Google Scholar]
- 30.Chiu TL, Mulakala C, Lambris JD, Kaznessis YN. Development of a new pharmacophore model that discriminates active compstatin analogs. Chem Biol Drug Des. 2008;72:249–256. doi: 10.1111/j.1747-0285.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morikis D, Lambris JD. Structural aspects and design of low-molecular-mass complement inhibitors. Biochem Soc Trans. 2002;30:1026–1036. doi: 10.1042/bst0301026. [DOI] [PubMed] [Google Scholar]
- 32.Klepeis JL, Floudas CA, Morikis D, Tsokos CG, Lambris JD. Design of peptide analogs with improved activity using a de novo protein design approach. Ind Eng Chem Res. 2004;43:3817–3826. [Google Scholar]
- 33.Holland MCH, Morikis D, Lambris JD. Synthetic small molecule complement inhibitors. Curr Opin Invest Dr. 2004;5:1164–1173. [PubMed] [Google Scholar]
- 34.Morikis D, Floudas CA, Lambris JD. Structure-based integrative computational and experimental approach for the optimization of drug design. In: Sunderam VS, van Albada GD, Sloot PMA, Dongarra JJ, editors. ICCS Lecture Notes in Computer Science: Computational Science. Springer-Verlag; Berlin-Heidelberg, Atlanta, GA, USA: 2005. pp. 680–688. [Google Scholar]
- 35.Morikis D, Mallik B, Zhang L. Biophysical and bioengineering methods for the study of the complement system at atomic resolution. WSEAS Trans Biol Biomed. 2006;6:408–413. [Google Scholar]
- 36.Qu H, Magotti P, Ricklin D, Wu EL, Kourtzelis I, Wu Y-Q, Kaznessis YN, Lambris JD. Novel analogues of the therapeutic complement inhibitor compstatin with significantly improved affinity and potency. Mol Immunol. 2011;48:481–489. doi: 10.1016/j.molimm.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamamis P, Archontis G. Solution conformational properties of the potential therapeutic complement inhibitor compstatin and selected analogs, investigated by MD simulations in implicit- and explicit-water. Biomedical Engineering Research. 1:14–21. [Google Scholar]
- 38.Rajgaria R, McAllister SR, Floudas CA. A Novel High Resolution Cα-Cα Distance Dependent Force Field Based on a High Quality Decoy Set. Proteins: Structure, Function, and Bioinformatics. 2006;65:726–741. doi: 10.1002/prot.21149. [DOI] [PubMed] [Google Scholar]
- 39.Rajgaria R, McAllister SR, Floudas CA. Distance Dependent Centroid to Centroid Force Fields Using High Resolution Decoys. Proteins. 2008;70:950–970. doi: 10.1002/prot.21561. [DOI] [PubMed] [Google Scholar]
- 40.Fung HK, Taylor MS, Floudas CA. Novel Formulations for the Sequence Selection Problem in De Novo Protein Design with Flexible Templates. Optim Methods & Software. 2007;22:51–71. [Google Scholar]
- 41.Fung HK, Floudas CA, Taylor MS, Zhang L, Morikis D. Toward Full Sequence De Novo Protein Design with Flexible Templates for Human Beta- Defensin-2. Biophys J. 2008;94:584–599. doi: 10.1529/biophysj.107.110627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellows ML, Taylor MS, Cole PA, Shen L, Siliciano RF, Fung HK, Floudas CA. Discovery of entry inhibitors for HIV-1 via a new de novo protein design framework. Biophys J. 2010;99:3445–3453. doi: 10.1016/j.bpj.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee MR, Baker D, Kollman PA. 2.1 and 1.8 Å Cα RMSD Structure Predictions on Two Small Proteins, HP-36 and S15. J Am Chem Soc. 2001;123:1040–1046. doi: 10.1021/ja003150i. [DOI] [PubMed] [Google Scholar]
- 44.Rohl CA, Baker D. De Novo Determination of Protein Backbone Structure from Residual Dipolar Couplings Using Rosetta. J Am Chem Soc. 2002;124:2723–2729. doi: 10.1021/ja016880e. [DOI] [PubMed] [Google Scholar]
- 45.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein Structure Prediction Using Rosetta. Methods in Enzymology. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 46.DiMaggio PA, McAllister SR, Floudas CA, Feng XJ, Rabinowitz JD, Rabitz HA. Biclustering via optimal re-ordering of data matrices in systems biology: rigorous methods and comparative studies. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiMaggio PA, McAllister SR, Floudas CA, Feng XJ, Rabinowitz JD, Rabitz HA. A network flow model for biclustering via optimal re-ordering of data matrices. J Global Optimization. 2010;47(3):343–354. [Google Scholar]
- 48.Daily MD, Masica D, Sivasubramanian A, Somarouthu S, Gray JJ. CAPRI Rounds 3–5 Reveal Promising Successes and Future Challenges for Rosetta- Dock. Proteins: Structure, Function, and Bioinformatics. 2005;60:181–186. doi: 10.1002/prot.20555. [DOI] [PubMed] [Google Scholar]
- 49.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-Protein Docking with Simultaneous Optimization of Rigid-body Displacement and Side-chain Conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 50.Gray JJ, Moughon SE, Kortemme T, Schueler-Furman O, Misura KMS, Morozov AV, Baker D. Protein-Protein Docking Predictions for the CAPRI Experiment. Proteins: Structure, Function, and Genetics. 2003;52:118–122. doi: 10.1002/prot.10384. [DOI] [PubMed] [Google Scholar]
- 51.Mackerell AD, et al. An all-atom empirical potential for molecular modeling and dynamics study of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 52.Mackerell AD, Jr, Feig M, Brooks CLIII. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2003;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 53.Macias AT, Mackerell AD., Jr CH/pi interactions involving aromatic amino acids: refinement of the CHARMM tryptophan force field. J Comput Chem. 2005;26:1452–1463. doi: 10.1002/jcc.20281. [DOI] [PubMed] [Google Scholar]
- 54.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Current Protocols in Bioinformatics. Supplement. Vol. 15. John Wiley & Sons, Inc; 2006. Comparative protein structure modeling with MODELLER; pp. 5.6.1–5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A. Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
- 58.Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov. 2000;18:113–135. [Google Scholar]
- 59.Im W, Lee MS, Brooks CLIII. Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 60.Chen J, Im W, Brooks CLIII. Balancing solvation and intramolecular interactions: toward a consistent generalized born force field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gohlke H, Case D. Converging Free Energy Estimates: MM-PB(GB)SA Studies on the Protein–Protein Complex Ras–Raf. J Comput Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 62.Page CS, Bates PA. Can MM-PBSA Calculations Predict the Specificities of Protein Kinase Inhibitors? J Comput Chem. 2006;27:1990–2007. doi: 10.1002/jcc.20534. [DOI] [PubMed] [Google Scholar]
- 63.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Bioph Biom. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 64.Zoete V, Irving MB, Michielin O. MM-GBSA binding free energy decomposition and T cell receptor engineering. J Mol Recognit. 2010;23:142–152. doi: 10.1002/jmr.1005. [DOI] [PubMed] [Google Scholar]
- 65.Singh N, Warshel A. Absolute binding free energy calculations: On the accuracy of computational scoring of protein–ligand interactions. Proteins: Structure, Function, and Bioinformatics. 2010;78:1705–1723. doi: 10.1002/prot.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lazaridis T, Massunov A, Gandolfo F. Contributions to the Binding Free Energy of Ligands to Avidin and Streptavidin. Proteins: Structure, Function and Genetics. 2002;47:194–198. doi: 10.1002/prot.10086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.