Abstract
Protein misfolding has been proposed to be a common pathogenic mechanism in many inborn errors of metabolism including cystathionine β-synthase (CBS) deficiency. In this work, we describe the structural properties of nine CBS mutants that represent a common molecular pathology in the CBS gene. Using thermolysin in two proteolytic techniques, we examined conformation of these mutants directly in crude cell extracts after expression in E. coli. Proteolysis with thermolysin under native conditions appeared to be a useful technique even for very unstable mutant proteins, whereas pulse proteolysis in a urea gradient had limited values for the study of the majority of CBS mutants due to their instability. Mutants in the active core had either slightly increased unfolding (p.A114V, p.E302K and p.G307S) or extensive unfolding with decreased stability (p.H65R, p.T191M, p.I278T and p.R369C). The extent of the unfolding inversely correlated with the previously determined degree of tetrameric assembly and with the catalytic activity. In contrast, mutants bearing aminoacid substitutions in the C-terminal regulatory domain (p.R439Q and p.D444N) had increased global stability with decreased flexibility. This study shows that proteolytic techniques can reveal conformational abnormalities even for CBS mutants that have activity and/or a degree of assembly similar to the wild-type enzyme. We present here a methodological strategy that may be used in cell lysates to evaluate properties of proteins that tend to misfold and aggregate and that may be important for conformational studies of disease-causing mutations in the field of inborn errors of metabolism.
Electronic supplementary material
The online version of this article (doi:10.1007/s10545-011-9407-4) contains supplementary material, which is available to authorized users.
Introduction
Protein misfolding plays a role in the pathogenesis of many diseases, including inborn errors of metabolism, such as phenylketonuria due to phenylalanine hydroxylase deficiency, lysosomal storage disorders and various acyl-CoA deficiencies (Muntau and Gersting 2010). Aberrant folding of mutant proteins may be repaired by treatment of patients with chaperones, and the possible rescue of the mutants represents an interesting option for successful therapy of conformational disorders (Leandro and Gomes 2008). The efficacy and specificity of the search for novel compounds with chaperone activity may be significantly increased if detailed knowledge of the structural mechanisms underlying the misfolding of the mutants is available. This has been demonstrated by the implementation of a novel therapy for phenylketonuria using sapropterin (Muntau et al. 2002), which possibly acts a pharmacological chaperone (Erlandsen et al. 2004; Pey et al. 2004; Perez et al. 2005).
Homocystinuria due to cystathionine beta-synthase (CBS) deficiency is a common enzymopathy that affects the metabolism of sulfur amino acids and manifests clinically as a multisystemic disease (Mudd et al. 2001; Skovby et al. 2010). It has been proposed that many missense mutations in the CBS gene disrupt correct protein folding. This notion was supported by several lines of evidence, including the abnormal migration of mutant proteins in native electrophoresis (Janosik et al. 2001b; Kozich et al. 2010), the decreased solubility of mutants in SDS (Kozich et al. 2010) and the increased rate of proteasome-dependent proteolysis (Singh and Kruger 2009). The misfolding of CBS mutants was also inferred from the rescue of mutant enzymes by chemical chaperones (Singh et al. 2007; Majtan et al. 2010; Kopecka et al. 2011), CBS ligands (Kopecka et al. 2011), proteasome inhibitors (Singh et al. 2010), or by expression under temperature permissive to folding (Kozich et al. 2010). Taken together, all these experiments strongly indicate that misfolding plays an important role in CBS deficiency; however, the conformational changes underlying the abnormal folding have not been studied in detail.
Biophysical techniques used for conformational studies require relatively large amounts of purified proteins. However, only a few CBS mutants have been successfully produced as recombinant proteins in E. coli, purified to homogeneity in sufficient yields and structurally characterized, including the C-terminal mutants p.I435T, p.S466L (Janosik et al. 2001a) and p.D444N (Evande et al. 2002; Sen et al. 2005) and a double-linked mutant p.P78R/K102N (Sen and Banerjee 2007). Six additional CBS mutants (p.P49L, p.P78R, p.A114V, p.R125Q, p.E176K, p.P422L) were successfully purified in the presence of chemical chaperones in the culture media (Majtan et al. 2010). Because chaperones facilitate correct folding, it is not surprising that the chaperoned mutants did not exhibit gross structural abnormalities. It should be also noted that during the purification procedure, an unknown portion of misfolded and aggregated proteins may be lost and, consequently, the structural information about this fraction may not be obtained from studying purified proteins.
To study mutants that are not amenable to purification, proteolytic techniques in crude cell extracts may be used. It has been demonstrated that protein folding may be studied quantitatively by employing thermolysin, even in crude cell extracts (Chang and Park 2009; Kim et al. 2009). The methodology comprises two techniques: proteolysis under native conditions and pulse proteolysis in a urea gradient. The rate of proteolysis under native conditions reveals the extent of unfolding in the conformational ensemble of studied proteins because proteases can only cleave flexible regions and partially or globally unfolded structures (Young et al. 2007). Pulse proteolysis is a technique that uses thermolysin to monitore urea-induced unfolding. After a short proteolytic pulse, the fraction of folded proteins remains uncleaved and the unfolded species are digested. Using this premise, the c m value, a measure of protein thermodynamic stability in the urea gradient, may be determined (Park and Marqusee 2005).
In the present work, we applied these recently developed techniques, whose feasibility for the conformational study of CBS was shown using purified wild-type proteins (Prudova et al. 2006; Hnizda et al. 2010), to study CBS mutants expressed in E. coli. For these analyses, we selected the three most prevalent mutants and six additional mutants at different locations within the CBS molecule, and we examined if the conformation of the CBS mutant differs from the wild-type enzyme.
Results
Selection of CBS mutants
The mutations selected for this study are located in different domains of the CBS molecule (Meier et al. 2003; Yamanishi et al. 2006; Hnizda et al. 2010) (see Table 1). Advanced molecular modelling of the impact of mutations on enzyme structure could not be performed with sufficient reliability because the 3-D structure of human full-length wild-type CBS has not been experimentally solved at atomic resolution. As reported previously, these mutants vary in the degree of correct assembly and subsequent tetramer formation as well as in the levels of their catalytic activity (Kozich et al. 2010).
Table 1.
List of CBS mutants analyzed in this study.
| Protein | Location of the mutated residue | The number of tetramers (% of wild-type) | Relative enzyme activity (% of wild-type activity) | Reference for the location of the mutation |
|---|---|---|---|---|
| p.H65R | heme-binding pocket | 23.1 | 3.9 | (Meier et al. 2003) |
| p.A114V | dimer interface of the active core | 73.7 | 76.9 | (Meier et al. 2003) |
| p.T191M | solvent-exposed at the periphery of the active core | 1.7 | 0.3 | (Kozich et al. 2010) |
| p.I278T | buried in the active core | 1.0 | 0.3 | (Meier et al. 2003) |
| p.E302K | regulatory interface | 90.7 | 95.4 | (Meier et al. 2003; Hnizda et al. 2010) |
| p.G307S | catalytic site | 111.7 | 0.2 | (Meier et al. 2003) |
| p.R369C | regulatory interface | 7.8 | 1.8 | (Yamanishi et al. 2006) |
| p.R439Q | regulatory domain | 117.6 | 117.2 | (Kozich et al. 2010) |
| p.D444N | regulatory domain - AdoMet binding site | 120.2 | 163.8 | (Hnizda et al. 2010; Kozich et al. 2010) |
Relative enzyme activities and the number of tetramers are expressed as a mean value from data published elsewhere (Kozich et al. 2010).
Determination of proteolytic kinetics under native conditions
We optimized the previously described procedure (Chang and Park 2009) and final experimental conditions designed to evaluate the proteolytic cleavage of wild-type CBS in crude extracts over a time course of 20 min (for details, see Methods). The analysis of proteolytic cleavage using SDS-PAGE (Fig. 1) and native electrophoresis (Fig. S1 available in Supplementary_Info.pdf) did not reveal the presence of any partially cleaved fragments or unfolded intermediates, respectively. Therefore, the assumption for a model to describe protein unfolding and consequent proteolysis of CBS by thermolysin in which there is an equilibrium between folded tetramers and unfolded monomers (Park and Marqusee 2004a) was satisfied. Because the opening of the folded structure is necessary for sufficient proteolysis (Park and Marqusee 2004b; Chang and Park 2009), the determination of proteolytic kinetics provides data on the extent of the unfolding of the conformational ensemble.
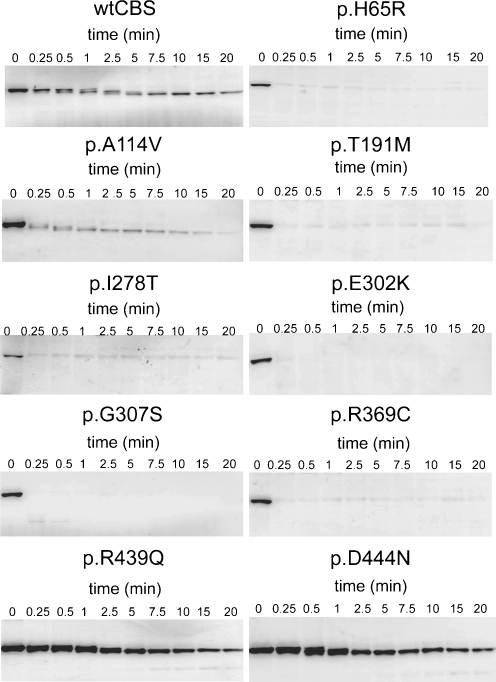
Fig. 1.
Proteolytic kinetics under native conditions for wild-type and mutant CBS proteins. Cell lysates (1 mg/ml protein) were cleaved by thermolysin (0.2 mg/ml). The amount of uncleaved protein was determined by Western blot followed by immunodetection with CBS antibody. Time points are depicted in minutes
The quantitative analysis of proteolysis under native conditions revealed that the CBS protein in crude cell extract comprises two different fractions. The major fraction of wild type CBS (≈80%) was cleaved via the first-order kinetics while the minor portion (≈20%) remained uncleaved at the end of proteolytic experiment. The data from the proteolytic kinetics of wild-type CBS were consistent with Eq. 1 (see Methods; r 2 = 0.9066; see Fig. S2A available in Supplementary_Info.pdf) and they show that the proteolysis with thermolysin under native conditions provides two parameters about the CBS proteins: the proteolytic rate of the major fraction and the relative abundance of the fraction that resists proteolysis during experiments.
Analysis of CBS mutants by native proteolysis
The kinetics of proteolysis under native conditions was successfully determined for all mutants. We show here that this method is robust and that it can be used for the analysis of even extremely unstable proteins.
All mutant proteins possessing an aminoacid substitution in the active core were cleaved too rapidly using the same default conditions for the capture of their proteolytic courses (Fig. 1). Therefore, the unstable mutants were exposed to lower and varying concentrations of thermolysin, which enabled the observation of the proteolytic time course and subsequent determination of its kinetics (Fig. S3, available in Supporting_Info.pdf). Because the dependence of the kinetic constant k obs on the thermolysin concentration appeared to be linear (as shown for wild-type CBS in Fig. S2B, available in Supplementary_Info.pdf), the proteolytic susceptibility of CBS mutants was assessed by comparing the k p values expressed in M−1 min−1; that is, k obs divided by the molar concentration of thermolysin in the proteolytic reaction (Park and Marqusee 2004b).
A significant increase in the proteolytic rates was observed for all mutants located in the active core, Fig. 1 illustrates the proteolysis of the mutants under the same conditions, and Table 2 provides the quantitative comparisons of the kinetic constant k p for the analyzed proteins. Unfortunately, faster cleavage of the active core mutant p.I278T could not be reliably quantified due to poor reproducibility of the proteolytic cleavage of this mutant at lower thermolysin concentration. In contrast, a decreased value of k p was observed for mutations located in the regulatory domain, suggesting a rigidification of their conformation.
Table 2.
Proteolytic kinetics of the CBS mutants under native conditions and pulse proteolysis in a urea gradient
| Proteolysis under native conditions | Pulse proteolysis | ||
|---|---|---|---|
| Protein | kp (106. M−1.min−1) | B (%) | Cm [M] |
| Wild-type | 0.30 ± 0.04 | 24.4 ± 1.7 | 3.05 ± 0.03 |
| p.H65R | 38.4 ± 5.1 | 8.2 ± 2.1 | N.D |
| p.A114V | 2.2 ± 0.4 | 24.1 ± 2.5 | 2.84 ± 0.04* |
| p.T191M | 44.3 ± 8.8 | 22.9 ± 3.0 | 2.4** |
| p.I278T | <2, 64.5>*** | 19.5 ± 1.2 | 2.42 ± 0.01 |
| p.E302K | 5.0 ± 0.6 | 0 ± 3.3 | N.D |
| p.G307S | 4.6 ± 0.4 | 0 ± 0.1 | N.D |
| p.R369C | 24.0 ± 3.5 | 23.4 ± 1.9 | 2.59 ± 0.09 |
| p.R439Q | 0.02 ± 0.01 | 25.7 ± 9.6 | 3.49 ± 0.04 |
| p.D444N | 0.03 ± 0.01 | 28.4 ± 6.2 | 3.32 ± 0.04 |
k p, B and c m values were determined by nonlinear data fitting from four different measurements. Representative gels with corresponding plots are shown in the Supplementary materials.
*The determination of c m for p.A114V neglected the cleavage of partially folded forms.
**Values were determined by visual inspection because these data could not be evaluated by nonlinear data fitting.
***k p could not be exactly determined due to low reproducibility at lower concentrations of thermolysin.
N.D. not determined due to the rapid proteolysis in the absence of urea.
In the next step, we compared the B values, which give the relative abundance of proteolytically resistant fractions of CBS proteins revealed from the determination of proteolytic kinetics. It should be noted that cleavage with lower concentrations of thermolysin might lead to an overestimation of the uncleaved protein fraction, as shown for wild-type CBS in Fig. S2C (available in the Supplementary_Info.pdf). However, an increased B value was not observed for any of the mutants analyzed in this study (Table 2). In contrast to wild-type CBS, several mutants (p.H65R, p.I278T, p.E302K and p.G307S) exhibited a decreased B value an increased k p. These results indicate that the conformer distribution in the structural ensemble of these mutants may possibly be shifted towards the unfolded intermediates.
In summary, proteolysis under native conditions revealed that all mutants had a different degree of structural flexibility. Mutations in the active core had an increased extent of protein unfolding, whereas mutations in the regulatory domain tended to form more rigid protein structures.
Pulse proteolysis
A previously developed procedure for pulse proteolysis of purified CBS (Hnizda et al. 2010) was successfully adopted for crude cell extracts and the data for wild-type CBS followed the two-state unfolding model (r 2 = 0.9562; Fig. S2D available in Supplementary_Info.pdf). Pulse proteolysis therefore describes an equilibrium between folded tetramers and unfolded monomers in a urea gradient; the validity of this model is further supported by a previous study (Kery et al. 1998) reporting that higher concentrations of urea lead to the dissociation of CBS tetramers.
Because a proportion of folded wild-type CBS was cleaved during the proteolytic pulse even in the absence of urea (see proteolysis under native conditions), a possible systematic error in the determination of c m was assessed according to a previously described procedure (Park and Marqusee 2005, 2006). The kinetic constant for proteolysis in 2.5 M urea was determined to be 0.13 ± 0.01 min−1 (see Fig. S4, available in Supplementary_Info.pdf), indicating negligible cleavage of folded protein during the proteolytic pulse at the beginning of the transition zone and, consequently, the absence of a systematic error (Park and Marqusee 2006).
It should also be noted that the c m calculated from the pulse proteolysis of multimeric protein does not simply represent global protein stability (ΔGunf°) and that the structural stability of the protein is dependent on its concentration (Park and Marqusee 2004a). However, the amount of CBS mutants in cell extracts was similar to or slightly less than the wild-type protein (Fig. S5, available in Supplementary_Info.pdf). A significant decrease in protein level to ~20% of wild-type enzyme was observed only for p.I278T. Although the decreased amount of p.I278T might lead to an underestimation of its stability, the experiments with a decreased amount of wild-type CBS (described in Materials and methods) did not reveal any changes in the apparent c m values. It was also proposed that, in spite of some limitations, c m values are still valuable for comparing protein stability under the same conditions (Schlebach et al. 2010). Consequently, we employed pulse proteolysis in a urea gradient for conformational studies of CBS mutants in crude cell extracts.
Analysis of CBS mutants using pulse proteolysis in a urea gradient
Pulse proteolysis in a urea gradient was used for six mutant proteins, whereas three mutants (p.H65R, p.E302K and p.G307S) were cleaved rapidly during the proteolytic pulse in the absence of urea, and less than 10% of the CBS protein remained uncleaved. It has been shown that pulse proteolysis in a urea gradient cannot be used for rapidly cleaved proteins (Chang and Park 2009).
Analysis of the mutantions in the regulatory-domain revealed higher c m values as a measure of increased protein stability (Table 2, Figs. S4 and S6, available in Supplementary_Info.pdf). However, we experienced several difficulties during the evaluation of mutants with an increased rate of proteolysis observed under native conditions (p.A114V, p.T191M, p.I278T and p.R369C). In the case of the slightly unfolded mutant p.A114V, we observed a significant proteolytic susceptibility at the beginning of the apparent transition zone (Fig. S4 in Supplemetary_Info.pdf), and the requirement for the kinetic constant to be less than 0.2 min−1 for an accurate determination of c m was therefore not satisfied. Despite this, pulse proteolysis can yield at least partial knowledge about the urea-induced transition between the partially and globally unfolded fractions. An analysis accounting for a systematic error (see Fig. S7, available in Supplementary_Info.pdf) showed that the correct value of c m could in fact be lower that the apparent value. Taken together, both the corrected and uncorrected value of c m indicated that the protein stability of the mutant p.A114V was at least slightly decreased compared to wild-type CBS.
Another type of obstacle arose during the pulse proteolysis of severely unfolded proteins, p.T191M, p.I278T and p.R369C. The amount of CBS mutants in cell lysates decreased after the overnight incubation even in the absence of urea in the uncleaved control samples, perhaps due to the formation of precipitates that did not enter into the electrophoretic gel (a representative gel together with a quantitative comparison of the amount of soluble CBS antigen before and after overnight incubation is shown in Fig. S8, available in Supplementary_Info.pdf).Therefore, pulse proteolysis could only be assessed for the soluble fraction of these mutants. Interestingly, the proteins were not significantly cleaved at the beginning of their apparent transition zone, indicating the reliable determination of these values (Fig. S4 in Supplementary_Info.pdf). The analyses showed a significant decrease in the stability of these mutant proteins, which is consistent with the higher extent of unfolding revealed by native proteolysis. However, these results should be interpreted with great care because we analyzed only the limited fraction of the conformational ensemble that did not precipitate.
It should be noted here that the reported difficulties in pulse proteolysis are most likely caused by the instability of the mutants. Various modifications of experimental procedures, such as a lower amount of thermolysin in the assay or a shorter equilibration time of cell lysates in urea, did not improve the outcome of the experiments (data not shown). This suggests that pulse proteolysis in a urea gradient for the conformational analysis of highly unstable mutant proteins may be of limited value. In spite of these limitations, pulse proteolysis showed that the different extent of the unfolding of CBS mutants is associated with corresponding changes in protein stability: unfolded mutant proteins had lower stability, whereas rigidified mutants were more stable than wild-type CBS.
Correlation of protein unfolding with the degree of tetramer assembly and with enzyme activity of CBS mutants
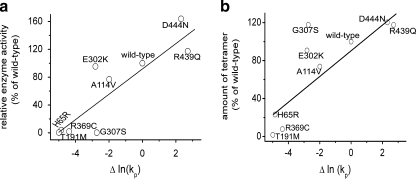
The data from proteolysis under native conditions were correlated with the number of correctly assembled tetramers and their catalytic activity, which was reported in a previous study (Kozich et al. 2010). The relative positions of the mutations are similar in both plots, with the exception of the catalytic site mutant p.G307S (compare Fig. 2a and b). The plots show that hypoactive and incorrectly assembled mutants had a higher extent of protein unfolding, whereas hyperactive and properly assembled mutants were more rigidified that wild-type CBS. This analysis indicates a trend between the proteolytic resistance, k p, and the number of tetramers (R 2 = 0.6646; p = 0.0074) and enzyme activity (R 2 = 0.7566; p = 0.0015) (Fig. 2). These data may imply that an optimal degree of conformational flexibility is a necessary condition for correct tetrameric assembly which further determines the catalytic efficiency of CBS mutants.
Fig. 2.
Conformational properties of CBS mutants: proteolytic data correlated with enzyme activity and with the degree of tetrametric assembly. The dependence of the proteolytic kinetics with thermolysin under native conditions (ln (k p)) on the enzyme activity of the CBS mutants (A) and on the number of assembled tetramers (B) – the correlation analysis indicates a possible trend between the proteolytic resistance and the number of tetramers (R 2 = 0.6646; p = 0.0074) and enzyme activity (R 2 = 0.7566; p = 0.0015). All values for the mutants are normalized to the number of tetramers or activity of the wild-type as appropriate
Discussion
In this study, we describe conformational properties of nine disease-causing CBS mutants that previously resisted purification and further characterization. By using proteolysis of mutants in crude cell extracts under native conditions and pulse proteolysis in a urea gradient, we demonstrated that the conformational properties of all analyzed mutants differ from the wild-type protein. Based on these data, we propose a possible molecular mechanism for the misfolding and misassembly of CBS mutants. Using a modified mathematical model for the folded tetramers-unfolded monomers equilibrium (Park and Marqusee 2004a), mutants may be divided into three major groups:
-
i)
Severely misassembled mutants with low residual activity and extensive unfolding (p.H65R, p.T191M, p.I278T and p.R369C). These mutants predominantly form aggregates that are likely composed of weakly interacting subunits with unfolded regions of high sensitivity to proteolytic cleavage.
-
ii)
Correctly assembled mutants that are only slightly unfolded (catalytically active p.A114V and p.E302K and the inactive p.G307S located in the active site). These proteins are characterized by increased unfolding through kinetically and/or thermodynamically more favourable formation of unfolded intermediates.
-
iii)
Correctly assembled hyperactive mutants that are more stable and more tightly folded (p.R439Q and p.D444N) due to a decreased formation of unfolded species.
Altered conformational properties of CBS mutants may affect protein turnover in vivo
The higher proteolytic susceptibility of the majority of CBS mutants in this study indicates their inclination to unfolding. This observation is consistent with previous work by Singh and Kruger demonstrating that the most common mutant, p.I278T, exhibited accelerated ubiquitin-dependent proteolysis in S. cerevisiae and that turnover of the mutated protein could have been altered by a manipulation of the levels of heat shock proteins (Singh and Kruger 2009). Furthermore, several mutants were rescued by treatment with the proteasome inhibitor bortezomib in yeast or by MG132 in patient-derived fibroblasts (Singh et al. 2010), indicating that more rapid degradation plays an important role in the pathophysiology of CBS deficiency.
All these data are consistent with a hypothesis that the unfolded intermediates of CBS mutants may be rapidly degraded by proteasome-dependent proteolysis. Increased protein unfolding was reported for oligomeric enzymes that are deficient in other inherited metabolic diseases such as S-adenosylhomocysteine hydrolase (Beluzic et al. 2006), medium chain acyl-CoA dehydrogenase (Maier et al. 2009) and electron transfer flavoprotein (Schiff et al. 2006). Furthermore, the relationship between increased proteolytic susceptibility in vitro and accelerated proteasome-dependent turnover in vivo was also demonstrated for misfolded mutants of alanine:glyoxylate aminotransferase (Coulter-Mackie and Lian 2006, 2008) and phenylalanine hydroxylase (Waters et al. 1999; Gersting et al. 2008).
Conversely, mutations in the regulatory domain of CBS led to an increase in proteolytic stability. This indicates that the conformation of these mutants is less flexible than of the wild-type protein. It remains to be addressed how the increased conformational rigidity and hyperactivity of the C-terminal mutants could result in their pathogenicity in vivo. In previous studies, the representative C-terminal mutants, p.I435T, p.D444N and p.S466L, had higher activity than the wild-type enzyme together and a lower affinity towards an allosteric activator of CBS, S-adenosylmethionine (AdoMet), and they failed to change conformation upon AdoMet binding (Janosik et al. 2001a; Evande et al. 2002; Sen et al. 2005). Based on these observations, it was proposed that the altered response to AdoMet may play a role in the pathogenicity of these C-terminal mutations (Yamanishi et al. 2006; Kozich et al. 2010). Interestingly, other work showed that the expression of the C-terminal mutant p.S466L in a transgenic mouse led to a lower steady-state protein level and to decreased catalytic efficiency in vivo (Gupta et al. 2008). Decreased amounts of hyperactive mutants were also observed in crude cell extracts for two patient-derived fibroblast cell lines carrying the mutations p.I435T/del ex8 (Maclean et al. 2002) or p.D444N/p.D444N (Evande et al. 2002). Based on these observations, we speculate that the more tightly folded mutants may be recognized by the cellular quality control machinery, thereby leading to their accelerated degradation.
It should be noted that our study revealed several limitations of proteolytic experiments for conformational studies in crude cell extracts. Pulse proteolysis could not be applied for three of the nine mutant proteins (p.H65R, p.E302K and p.G307S) because these mutants were substantially cleaved during the proteolytic pulse, even in the absence of urea. In addition, we experienced difficulties with the evaluation of the other unfolded mutants (p.A114V, p.T191M, p.I278T and p.R369C) due to significant cleavage at the beginning of the apparent transition zone or precipitation events during the overnight equilibration in the urea gradient. In this case, the determination of c m values and subsequent data interpretation must be performed carefully and pulse proteolysis in the urea gradient may indicate changes in thermodynamic stability of unfolded mutant proteins.
On the other hand, the determination of the rate of proteolysis with thermolysin under native conditions appeared to be robust and provided conformational data about all mutants studied. Despite this, there were some obstacles while performing this technique: a) the relatively demanding optimalization of the procedure in order to find an appropriate ratio of the total protein in lysates to the thermolysin concentration (w/w) to accurately determine proteolysis kinetics; b) the presence of a small protein fraction that was not cleaved according to first-order kinetics and that remained intact and catalytically active at the end of experiments (data not shown). Because the small uncleaved fraction was also observed in mammalian HepG and HeK cell-lines with endogenous CBS expression (our unpublished data) but not with purified CBS proteins (Hnizda et al. 2010), it is possible that the minor fraction of CBS in cell lysates may result from intermolecular interactions with small ligands or high molecular weight molecules such as other proteins or nucleic acids. This uncleaved fraction is most likely not caused by binding of a small compound because the dialysis of the bacterial lysate containing wild-type CBS prior to the assay did not significantly change the proteolytic time-course (data not shown). In spite of these limitations, the proteolytic techniques used in crude cell extracts enabled the expansion of our knowledge about the conformational properties of the CBS mutants that are not amenable for purification and consequently not available for detailed studies by more conventional methods.
We present here a general methodological strategy that may be used directly in crude cell extracts to evaluate properties of proteins that tend to misfold and aggregate and that may be important for conformational studies of disease-causing mutations in the field of inborn errors of metabolism. This approach may benefit the conformational analysis of mutants after expression without affinity tags or after endogenous expression in patient-derived fibroblasts, other available cell lines or tissues. Proteolytic techniques are able to reveal conformational abnormalities even in mutant enzymes with normal catalytic activity and may be applied to the mutational analysis of non-enzymatic proteins whose functional assays are lacking or are difficult to employ.
Materials and methods
Materials
If not specified otherwise, all chemicals were purchased from Sigma-Aldrich.
Expression of CBS enzymes and preparation of crude cell extract
The CBS proteins were expressed at 37°C in E. coli transformed with pHCS3 plasmids that express untagged full-length wild-type or mutant CBS proteins (Kozich and Kraus 1992). Crude cell-extracts were prepared as described previously (Kozich et al. 2010). The protein concentration was determined using Bradford reagent with bovine serum albumin as a standard.
Proteolysis with thermolysin under native conditions
Crude cell extracts (final protein concentration 1 mg/ml; total volume 25 μl) were diluted in 20 mM Tris-HCl (pH 8.0) containing 10 mM CaCl2, equilibrated at 25°C for 10 minutes and then digested with thermolysin. The concentration of thermolysin ranged from 0.4 to 200 μg/ml as described in the Results. At the selected time points, proteolysis was quenched in 20 mM EDTA.
Pulse proteolysis
Pulse proteolysis was performed according to a previously reported procedure (Hnizda et al. 2010) with some modifications. Cell lysates (final protein concentration 1 mg/ml; total volume 25 μl) were incubated overnight at 4°C in 20 mM Tris-HCl (pH 8.0) containing 10 mM CaCl2 and urea (0–7 M). After subsequent equilibration at 25°C for 10 minutes, the samples were digested by thermolysin from Bacillus thermoproteolyticus (0.2 mg/ml) at 25°C. The proteolytic pulse (60 s) was quenched in 20 mM EDTA.
Because the thermodynamic stability of multimers may be dependent on protein concentration, we assessed the effect of different amounts of wild-type CBS in the assay on the determined c m values. Pulse proteolysis of the wild-type CBS was performed for lower concentration of total protein (0.5 or 0.25 mg/ml) that was digested by the corresponding amount of thermolysin (100 or 50 μg/ml). The lower protein concentration in pulse proteolysis did not change the apparent c m values (data not shown). These experiments showed that lower amounts of CBS proteins in the assay did not have an effect on the accurate determination of their thermodynamic stability implying that this approach is also useful for mutant proteins with decreased steady-state levels.
Western blotting and data fitting
The amount of uncleaved CBS was analyzed under denaturing or native conditions by the Western blot procedure described previously (Kopecka et al. 2011). The CBS antigen was detected using purified chicken anti-hCBS serum H19 (HenA, Czech Republic) diluted 1:500 in 3% non-fat dry milk in 10 mM Tris-HCl (pH 7.4) containing 150 mM NaCl. Secondary rabbit anti-chicken horseradish-peroxidase-cojugated antibody (ThermoFischer Scientific) was diluted 1:15,000 in PBS containing 3% non-fat dry milk. The signal was quantified, and the data were fitted to non-linear curves as reported previously (Hnizda et al. 2010; Kozich et al. 2010). Each experiment was carried out twice, each in duplicate; data shown in figures and tables are therefore the means and standard deviations of four determinations.
Proteolysis under native conditions was evaluated using the first-order kinetics (Park and Marqusee 2004b). Data were fitted into the equation as follows:
| 1 |
where N and N 0 are the amounts of intact protein, k obs is the first-rate kinetics constant, t is time and B is the fraction of protein that was not cleaved via the first-order kinetics and that remained uncleaved at the end of proteolytic experiment. Data from the pulse proteolysis were fitted into the sigmoidal equation as follows:
| 2 |
where f fold is a fraction of folded proteins remaining intact after the proteolytic pulse and is related to the amount of uncleaved protein after the pulse in absence of urea, c m is the urea concentration at which f fold is 0.5 and c is the urea concentration. p is a slope of the curve at c m and reflects unfolding cooperativity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 3.30 mb)
Acknowledgements
We thank Dr. Jakub Krijt for determination of CBS activity in the proteolytically resistant fraction.
Funding
This work was supported by the Welcome Trust International Senior Research Fellowship in Biomedical Science in Central Europe (reg. No 070255/Z/03/Z), by the grant No. 7310 from Grant Agency of Charles University in Prague and by the grant SVV262502 from the Charles University in Prague. Institutional support was provided by the Research Project from the Ministry of Education of the Czech Republic No. MSM0021620806.
Authors confirm independence from the sponsors; the content of the article has not been influenced by the sponsors.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Competing interests: None declared.
References
- Beluzic R, Cuk M, Pavkov T, et al. A single mutation at Tyr143 of human S-adenosylhomocysteine hydrolase renders the enzyme thermosensitive and affects the oxidation state of bound cofactor nicotinamide-adenine dinucleotide. Biochem J. 2006;400:245–253. doi: 10.1042/BJ20060749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Park C. Mapping transient partial unfolding by protein engineering and native-state proteolysis. J Mol Biol. 2009;393:543–556. doi: 10.1016/j.jmb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Coulter-Mackie MB, Lian Q. Consequences of missense mutations for dimerization and turnover of alanine:glyoxylate aminotransferase: study of a spectrum of mutations. Mol Genet Metab. 2006;89:349–359. doi: 10.1016/j.ymgme.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Coulter-Mackie MB, Lian Q. Partial trypsin digestion as an indicator of mis-folding of mutant alanine:glyoxylate aminotransferase and chaperone effects of specific ligands. Study of a spectrum of missense mutants. Mol Genet Metab. 2008;94:368–374. doi: 10.1016/j.ymgme.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Erlandsen H, Pey AL, Gamez A, et al. Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations. Proc Natl Acad Sci USA. 2004;101:16903–16908. doi: 10.1073/pnas.0407256101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evande R, Blom H, Boers GH, Banerjee R. Alleviation of intrasteric inhibition by the pathogenic activation domain mutation, D444N, in human cystathionine beta-synthase. Biochemistry. 2002;41:11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- Gersting SW, Kemter KF, Staudigl M, et al. Loss of function in phenylketonuria is caused by impaired molecular motions and conformational instability. Am J Hum Genet. 2008;83:5–17. doi: 10.1016/j.ajhg.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wang L, Hua X, Krijt J, Kozich V, Kruger WD. Cystathionine beta-synthase p.S466L mutation causes hyperhomocysteinemia in mice. Hum Mutat. 2008;29:1048–1054. doi: 10.1002/humu.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnizda A, Spiwok V, Jurga V, Kozich V, Kodicek M, Kraus JP. Cross-talk between the catalytic core and the regulatory domain in cystathionine beta-synthase: Study by differential covalent labeling and computational modelling. Biochemistry. 2010;49:10526–10534. doi: 10.1021/bi101384m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- Janosik M, Oliveriusova J, Janosikova B, et al. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kery V, Poneleit L, Kraus JP. Trypsin cleavage of human cystathionine beta-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- Kim MS, Song J, Park C. Determining protein stability in cell lysates by pulse proteolysis and Western blotting. Protein Sci. 2009;18:1051–1059. doi: 10.1002/pro.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka J, Krijt J, Rakova K, Kozich V. Restoring assembly and activity of cystathionine beta-synthase mutants by ligands and chemical chaperones. J Inherit Metab Dis. 2011;34:39–48. doi: 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich V, Kraus JP. Screening for mutations by expressing patient cDNA segments in E. coli: homocystinuria due to cystathionine beta-synthase deficiency. Hum Mutat. 1992;1:113–123. doi: 10.1002/humu.1380010206. [DOI] [PubMed] [Google Scholar]
- Kozich V, Sokolova J, Klatovska V, et al. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010;31:809–819. doi: 10.1002/humu.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro P, Gomes CM. Protein misfolding in conformational disorders: rescue of folding defects and chemical chaperoning. Mini Rev Med Chem. 2008;8:901–911. doi: 10.2174/138955708785132783. [DOI] [PubMed] [Google Scholar]
- Maclean KN, Gaustadnes M, Oliveriusova J, et al. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum Mutat. 2002;19:641–655. doi: 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- Maier EM, Gersting SW, Kemter KF, et al. Protein misfolding is the molecular mechanism underlying MCADD identified in newborn screening. Hum Mol Genet. 2009;18:1612–1623. doi: 10.1093/hmg/ddp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. J Biol Chem. 2010;285:15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003;1647:206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- Mudd SH, Levy LH, Kraus JP (2001) Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, Vale D, Childs B, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 7th edn. McGraw-Hill, 1007–1056
- Muntau AC, Gersting SW. Phenylketonuria as a model for protein misfolding diseases and for the development of next generation orphan drugs for patients with inborn errors of metabolism. J Inherit Metab Dis. 2010;33:649–658. doi: 10.1007/s10545-010-9185-4. [DOI] [PubMed] [Google Scholar]
- Muntau AC, Roschinger W, Habich M, et al. Tetrahydrobiopterin as an alternative treatment for mild phenylketonuria. N Engl J Med. 2002;347:2122–2132. doi: 10.1056/NEJMoa021654. [DOI] [PubMed] [Google Scholar]
- Park C, Marqusee S. Analysis of the stability of multimeric proteins by effective DeltaG and effective m-values. Protein Sci. 2004;13:2553–2558. doi: 10.1110/ps.04811004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. J Mol Biol. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- Park C, Marqusee S. Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- Park C, Marqusee S. Quantitative determination of protein stability and ligand binding by pulse proteolysis. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT, Ploegh HL, editors. Current protocols in protein sciences. Hoboken: Wiley; 2006. pp. 20.11.1–20.11.14. [DOI] [PubMed] [Google Scholar]
- Perez B, Desviat LR, Gomez-Puertas P, Martinez A, Stevens RC, Ugarte M. Kinetic and stability analysis of PKU mutations identified in BH4-responsive patients. Mol Genet Metab. 2005;86(Suppl 1):S11–S16. doi: 10.1016/j.ymgme.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Pey AL, Perez B, Desviat LR, et al. Mechanisms underlying responsiveness to tetrahydrobiopterin in mild phenylketonuria mutations. Hum Mutat. 2004;24:388–399. doi: 10.1002/humu.20097. [DOI] [PubMed] [Google Scholar]
- Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci USA. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff M, Froissart R, Olsen RK, Acquaviva C, Vianey-Saban C. Electron transfer flavoprotein deficiency: functional and molecular aspects. Mol Genet Metab. 2006;88:153–158. doi: 10.1016/j.ymgme.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Schlebach JP, Kim MS, Joh NH, Bowie JU, Park C. Probing membrane protein unfolding with pulse proteolysis. J Mol Biol. 2010;406:545–551. doi: 10.1016/j.jmb.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Banerjee R. A pathogenic linked mutation in the catalytic core of human cystathionine beta-synthase disrupts allosteric regulation and allows kinetic characterization of a full-length dimer. Biochemistry. 2007;46:4110–4116. doi: 10.1021/bi602617f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Yu J, Yamanishi M, Schellhorn D, Banerjee R. Mapping peptides correlated with transmission of intrasteric inhibition and allosteric activation in human cystathionine beta-synthase. Biochemistry. 2005;44:14210–14216. doi: 10.1021/bi051046d. [DOI] [PubMed] [Google Scholar]
- Singh LR, Kruger WD. Functional rescue of mutant human cystathionine beta-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae. J Biol Chem. 2009;284:4238–4245. doi: 10.1074/jbc.M806387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Chen X, Kozich V, Kruger WD. Chemical chaperone rescue of mutant human cystathionine beta-synthase. Mol Genet Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Gupta S, Honig NH, Kraus JP, Kruger WD. Activation of mutant enzyme function in vivo by proteasome inhibitors and treatments that induce Hsp70. PLoS Genet. 2010;6:e1000807. doi: 10.1371/journal.pgen.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovby F, Gaustadnes M, Mudd SH. A revisit to the natural history of homocystinuria due to cystathionine beta-synthase deficiency. Mol Genet Metab. 2010;99:1–3. doi: 10.1016/j.ymgme.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PJ, Parniak MA, Akerman BR, Jones AO, Scriver CR. Missense mutations in the phenylalanine hydroxylase gene (PAH) can cause accelerated proteolytic turnover of PAH enzyme: a mechanism underlying phenylketonuria. J Inherit Metab Dis. 1999;22:208–212. doi: 10.1023/A:1005533825980. [DOI] [PubMed] [Google Scholar]
- Yamanishi M, Kabil O, Sen S, Banerjee R. Structural insights into pathogenic mutations in heme-dependent cystathionine-beta-synthase. J Inorg Biochem. 2006;100:1988–1995. doi: 10.1016/j.jinorgbio.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Young TA, Skordalakes E, Marqusee S. Comparison of proteolytic susceptibility in phosphoglycerate kinases from yeast and E. coli: modulation of conformational ensembles without altering structure or stability. J Mol Biol. 2007;368:1438–1447. doi: 10.1016/j.jmb.2007.02.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 3.30 mb)