Abstract
Fyn is a non-receptor protein tyrosine kinase that belongs to a highly conserved kinase family, Src family kinases (SFKs). Fyn plays an important role in inflammatory processes and neuronal functions. To generate a synthetic affinity reagent that can be used to probe Fyn, a phage-display library of fibronectin type III (FN3) monobodies was affinity selected with the SH3 domain of Fyn and three binders were isolated. One of the three binders, G9, is specific in binding to the SH3 domain of Fyn, but not to the other members of the Src family (i.e., Blk, Fgr, Hck, Lck, Lyn, Src, Yes), even though they share 51-81% amino acid identity. The other two bind principally to the Fyn SH3 domain, with some cross-reactivity to the Yes SH3 domain. The G9 binder has a dissociation constant of 166 ± 6 nM, as measured by isothermal titration calorimetry, and binds only to the Fyn SH3 domain out of 150 human SH3 domains examined in an array. Interestingly, although the G9 monobody lacks proline in its randomized BC and FG loops, it binds at the same site on the SH3 domain as proline-rich ligands, as revealed by competition assays. The G9 monobody, identified in this study, may be used as a highly selective probe for detecting and purifying cellular Fyn kinase.
Keywords: affinity selection, dissociation constant, engineered scaffold, fibronectin domain, phage display, Src family kinases
Introduction
While recombinant antibodies continue to be a fruitful source of affinity reagents, there has been a great deal of interest in engineering useful affinity reagents from other scaffolds. Several other scaffold proteins have been engineered into affinity reagents [1]. These include three α-helix bundles of the Z domain of protein A, called affibodies [2], avimers [3], cystine-knot peptides [4], green fluorescent protein (GFP) [5], lipocalin [6], camelid VHH [7], and designed ankyrin repeat proteins (DARPins) [8], to name a few. The scaffold we have exploited is the fibronectin type III (FN3) domain, which is 94 amino acids in length, and has a three-dimensional structure similar to that of the immunoglobulin fold. There are over 8000 examples of this domain in GenBank, making it one of the most prevalent domains known. It should be pointed out that the tenth repeat of the domain in fibronectin contains the Arg-Gly-Asp (RGD) motif in one loop, which interacts with the fibronectin receptor on cell surfaces [9]. Protein engineering experiments have shown it is possible to substitute amino acids within five of the six loops without loss of stability [10]; in particular, from libraries of variants with combinatorial peptides within two loops on one side of the domain, one generates libraries of synthetic affinity reagents, termed “monobodies”. Through phage display, RNA display, or yeast display, binding FN3 monobodies have been generated for a variety of targets, such as the estrogen receptor [11], integrin [12], lysozyme [13], a phosphorylated IκBα peptide [14], small ubiquitin-like modifier 4 (SUMO4) [15], streptavidin [16], tumor necrosis factor [17], ubiquitin [18], and vascular endothelial growth factor receptor 2 [19]. In addition, the FN3 domain can be expressed inside eukaryotic cells, where it can fold properly and bind to its target [11]. It is also possible to combine a monobody with a protein interaction module to create affinity reagents that can “clamp” short peptides with picomolar affinity [20, 21]. Three advantages of the FN3 domain are that it lacks disulfide bonds, it can be highly overexpressed (≥ 50 mg/L culture) in E. coli, and it is thermally stable (Tm = 88°C).
Recently, we have screened a phage-display library of monobodies for variants that bind to the Src Homology 3 (SH3) domain [16]. This 60 amino acid domain is involved in protein-protein interactions [22], and is present in many different eukaryotic proteins that are involved in signal transduction [23], cytoskeleton assembly [24], and endocytosis [25]. In mammals, the Src family of protein tyrosine kinases (Fig. 1) plays an important role in cell signaling pathways, by transferring signals from activated receptors to downstream signaling proteins. This family of kinases shares a common architecture that consists of an N-terminal myristic acid, a unique region, a Src Homology 3 (SH3) domain, a Src homology 2 (SH2) domain, a linker, and a C-terminal catalytic kinase domain. Generally, these proteins have two conformations: a catalytically inactive conformation, in which the protein is folded into a ‘closed’ state, due to intramolecular interactions, and a catalytically active conformation, in which the protein is in an extended, ‘open’ state [26]. The Fyn protein has been proposed to play a role in T-cell receptor activation [27], lipid utilization [28], exit from meiotic and mitotic metaphases [29], mast cell signaling [30], and carcinogenesis [31, 32].
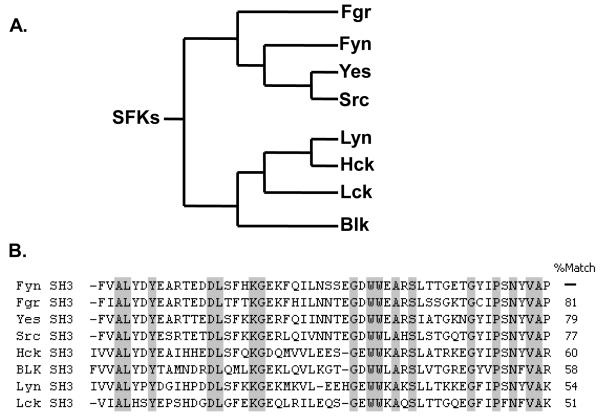
FIGURE 1.
Cladogram of human Src family kinases (SFKs) and sequence alignment of their SH3 domains. A. A phylogenetic tree of human SFKs, which is based on the similarity of their catalytic domains, and adapted from another publication [41]. B. The sequences of SH3 domains of SFKs were aligned to reveal their amino acid identity to the Fyn SH3 domain; conserved residues are highlighted in gray columns, and dashes are introduced to maximize alignment.
This work extends prior efforts in which we identified monobodies to the Src SH3 domain [16], and one of the isolated monobodies was converted into a biosensor of in situ activation of Src in cultured cells [33]. We were interested in extending this approach to other members of the Src family, which consists of Blk, Fgr, Fyn, Hck, Lck, Lyn, Src, and Yes [34]. We report the isolation of monobodies that are specific in binding to the Fyn SH3 domain. One of the isolated monobodies, G9, is highly selective and can be potentially used as a biosensor of Fyn kinase activation.
Materials & Methods
Construction of phage display library
A library of FN3 monobodies was constructed by oligonucleotide-directed mutagenesis [35, 36]. Briefly, the FN3 coding region was subcloned into a drop-out phagemid vector [37], which encodes a truncated form of gene III of the M13 bacteriophage and the DsbA signal sequence [38]. The recombinant phagemid construct was transformed into the bacterial strain CJ236 (New England BioLabs, Ipswich, MA), an Escherichia coli host used for producing uracil-containing single-stranded DNA (ssDNA). The ssDNA was annealed to two mutagenic oligonucleotides (IDT DNA, Coralville, Iowa), which encoded five NNK codons in the BC loop and FG loop (Fig. 2A). (N is an equimolar distribution of A, G, C, or T, whereas K is G or T. NNK allows incorporation of all 20 amino acids, plus the TAG amber codon.) With T7 DNA polymerase (New England BioLabs) and T4 DNA ligase (New England BioLabs), the ssDNA was converted into the double-stranded DNA (dsDNA) with the two annealed degenerate oligonucleotides. Synthesized dsDNA was recovered with a PCR clean-up kit (Qiagen, Valencia, CA), and electroporated into TG1 E. coli cells (Lucigen, Madison, WI). Transformed cells were immediately transferred to pre-warmed recovery medium (Lucigen), incubated at 37°C at 200 rpm for 35 min, and then aliquots were taken out for determining the transformation efficiency. To ensure optimal recovery, cells were allowed to recover for an additional 25 min, before being spun down and plated for overnight growth at 30°C. The output of 85 electroporations were pooled to generate a library containing 2.8×1010 transformants. The library DNA sequencing of 60 clones showed that both BC and FG loops were replaced by mutagenic sequences in 46% of the transformants. The library was estimated to have a diversity of 1.3×1010 recombinant clones (46% of 2.8×1010).
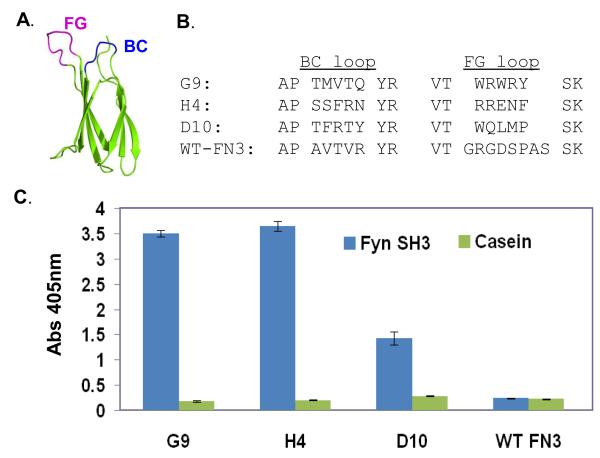
FIGURE 2.
Characterization of three FN3 monobodies that bind to the Fyn SH3 domain. A. Cartoon of the FN3 scaffold with its randomized BC and FG loops labeled. The beta-sheet secondary structures of FN3 are shown as arrows in the figure, which was generated with the PyMol program [59], using published coordinates (PDB: 1TTG structure) [60]. B. Sequences of the BC and FG loops of the three isolated monobodies and the wild-type domain. The length of FG loop of wild-type (WT) FN3 was shortened from eight residues to five residues in the library design. C. Phage ELISA of the three binders and the wild-type monobody (WT FN3). Casein served as the negative control target.
To amplify the phage particles displaying the recombinant FN3 monobodies, approximately 1.3 × 1011 cells were diluted into Luria Broth (LB; 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) - 50 μg/mL carbenicillin to a final density of OD600nm= 0.05. After 2 h incubation at 37°C at 250rpm, cells were infected with M13KO7 helper phage (New England BioLabs) at a multiplicity of infection (MOI) of 10, and then incubated at 37°C for 2 h at 150 rpm. Infected cells were recovered by centrifugation, resuspended in fresh LB (supplemented with 50 μg/mL carbenicillin and 100 μg/mL kanamycin), and incubated overnight at 30°C. The next day, secreted phage particles were separated from bacterial cells by centrifugation, followed by precipitation with 5% PEG8000 - 300 mM NaCl (final concentrations). The phage particle pellet was dissolved in phosphate buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4), and stored in 16% glycerol (with PBS) at −80°C.
Gene synthesis, plasmid preparation, protein overexpression, and purification
The coding sequences of FN3 monobodies and the Fyn SH3 domain were amplified by polymerase chain reaction (PCR), and subcloned into a modified form of the pET-14b expression vector (A gift from Dr. Arnon Lavie, University of Illinois at Chicago), which carries a His6-tag that is fused in frame to the small ubiquitin-like modifier (SUMO) at the N-terminus. The coding sequences of the other seven human SH3 domains of the SFKs were synthesized commercially (GenScript, Piscataway, NJ) and subcloned into the same vector.
All expression constructs were transformed into the BL21-DE3 (Novagen, Rockland, MA) or C41-DE3 (A gift from Dr. Arnon Lavie, University of Illinois at Chicago) strains of E. coli. Cells were inoculated into 10 mL LB with 50 μg/mL carbenicillin, and incubated at 37°C overnight. After a 30:1 dilution in auto-induction medium (Novagen), the cells were grown for 20-24 h at 30°C. The cell pellet was lysed with Bugbuster detergent (Novagen), which was supplemented with rLysosme (Novagen) and ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor (Roche). After 10 min of sonication (Thermo Fisher Scientific), the clarified lysate was incubated with nickel-nitriloacetic acid (Ni-NTA) resin (Qiagen) for immobilized metal affinity chromatography of the His6-tagged proteins. The SUMO fusion proteins (SH3 domain and monobody) were subsequently chromatographed through a Superdex 200 column (GE Healthcare, Piscataway, NJ) to attain >95% purity, with yields of 50-200 mg/L of cell culture.
Affinity selection
Three rounds of affinity selection were used to select monobodies binding to the Fyn SH3 domain. A Fyn SH3 domain fusion to glutathione-S-transferase (GST) was purified with GST bind resin (GE Healthcare) and chemically biotinylated with the EZ-link NHS biotinylation kit (Thermo Fisher Scientific). Neutravidin (Thermo Fisher Scientific) was diluted in PBS (25 ng/μL) and incubated in Nunc microtiter plate wells (Thermo Fisher Scientific) overnight at 4°C. The next day, after washing the wells, biotinylated Fyn SH3 domain-GST fusion protein (25 ng/μL) was added to microtiter plate wells for 20min at room temperature, followed by blocking of non-specific binding sites with casein (Thermo Fisher Scientific), which was supplemented with 10 μM free biotin. In the affinity selection experiment, ~5×1012 phage particles, which displayed variants of the FN3 monobody, were incubated in microtiter plate wells for 2 h, followed by three washes with PBS-0.1% Tween (PBST) and three washes with PBS. Bound phage particles were eluted with 50 mM glycine (pH 2.0). Eluted phages were neutralized with Tris-HCl (pH 10) and used to infect TG1 cells (OD600nm=0.4-0.6) for 1 h at 37°C at 150 rpm. Infected cells were spread onto Petri plates containing 1.5% agar, LB, and 50 μg/mL carbenicillin, and incubated overnight at 30°C. The next day, colonies were scraped from plates and ~5 × 108 cells were inoculated into 50 mL of LB + 50 μg/mL carbenicillin and grown for 2-3 h at 37°C at 250 rpm. When the culture reached an OD600nm =0.4-0.6, the cells were infected with M13K07 helper phage (MOI = 10) for 1 h, transferred to fresh LB + 50 μg/mL carbenicillin and 100 μg/mL kanamycin, and incubated overnight at 30°C. The next day, phage particles were harvested from the culture supernatant, precipitated with 5% PEG8000 and 300 mM NaCl (final concentrations), dissolved in PBS, and used for the next round of affinity selection. The second and third rounds of affinity selection were conducted in the same manner, except that PBS-dissolved phage particles was supplemented with soluble GST for binding to the Fyn SH3 domain in the microtiter plate wells, as a means of eliminating recovery of FN3 monobodies that bind to the GST portion of the fusion protein. After the third round of affinity selection, individual clones were picked for phage amplification, followed by enzyme-linked immunosorbent assay (ELISA) to identify binders to the Fyn SH3 domain. Subsequently, positive binding clones were sequenced.
Phage ELISA and mapping of binding location
Phage ELISA assays were performed as described in previous work [16]. Briefly, the SH3 domain fusion proteins were directly immobilized on Nunc microtiter plate (Thermo Fisher Scientific), by aliquoting 5 μg/mL solutions (in PBS) into triplicate wells, and incubating the plates overnight at 4°C. The following day, non-specific binding sites in the wells were blocked with excess casein (in PBS) for 1 h. After washing the wells with PBST, culture supernatants, which contained phage particles, were added to wells for 1 h incubation, followed by washes of microtiter plate and incubation with an anti-phage antibody that is conjugated to horseradish peroxidase (GE Healthcare). After 1 h incubation, the wells were washed, and the chromogenic substrate, 2,2′-Azino-bis(3-Ethylbenzthiazoline-6-Sulfonic Acid (ABTS), in the presence of hydrogen peroxide, was added and the resulting absorbance was determined at 405 nm with a microtiter plate spectrophotometer (BMG Labtech, Germany).
To determine if the isolated monobodies bind to the Fyn SH3 domain in the same site used for binding to proline-rich peptides, a competition binding assay was devised. First, we optimized the phage ELISA assay, with respect to signal generation for the lowest amount of phage binding particles. Second, a proline-rich peptide (VSLARRPLPPLPGGK), which is a peptide ligand for the Fyn SH3 domain [39], was added over a range of concentrations, from 0.1 μM to 512 μM, along with the phage particles to the wells. After 1 h of incubation, the wells were washed with PBST, and phage particles that were retained in the wells were detected by ELISA, as described above. A second peptide (VSAARAALAPLAGGK), in which several residues were substituted with alanine, served as a negative control. In a separate experiment (data not shown), we confirmed that the VSLARRPLPPLPGGK peptide bound to the Fyn SH3 domain and the VSAARAALAPLAGGK control peptide did not. In a parallel set of experiments, the binding of G9 to the Fyn SH3 was competed over a range of concentrations with the 1F11 monobody, a ligand with proline-rich binding motif.
Probing of SH3 domain array
Four PVDF membranes, spotted with a total of 150 human SH3 domains, were purchased from Panomics (Freemont, CA). The membranes were probed according to the manufacturer’s instructions. Briefly, the four membranes were soaked in the wash buffer, provided by the manufacturer, for 1 h until the membrane was completely wet. The soaked membranes were blocked for 2 h, prior to the addition of the His6-tagged SUMO-G9 monobody fusion at a final concentration of 1 nM. After overnight incubation at 4°C, the membranes were washed four times with PBST, followed by the addition of anti-his6-tag antibody, conjugated to HRP (diluted 1:1200). After 1 h incubation, the membranes were washed 10 times with PBST and then incubated with a substrate for enhanced chemiluminescence (ECL-Plus, GE Healthcare). The four membranes were scanned simultaneously with a blue fluorescence filter on the Storm 860 PhosphorImager (GE Healthcare).
Isothermal titration calorimetry
His6-tagged SUMO-FN3 monobody fusions and His6-tagged SUMO-SH3 domain fusions were purified to homogeneity of >95% and dialyzed together against 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 100 mM imidazole. After overnight dialysis, their concentrations were determined with a NanoDrop ND-1000 spectrophotometer. Degassed samples were added to the loading cell (1.4 mL) and syringe (300 μL) of a VP-ITC (GE Healthcare). FN3 monobodies were loaded into the syringe at 200 μM and SH3 domains were loaded into the cell at 22 μM. The reference well was loaded with water. FN3 monobodies were injected into the cell with a volume of 10 μL per injection at 25°C, with a reference power of 10 μcal/s. The heat change of each injection was recorded, and analyzed with Origin software (GE Healthcare).
Results & Discussion
One of the criteria of an affinity reagent’s usefulness is its specificity. This is especially difficult to achieve when generating affinity reagents to members of a closely related protein family. Members of human SFKs are evolved from the same kinase, Src64, and thus share a highly conserved sequence with one another [40]. Based on the sequence identity of the catalytic domain [41], the SFKs are divided into two groups (Fig. 1A): the SrcA group (Fgr, Fyn, Src, and Yes) and the SrcB group (Blk, Hck, Lck, and Lyn). In general, the Fyn, Src, and Yes protein kinases are ubiquitously expressed in different types of tissues while the other members are expressed mostly in myeloid-lineage cells [42]. Like the catalytic domain, the SH3 domain is highly conserved among the SFKs; for example, the SH3 domain of Fyn shares 51-81% identity with the SH3 domains of the other SFKs (Fig. 1B). As recombinant antibodies have the prospect of being more specific than natural antibodies, due to the ability to control the selection process in vitro [43], we decided to engineer a synthetic affinity reagent to the SH3 domain of Fyn.
Isolation of FN3 monobodies to Fyn SH3 domain
The coding sequence of FN3 monobody was subcloned into a new phagemid vector [37], fusing it with the truncated gene III of M13 bacteriophage, which allows monovalent display of the monobody protein on the phage surface. Diversifying the 5 residues of both the BC and FG loops of FN3 scaffold (Fig. 2A), by a modified form of Kunkel mutagenesis [36, 44], generated a library containing 1.3 × 1010 variants. The library was affinity selected for binders to the Fyn SH3 domain. The first experiment yielded one binding FN3 monobody, D10, and when repeated, yielded two more, G9 and H4. A follow-up ELISA of all three clones confirmed binding to the Fyn SH3 domain (Fig. 2C). The sequencing revealed that all three clones were different, with four residues shared between clones G9 and D10 (Fig. 2B). Interestingly, two of the clones, G9 and H4, contained no proline residues in either BC or FG loops, while the third clone, D10, had a single proline residue in the FG loop. The absence of proline is unexpected, as most cellular ligands for SH3 domains contain a proline-rich (i.e., PxxP) motif [22, 39, 45, 46]. However, the G9 clone contains an RxxK motif in its FG loop (RY-SK), which has been shown to be a non-canonical class of SH3-binding ligand [47]. Mutagenesis experiments and three-dimensional structural determinations will ultimately be necessary to learn the finer details of binding.
Characterization of binding specificity
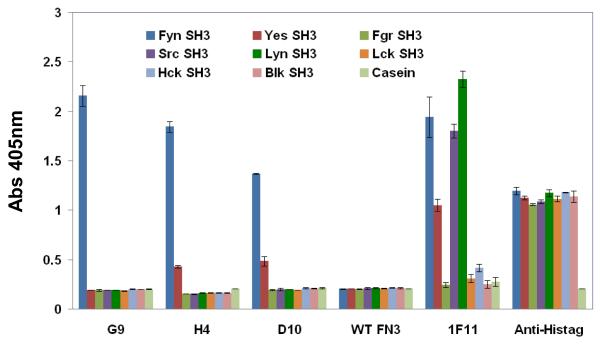
As the SH3 domains of SFKs are highly conserved in sequence, cross-reactivity is an important parameter to evaluate. To assess the binding specificity of the three monobodies, the SH3 domains of all eight SFKs were used in an ELISA (Fig. 3). Phage particles that displayed the G9 monobody bound exclusively to the Fyn SH3 domain with no cross-reactivity to the other seven SH3 domains, even though they share a common three-dimensional structure and are 51-81% identical in amino acid sequence to the Fyn SH3 domain. The same binding specificity was observed in the binding assays performed with purified protein of G9 monobody. The other two monobodies, H4 and D10, bound principally to the Fyn SH3 domain, with some cross-reactivity to the Yes SH3 domain. The binding of the D10 clone was improved through mutagenesis and affinity selection, resulting in 25-fold tighter binding (measured by Isothermal titration calorimetry, data not shown) to the Fyn SH3 domain and loss of binding to the Yes SH3 domain (data not shown).
FIGURE 3.
Phage ELISA of monobodies to the SH3 domains of all eight human SFKs. Microtiter plate wells were coated with equal amounts of target or casein (negative control), and probed with equivalent amounts of phage particles displaying the various binding monobodies. Phage particles displaying wild-type monobody (WT FN3) served as the non-binding negative control. 1F11 is a monobody that binds to several, but not all, SFKs SH3 domains. An anti-his6 tag antibody, conjugated to horseradish peroxidase (HRP), was used to normalize the amount of SH3 domain protein immobilized in the microtiter plate wells. Error bars correspond to standard deviation of triplicate measurements.
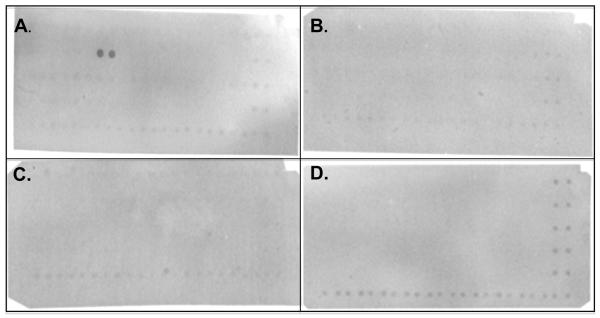
As there are 297 SH3 domains encoded in the human genome [48], it would be desirable to profile the cross-reactivity of the isolated monobody to all of them. Toward that end, a protein array containing 150 human SH3 domains was probed with the G9 monobody. Among the 150 SH3 domains arrayed on the four membranes, G9 bound exclusively with the Fyn SH3 domain but not to the other 149 (Fig. 4). This level of selectivity is remarkable given the relatively small size (i.e., 60 amino acids) and the highly conserved primary structure of SH3 domains. It should be noted that when G9 was fused to bacterial alkaline phosphatase, it bound predominantly to the Fyn SH3 domain, but showed some binding to three other SH3 domains in the array (i.e. Yes, Src, Grap-2-D2; data not shown). We interpret the slightly broadened specificity of the fusion protein to be caused by avidity effect, due to dimerization of bacterial alkaline phosphatase [49].
FIGURE 4.
Probing of SH3 domain array with the G9 monobody. Four membranes (purchased from Panomics) were incubated with His6-tagged SUMO fusion of the G9 monobody, washed, incubated with anti-His6-tag antibody conjugated to HRP, followed by detection with Enhanced Chemiluminescence-Plus (ECL-Plus). Panels A-D correspond to Panomics Arrays I-IV, respectively. Of the 150 SH3 domains tested, only the Fyn SH3 domain (duplicate, adjacent spots in panel A) bound to the G9 monobody. A His6-tagged ligand has been spotted in duplicate along the right side and the bottom of each membrane; these positive control spots are intended for alignment.
Measurement of dissociation constant
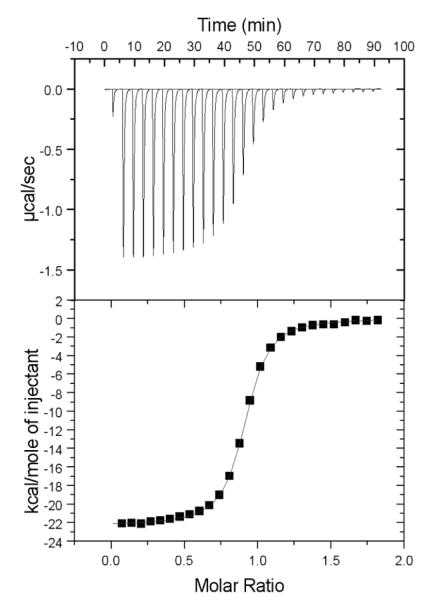
To determine the binding strength of the monobodies to the Fyn SH3 domain, isothermal titration calorimetry (ITC) was performed, in which the amount of heat change upon binding was plotted as a titration curve for calculating the dissociation constant (KD). Purified monobodies were injected into the sample cell loaded with Fyn SH3 domain, and the KD of G9 to Fyn SH3 domain was determined to be 166 ± 6 nM (Fig. 5). This value is approximately 10-fold lower than the KDs of most peptide ligands [39]. The clone D10 bound to the Fyn SH3 domain with a KD = 8.2 ± 0.5 μM, making it less attractive than G9 as an affinity reagent. However, the dissociation constant of an affinity-matured derivative of D10 could be lowered 25-fold to 325 ± 21 nM (data not shown), demonstrating that enhanced binding can be obtained through second round of protein engineering. To verify the selectivity exhibited in the ELISA and the array probing experiment, ITC was carried out with the SH3 domains of Fgr and Yes, the two closest relatives of Fyn. However, due to the small amount of heat released during the titrations, we could not determine dissociation constants of G9 binding to these two SH3 domains (data not shown).
FIGURE 5.
Isothermal titration calorimetry (ITC) measurements of the G9 monobody binding to the Fyn SH3 domain. The thermogram (top panel) and the plotted titration curve (bottom panel) were obtained with a Microcal VP-ITC. The observed N, ΔH, ΔS, and dissociation constant for this interaction are 0.89, −2.22×104 cal/mole, −43.37 Joule/°K, and 160 nM, respectively.
Mapping the binding location on the SH3 domain
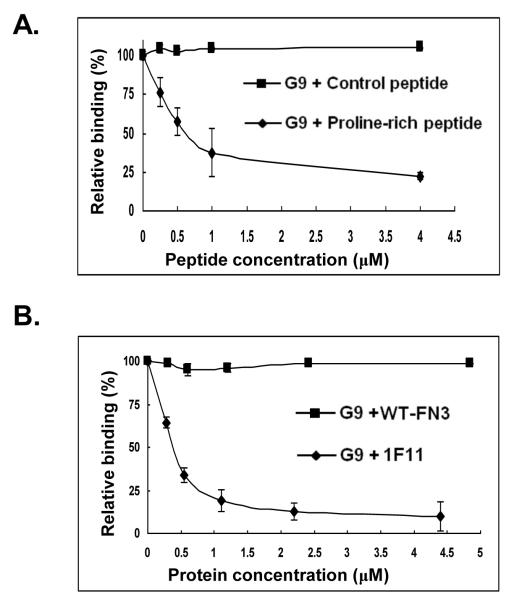
SH3 domains of SFKs contain a shallow groove that, upon kinase activation [50-53], binds to proline-rich peptides [46, 54-56]. Thus, ligands to this region of SH3 domain can be potentially used as biosensors for SFKs activation. As the monobody G9 does not contain such a proline-rich motif, it is of interest to learn if it binds to the Fyn SH3 domain in the same location or not. To that aim, a class I proline-rich peptide, VSLARRPLPPLPGGK, which binds to the Fyn SH3 with a KD of 0.6 μM [39], and a negative control peptide, VSAARAALAPLAGGK, were tested for their ability to compete with G9 for binding to the Fyn SH3 domain. In Figure 6A, it is shown that the Fyn SH3-binding peptide competed with G9 for binding to the SH3 domain in a dose-dependent manner, with an IC50 value of ~440 nM, while the negative control peptide did not compete at all.
FIGURE 6.
Mapping the binding location of the G9 monobody. A. Competition binding of the G9 monobody to the Fyn SH3 domain in the presence of a proline-rich peptide (VSLARRPLPPLPGGK) or a control peptide, a less proline-rich form of the same peptide (VSAARAALAPLAGGK). B. Competition binding of G9 monobody to the Fyn SH3 domain by the 1F11 monobody, which contains a proline-rich motif in its FG loop [16]. Wild type monobody (WT-FN3) served as the non-competitive control.
To confirm the above finding, 1F11, a monobody with a sequence of GISQG (BC loop) and SRPLP (FG loop) [16], was also used in the competition experiment. The 1F11 monobody binds to several SH3 domains of SFKs, including Src and Fyn. The chemical shift of residues in Src SH3 upon binding to 1F11 was mapped by NMR, which exhibited the same profile upon binding to a proline-rich peptide [16]. In the competition assay, 1F11 competed with G9 for binding to the Fyn SH3 domain with an IC50 value of ~210 nM (Fig. 6B). The wild-type FN3 (WT FN3) didn’t compete with the G9 for binding to the SH3 domain and served as the non-competitive control.
To determine if the G9 monobody can bind to full-length, active Fyn kinase, the biotinylated G9 monobody has been used in pull-down experiments. G9 monobody was capable of pulling down recombinant active Fyn kinase (data not shown). Thus, based on the above competition assays and pull-down experiment, we suggest that G9 can serve as a biosensor for monitoring the activation of Fyn kinase inside cells.
Recently with the advancement of phage display technology, highly selective reagents have been generated that distinguish closely related proteins. McCafferty and his colleagues [57] have isolated antibodies that can differentiate SH2 domains of Abl1 and Abl2, which share 89% identity. In addition, Koide and his coworkers [58] have identified a FN3 monobody that is capable of discriminating between the SH2 domains of Abl (1 and 2) and the other 82 SH2 domains, demonstrating an impressive level of selectivity. Here, we report the discovery of a FN3 monobody, which not only discriminated between Fyn SH3 domain and its closest relative, Fgr SH3 domain (81% sequence identity), but also exhibited exquisite selectivity to the SH3 domain of Fyn out of 150 SH3 domains in the human genome. In an unpublished work, we isolated monobodies that could differentiate between the SH3 domains of Abl1 and Abl2, which have 91% sequence identity. All of the above evidence suggests that FN3 monobody, even with a relatively small binding face due to its small molecular size (10 kDa), is a valuable scaffold for generating affinity reagents with high selectivity.
Acknowledgements
This work was funded by a research grant (GM082288) from the National Institutes of Health (NIH). We are grateful for the pET14-b-SUMO plasmid and C41-DE3 E. coli strain from Dr. Arnon Lavie at University of Illinois at Chicago. We appreciate the editorial comments of Ms. Kritika Pershad, Dr. Sujatha Koduvayur, and Mr. Michael Kierny.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gebauer M, Skerra A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr. Opin. Chem. Biol. 2009;13:245–255. doi: 10.1016/j.cbpa.2009.04.627. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson FY, Tolmachev V. Affibody molecules: new protein domains for molecular imaging and targeted tumor therapy. Curr. Opin. Drug Discov. Devel. 2007;10:167–175. [PubMed] [Google Scholar]
- 3.Silverman J, Liu Q, Bakker A, et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 4.Kolmar H. Alternative binding proteins: Biological activity and therapeutic potential of cystine-knot miniproteins. FEBS J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 5.Dai M, Temirov J, Pesavento E, et al. Using T7 phage display to select GFP-based binders. Protein Eng. Des. Sel. 2008;21:413–424. doi: 10.1093/protein/gzn016. [DOI] [PubMed] [Google Scholar]
- 6.Skerra A. Alternative binding proteins: anticalins - harnessing the structural plasticity of the lipocalin ligand pocket to engineer novel binding activities. FEBS J. 2008;275:2677–2683. doi: 10.1111/j.1742-4658.2008.06439.x. [DOI] [PubMed] [Google Scholar]
- 7.Muyldermans S, Baral TN, Retamozzo VC, et al. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009;128:178–183. doi: 10.1016/j.vetimm.2008.10.299. [DOI] [PubMed] [Google Scholar]
- 8.Boersma YL, Pluckthun A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr. Opin. Biotechnol. 2011 doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Xiong JP, Stehle T, Zhang R, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 10.Batori V, Koide A, Koide S. Exploring the potential of the monobody scaffold: effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002;15:1015–1020. doi: 10.1093/protein/15.12.1015. [DOI] [PubMed] [Google Scholar]
- 11.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc. Natl. Acad. Sci. U S A. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards J, Miller M, Abend J, et al. Engineered fibronectin type III domain with a RGDWXE sequence binds with enhanced affinity and specificity to human alphavbeta3 integrin. J. Mol. Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 13.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J. Mol. Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson CA, Liao HI, Sun R, Roberts RW. mRNA display selection of a high-affinity, modification-specific phospho-IkappaBalpha-binding fibronectin. ACS Chem. Biol. 2008;3:480–485. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbreth RN, Truong K, Madu I, et al. Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design. Proc. Natl. Acad. Sci. U S A. 2011;108:7751–7756. doi: 10.1073/pnas.1102294108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karatan E, Merguerian M, Han Z, et al. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem. Biol. 2004;11:835–844. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Aha P, Gu K, et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 18.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 19.Parker MH, Chen Y, Danehy F, et al. Antibody mimics based on human fibronectin type three domain engineered for thermostability and high-affinity binding to vascular endothelial growth factor receptor two. Protein Eng. Des. Sel. 2005;18:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl. Acad. Sci. U S A. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Makabe K, Biancalana M, et al. Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution. J. Mol. Biol. 2009;392:1221–1231. doi: 10.1016/j.jmb.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer BJ. SH3 domains: complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 23.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell. Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 25.McPherson PS. Regulatory role of SH3 domain-mediated protein-protein interactions in synaptic vesicle endocytosis. Cell. Signal. 1999;11:229–238. doi: 10.1016/s0898-6568(98)00059-x. [DOI] [PubMed] [Google Scholar]
- 26.Engen JR, Wales TE, Hochrein JM, et al. Structure and dynamic regulation of Src-family kinases. Cell. Mol. Life Sci. 2008;65:3058–3073. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmond RJ, Filby A, Qureshi I, et al. T-cell receptor proximal signaling via the Src-family kinases, Lck and Fyn, influences T-cell activation, differentiation, and tolerance. Immunol. Rev. 2009;228:9–22. doi: 10.1111/j.1600-065X.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- 28.Vatish M, Yamada E, Pessin JE, Bastie CC. Fyn kinase function in lipid utilization: a new upstream regulator of AMPK activity? Arch. Physiol. Biochem. 2009;115:191–198. doi: 10.1080/13813450903164348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levi M, Shalgi R. The role of Fyn kinase in the release from metaphase in mammalian oocytes. Mol. Cell. Endocrinol. 2010;314:228–233. doi: 10.1016/j.mce.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Siraganian RP, de Castro RO, Barbu EA, Zhang J. Mast cell signaling: the role of protein tyrosine kinase Syk, its activation and screening methods for new pathway participants. FEBS Lett. 2010;584:4933–4940. doi: 10.1016/j.febslet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116:1629–1637. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenone S, Brullo C, Musumeci F, et al. Fyn Kinase in Brain Diseases and Cancer: The Search for Inhibitors. Curr. Med. Chem. 2011 doi: 10.2174/092986711796150531. [DOI] [PubMed] [Google Scholar]
- 33.Gulyani A, Vitriol E, Allen R, et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat. Chem. Biol. 2011;7:437–444. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U S A. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss GA, Watanabe CK, Zhong A, et al. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. U S A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pershad K, Sullivan MA, Kay BK. Drop-out phagemid vector for switching from phage displayed affinity reagents to expression formats. Anal. Biochem. 2011;412:210–216. doi: 10.1016/j.ab.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner D, Forrer P, Stumpp MT, Pluckthun A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat. Biotechnol. 2006;24:823–831. doi: 10.1038/nbt1218. [DOI] [PubMed] [Google Scholar]
- 39.Rickles RJ, Botfield MC, Zhou XM, et al. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc. Natl. Acad. Sci. U S A. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu B, Shah B, Jablonowski B, et al. The SH2 domain-containing proteins in 21 extant species establish the provenance and scope of phosphotyrosine signaling in Eukaryotes. Science Signaling. doi: 10.1126/scisignal.2002105. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 42.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 43.Parsons HL, Earnshaw JC, Wilton J, et al. Directing phage selections towards specific epitopes. Protein Eng. 1996;9:1043–1049. doi: 10.1093/protein/9.11.1043. [DOI] [PubMed] [Google Scholar]
- 44.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2007;2:1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 45.Sparks AB, Rider JE, Kay BK. Mapping the specificity of SH3 domains with phage-displayed random- peptide libraries. Methods Mol. Biol. 1998;84:87–103. doi: 10.1385/0-89603-488-7:87. [DOI] [PubMed] [Google Scholar]
- 46.Lim WA, Richards FM. Critical residues in an SH3 domain from Sem-5 suggest a mechanism for proline-rich peptide recognition. Nat. Struct. Biol. 1994;1:221–225. doi: 10.1038/nsb0494-221. [DOI] [PubMed] [Google Scholar]
- 47.Berry DM, Nash P, Liu SK, et al. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr. Biol. 2002;12:1336–1341. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- 48.Karkkainen S, Hiipakka M, Wang JH, et al. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 2006;7:186–191. doi: 10.1038/sj.embor.7400596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlesinger MJ, Andersen L. Multiple molecular forms of the alkaline phosphatase of Escherichia coli. Ann. N. Y. Acad. Sci. 1968;151:159–170. doi: 10.1111/j.1749-6632.1968.tb11886.x. [DOI] [PubMed] [Google Scholar]
- 50.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 51.Williams JC, Weijland A, Gonfloni S, et al. The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions. J. Mol. Biol. 1997;274:757–775. doi: 10.1006/jmbi.1997.1426. [DOI] [PubMed] [Google Scholar]
- 52.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi H, Hendrickson WA. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 54.Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 55.Yu H, Chen JK, Feng S, et al. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 56.Musacchio A, Saraste M, Wilmanns M. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat. Struct. Biol. 1994;1:546–551. doi: 10.1038/nsb0894-546. [DOI] [PubMed] [Google Scholar]
- 57.Pershad K, Pavlovic JD, Graslund S, et al. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng. Des. Sel. 2010;23:279–288. doi: 10.1093/protein/gzq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wojcik J, Hantschel O, Grebien F, et al. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 2010;17:519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLano W. The PYMOL molecular graphics system. 2002.
- 60.Main AL, Harvey TS, Baron M, et al. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]