Abstract
Structural DNA nanotechnology utilizes DNA molecules as programmable information-coding polymers to create higher order structures at the nanometer scale. An important milestone in structural DNA nanotechnology was the development of scaffolded DNA origami in which a long single-stranded viral genome (scaffold strand) is folded into arbitrary shapes by hundreds of short synthetic oligonucleotides (staple strands). The achievable dimensions of the DNA origami tiles units are currently limited by the length of the scaffold strand. Here we demonstrate a strategy referred to as ‘super-origami’ or ‘origami of origami’ to scale up DNA origami technology. First, this method uses a collection of bridge strands to pre-fold a single stranded DNA scaffold into a loose framework. Subsequently, pre-formed individual DNA origami tiles are directed onto the loose framework so that each origami tile serves as a large staple. Using this strategy, we demonstrate the ability to organize DNA origami nanostructures into larger spatially addressable architectures.
Keywords: Structural DNA nanotechnology, self-assembly, DNA origami, scale up
Since the introduction of scaffolded DNA origami1 the technology has been extensively applied to engineer a variety of two-dimensional (2D)1–4 and three-dimensional (3D)5–14 nanostructures with a broad range of geometric complexities. One of the challenges to the functional development of DNA origami technology is to expand and adjust the size of the assemblies. Thus far the size has been restricted by the limited lengths of available single stranded DNA (ssDNA) scaffolds, where a 7 kilobase single stranded genome from the bacteriophage M13mp18 has become the standard. Two methods have recently been developed to address this problem. In the first approach, Shih and coworkers 15 utilized a one-pot assembly strategy to produce two different origami structures from a single double stranded scaffold (7560 bps). To achieve this, the initial double stranded DNA scaffold was denatured by a combination of heat and formamide to get complete separation of the forward and reverse scaffold strands. While denatured, the mixture was quickly cooled to room temperature to promote the faster hybridization of the staple strands and kinetically trap the scaffold-staple complexes. The remaining formation of the structures was achieved by gradually removing the formamide by dialysis. In the second method Woolley and co-workers4 used biotinylated primers in a PCR reaction to obtain single stranded DNA (ssDNA) products to be used as scaffolds for the assembly of origami structures. Using this approach they generated several different DNA origamis with sizes ranging from 756 to 4808 bps. Although large double stranded genomes are a promising source for longer DNA origami scaffolds, it is still not known how to optimize the assembly of larger structures. As the scaffold strand gets significantly longer the number of staple strands required to fold the scaffold will also drastically increase, which may result in considerable sequence mismatches. Furthermore, shear forces applied to longer scaffolds may lead to DNA breaks and only partial assembly of the target structures.
Another strategy to create large DNA origami superstructures is to connect individual origami tiles through sticky end associations. Recently, a periodic 2D lattice of DNA origami tiles was achieved by Seeman and co-workers16. They used a symmetric cross-like design with the helical axes of the component DNA propagating in two perpendicular directions to avoid nonspecific polymerization. This design strategy led to large periodic DNA origami lattices with dimensions up to 2 µm × 3 µm. This design has not been applied to create large discrete architectures with multiple units. In another effort, Sugiyama and coworkers17 employed a ‘JigSaw puzzle strategy’ which relied on shape complementarity and sticky end association to create a large, discrete DNA origami structure composed of 9 different DNA origami tiles with overall assembly efficiency of ~35%.
An alternative way to assemble larger DNA origami structures is to use more complex staples. We recently reported the use of 8-helix tiles (20 nm × 20 nm × 2nm), rather than single stranded oligonucleotides, as staples and demonstrated that DNA origami assemblies of more than 30000 bps can be constructed.18 Herein, we aim to determine whether the ‘tile staple’ concept can be applied to large DNA origami tiles (e.g. equilateral triangle shaped DNA origami tiles with 120 nm edges and 2 nm thickness) to create ‘origami of origami’ and, if successful, what are the key factors to achieve high assembly efficiency.
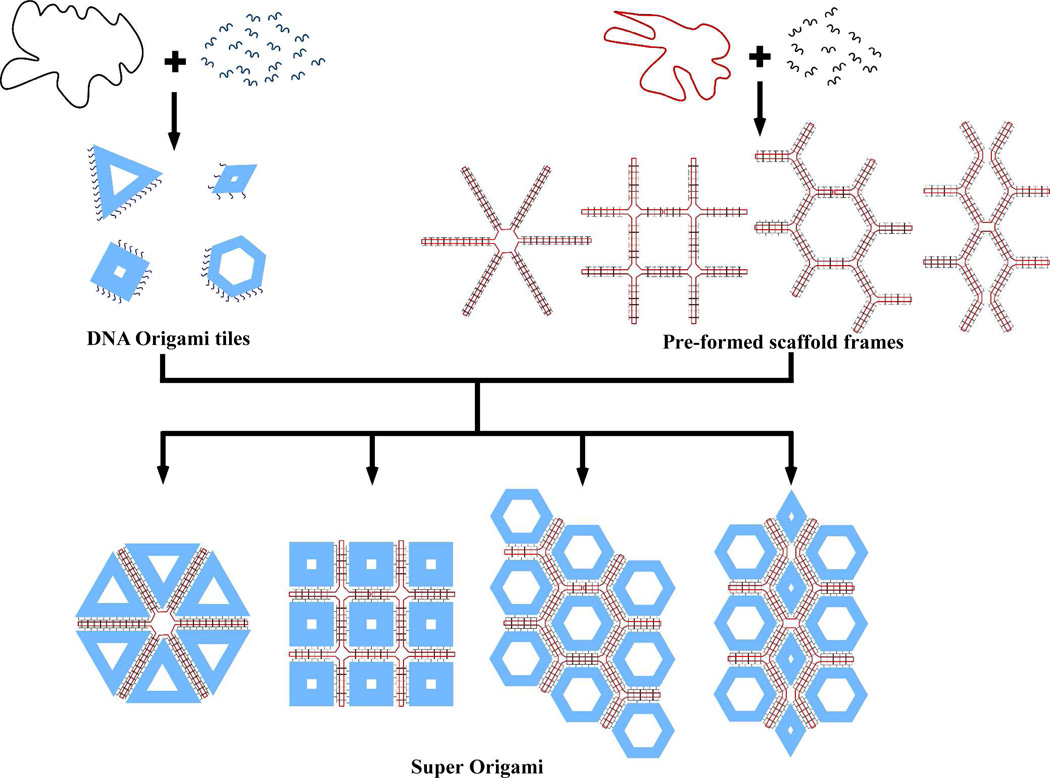
As illustrated in Figure 1, a multi-step folding procedure is necessary to implement the ‘origami of origami’ strategy. First a series of DNA origami tiles, each with a unique set of single stranded extensions (probes) is assembled in separate tubes. Concurrently, a loose framework is constructed by folding a different single stranded DNA scaffold with a separate group of bridge strands. Finally, the loose framework is folded further by the large, pre-formed origami tile staples through hybridization between the probes of the staple origami and the complementary sites within the loose framework. We demonstrated that very high assembly efficiencies (up to 85%) can be achieved by optimizing the formation of the loose framework and that the ‘origami of origami’ approach is a highly programmable approach to organize DNA origami tiles into larger complexes.
Figure 1.
Schematic illustrating the ‘origami of origami’. Top left: An M13 scaffold (black circular strand) is folded by a set of short DNA staples (blue strands) to form various individual DNA origami tiles. Each individual origami tile displays a group of single stranded extensions that are subsequently used as sticky-points to interact with the pre-formed scaffold frames shown on the right. Top right: A PhiX174 scaffold (red circular strand) is folded by a set of bridge strands (black strands) to form the loose frameworks that interact with the individual pre-formed origami tiles to create the various super-origami structures shown at the bottom.
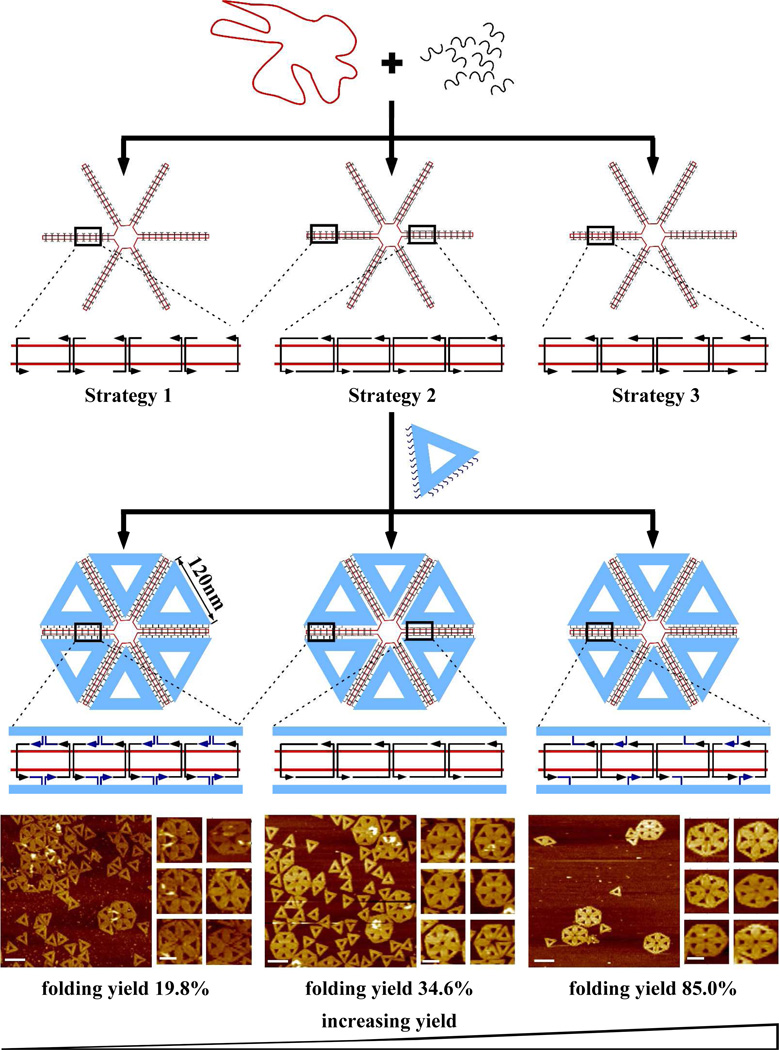
For our initial design (Figure 2) we used six triangular origami tiles (M13 scaffold; 120 nm × 120 nm × 120 nm) as the preassembled staple tiles (shown in blue) and single stranded PhiX174 as the scaffold forming the loose framework (shown in red) to assemble hexagonally shaped super-origami structures. The PhiX174 scaffold was partitioned into six equivalent loops; half of each loop was designed to interact with probes from a specific side of the triangular origami and the other half with a different side. The final side of the triangular origami tile remained unmodified. Three different strategies of association between the staple tiles and the framework scaffold were investigated with various yields of the final hexagonal super-structure. It should be noted that the single stranded PhiX174 scaffold shares little sequence similarity with the M13 scaffold so that any sequence overlap is minimal and can be neglected.
Figure 2.
Hexagonal shaped super-origami assembled from six individual triangular origami tiles. Three different strategies for the association between the origami tiles (blue) and the framework scaffold strand (red) are shown. Probe strands (dark blue, arrow points to 3‘ end) were extended from two sides of each of the individual origami tile and were designed to hybridize to specific positions within the framework scaffold strand. Periodic bridge strands (black) were also designed to assist the folding of the framework scaffold. AFM images of the final super-structures reveal varying efficiency among the designs, increasing from strategy 1 to strategy 3 (scale bar: 200 nm for zoom out images and 100 nm for zoom in images).
For strategy 1 (Figure 2, left), 22 probes were extended from two sides of the triangular origami staple tiles. Each ssDNA probe consisted of 8-nucleotides (shown in blue) that were designed to hybridize directly to the PhiX174 scaffold at the corresponding positions. Bridge strands (~ 16 nts long, shown in black) were designed to hybridize to the remaining portions of the scaffold framework, holding the framework in place and maintaining the correct spacing. Each crossover point (junction) between the individual triangular origami and the scaffold framework is formed from the participation of two probe strands. The distance between the neighboring crossovers of adjacent helices was kept at 32 bp, approximately three full turns.
For strategy 2, in contrast to strategy 1, only 12 probes were extended from the sides of the triangular origami tiles. In this design only the 12 probes corresponding to positions farthest from the center (with respect to the hexagonal super-structure) were kept, while the 10 probes nearest the center were deleted. The bridge strands (32 nt each) were extended to include the deleted positions and designed to hybridize to the available portions of scaffold strand at those locations. In this way, the potential twisting and structural tension in the super-structure that might occur due to inclusion of non B-form DNA conformations could be partially relaxed.
Strategy 3 involved the use of 11 probes spaced evenly along two arms of the triangular origami tiles. Unlike the first two strategies which contain reciprocal crossovers at the junctions between the individual triangular origami and the scaffold framework, each crossover point for strategy 3 is formed by a single probe strand. This design can more effectively relax any structural tension.
For all three design strategies, the formation of the hexagonal shaped super-origami was carried out in a two-step annealing procedure: 1) six individual triangular DNA origami tiles, with unique single stranded probes extended from two arms at selected positions, were annealed in separate tubes from 90 °C to 4 °C over 10 h in 1×TAE-Mg2+ buffer (pH 8.0, 20 mM Tris acetate, 1 mM EDTA, and 12.5 mM Mg(OAc)2), with a 1:10 molar ratio of the M13 scaffold strand to the staple strands. The annealed structures were purified with 100 KD MWCO Microcon centrifugal filters to remove any excess staple strands. Concurrently, the PhiX174 framework scaffold strand and the entire set of bridge strands were mixed, with a 1:10 molar ratio of the PhiX174 scaffold strand to the bridge strands, in a separate tube and annealed from 90 °C to 4 °C in 1×TAE-Mg2+ buffer. 2) The two solutions were subsequently mixed together (with a 1.5:1 or 2:1 molar ratio of individual origami tiles to framework scaffold) and annealed from 45 °C to 4 °C with a temperature gradient of 2°C per hour. The annealing program was repeated 10 times and in each consecutive cycle the starting temperature of the program was decreased by 0.5 °C. The entire annealing process lasted approximately 100 hrs.
In this design strategy each individual triangular DNA origami tile contains the same scaffold and most of the same staple strands, differing only in the locations and sequences of the probes; thus, it necessary to form each of the individual origami tiles separately in the first step. This prevents the individual origami tiles from forming incorrect associations with the PhiX174 scaffold strand. Assembly of the PhiX174 scaffold with the bridge strands pre-folds the scaffold framework into approximately the desired shape so that the subsequent addition of the pre-formed individual tiles will proceed efficiently, with each individual origami tile fitting into the evenly spaced cavities along the scaffold. This process is analogous to protein folding in which stepwise folding provides fast, pre-determined kinetic pathways to efficiently achieve the most thermodynamically stable folded structure.
The AFM images shown in Figure 2 reveal that strategy 3 has the best assembly efficiency, with approximately 85.0% complete (all six individual tiles) super-origami formation. The efficiency is calculated by multiplying the number of the complete hexagons by 6 and dividing the result by the total number of origami tiles. Strategy 2, which relieved some of the structural tension at the core of the hexagonal super-structure, resulted in ~ 34.6% assembly efficiency. Meanwhile, strategy 1 achieved only ~19.8% assembly efficiency. Furthermore, from the AFM images it is evident that the superstructure assembled by strategy 1 does not always form correctly; occasionally the individual origami tiles do not fit perfectly within the framework and the hexagonal superstructure often appears twisted or partially broken. The super-structures assembled by strategy 2 displayed improved morphology and those assembled by strategy 3 appear nearly perfect. These results indicate that relaxing the structural tension within the superstructure, either by deliberate probe placement or through single stranded crossovers (rather than reciprocal crossovers) between the individual tiles and the scaffold, can significantly improve the efficiency of super-structure assembly. In Rothemund’s original DNA origami report he attempted to utilize complementary sticky end association to organize six triangular DNA origami tiles into the same hexagonal structure.1 However, the reported assembly efficiency was only ~2%, lower than the efficiency achieved using all three strategies reported here, and much lower than what was achieved by strategy 3.
To test the versatility of our super-origami method we designed several other unique DNA origami staple tiles including square, hexagonal and diamond shaped tiles. For each of the additional staple tile systems we assembled the super-origami structures using the optimized folding strategy 3.
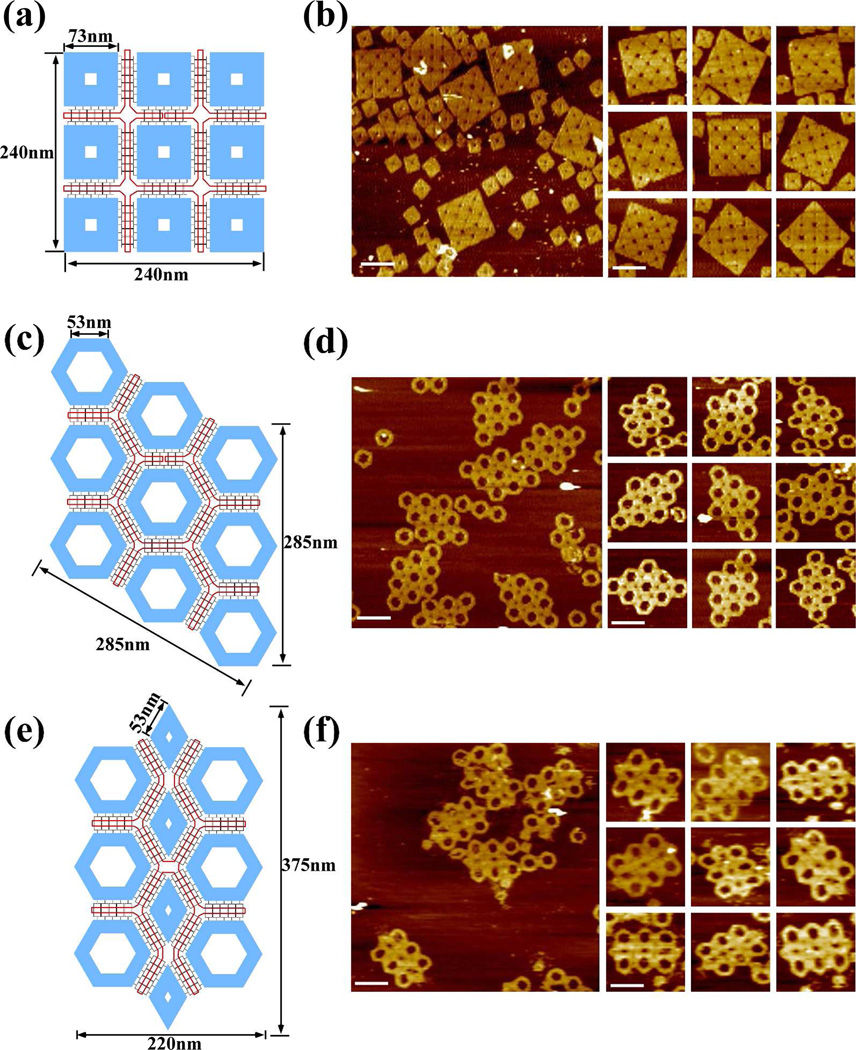
The square shaped staple tile14 has four equivalent sides, with each side consisting of nine parallel helices decreasing in length from the outermost to innermost layer (Figure 3). The longest helix is 224 base-pairs (bps), or 73 nm, in length. To form perfect 90 degree angles at each of the four corners, the length of helix n is designed to be 16 bps greater than the immediate neighboring helix n-1 (corresponds the relative outer helical layer). This is based on the consideration that an 8 bp DNA duplex has a length of ~ 2.5 nm; 2.5 nm is equal to the diameter of a single DNA double helix (2.0 nm) plus the estimated gap between two neighboring parallel double helices (0.5 nm). Nondenaturing gel electrophoresis (Figure S9) and AFM analysis (Figure S8) revealed that the square origami tiles formed properly with very high yield (>95%).
Figure 3.
Square, hexagonal and diamond shaped DNA origami staple tiles assembled into super-structures using the design strategy depicted in Figure 2. (a), (b) Design and AFM images, respectively, of 3 × 3 square staple tiles assembled into a super-structure. The length of each side of the individual square tiles is 73 nm; the length of each side of the super- structure is 240 nm. (c), (d) Design and AFM images, respectively, of 3 × 3 hexagonal staple tiles assembled into a super-structure. The length of each side of the hexagonal tiles is 53 nm; the length of each side of the super-structure is 285 nm. (e), (f) Design and AFM images, respectively, for mixed hexagonal and diamond staple tiles assembled into a super-structure. The length of each side of the diamond tiles is 53 nm; the dimensions of the super-structure are 220 nm × 375 nm. (scale bars:200 nm)
PhiX174 scaffold framework was pre-formed to accommodate nine square origami tiles, ultimately arranged in a 3×3 pattern within the super-structure. The super-structure was assembled following the same annealing procedure as described above, with 1:10:2 molar ratios between the PhiX174 scaffold strand, the bridges strands, and the pre-assembled square tiles. AFM images (Figure 3b) reveal that the super-structure is assembled with ~ 49% efficiency, somewhat lower than the folding efficiency for the triangle staple tile system.
The lower efficiency may have several causes: 1) 9 origami tiles were used in the square staple tile super-structure, while only 6 tiles were used in the triangle staple tile super-structure. It is possible that as the final assembly grows larger there is a requirement for more units to simultaneously associate with the correct stoichiometry resulting in a less favorable kinetic situation. 2) Although the total number of probe-scaffold framework connections is slightly more in the square staple tile superstructure than the triangle staple tile super-structure, 144 vs. 132, the number of probes per origami unit (on average) is fewer, 16 vs. 22. This is especially relevant to the 4 square tiles located in the corners of the square super-structure which are only linked to the scaffold framework by 12 probes, far fewer than the 22 probes per triangular origami in the hexagonal super-structure. Thus, the total enthalpy gain per origami unit tile is lower for the square tile than the triangular tile.
The hexagonal shaped staple tile (Figure S11) was designed with similar principles as the square staple tile. Each side contains nine parallel helices decreasing in length from the outermost (160 bps, or ~52 nm) to the innermost layer. To achieve the 120 degree angle at each corner, the length of helix n is designed to be 8 bps greater than the immediate neighboring helix n-1 (n corresponds the relative outer helical layer). Non-denaturing gel electrophoresis (Figure S13) and AFM analysis (Figure S12) confirm that the hexagonal tiles form as designed with >95% yield. PhiX174 scaffold framework was preformed to accommodate nine hexagonal origami tiles assemble in the same manner as described above, with 1:10:1.5 molar ratios between the PhiX174 scaffold, the bridge strands, and the individual origami tiles. AFM images (Figure 3d) reveal that this super-structure forms with efficiency ~55%, similar to the square staple tile system. The total number of probe-scaffold framework connections is ~ 160 and the average number of probe strands per hexagonal origami unit is 17.8, both of which are similar to the square super-origami structure.
The square and hexagonal staple tile super-structure assemblies demonstrate that nine individual origami unit tiles can be co-assembled with a PhiX174 scaffold framework with relatively high efficiency. Furthermore, each of the staple tile units share the same core strands, differing only in the sequences of the probe extensions which keeps the cost of super-structure assembly relatively low. Even when you consider the need for a second scaffold strand (PhiX174 to form the scaffold framework, the cost to assemble a large super-structure increases by less than 1 fold compared to an individual tile.
Finally, we designed a diamond shaped staple tile (Figure S15) and assembled it with the hexagonal staple tile and PhiX174 scaffold framework to form a super-structure with mixed staple tiles. The pattern of the final structure is similar to a tessellation pattern; the gaps between the hexagonal tiles are filled in by the smaller diamond shaped tiles (Figure 3e).
The diamond shaped staple tile was also designed with similar principles as the hexagon and square staple tiles. Each side is composed of 9 parallel helices and the length of the outermost helix is 160 bps, or 53 nm, the same length as in the hexagonal tile. One end of each side forms a 120 angle with the adjacent side, and the other end forms a 60 degree angle with the other adjacent side. The same strategy employed for the hexagonal staple tiles was used to create the 120 degree angles, i.e. 4 bps were deleted from each helix n-1 compared to the outer neighboring helix (n); 13 bps were deleted to make the 60 degree angles. The formation of the diamond shaped staple tiles was confirmed by nondenaturing gel electrophoresis (Figure S17) and AFM analysis (Figure S16). The entire M13 scaffold strand was not utilized to assemble the individual staple tiles; the unused portion was left as an unpaired loop in the inner cavity of the diamond. The single stranded loops can be observed in the background behind the super-structures in the AFM images.
Again the PhiX174 scaffold framework was pre-formed to accommodate the hexagonal and diamond shaped origami staple tiles and assembled in the same manner as described above, with 1:10:2:1.5 molar ratios between the PhiX174 scaffold, the bridge strands, the diamond shaped staple tiles and the hexagonal tiles. AFM results showed that the corresponding super-structure forms with ~ 41% efficiency. The lower efficiency may be related to the unique size and shape of the two origami staple tiles; notably, the diffusion and rotational dynamics of each of the tiles is expected to differ. In addition, the closely-packed design of the super-structure may impose considerable structural strain with the unit tiles experiencing increased steric hindrance. The unpaired region of the M13 scaffold within each staple tile may also interfere with the super-structure formation, ultimately reducing the overall yield.
In summary, we have improved and expanded upon the super-origami method that connects preassembled DNA origami tiles together to generate complex DNA super-structures. Uniquely shaped, geometric origami structures were designed and used as unit tiles to further assemble into large superstructures demonstrating the versatility of the method described here. The super-structures were assembled with high efficiency and exhibit an order of magnitude increase in size compared to the individual origami tile units. Super origami architectures formed from the triangular, square, hexagonal, hexagonal plus diamond origami unit tiles have molecular weights of 31.8 MD (96430 nt), 44.5 MD (134745 nt), 45.6 MD (138204 nt), 45.5 MD (137962 nt), respectively. The dimensions of the origami super-structures are close to the size domain of patterns generated by top-down photolithography, thus it may provide a viable approach to bridge bottom-up self-assembly with top-down methods and open up opportunities to build functional nanodevices. Future efforts are needed to establish protocols to purify the super-origami DNA structures from excessive unit origami tiles. Conventional DNA nanostructure purification methods such as agarose gel electrophoresis or semi-permeable membrane separation currently did not provide satisfying separation. It is also interesting to test whether the super-origami strategy can be applied to organize DNA origami tiles into 3D architectures, which may require more rigid frames as scaffolds. In this regard, DNA junction formed with a transition metal at the branch point19 may be used to create a rigid frame for 3D super-origami.
Supplementary Material
ACKNOWLEDGMENT
This work was partly supported by grants from the ONR, NSF, ARO, DOE, and NIH to H. Y., and from the Sloan Research Foundation to H.Y.; H.Y. and Y. L. were supported by the Center for Bio-Inspired Solar Fuel Production, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001016. H. Y. also thank grant from NSFC (21028005) and Wong Kwan Cheng education foundation scholarship. We thank Dongran Han and Zhe Li for helpful discussions and Jeanette Nangreave for proof-reading the manuscript.
Footnotes
Supporting Information Available. DNA sequences, materials, methods and additional AFM results. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Qian L, et al. Chin. Sci. Bull. 2006;51:2973–2976. [Google Scholar]
- 3.Anderson ES, et al. ACS Nano. 2008;2:1213–1218. doi: 10.1021/nn800215j. [DOI] [PubMed] [Google Scholar]
- 4.Pound E, Ashton JR, Becerril HA, Woolley AT. Nano Letter. 2009;9:4302–4305. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]
- 5.Anderson ES, et al. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 6.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shis WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzuya A, Komiyama M. Chem Comm. 2009;28:4182–4184. doi: 10.1039/b907800b. [DOI] [PubMed] [Google Scholar]
- 9.Endo M, Hidaka K, Kato T, Namba K, Sugiyama H. J. Am. Chem. Soc. 2009;131:15570–15571. doi: 10.1021/ja904252e. [DOI] [PubMed] [Google Scholar]
- 10.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Nano Letter. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 12.Liedl T, Högberg B, Tytell J, Ingber DE, Shih WM. Nature Nanotechnology. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han D, Pal S, Liu Y, Yan H. Nature Nanotechnology. 2010;5:712–717. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 15.Högberg B, Liedl T, Shih WM. J. Am. Chem. Soc. 2009;131:9154–9155. doi: 10.1021/ja902569x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Zhong H, Wang S, Seeman NC. Angew Chem Int Ed. 2010;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajendran A, Endo M, Katsuda Y, Hidaka K, Sugiyama H. ACS Nano. 2011;5:665–671. doi: 10.1021/nn1031627. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Yan H, Liu Y. Angew Chem Int Ed. 2010;49:1414–1417. doi: 10.1002/anie.200906225. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Alvater F, de Bruijin D, McLaughlin CK, Lo PK, Sleiman HF. Angew Chem Int Ed. 2011;50:4620–4623. doi: 10.1002/anie.201007403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.