Abstract
Dicer is a multi-domain enzyme that generates small RNAs for gene silencing in eukaryotes. Current understanding of Dicer structure is restricted to simple forms of the enzyme, while that of the large and complex Dicer, widespread in eukarya, is unknown. Here, we describe a novel domain localization strategy developed to determine the structure of human Dicer by electron microscopy. A rearrangement of the nuclease core, compared to the archetypal Giardia Dicer, explains how metazoan Dicers generate 21–23 nucleotide products. The helicase domains form a clamp-like structure adjacent to the RNase III active site, facilitating recognition of pre-miRNA loops or translocation on long dsRNAs. Drosophila Dicer-2 displays similar features, revealing that the three-dimensional architecture is conserved. These results illuminate the structural basis for small RNA production in eukaryotes and provide a versatile new tool for determining structures of large molecular machines.
Introduction
Small regulatory RNAs, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs), are involved in a myriad of biological processes ranging from viral defense to brain development1. The RNase III enzyme, Dicer, plays a fundamental role in small RNA biogenesis by cleaving dsRNA substrates into functional small RNAs of a discrete size (typically 21–23 nt)2,3. Small RNA products of Dicer are incorporated into large multiprotein complexes termed RNA-induced silencing complexes (RISC). RISC and RISC-like complexes use the small RNAs as guides for the sequence-specific silencing of cognate genes through mRNA degradation4, translational repression5 and heterochromatin formation6.
In living cells, Dicer enzymes often function within larger protein complexes required for initiation of RNA silencing pathways. In Tetrahymena, Dcr2 is physically coupled to the RNA-dependent RNA polymerase Rdr1 for the biogenesis of siRNAs7. In Drosophila, Dcr-2 interacts with the protein R2D2 and facilitates siRNA loading into Ago28. In C. elegans, Dcr-1 associates with more than 20 other protein factors and exists in at least two different functional complexes that are distinctly responsible for initiating the endo- and exo- arms of the RNA interference (RNAi) pathway9,10. In addition to generating small RNA duplexes, Dicer itself functions as a molecular scaffold in all of these complexes.
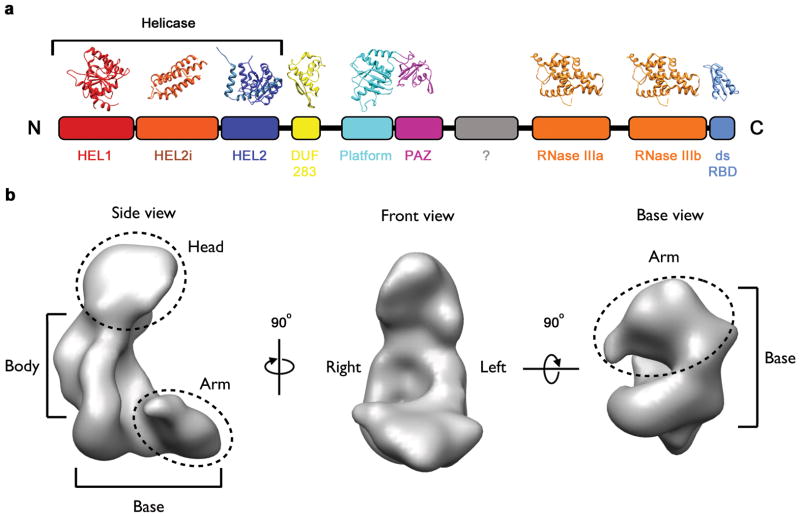
Central to the activity of Dicer is its ability to recognize dsRNA, to generate precisely sized products and, in some cases, to translocate along a long substrate. Structural and mechanistic insights into these activities have proved difficult because the metazoan Dicers are large and complicated proteins, recalcitrant to crystallization. Previous studies have primarily focused on simple forms of the enzyme, either Dicers from lower eukaryotes or isolated domains from higher eukaryotic Dicers11–15. Consequently, although structures of many of the individual domain components of Dicer have been established (Fig. 1a), the overall architecture of the enzyme remains unknown.
Fig. 1. The conserved domain structure of metazoan Dicer.
a, Schematic of the 2D domain structure of human Dicer with crystal structures homologous to each module. b, The EM map of Dicer (EMD-1646) shown in three orientations.
To date, two electron microscopy (EM) reconstructions of human Dicer have been reported16,17. Both described an “L” shaped particle composed of several morphologically discrete regions (Fig. 1b), and a working model for the domain architecture of Dicer has emerged17–22. However, this model has not been examined rigorously and seemingly contradicts several observed biochemical properties of the enzyme23–25. The difficulty in testing the model stems from technical challenges associated with localizing individual domains in EM maps—this is particularly difficult in relatively small, asymmetric particles like Dicer, which is less than 250 kDa. To overcome this issue we devised a versatile, site-specific tagging strategy compatible with single-particle analysis, allowing us to establish the first experimentally validated structure of this essential enzyme. Our structure differs significantly from the model proposed previously17, but is well aligned with known biochemical properties of Dicer.
Results
RNA recognition by PAZ occurs in the head of the Dicer
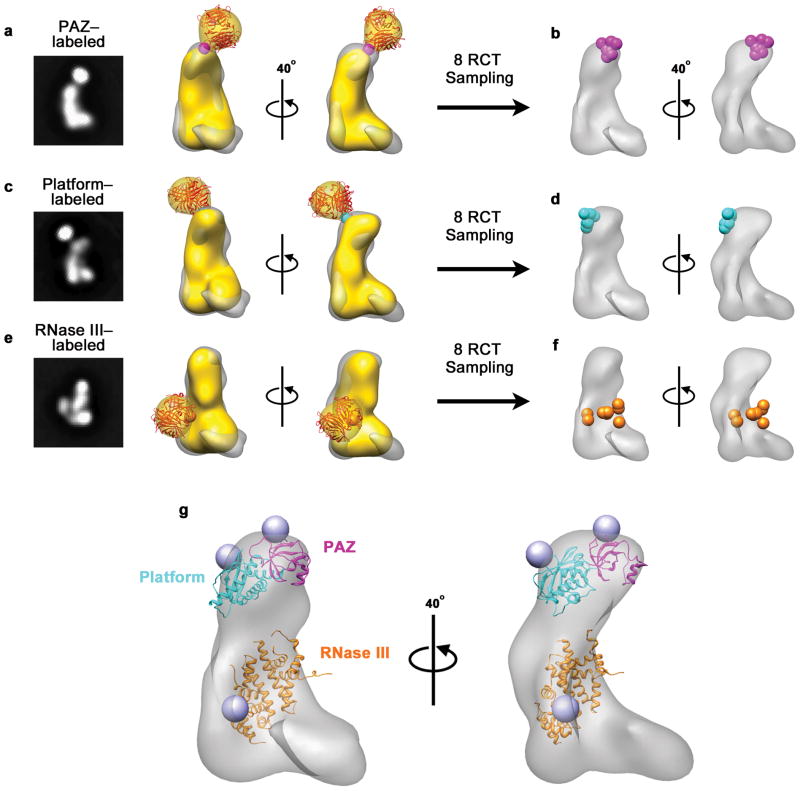
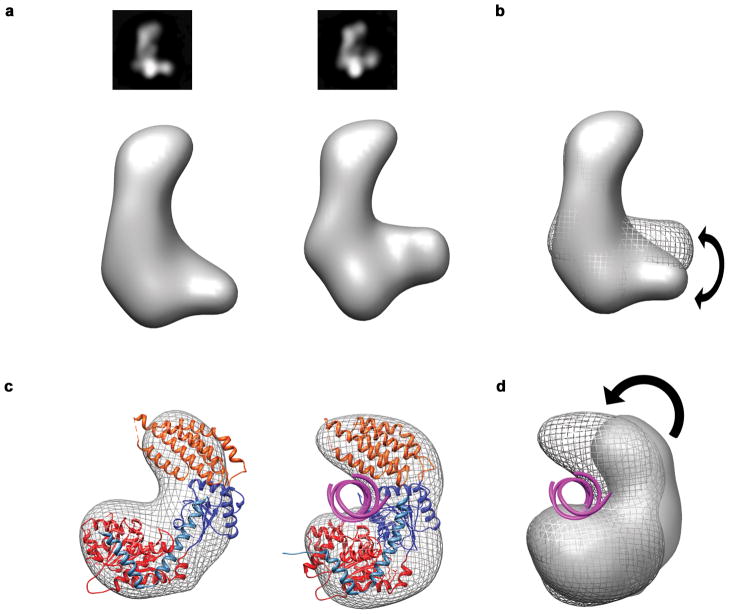
Dicer generates small RNAs by cleaving dsRNAs ~22 base pairs from their open helical ends25–28. DsRNA end recognition is directly mediated by an RNA-binding domain called PAZ26,29. We identified the position of the PAZ domain in the EM map of human Dicer by inserting the 15-amino acid AviTag sequence30, a substrate for biotin-protein ligase, into a surface loop in the PAZ domain (between residues K916 and E917) and, after subsequent biotinylation, tagging with a monovalent form of streptavidin31. Tagged proteins were purified and visualized by negative stain EM. Extra density with the size and shape of streptavidin extending from the head of the L was observed in many unsupervised 2D class averages (Fig. 2a and Supplementary Fig. 1). Eight independent 3D reconstructions of the tagged Dicer were generated by the random conical tilt (RCT) method and the L-shaped portion of each reconstruction was aligned with the refined Dicer structure. The point of streptavidin attachment (estimated as the central point in the bridging region between Dicer and streptavidin densities) was then mapped onto the refined structure. The estimated attachment points lie within a 10 Å-radius located at the front of the head region (Fig. 2d), revealing that the PAZ domain—and thus the site of dsRNA end recognition—is in the very top of the molecule, in the front of Dicer’s head.
Fig. 2. Mapping the nuclease core of human Dicer.
a–c, 2D class averages of Dicer labeled with streptavidin in the PAZ, platform or RNase IIIb domain (left) and corresponding RCT reconstruction (yellow) superimposed on the unlabeled Dicer map (gray). Streptavidin (red) is shown docked into the additional density. Estimated streptavidin attachment sites are indicated with a sphere. d–f, Estimated streptavidin attachment sites from 8 RCT reconstructions on the refined Dicer map g, Crystal structures of the PAZ-Platform and RNase III modules docked into the EM map of Dicer based on streptavidin labeling results. Blue spheres indicate positions of labeled sites in the crystal structures.
The platform domain is tightly associated with the PAZ domain
Upon recognition of dsRNA by PAZ, Dicer then cleaves the substrate ~22 nucleotides from the open helical end. In the simple Dicer enzyme from the protozoan Giardia lamblia, a “platform” domain separates PAZ from the RNase III catalytic site by a distance of ~70 Å, thereby providing the structural basis for production of RNAs 25–27 nt in length32. Human Dicer has been proposed to use a similar measuring mechanism26—although its products are 4 nt shorter. We identified the position of the platform domain in the human Dicer EM map by inserting the AviTag between residues D886 and S887. For platform-labeled particles, streptavidin density extended from the back of the head in 2D class averages (Fig. 2b and Supplementary Fig. 1), and 3D reconstructions from eight class averages mapped the point of attachment to a 10 Å radius region in the back of the Dicer head (Fig. 2e). The position of platform suggests that both PAZ and Platform are tightly associated as in the case of Giardia Dicer.
dsRNA cleavage by the RNase III domains occurs in the body
Zhang and coworkers first proposed that a fixed spacing between the PAZ and RNase III domains in human Dicer could lead to cleavage of dsRNA ~22 nt from the open helical end26. To test this model directly, we tagged the RNase IIIb domain with streptavidin (residues N1780-E1800 were replaced with the AviTag). Streptavidin density was apparent in 2D class averages and extended from the body of the L, approximately 55 Å from the PAZ domain (Fig. 2c and Supplementary Fig. 1). The tagged RNase IIIb loop appeared to be more mobile than the loops labeled in the PAZ and platform domains, as the estimated points of streptavidin attachment for eight different RCT models lie within a 20 Å radius, with the labeled loop extending out from the right side of the body (Fig. 2f). Consistent with this arrangement, a reconstruction of Dicer lacking the C-terminal dsRBD, which lies adjacent to the tagged loop in the RNase IIIb domain12, is missing density from the right side of the body (Supplementary Fig. 2).
The human Dicer nuclease core is rearranged relative to Giardia Dicer
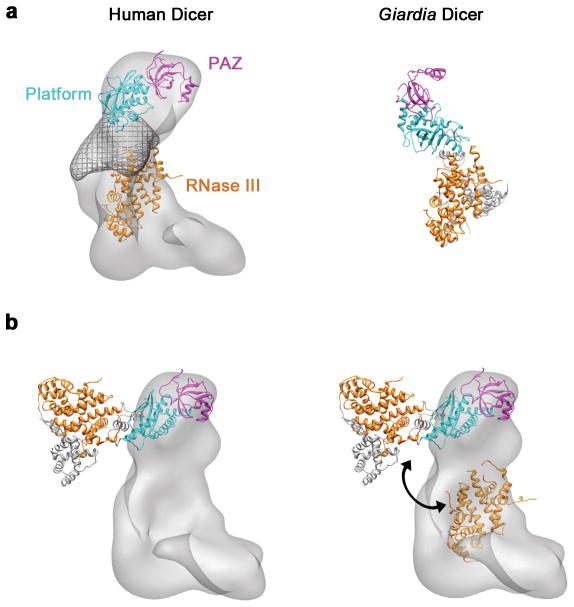
Based on the positions of the streptavidin tags, we docked the platform-PAZ module and RNase III domains from the Giardia Dicer crystal structure into the EM map of human Dicer (Fig. 2g). While the head easily accommodated the platform-PAZ module, the RNase III domains of Giardia Dicer could not be fit into the body of the EM map without a major rearrangement relative to the platform (Fig. 3). Rearranging these domains established a 3D model for the architecture of the human Dicer nuclease core. Conceptually, the core of the human enzyme is similar to Giardia Dicer—both have PAZ and RNase III domains separated by a specific distance. However, relative to their PAZ domains, the RNase III active sites of human and Giardia Dicer are offset from each other by a rotation of roughly 120 around the long axis of the human enzyme. Moreover, in human Dicer, the platform domain does not lie directly between PAZ and RNase III. Instead, a structurally undefined “ruler domain” physically separates the two functional domains (Fig. 3a and Supplementary Fig. 3). These large-scale differences in 3D architecture likely reflect the fact that small RNAs in humans are 4 nt (one third of a dsRNA helical turn) shorter than in Giardia—the human enzyme must attack a completely different face of its dsRNA substrates relative to their helical ends.
Fig. 3. Comparison of the human Dicer and Giardia Dicer nuclease cores.
a, Modeled positions of the PAZ, platform and RNase III domains in human Dicer (left) compared to the Giardia Dicer crystal structure (right). The proposed Ruler domain in human Dicer, estimated by segmentation of the refined map, is shown as a wire mesh. b, The full-length crystal structure of Giardia Dicer docked onto the positions of the PAZ and platform domains identified in the human Dicer EM map (left). The RNase III domains modeled in to the human Dicer EM map, compared to the RNase III domains in the Giardia Dicer crystal structure (right). Double-headed arrow indicates the rearrangement required to align the RNase III domains in the two models.
The helicase forms a clamp-shaped structure at the base of Dicer
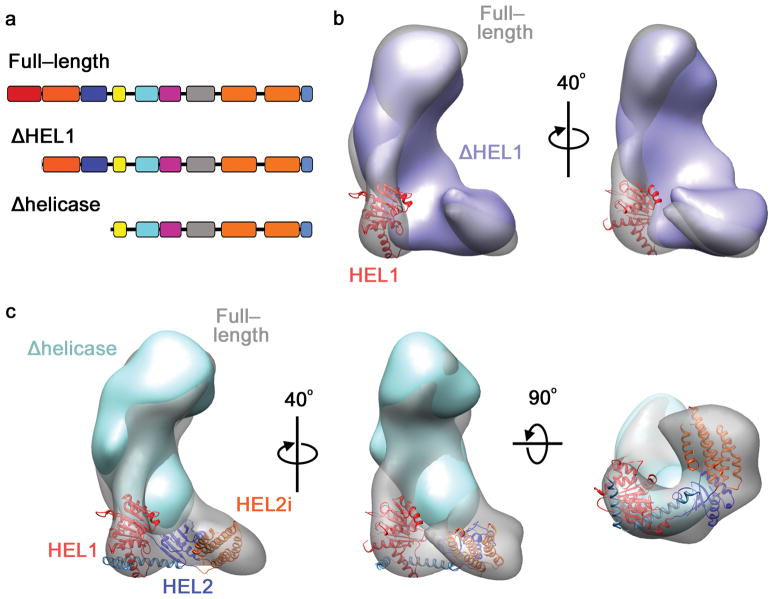
In addition to differences in product length, human Dicer further differs from Giardia in the complexity of its accessory domains. Human Dicer contains an N-terminal helicase, which itself is composed of three predicted globular domains (HEL1, HEL2i, and HEL2 domains) (Fig. 1a). Previously, after comparing 2D class averages of full-length and helicase-deleted Dicer proteins, Wang et al. proposed that the helicase resides within the arm, or “base-branch”, of the L17. However, we note that the volume of the arm is too small to accommodate all three globular domains of the Dicer helicase. Indeed, the crystal structure of DDX3X, which was used previously as a model for the Dicer helicase17, is composed of only two globular domains and lacks any structure analogous to the HEL2i domain that is observed in the Dicer primary sequence. To probe this issue, we generated 3D reconstructions of a truncated Dicer in which the three helicase domains (residues 1–604) were deleted (Fig. 4a). RCT reconstruction of Δhelicase-Dicer produced an oblong structure with dimensions similar to the head and body portion of Dicer. Likewise, a variety of projection-matching refinements of Δhelicase particles consistently produced structures resembling the head and body (Supplementary Fig. 4). We conclude that the deleted helicase forms not just the arm, but the entire base of full-length Dicer.
Fig. 4. The helicase forms a clamp-like structure in the base.
a, Schematic of full-length, ΔHEL1 and Δhelicase Dicers. b, Reconstruction of ΔHEL1 Dicer (blue) overlaid on the full length Dicer (transparent). HEL1 domain of the RIG-I helicase (red) is modeled into the major difference density. c, 3D reconstruction of Δhelicase Dicer (cyan) overlaid on the full length Dicer. The RIG-I helicase crystal structure is modeled into the base of the full-length map.
Sequence homology suggests the Dicer helicase belongs to the RIG-I family of RNA helicases33. Indeed, the core helicase domains of RIG-I form a “C” shape similar to the base of the Dicer L (Supplementary Fig. 5)34. To establish the orientation of the helicase domains, we determined the structure of a truncated Dicer protein with only the HEL1 domain (residues 1–211) deleted (Fig. 4a). The ΔHEL1 reconstruction lacks density in the bottom corner of the L, corresponding to a mass of approximately 25 kDa, in agreement with the mass of the deleted HEL1 domain (Fig. 4b).
Based on the position of the HEL1 domain, we docked the crystal structure of duck RIG-I helicase34 into the base of the L (Fig. 4c). Importantly, the RIG-I helicase and Δhelicase-Dicer reconstruction together accounted for the total density of the full-length Dicer map. The interior channel of the helicase, which constitutes the dsRNA binding site in RIG-I, is aligned with the central channel that runs up the body of the L (Supplementary Fig. 6). Thus, a single continuous channel runs through clamp of the helicase, past the RNase III active site, and ends with the RNA-binding pocket of the PAZ domain (Supplementary Fig. 6). We propose that this channel is the major surface used by the enzyme for processing dsRNA.
The Dicer helicase adopts conformations reminiscent of the RIG-I helicase
Crystal structures of RIG-I suggest that, upon dsRNA binding, the helicase domains shift to clamp down on dsRNA substrates34. We looked for similar conformational changes in the helicase of human Dicer, by examining samples of Dicer negatively stained in the presence of a dsRNA substrate. Despite the challenge in detecting nucleic acids in negative stain EM, we observed two discrete conformations of the helicase relative to the body and head in RCT reconstructions (Fig. 5a). Subjecting these reconstructions to multi-model projection matching refinement confirmed the presence of at least two discrete conformations in the base. The two conformations resembled the apo and dsRNA-bound crystal structures of RIG-I, suggesting the Dicer helicase may shift to clamp down on dsRNA in a manner structurally analogous to dsRNA binding by RIG-I (Fig. 5).
Fig. 5. Conformational states of the Dicer helicase.
a, 2D class averages and corresponding RCT reconstructions of two distinct conformations of human Dicer observed when stained the presence of a dsRNA substrate. b, Alignment of the RCT maps showing the conformational differences between the two reconstructions. c, Docking RIG-I into the base of the L reveals that the two observed conformations of the Dicer helicase resemble RIG-I in its apo (PDB code: 4A2P) and dsRNA-bound bound forms (PDB code: 4A36). d, Overlay of the EM density maps of each helicase conformation shows a large scale rearrangement similar to that observed in the RIG-I crystal structures.
Dicer architecture is conserved between human and Drosophila
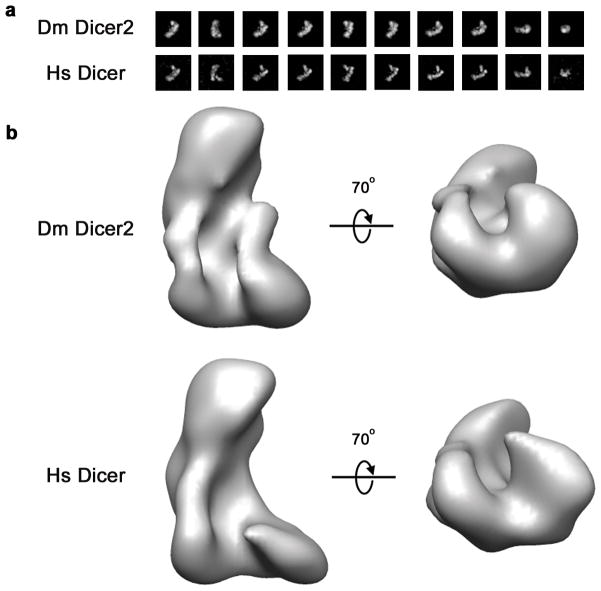
The function of the Dicer helicase has been enigmatic—it has been suggested to contribute to substrate binding in human Dicer35,36, facilitate pre-miRNA recognition in Drosophila Dcr-124, and catalyze translocation on long dsRNA substrates in C. elegans Dcr-1 and Drosophila Dcr-223,25. Considering the divergent functions reported for the Dicer helicase, we wondered how the architecture of Dicer varies between different species. To explore this issue, we extended our EM analysis to a sample of Drosophila Dcr-2. Dcr-2 is one of the best studied Dicer enzymes and differs from human Dicer in that it requires ATP to cleave dsRNA and is believed to couple ATP hydrolysis to translocation on long dsRNA23,37. 2D class averages of Dcr-2 contained many L-shaped particles similar to those observed in class averages of human Dicer (Fig. 6a). Furthermore, projection-matching using Dcr-2 particles led to an L-shaped reconstruction with dimensions strikingly similar to those of the human enzyme (Fig. 6b). We therefore conclude that, despite functional clear differences among various forms of the enzyme, the overall three-dimensional architecture of Dicer is well conserved.
Fig. 6. Comparison of Human and Drosophila Dicer structures.
a, Class averages of corresponding views of Drosophila (Dm) Dicer2 and Human (Hs) Dicer particles. b, Reconstructions of Dm Dicer2 and Hs Dicer reveal that the two proteins share a common overall shape and many 3D features.
Discussion
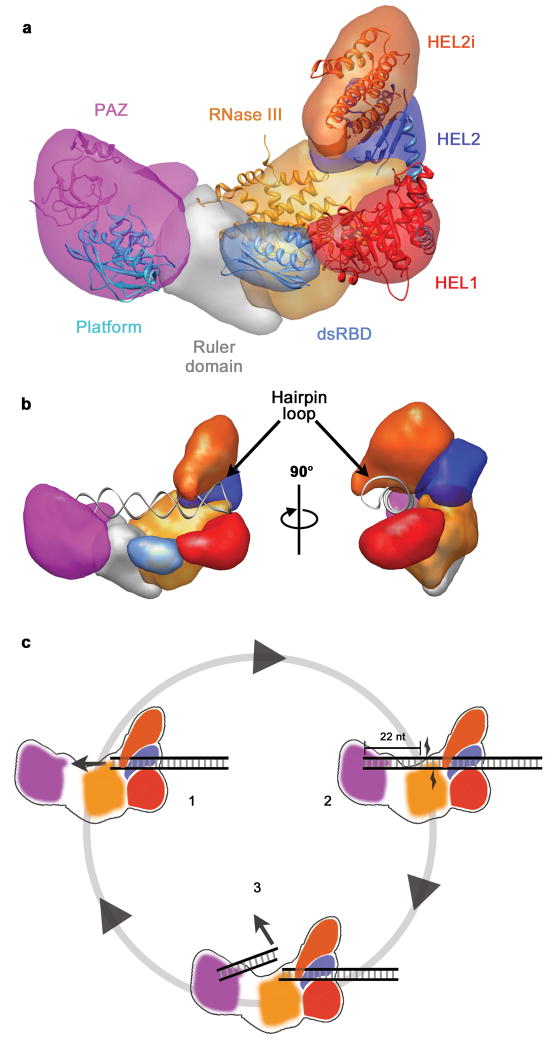
The structural analysis of Dicer presented here allows reconciliation of the seemly disparate functions of the helicase observed in different Dicer homologs. Adjacent to the RNase III domains, the helicase is positioned to bind the stem loops of pre-miRNAs (Fig. 7a, b). This explains how the helicase contributes to pre-miRNA binding in human Dicer36 and selective processing of pre-miRNAs in Drosophila Dcr-124. For processive Dicers23,25 the helicase could use ATP hydrolysis to translocate dsRNA into the nuclease core of the enzyme. The helicase is positioned to remain bound to long dsRNAs after cleavage and formation of each siRNA product, providing a structural basis for processivity on long substrates (Fig. 7c).
Fig. 7. Architecture and Mechanism of Dicer.
a, Segmented map of human Dicer with crystal structures of homologous domains docked. b, Model for pre-miRNA recognition. A pre-miRNA hairpin is modeled into the proposed binding channel of Dicer with the stem loop fit in the RNA-binding cleft of the helicase. c, Schematic for processive dicing: (1) The helicase translocates dsRNA into the nuclease core. (2) The PAZ domain (purple) recognizes the dsRNA end, positioning RNase III (orange) for cleavage. (3) The siRNA product is released while the dsRNA substrate remains bound to the helicase.
The previously proposed structural model for Dicer suggested the opposite orientation for the nuclease core—with the PAZ domain in the body, adjacent to the helicase, and RNase III domains in the head17–20. This model implies that the end of dsRNA substrates bind within the body and extend out past the head, never directly interacting with the helicase. It has thus been difficult to rationalize how the helicase could facilitate processivity or substrate binding if it is positioned on the opposite side of the molecule from where the dsRNA feeds in. We have now experimentally determined the 3D positions of the PAZ, RNase III and helicase domains, allowing us to exclude this model, and cohesively integrate a structural view of the enzyme with its known biochemical functions. Moreover, Dicer enzymes typically function as central components in larger complexes required for initiation of diverse RNA silencing pathways7–10. Therefore, establishing the correct structure of Dicer is necessary for understanding the mechanisms of the multi-protein assemblies required for a wide range of gene silencing processes.
Methods
Generation of Avitag Dicer constructs and truncated Dicer constructs
DNA encoding the Avitag sequence (LNDILEAQKIEWHEG) was cloned into specific positions corresponding to surface loops in a cDNA clone of human Dicer (NM_030621) in the plasmid pFastBac HT A (Invitrogen) within the PAZ (between residues K916 and E917), Platform (between residues D886 and S887) and RNase IIIb domains (replacing residues N1780-E1800).
Deletion mutants of human Dicer were amplified by PCR using the full-length cDNA clone as a template. The ΔHEL1 Dicer lacks amino acids 1 – 211, while the ΔHelicase constructs lacks amino acids 1 – 604. The cloning details are described in the Supplementary Methods.
Dicer expression and purification
All the Dicer constructs were produced and purified from Sf9 cells using the Bac-to-Bac system (Invitrogen) as described38. Recombinant proteins were purified using a His6 tag, which was cleaved by treatment with TEV protease. The samples were then passed through a 5mL HisTrap FF column, with the flow through being collected. Samples were then concentrated and applied to size exclusion chromatography. Further details can be found in the Supplementary Methods.
Biotinylation and streptavadin labeling of Avitag Dicer
Partially purified AviTag Dicers were biotinylated after the His-Trap FF step (see above). Flow though from the His-Trap column was concentrated and exchanged into a buffer containing 250 mM potassium glutamate with 25 mM Tris, pH 8.0—biotin ligase (BirA) is inhibited at high concentrations of sodium chloride. Biotinylation reactions were carried out in a reaction volume of 0.5 to 1 ml. One tenth the volume of a 10X buffer containing 100 mM ATP, 100 mM magnesium acetate and 500 μM d-biotin, pH 8 was added to the sample together with the purified BirA enzyme (20uL of 5mg/mL). Biotinylation reactions were incubated for 1 hour at 37°C.
Following the biotinylation reaction, the samples were dialyzed extensively (against a low imidazol buffer) to remove excess biotin. The biotinylated Dicers were then bound to purified monovalent streptavidin31. Biotinylation efficiency was typically about 60%. The streptavidin-tagged Dicer samples were purified from untagged proteins by applying the sample to a 1 mL His-Trap FF column. As only the streptavidin contained a His6 tag, non-biotinylated Dicers did not bind the column. The retained samples were eluted by increasing imidazole concentration and then concentrated and passed through a HiLoad 16/60 Superdex 200 column (GE Healthcare Life Sciences) to separate the Dicer-Streptavidin complex from free streptavidin.
Negative staining and electron microscopy
Samples were adhered to C-flat grids coated with a layer of thin carbon. Heavy metal uranyl solution was used to embed and fix the sample. All data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 keV, using a dose of ~20 e-/Å2 and a nominal defocus range of −1 to −3 μm; Images were automatically collected during multiple sessions at a nominal magnification of between 50,000× and 62,000× at a pixel size at the specimen level of 0.151 nm and 0.131 nm respectively. Images were recorded using either a Tietz F415 4 × 4 K pixel CCD camera (15 μm pixel) or Gatan 4 × 4 K pixel CCD camera utilizing the Leginon data collection software39. Random Conical Tilt (RCT) experiments were carried out using the RCT node of Leginon40, with image pairs taken at 0 and (−) 50 degrees. Additional details are available in the Supplementary Methods.
Image processing and model reconstructions
Image processing and model reconstructions were performed using the Appion software package41. Particles were extracted from the raw micrographs using a reference-free method42. For alignments and classifications, a combination of Xmipp 43 and Spider44 protocols were used. The 3D reconstruction was performed using a combination of SPIDER and EMAN reconstruction packages44,45. Additional details are available in the Supplementary Methods.
Model fitting
All model fitting was done using Chimera visualization software46. Map segmentations was performed using Segger47 available in Chimera. Additional details on modeling are available in the Supplementary Methods.
Supplementary Material
Acknowledgments
We are grateful to D.K. Simanshu, Y. Tian and D.J. Patel for sharing the human Dicer PAZ-platform structure prior to publication. EM imaging and reconstruction was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the P41 program of the National Center for Research Resources (RR017573) and the National Institute of General Medical Sciences (GM103310). This work was also supported by the NIH grant R01 GM086701 to I.J.M. P.W.L is a pre-doctoral fellow of the American Heart Association. I.J.M. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Author Contributions. C.S.P., B.C. and I.J.M. conceived of the project. P.W.L, K.G. and N.D. prepared the samples. P.W.L. performed data collection. P.W.L., C.S.P., B.C. and I.J.M. analyzed the data. P.W.L. and I.J.M. wrote the manuscript.
References
- 1.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 5.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 6.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 7.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–10. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 9.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–54. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 10.Thivierge C, et al. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol. 2011;19:90–7. doi: 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci U S A. 2008;105:2391–6. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita D, et al. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106–20. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 14.Qin H, et al. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA. 2010;16:474–81. doi: 10.1261/rna.1965310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg DE, Nakanishi K, Patel DJ, Bartel DP. The inside-out mechanism of dicers from budding yeasts. Cell. 2011;146:262–76. doi: 10.1016/j.cell.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–32. doi: 10.1016/j.str.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HW, et al. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–53. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43:110–21. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr Opin Struct Biol. 2010;20:90–7. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartig JV, Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2010;39:3836–51. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snead NM, Rossi JJ. Biogenesis and function of endogenous and exogenous siRNAs. Wiley Interdiscip Rev RNA. 2010;1:117–31. doi: 10.1002/wrna.14. [DOI] [PubMed] [Google Scholar]
- 23.Cenik ES, et al. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42:172–84. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 25.Welker NC, et al. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–99. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–21. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–8. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- 29.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–22. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–43. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 31.Howarth M, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–73. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–40. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 33.Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol. 2009;9:85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Soifer HS, et al. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–22. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–43. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–7. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Yoshioka C, et al. Automation of random conical tilt and orthogonal tilt data collection using feature-based correlation. J Struct Biol. 2007;159:335–46. doi: 10.1016/j.jsb.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–13. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres SH, et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol. 2005;348:139–49. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 44.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 45.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–38. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.