Abstract
Background
Severe trauma, including burns, triggers a systemic response that significantly impacts on the liver, which plays a key role in the metabolic and immune responses aimed at restoring homeostasis. While many of these changes are likely regulated at the gene expression level, there is a need to better understand the dynamics and expression patterns of burn injury-induced genes in order to identify potential regulatory targets in the liver. Herein we characterized the response within the first 24 h in a standard animal model of burn injury using a time series of microarray gene expression data.
Methods
Rats were subjected to a full thickness dorsal scald burn injury covering 20% of their total body surface area while under general anesthesia. Animals were saline resuscitated and sacrificed at defined time points (0, 2, 4, 8, 16, and 24 h). Liver tissues were explanted and analyzed for their gene expression profiles using microarray technology. Sham controls consisted of animals handled similarly but not burned. After identifying differentially expressed probesets between sham and burn conditions over time, the concatenated data sets corresponding to these differentially expressed probesets in burn and sham groups were combined and analyzed using a “consensus clustering” approach.
Results
The clustering method of expression data identified 621 burn-responsive probesets in 4 different co-expressed clusters. Functional characterization revealed that these 4 clusters are mainly associated with pro-inflammatory response, anti-inflammatory response, lipid biosynthesis, and insulin-regulated metabolism. Cluster 1 pro-inflammatory response is rapidly up-regulated (within the first 2 h) following burn injury, while Cluster 2 anti-inflammatory response is activated later on (around 8 h post burn). Cluster 3 lipid biosynthesis is downregulated rapidly following burn, possibly indicating a shift in the utilization of energy sources to produce acute phase proteins which serve the anti-inflammatory response. Cluster 4 insulin-regulated metabolism was down-regulated late in the observation window (around 16 h postburn), which suggests a potential mechanism to explain the onset of hypermetabolism, a delayed but well-known response that is characteristic of severe burns and trauma with potential adverse outcome.
Conclusions
Simultaneous analysis and comparison of gene expression profiles for both burn and sham control groups provided a more accurate estimation of the activation time, expression patterns, and characteristics of a certain burn-induced response based on which the cause-effect relationship among responses were revealed.
Keywords: Burn, gene expression, microarray, inflammation, liver
Introduction
Thermal injury, one of the most severe forms of trauma, triggers a number of physiological responses including local and systemic inflammation, hyper-metabolism, immune-suppression, and eventually organ dysfunction (1). Clinical studies have shown that an uncontrolled and prolonged action of inflammatory cytokines, which is evidenced by a sustained release of acute phase proteins, may contribute to detrimental complications (2). Liver is an important player in the modulation of the inflammatory response since it largely controls circulating levels of metabolites and the production of acute phase proteins. It is known that inflammatory mediators as well as metabolic changes in the circulation result in alterations in gene expression levels in the liver (1, 3). Therefore, understanding the liver response at the molecular level is critical to understanding the systemic inflammatory disease, as well as its potential as a target for therapeutic approaches.
Prior studies using classical RT-PCR to analyze gene expression in liver have shown that inflammation upregulated specific receptors (such as CD14 receptors, protease activated receptors, histamine H-1 and H-2 receptors), transcription factors (NF-κβ, Stat3, and C/EBP-β) and other proteins or kinases (such as ERK, JNK, and p38) involved in the MAPK, Jac/STAT, and Ik-B/NF-kB signaling pathways (4–12). Recently, microarray technology and transcriptional profiling have been used to elucidate genome-wide changes in the liver following the burn injury (1, 3, 13). Vemula and co-workers (3) analyzed gene expression changes in rat livers during the first 24 h following the burn injury. Functional analysis of differentially expressed genes revealed that metabolism and inflammation accounted for the majority of the differentially expressed genes. Altered inflammatory genes included several classic acute phase response markers, and other genes involved in the complement, kinin, clotting, and fibrinolytic protein systems. On the other hand, metabolic genes showed that fatty acid oxidation increased after burn presumably to meet the enhanced energy demands. Dasu et al. (1) also analyzed the gene expression profiles in rat livers at different time points (2h, 6h, 24h, 240h) after a more severe burn than in the prior studies.
In general, unsupervised hierarchical clustering was applied in order to identify specific patterns of gene expression in the liver associated with burn injury. A limitation of the aforementioned studies is that the control sham-burn group was defined as the initial pre-burn condition corresponding to the 0 h time point. However, gene expression in a healthy animal liver naturally fluctuates over time due to circadian rhythms (14). In order to obtain a better resolution of the dynamics of the injury response, it is therefore necessary to account for the dynamics of the sham group as well.
In this study, we used a standard burn injury model of the rat to compare the dynamics of gene expression in liver in burn vs. sham conditions during the first 24 h. The differentially expressed genes between sham and burn condition over time whose expression patterns were significantly altered following burn were identified and clustered. Simultaneous analysis of both burn and sham-burn groups’ expression profiles enabled to characterize the dynamic patterns of both groups and reveal a comprehensive picture regarding the temporally coordinated inflammatory and metabolic changes in the liver following burn injury.
Material and Methods
Animal Model
Male Sprague-Dawley rats (Charles River Labs, Wilmington, MA) weighing between 150 and 200 g were used. The animals were housed in a temperature-controlled environment (25°C) with a 12-hour light-dark cycle and provided water and standard chow ad libitum. All experimental procedures were carried out in accordance with National Research Council guidelines and approved by the Rutgers University Animal Care and Facilities Committee.
A systemic hypermetabolic response was induced by applying a full-thickness burn on an area of the dorsal skin corresponding to 20% of the total body surface area (TBSA) as described elsewhere (15). This model was chosen because it has nearly 100% long-term survival, no evidence of systemic hypoperfusion, and no significant alterations on feeding patterns (13). Rats were first randomized into two groups: burn and sham burn (control group). Rats were anesthetized by intraperitoneal injection of 80 to 100 mg/kg ketamine + 12 to 10 mg/kg xylazine, and all hair removed from the dorsal abdominal area using electric clippers. The animal's back was immersed in water at 100°C for 10 s to produce a full-thickness scald injury covering 20% TBSA. Immediately after burns, the animals were resuscitated with 50 mL/kg of saline injected intraperitoneally. Negative controls (sham burn) consisted of animals treated identically but immersed in lukewarm water (37°C). Rats were single caged after burn or sham burn and given standard rat chow and water ad libitum until sacrifice. No post-burn analgesics were administered, consistent with other studies with this full thickness burn model since the nerve endings in the skin are destroyed and the skin becomes insensate (16). Furthermore, after animals woke up, they ate, drank and moved freely around the cage, responded to external stimuli, and did not show clinical signs of pain or distress. Animal body weights were monitored daily and found to increase at the same rate in both groups.
Microarray experiments to generate liver gene expression data have been explained elsewhere (3). Briefly, animals were sacrificed (starting at 9am) at different time points (0, 2, 4, 8, 16 and 24hr post-treatment, i.e., sham burn and burn) and liver tissues were collected, snap frozen in liquid nitrogen and stored at −80°C (n=3 per time point per group). The tissues were lysed and homogenized using Trizol, and the RNAs were further purified and treated with DNase using RNeasy columns (Qiagen). Then cRNAs prepared from the RNAs of liver tissues using protocols provided by Affymetrix were utilized to hybridize Rat Genome 230 2.0 Array (GeneChip, Affymetrix) comprised of more than 31,000 probe sets.
Data Analysis
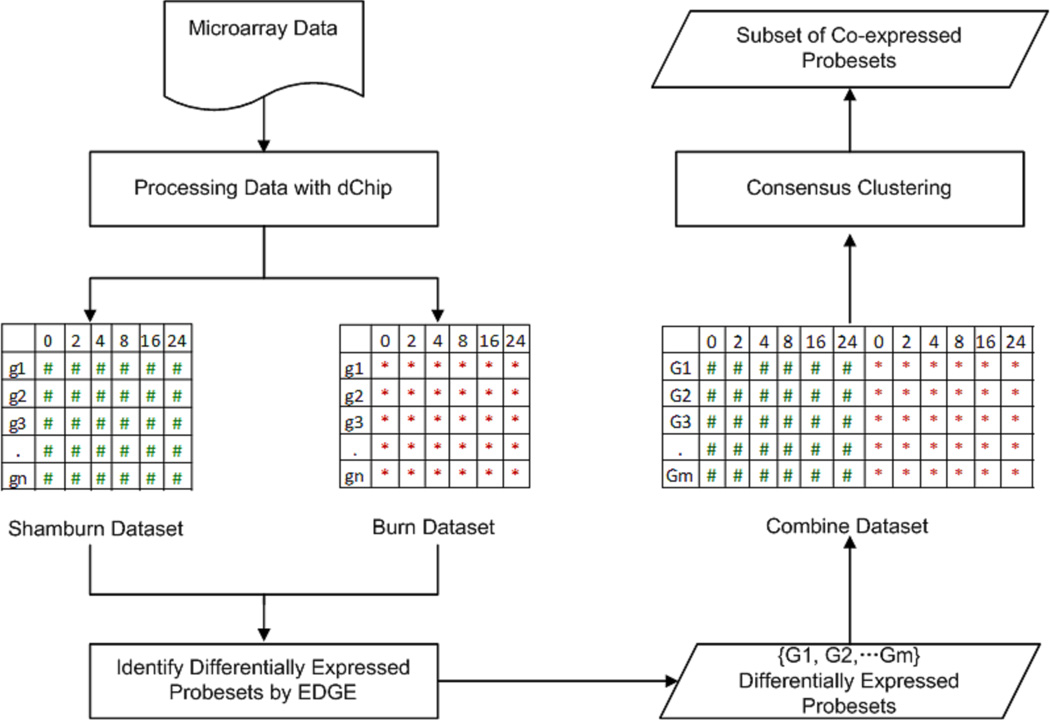
In this study gene expression data analysis includes data preprocessing, filtering for “between class temporal differential expressions”, combining the datasets and clustering as seen in Figure 1. First, DNA chip analyzer (dChip) software (17) was used with invariant-set normalization and perfect match (PM) model to generate expression values. Microarray outlier filter analysis (18) identified that there were approximately 10% outliers in sham and burn gene expression data, which is typically observed in a microarray data. The outliers were replaced by the mean of the replicates (19). Then the data sets corresponding to burn and sham groups were investigated to identify the differentially expressed probesets by using the method (EDGE) proposed by Storey et al. (20). The statistical test used is analogue to an F statistics which compares the goodness of fit of the model under the null hypothesis to that under the alternative hypothesis. The null hypothesis model is obtained by fitting a time-dependent curve to the two or more groups combined, and the alternative hypothesis model is obtained by fitting a separate curve to each group. The significance threshold for this analysis was set as q-value <0.01 and p-value<0.01. This step determined a set of probesets whose expression patterns were significantly altered following the treatment considering the temporal differences between the control and treatment groups. Finally the data sets corresponding to those differentially expressed probesets in either burn and/ or sham groups were combined to form one single matrix, which was then clustered using the a approach “consensus clustering” (21), in an unsupervised manner. This provided a set of burn responsive genes, which is significantly different than that of control group. We further applied one-way ANOVA test (p<0.01) independently for each gene in each cluster and animal group in order to verify if the gene has been differentially expressed across the time only. Moreover, t-test was used additionally for pair-wise comparison of burn and sham genes identified in the clusters at each time point in order to estimate the activation time of a certain response related to burn injury. We characterized the biological relevance of the intrinsic responses by evaluating the enrichment of the corresponding gene subsets using the KEGG database through ARRAYTRACK (22) as well as analyzing the functions of each individual gene (23).
Figure 1. Schematic overview of the microarray data analysis.
Microarray data was preprocessed by using dChip. Then, two data sets corresponding to burn and sham groups, respectively, were analyzed to identify the differentially expressed probesets by using EDGE with ‘between classes’ option under the statistical threshold q<0.01, p<0.01. Finally, the data sets corresponding to those differentially expressed probesets in burn and sham groups were combined to form one single matrix, which was then clustered using the approach of “consensus clustering” with threshold p<0.01.
Results and Discussion
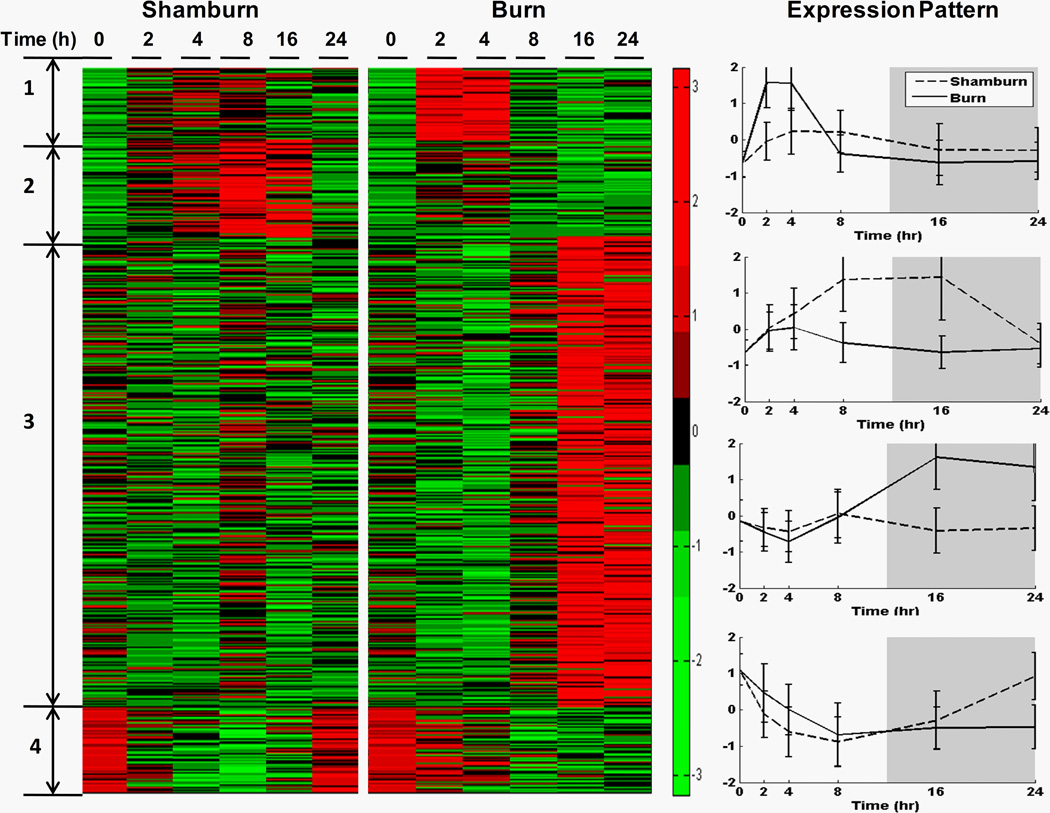
We examined liver-specific gene expression levels at 0, 2, 4, 8, 16, and 24 h post-treatment consisting of a 20% total body surface area (TBSA) burn or sham-burn. 1534 temporally differentially expressed, i.e., burn responsive, were identified and subsequently clustered, using a consensus clustering approach, leading to identification of sub-set of genes assigned to 4 dominant expression patterns composed of 62, 82, 404, and 73 probe sets respectively. The average expression patterns of each dominant cluster are depicted in Figure 2 (right panel) while a heat map of all probe sets is shown in the left panel. A subset of critical genes is listed in Table 1, while a complete description for all 621 probe sets can be found in “Supplementary Materials”. ArrayTrack, as well as single gene ontology analysis, was used to further elaborate the functional annotations of burn injury responsive genes.
Figure 2. Gene expression profiles of rat livers in response to sham-burn or burn injury.
Left Panel, expressions of 62, 82, 404 and 73 probesets in 4 clusters in sham-burn rats and burn rats at 0, 2, 4, 8, 16, 24 h post-treatment are exhibited in a heatmap.
Right Panel, the expression patterns are shown by plotting the average normalized (z-score) expression values of 62, 82, 404 and 73 probe sets in 4 clusters in sham-burn and burn groups (displayed as the means ± SEM).
Table 1.
Information of critical genes in each of the four clusters
| Cluster number |
Function | Gene name |
|---|---|---|
| 1 | Pro-inflammatory Cytokine | IL1a |
| Chemokine | Cxcl16, Ccl11, Ccl9 | |
| Adaptive immune response regulation | Ceacam1 | |
| 2 | Unsaturated fatty acid biosynthesis | Acot1, Acot2, Acot3 |
| Fatty acid metabolism | Acaa2, Cpt1a, Dci | |
| Synthesis of ketone bodies | Hmgcs2 | |
| Lipid metabolism and lipid transport | Eci1, Pigo, cyp4b1, Adfp, Pnpla8, Pank4, Crot, Etfdh | |
| Cell-cell junctions | Ablim3, Acer2, Cdh17 | |
| 3 | Complement and coagulation cascade | C2, C4bpa, C8a, Cfh, Masp1, Serping1 |
| N-glycan biosynthesis | B4galt1, Dad1, Ddost, Dpagt1, Ganab, Man1b1 | |
| Ribosome | Rps25, Rps2 | |
| Jak-STAT signaling | Il13, Il4, Il7, Jak3 | |
| TLR4 signaling | IRAK1, LBP, TRAF3 | |
| Anti-inflammatory cytokine | Il13, Il4 | |
| Transcription | Brca1, Mcm7, Tcf25, Kdm1, Nfyb, Tef, Cited4, Otx1, Sox4, Acvr1, Tbx2, Zfhx2, Dmrt2, Tsc22d1, Ccdc 101 | |
| Translation | Atpif1, Mrps21, Rps25, Rps2, Mrps11 | |
| Protein folding | Dnajb11, Ppib, Hyou1, Edem2, Sep15, Pdia6, Dnajc3, Pdia3, Ugcgl1, Sec63, Mlec, Mecp2 | |
| Protein degradation | Pcolce, Cpn1, caspase 12, Cdc34, Spink3, Hspa5, RGD1306508, Erlec1, Aph1a, Derl2, Os9, Rnf20, Ppp2r5c, Aurkaip1, Prss32, Prepl, Tbl1xr1, St14, P4hb | |
| Protein target | Tmed3, Ssr4, Tram1, Sec61a1, Rrbp1, Arfgap2, Gabarap, Erp29, Ssr3, Rrbp1, Sec61a1, Derl1, Gosr2, Cope, Tmed2, Copz1, Copg, Sec13, Abcb10, Kdelr1, Kdelr2, Tram1, Serp1, Ssr3, Atp6v0d1, Rabac1, Vps28 | |
| Bile acid production | Idi1, tmem97, Npc2, Hsd17b4 | |
| 4 | Insulin signaling pathway | Gck, Irs1, Mknk2, Trip10 |
| Phenylalanine metabolism | Ddc | |
| Glycine, serine and threonine metabolism | Bhmt | |
| Galactose metabolism | Gck |
Functional Characterization of major clusters
Cluster 1 (Figure 2) exhibits an early up-regulation during the first two hours following thermal injury. One-way ANOVA (p<0.01) indicates that the majority of the probesets in this cluster are not differentially expressed in sham-burn, clearly indicating that their activity is the result of the burn injury. Burn injury induces a rapid, but transient, up-regulation of the genes in this cluster which resolves within 8 hrs post-burn. Functional annotation and characterization of cluster 1 identifies cytokines, chemokines and chemokine receptors as well as genes related to the modulation of innate and adaptive immune responses, including: IL-1α, a pro-inflammatory cytokine playing a central role in the regulation of the immune response by binding to the IL-1 receptor (25) known to exhibit increased activity in the early stage of inflammatory response (26); chemokine (C-X-C motif) ligand 16 (CXCL16), a chemoattractant belonging to the CXC chemokine family whose expression is induced by inflammatory cytokines, such as IFN-γ and TNF-γ (27); chemokine (C-C motif) ligand 11 (CCL11), an inflammatory mediator belonging to the CC chemokine family that is known as Eotaxin-1; chemokine (C-C motif) ligand 9 known as macrophage inflammatory protein (MIP)-1γ which is constitutively expressed in macrophages (28). The release of the pro-inflammatory cytokines of Cluster 1 postburn are hypothesized to trigger and enhance the inflammatory response and to mediate catabolic effects (24). A list of representative genes is depicted in Table 1, while a detailed account of the gene information is provided in the supplementary material accompanying the manuscript.
Cluster 2 (Figure 2) exhibits characteristics of a persistent down regulation following burn injury, beginning at about 2hr post-burn, compared to the temporal differential expression under sham (ANOVA, p<0.01), thus indicating suppression of expression in burn. A Student t-test (p<0.01) reveals that the most significant suppression occurred at 8 h and 16 h post-burn. The functional characterization of Cluster 2 revealed down-regulation of genes involved primarily in unsaturated fatty acid biosynthesis, fatty acid metabolism, synthesis of ketone bodies and lipid metabolism and transport, consistent with earlier indications. The decrease in fatty acid biosynthesis as well as increase in fatty acid oxidation during the first 24h suggests that fatty acid is utilized in the liver during the first 24h postburn as the early energy source (3). Thus, down-regulation of fatty acid biosynthesis associated enzymes is possibly implying an enhanced energy demand. Prior studies elucidating the circadian rhythmicity of gene expression in rat indicated that fatty acid biosynthesis is up-regulated in the late afternoon and early evening hours (32). Our sham results, consistent with this observation, indicate the possibility of circadian rhythmicity in Cluster 2 with a return to base-line values within 24 hr. This observation leads us to speculate the possibility of circadian disruption following burn injury. Furthermore, genes related to cell-cell junctions are also identified in Cluster 2 including: ABLIM3, a molecular component of adherence junctions (AJs) and possibly a novel component of adherens junctions with actin-binding activity (33); alkaline ceramidase 2 encoded by Acer2 playing an important role in regulating β1 maturation and cell adhesion mediated by β1 integrins (34); cadherin 17 is a Ca(2+)-dependent cell-cell adhesion molecule expressed in liver and intestine which plays a role in the morphological organization of liver and intestine (35). The products of those genes are associated with the integrity of the barrier function of hepatocytes linings. Given the many studies revealing that intestinal permeability is increased in burn patient shortly after the injury possibly due to the junction integrity alterations (36), the suppression of cell-cell junctions and membrane structural integrity may be indication of the liver damage caused by burn injury. Representative genes are depicted in Table 1, while a detailed account of the gene information is provided in the supplementary material accompanying the manuscript.
Cluster 3 (Figure 2) captures a dynamics response which, under burn, exhibits deviation from sham around 8 hr post injury. Beyond this point, the burn responses remain effectively suppressed (ANOVA, p<0.01). Functional annotation of Cluster 3 reveals gene products involved in complement and coagulation cascade, N-Glycan biosynthesis, ribosome and Jak-STAT signaling as well as involved in transcription/translation, protein synthesis/folding and targeting. All the above constitute processes critical in the production of acute phase proteins (APP) which are diffusible anti-inflammatory mediators (38). Furthermore, the anti-inflammatory response is induced by the suppressor of cytokine signaling proteins (SOCS) activated by Jak/STAT signaling pathway. Thus these gene encompassing Cluster 3 point to the activation of anti-inflammatory mechanisms resulting in an increase in the synthesis of the acute phase proteins and important anti-inflammatory cytokines. APPs produced by the liver is a prominent characteristic of the acute phase response following thermal injury, which is believed to be critical for the adaptation of the body to stress (39). In addition, the transcription of APPs is activated in the late phase starting around 8 h post-burn, consistent with previous observations that the level of amyloid A, a APP, is not increased until the concentration of IL-6, a late phase cytokine, increases (40). The requirement of the energy and amino acids (AA) to produce large amount of APP in liver are satisfied by the increased flux of amino acids from the periphery to the liver, especially from the accelerated breakdown of muscle proteins (41). The alterations in nitrogen and protein metabolism represent a major threat for the organism, as it leads to a debilitating loss of lean body mass (42). Thus, a sustained or exaggerated acute phase response has been shown to be an indicator of a potentially life threatening uncontrolled and prolonged action of pro-inflammatory cytokines leading to multiple-organ failure.
Critical cytokines in this cluster are well known anti-inflammatory cytokines such as IL-13 and IL-4. IL-13 inhibits the ability of host immune cells to destroy intracellular pathogens by recruiting a large number of Th2 cells while IL-4 induces differentiation of naive helper T cells (Th0 cells) to Th2 cells. IL-4 promotes the activation of macrophages into repair macrophages which is coupled with secretion of IL-10 and TGF-beta that result in the diminution of pathological inflammation. Anti-inflammatory cytokines, such as IL-4, are released later on in an attempt to counter-regulate the effects of the pro-inflammatory cytokines (43). Following the burn injury, a state of immunosuppression occurs whose intensity and duration is closely related to morbidity and mortality in burn patients (44). The inflammatory response after burn injury may play a role in the induction of adaptive immunosuppression. Both in vivo and in vitro studies manifest the altered adaptive immunity after burn which have shown that there is a decreased production of Th1- type cytokines (IL-2 and IFN-γ) and an increased production of Th2- type cytokines (IL-4 and IL-10) (45). In the current study, the gene expression of Th2- type cytokines, IL-4 and IL-13, is enhanced starting from 8 h post burn, which may imply the onset of the host immunosuppression. Our results reveal that the gene expression of critical proteins (IRAK1, LBP, and TRAF3) in the TLR4 signaling pathway is upregulated at 8 h post burn injury (Figure2, cluster 3). The TLR4 signaling pathway is critical for Gram-negative bacterial infections. It is well known that patients with severe burn injury are exceedingly susceptible to bacterial infections. Not only bacterial infection from the injured area but also bacterial translocation from the gut cause septic complications in the hosts. Mesenteric lymph nodes and liver indeed contain bacteria after burn injury in mice (46). It is generally accented that the decreased resistance to infection and enhanced secondary inflammatory response following serious injury is associated with abnormalities of both natural and adaptive immunity. Fang et al. (47) observed that thermal injury can markedly up-regulate lipopolysaccharide-binding protein (LBP) gene expression in various organs. LBP, a soluble acute-phase protein, binds to bacterial LPS to facilitate the immune response. Excessive LBP mRNA expression may be associated with enhanced synthesis and release of TNF-α stimulated by burn induced-endotoxin. Paterson et al. demonstrated that burn injury significantly increased TLR2- and TLR4-induced IL-1, IL-6, and TNF- α production by liver cells as early as 1 day after injury and they were found to be persistent for at least 7 days (48). Thus, the alteration of the TLR4 signaling pathway may imply that burn injury primes the innate immune system for enhanced TLR4-mediated responses to subsequent infection and provides evidence to suggest that an augmented Toll-like receptor signaling pathway might contribute to the development of increased systemic inflammation following severe burn injury.
Finally, a number of bile acid production related genes were also identified in this cluster. Bile acids are end products of cholesterol and the major driving force for bile formation, and the major excretory products of cholesterol. Bile acid production is expected to increase following burn injury (3). The main function of bile acids is to promote the formation of micelles, which facilitate fat digesting and absorption. Therefore, the enhanced production of bile acids may also reflect the demand of the energy from food intake. In fact, nutritional therapy is commonly used with burn patients (52) in an attempt to compensate for burn injury-induced metabolic abnormalities although it is limited given that it does not address the underlying mechanisms that are responsible for hypermetabolic and catabolic induction. Although nutritional supplements partially alleviate the hyper-catabolic condition, they seldom can cannot reverse or completely restore the nitrogen balance. Representative genes are listed in Table 1, and detailed information is provided in the supplementary material accompanying the manuscript.
Cluster 4 is encompasses genes which are downregulated long after the injury is induced (16 h post burn) (Figure 2, cluster 4). The probesets of this cluster in both sham and burn groups exhibit an early down-regulation. Although the control group to recover their expression within 24 h, persistent downregulation is observed in the burn group. The maximum deviation between the sham-burn and burn groups occurs at 24 h postburn (two sample t-test, p<0.01). The genes in this cluster are involved in the insulin signaling pathway, glycine, serine and threonine metabolism, and galactose metabolism. Consistent with prior observations, our data point to the possibility of impaired insulin signaling (55). Insulin is an anabolic hormone which promotes the storage of substrates in liver by stimulating lipogenesis, glycogen and protein synthesis (56). Thus, downregulation of the genes involved in the insulin signaling pathway suggests a potential mechanism to explain the onset of a hypercatabolic state which is characteristic of hypermetabolism. Furthermore, the expression of genes associated with amino acid metabolism are known to be under circadian regulation in rat liver (32). Consistent with this observation, the insulin and amino acid metabolism-related genes in the sham-burn group also exhibited characteristics reminiscent of daily oscillation reaching a nadir at the interface of the light and dark phases. However, this daily oscillation was disrupted and suppressed maximally 24 h postburn, as demonstrated by the dynamic gene expression profile of the burn group pointing again to the possibility of circadian rhythms disruption. Representative genes are listed in Table 1, and detailed information is provided in the supplementary material accompanying the manuscript.
Assessing and interpreting the differences in transcriptional dynamics between sham and burn
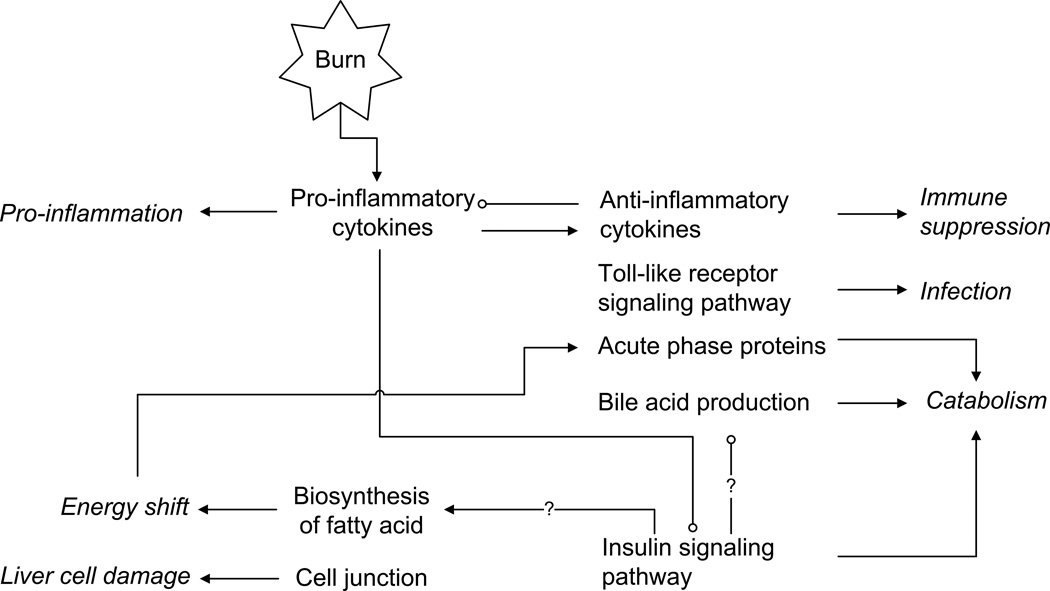
The richness of our data and the fact that we analyze in tandem the dynamics of sham and burn injuries allowed to identify not only the differentially expressed responses but also the critical turning points where deviations induced by the burn injury manifest themselves. We identify, therefore, that the release of pro-inflammatory cytokines (Cluster 1) is almost instantaneous, whereas the synthesis of APP is delayed (Cluster 3), fatty acid biosynthesis (Cluster 2) precedes impairment of insulin signaling (Cluster 4). In a manner analogous to (57) we hypothesize that the time dependence among the profiles, may imply putative causal relations, which are succinctly summarized in the putative network structure of Figure 3, where arrows indicate possibly activation and/or induction, and circles indicate inhibition. The relations are derived based on the time lag elucidated from the temporal dynamics of individual responses.
Figure 3. Proposed network of changes in the liver following burn injury.
Italics represent outcomes following burn-induced gene expression alterations. Arrows indicate activation and/or induction, and circles indicate inhibition.
The early upregulation of pro-inflammatory cytokines and chemokines, and their corresponding receptors in Cluster 1 indicates the activation of the immune system and a pro-inflammatory response, whereas the suppression of fatty acid biosynthesis associated genes in Cluster 2 implies an enhanced energy demand. In Cluster 3, the downregulation of the genes functioning as cell-cell junctions and providing membrane structural integrity indicate possible damage caused by the injury. Later activation of the expression of well-known anti-inflammatory cytokines may suggest the upcoming immune suppression. The activation of the Toll-like receptor signaling pathway, also in the Cluster 3, is possibly indicative of a priming effect to a subsequent secondary stimulus, i.e., infection. The most significant feature of Cluster 3 is the enhanced production of positive APPs, which is correlated to hyper-catabolism in muscle. In the same cluster, the enhanced expression of bile acid synthesis related genes may also be an indication of enhanced energy demand from nutrition supply. Finally, the late downregulation of the insulin signaling pathway-associated genes in Cluster 4 leads to the catabolism and insulin resistance. The dynamic picture which is assembled is indicative of the fact that once a pro-inflammatory response has been mounted there is a subsequent release of signals stimulating an anti-inflammatory response that inhibits the pro-inflammatory response, which drives the system back to homeostasis. The burn-induced response in Cluster 4, representing insulin-mediated metabolism, was characterized by an early and persistent downregulation. While prior work (58), indicated the possibility of an early downregulation the anabolic response in liver, our results indicate that the down-regulation is in fact delayed in time, given that the nature progression of the sham responses also points to an early down-regulation, although it recovers, pointing to the possibility that the burn-specific down-regulation occurs only later in time. Therefore, we argue that the onset of insulin resistance and the putative associated catabolic response (which is regulated by insulin) is not as immediate as previously thought (58). A delayed response is in fact more consistent with the observed dynamics of cytokine and chemokine activation, which presumably drive the molecular mechanisms leading to insulin resistance (59), because impaired insulin signaling should occur after and not before the release of cytokines. In addition, insulin is known to suppress bile acid synthesis in cultured rat hepatocytes by down-regulating the key enzymes in the synthesis of bile acids from cholesterol. Therefore, the impaired insulin signaling can also explain the increased bile acid excretion observed in humans with untreated diabetes mellitus and in experimental animals with insulin deficiency (60). However, in our study, the insulin signaling pathway was suppressed around 16 h postburn, which was later than the enhanced production of bile acids. Thus, our results suggest that the increased bile acid production in inflammation is more likely caused by mechanisms other than the impaired insulin signaling pathway. Moreover, insulin is a well-known critical anabolic hormone which promotes the storage of substrates in liver such as lipids (56). However, since the decreased synthesis of fatty acids (~2 h postburn) occurs earlier than the impaired insulin signaling (~16 h postburn), the suppression of fatty acid biosynthesis herein may not be caused by the insulin resistance either.
We observed that a significant number of positive APP genes were up-regulated, which requires an increased energy utilization. Thus, the biosynthesis of unsaturated fatty acid starts to be suppressed around 2 h post burn which may imply the preservation of the energy sources for the synthesis of positive APP activated later - around 8h post burn. This suggests that FA is utilized in the liver during the first 16 h after the burn injury. However, new energy sources and AA pool to produce positive APP are required after the exhaustion of available energy and AAs in the liver. It is well known that burn injury results in accelerated breakdown of muscle proteins which increase the AA concentrations in the circulation thus AA uptake-rates in the liver (41). We observed that the impaired insulin signaling pathway occurs later, starting around 16h postburn consistent with previous reports indicating that muscle protein breakdown is exacerbated in burn-injured patients as a result of insulin resistance (61). Thus, our results suggest the possibility that the impaired insulin signaling pathway may further intensify the catabolic response which has already been driven by the uncontrolled positive APP production. In addition, due to the suppression of fatty acid biosynthesis, it seems that the fatty acid oxidation might serve as the main source of energy in the liver following the burn injury.
Conclusions
We have shown a short-term liver gene expression profiling in response to thermal injury. This analysis characterizing the dynamic patterns of both burn and sham groups elucidated that temporal changes in the expression levels after the injury are mainly associated with the pro-inflammatory response, fatty acid biosynthesis, the anti-inflammatory response, and insulin-regulated metabolic responses. The network of dynamic changes in gene expression observed in this study revealed the possible links between the diverse burn-induced responses. Based on our results, the pro-inflammatory response is activated immediately around 2 h following burn treatment which triggers the anti-inflammatory response starting around 8 h postburn. The biosynthesis of unsaturated fatty acid starts to be suppressed around 2 h which may imply the preservation of the energy sources for the synthesis of APPs whose genes were activated later around 8 h post burn. In addition, the impaired insulin signaling pathway, starting from around 16 h postburn and putatively as a result of the alterations in inflammatory gene expression, is expected to further strengthen the catabolic response. A Suppression of fatty acid synthesis and enhanced production of bile acids were also observed, but were not likely due to the impaired insulin signaling because of the discrepancy in the dynamics of these responses. In conclusion, simultaneous analysis of both burn and sham-burn groups’ expression profiles enables to characterize the dynamic patterns of both groups. Our results reveal critical gene expression pattern changes triggered by burn injury which reflects host physiological and biological alterations and provides a more comprehensive understanding of the pathophysiology of the disease state.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support from NIH grant GM082974.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Dasu MRK, Cobb JP, Laramie JM, Chung TP, Spies M, et al. Gene expression profiles of livers from thermally injured rats. Gene. 2004;327:51–60. doi: 10.1016/j.gene.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Drost AC, Burleson DG, Cioffi WG, Jr., Jordan BS, Mason AD, Jr., et al. Plasma cytokines following thermal injury and their relationship with patient mortality, burn size, and time postburn. J Trauma. 1993;35:335–339. doi: 10.1097/00005373-199309000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Vemula M, Berthiaume F, Jayaraman A, Yarmush ML. Expression profiling analysis of the metabolic and inflammatory changes following burn injury in rats. Physiological Genomics. 2004;18:87–98. doi: 10.1152/physiolgenomics.00189.2003. [DOI] [PubMed] [Google Scholar]
- 4.Jeong J, Adamson LK, Hatam R, Greenhalgh DG, Cho K. Alterations in the expression and modification of histones in the liver after injury. Exp. Mol. Pathol. 2003;75:256–264. doi: 10.1016/s0014-4800(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 5.Cho K, Adamson LK, Jeong J, Crivello SD, Vanhook TG, et al. CD14-dependent alterations in c-Jun expression in the liver after burn injury. J. Surg. Res. 2004;122:36–42. doi: 10.1016/j.jss.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Jesmin S, Gando S, Zaedi S, Sakuraya F. Chronological expression of PAR isoforms in acute liver injury and its amelioration by PAR2 blockade in a rat model of sepsis. Thromb. Haemostasis. 2006;96:830–838. [PubMed] [Google Scholar]
- 7.Klein D, Einspanier R, Bolder U, Jeschke MG. Differences in the hepatic signal transcription pathway and cytokine expression between thermal injury and sepsis. Shock. 2003;20:536–543. doi: 10.1097/01.shk.0000093345.68755.98. [DOI] [PubMed] [Google Scholar]
- 8.Nishiura T, Nishimura T, deSerres S, Godfrey V, Bradham CA, et al. Gene expression and cytokine and enzyme activation in the liver after a burn injury. J. Burn Care Rehabil. 2000;21:135–141. doi: 10.1097/00004630-200021020-00009. [DOI] [PubMed] [Google Scholar]
- 9.Andrejko KM, Chen JD, Deutschman CS. Intrahepatic STAT-3 activation and acute phase gene expression predict outcome after CLP sepsis in the rat. Am. J. Physiol.-Gastroint. Liver Physiol. 1998;275:G1423–G1429. doi: 10.1152/ajpgi.1998.275.6.G1423. [DOI] [PubMed] [Google Scholar]
- 10.Cho K, Pham TN, Crivello SD, Jeong J, Green TL, et al. Involvement of CD14 and toll-like receptor 4 in the acute phase response of serum amyloid a proteins and serum amyloid P component in the liver after burn injury. Shock. 2004;21:144–150. doi: 10.1097/01.shk.0000108398.56565.ae. [DOI] [PubMed] [Google Scholar]
- 11.Masaki T, Chiba S, Tatsukawa H, Noguchi H, Kakuma T, et al. The role of histamine H-1 receptor and H-2 receptor in LPS-induced liver injury. FASEB J. 2005;19:1245–1252. doi: 10.1096/fj.04-3195com. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Dong LW, Tang CS, Liu MS. Transcriptional and posttranscriptional regulation of beta(2)-adrenergic receptor gene in rat liver during sepsis. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1999;277:R132–R139. doi: 10.1152/ajpregu.1999.277.1.R132. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman A, Maguire T, Vemula M, Kwon DW, Vannucci M, et al. Gene Expression Profiling of Long-Term Changes in Rat Liver Following Burn Injury. J. Surg. Res. 2009;152:3–17. doi: 10.1016/j.jss.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almon RR, Yang E, Lai W, Androulakis IP, DuBois DC, et al. Circadian variations in rat liver gene expression: relationships to drug actions. J Pharmacol Exp Ther. 2008;326:700–716. doi: 10.1124/jpet.108.140186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banta S, Vemula M, Yokoyama T, Jayaraman A, Berthiaume F, et al. Contribution of gene expression to metabolic fluxes in hypermetabolic livers induced through burn injury and cecal ligation and puncture in rats. Biotechnol Bioeng. 2007;97:118–137. doi: 10.1002/bit.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valenti LM, Mathieu J, Chancerelle Y, De Sousa M, Levacher M, et al. High levels of endogenous nitric oxide produced after burn injury in rats arrest activated T lymphocytes in the first G1 phase of the cell cycle and then induce their apoptosis. Exp. Cell Res. 2005;306:150–167. doi: 10.1016/j.yexcr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Guo X, Yang YC, Papcunik D, Heckman C, et al. Detecting outlier microarray arrays by correlation and percentage of outliers spots. Cancer Inform. 2007;2:351–360. [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson R, Gonye G, Schwaber J. Outliers in microarray data analysis. Methods of Microarray Data Analysis III. 2003:41–55. [Google Scholar]
- 20.Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen TT, Nowakowski RS, Androulakis IP. Unsupervised selection of highly coexpressed and noncoexpressed genes using a consensus clustering approach. Omics. 2009;13:219–237. doi: 10.1089/omi.2008.0074. [DOI] [PubMed] [Google Scholar]
- 22.Tong W, Cao X, Harris S, Sun H, Fang H, et al. ArrayTrack--supporting toxicogenomic research at the US Food and Drug Administration National Center for Toxicological Research. Environmental health perspectives. 2003;111:1819–1826. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twigger S, Lu J, Shimoyama M, Chen D, Pasko D, et al. Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res. 2002;30:125–128. doi: 10.1093/nar/30.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauglitz GG, Song J, Herndon DN, Finnerty CC, Boehning D, et al. Characterization of the inflammatory response during acute and post-acute phases after severe burn. Shock. 2008;30:503–507. doi: 10.1097/SHK.0b013e31816e3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinarello CA. Interleukin-1 and its biologically related cytokines. Adv Immunol. 1989;44:153–205. doi: 10.1016/s0065-2776(08)60642-2. [DOI] [PubMed] [Google Scholar]
- 26.Mester M, Carter EA, Tompkins RG, Gelfand JA, Dinarello CA, et al. Thermal injury induces very early production of interleukin-1 alpha in the rat by mechanisms other than endotoxemia. Surgery. 1994;115:588–596. [PubMed] [Google Scholar]
- 27.Abel S, Hundhausen C, Mentlein R, Schulte A, Berkhout TA, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol. 2004;172:6362–6372. doi: 10.4049/jimmunol.172.10.6362. [DOI] [PubMed] [Google Scholar]
- 28.Youn BS, Jang IK, Broxmeyer HE, Cooper S, Jenkins NA, et al. A novel chemokine, macrophage inflammatory protein-related protein-2, inhibits colony formation of bone marrow myeloid progenitors. J Immunol. 1995;155:2661–2667. [PubMed] [Google Scholar]
- 29.Motamed-Khorasani A, Jurisica I, Letarte M, Shaw PA, Parkes RK, et al. Differentially androgen-modulated genes in ovarian epithelial cells from BRCA mutation carriers and control patients predict ovarian cancer survival and disease progression. Oncogene. 2007;26:198–214. doi: 10.1038/sj.onc.1209773. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, et al. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem. 2005;280:42841–42847. doi: 10.1074/jbc.M506569200. [DOI] [PubMed] [Google Scholar]
- 31.Yoda E, Hachisu K, Taketomi Y, Yoshida K, Nakamura M, et al. Mitochondrial dysfunction and reduced prostaglandin synthesis in skeletal muscle of Group VIB Ca2+-independent phospholipase A2gamma-deficient mice. J Lipid Res. 2010;51:3003–3015. doi: 10.1194/jlr.M008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ovacik M, Sukumaran S, Almon R, DuBois D, Jusko W, et al. Circadian signatures in rat liver: from gene expression to pathways. BMC Bioinformatics. 2010;11:540. doi: 10.1186/1471-2105-11-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda M, Yamashita JK, Tsukita S, Furuse M. abLIM3 is a novel component of adherens junctions with actin-binding activity. Eur J Cell Biol. 89:807–816. doi: 10.1016/j.ejcb.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Sun W, Hu W, Xu R, Jin J, Szulc ZM, et al. Alkaline ceramidase 2 regulates beta1 integrin maturation and cell adhesion. Faseb J. 2009;23:656–666. doi: 10.1096/fj.08-115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berndorff D, Gessner R, Kreft B, Schnoy N, Lajous-Petter AM, et al. Liver-intestine cadherin: molecular cloning and characterization of a novel Ca(2+)-dependent cell adhesion molecule expressed in liver and intestine. J Cell Biol. 1994;125:1353–1369. doi: 10.1083/jcb.125.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjarnason I, MacPherson A, Hollander D. Intestinal permeability: an overview. Gastroenterology. 1995;108:1566–1581. doi: 10.1016/0016-5085(95)90708-4. [DOI] [PubMed] [Google Scholar]
- 37.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 38.Tracey KJ. The inflammatory reflex. Nature. 2002;420:853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 39.Kataranovski M, Magic Z, Pejnovic N. Early inflammatory cytokine and acute phase protein response under the stress of thermal injury in rats. Physiol Res. 1999;48:473–482. [PubMed] [Google Scholar]
- 40.Plackett TP, Colantoni A, Heinrich SA, Messingham KA, Gamelli RL, et al. The early acute phase response after burn injury in mice. J Burn Care Res. 2007;28:167–172. doi: 10.1097/BCR.0b013E31802CB84F. [DOI] [PubMed] [Google Scholar]
- 41.Hasselgren PO, Pedersen P, Sax HC, Warner BW, Fischer JE. Current concepts of protein turnover and amino acid transport in liver and skeletal muscle during sepsis. Arch Surg. 1988;123:992–999. doi: 10.1001/archsurg.1988.01400320078016. [DOI] [PubMed] [Google Scholar]
- 42.Windsor JA, Hill GL. Weight loss with physiologic impairment. A basic indicator of surgical risk. Ann Surg. 1988;207:290–296. doi: 10.1097/00000658-198803000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finnerty CC, Przkora R, Herndon DN, Jeschke MG. Cytokine expression profile over time in burned mice. Cytokine. 2009;45:20–25. doi: 10.1016/j.cyto.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran K, Munster AM. Alterations of the host defense mechanism in burned patients. Surg Clin North Am. 1987;67:47–56. doi: 10.1016/s0039-6109(16)44132-0. [DOI] [PubMed] [Google Scholar]
- 45.Lederer JA, Rodrick ML, Mannick JA. The effects of injury on the adaptive immune response. Shock. 1999;11:153–159. doi: 10.1097/00024382-199903000-00001. [DOI] [PubMed] [Google Scholar]
- 46.Maejima K, Deitch E, Berg R. Promotion by burn stress of the translocation of bacteria from the gastrointestinal tracts of mice. Arch Surg. 1984;119:166–172. doi: 10.1001/archsurg.1984.01390140032006. [DOI] [PubMed] [Google Scholar]
- 47.Fang CW, Yao YM, Shi ZG, Yu Y, Wu Y, et al. Lipopolysaccharide-binding protein and lipopolysaccharide receptor CD14 gene expression after thermal injury and its potential mechanism(s) J Trauma. 2002;53:957–967. doi: 10.1097/00005373-200211000-00024. [DOI] [PubMed] [Google Scholar]
- 48.Paterson HM, Murphy TJ, Purcell EJ, Shelley O, Kriynovich SJ, et al. Injury primes the innate immune system for enhanced Toll-like receptor reactivity. J Immunol. 2003;171:1473–1483. doi: 10.4049/jimmunol.171.3.1473. [DOI] [PubMed] [Google Scholar]
- 49.Bartz F, Kern L, Erz D, Zhu M, Gilbert D, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Klein A, Amigo L, Retamal MJ, Morales MG, Miquel JF, et al. NPC2 is expressed in human and murine liver and secreted into bile: potential implications for body cholesterol homeostasis. Hepatology. 2006;43:126–133. doi: 10.1002/hep.20985. [DOI] [PubMed] [Google Scholar]
- 51.Baes M, Huyghe S, Carmeliet P, Declercq PE, Collen D, et al. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J Biol Chem. 2000;275:16329–16336. doi: 10.1074/jbc.M001994200. [DOI] [PubMed] [Google Scholar]
- 52.Chan MM, Chan GM. Nutritional therapy for burns in children and adults. Nutrition. 2009;25:261–269. doi: 10.1016/j.nut.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 54.Jung UJ, Lee MK, Park YB, Jeon SM, Choi MS. Antihyperglycemic and antioxidant properties of caffeic acid in db/db mice. J Pharmacol Exp Ther. 2006;318:476–483. doi: 10.1124/jpet.106.105163. [DOI] [PubMed] [Google Scholar]
- 55.Gauglitz GG, Halder S, Boehning DF, Kulp GA, Herndon DN, et al. Post-Burn Hepatic Insulin Resistance Is Associated with Er Stress. Shock. 2009 doi: 10.1097/SHK.0b013e3181b2f439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 57.Qian J D-FM, Lin J, Yu H, Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J Mol Biol. 2001;314:1053–1066. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 58.Yang Q, Berthiaume F, Androulakis IP. A quantitative model of thermal injury-induced acute inflammation. Math Biosci. 2010;229:135–148. doi: 10.1016/j.mbs.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Twisk J, Hoekman MF, Lehmann EM, Meijer P, Mager WH, et al. Insulin suppresses bile acid synthesis in cultured rat hepatocytes by down-regulation of cholesterol 7 alpha-hydroxylase and sterol 27-hydroxylase gene transcription. Hepatology. 1995;21:501–510. [PubMed] [Google Scholar]
- 61.Gore DC, Chinkes DL, Hart DW, Wolf SE, Herndon DN, et al. Hyperglycemia exacerbates muscle protein catabolism in burn-injured patients. Crit Care Med. 2002;30:2438–2442. doi: 10.1097/00003246-200211000-00006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.