Abstract
This study aims to determine whether abdominal microbial profiles in early severe secondary peritonitis are associated with ongoing infection or death. The study is performed within a randomized study comparing two surgical treatment strategies in patients with severe secondary peritonitis (n = 229). The microbial profiles of cultures retrieved from initial emergency laparotomy were tested with logistic regression analysis for association with ‘ongoing infection needing relaparotomy’ and in-hospital death. No microbial profile or the presence of yeast or Pseudomonas spp. was related to the risk of ongoing infection needing relaparotomy. Resistance to empiric therapy for gram positive cocci and coliforms was moderately associated with ongoing abdominal infection (OR 3.43 95%CI 0.95–12.38 and OR 7.61, 95%CI 0.75–76.94). Presence of only gram positive cocci, predominantly Enterococcus spp, was borderline independently associated with in-hospital death (OR 3.69, 95%CI 0.99–13.80). In secondary peritonitis microbial profiles do not predict ongoing abdominal infection after initial emergency laparotomy. However, the moderate association of ongoing infection with resistance to the empiric therapy compels to more attention for resistance when selecting empiric antibiotic coverage.
Introduction
Abdominal sepsis is an often encountered, severe condition, treated by a multidisciplinary team of surgeons, intensive care specialists, radiologists and microbiologists. Surgical source control by elimination of the infectious focus is the main constituent of treatment. However, organ failure support and additional microbial therapy are indispensable features of treatment [1]. Especially very early antibiotic intervention is propagated as effective in reducing mortality in sepsis [2–4].
Previous studies have focused on identification of clinical and laboratory variables of value for identification of patients at high risk for ongoing infection [5–17]. In particular post-operative physiological parameters are helpful in identifying abdominal sepsis patients in need of a relaparotomy rather than peritonitis and operative characteristics [14]. However, the relation between the microbial profile of peritoneal infection and patient outcome has not been studied extensively in a prospective setting. If there is a relation, this could have consequences for the choice of the empiric broad spectrum antibiotic coverage aimed at possible pathogens in the intestinal flora [18, 19].
The clinical management consequences of abdominal fluid cultures obtained at initial emergency laparotomy is often questioned. Culture results including susceptibility patterns are first available after at least 48–72 hours, and the retrieved species may not vary that much. Furthermore, it is stated that the antibiotic treatment window really affecting patient outcome lies in the first few hours, stressing the importance of adequate empiric regimes [20]. Moreover, some secondary peritonitis patients require a relaparotomy because of clinical suspicion of ongoing infection. This decision is usually made before culture results become available. However, if index cultures are predictive of a complicated course with (multiple) relaparotomies or death, early identification of eventual microbial profile may influence treatment decision and thereby affect outcome. The aim of this study is to determine whether abdominal microbial profiles in early secondary peritonitis are predictive of the course of disease.
Methods
Design and eligibility
All patients from the RELAP trial (ISRCTN51729393) were enrolled in this study (n = 229). The RELAP trial was a randomized controlled clinical trial comparing two main surgical strategies for severe secondary peritonitis following the initial emergency laparotomy: on-demand relaparotomy (n = 114) versus planned relaparotomy (n = 115) [12]. Patients were eligible with an APACHE-II score >10 and diagnosed with abdominal sepsis, verified during surgery, caused by perforation or infection of a visceral organ or ischemia/necrosis of part of the gastrointestinal tract due to strangulation or postoperative peritoneal infection. Details on design and patient inclusion have been described elsewhere [12].
‘On-demand’ relaparotomy was performed only in case of no clinical improvement or in case of clinical deterioration, monitored by physiological, laboratory and radiology parameters. Planned relaparotomy was performed every 36 to 48 hours until the abdomen was macroscopically clean at the beginning of the final relaparotomy. All eligible patients were enrolled at two academic and seven major teaching hospitals. The study was approved by the Medical Ethics Committees of all participating hospitals.
Outcome
The aim of the present study was to evaluate whether specific microbial profiles are associated with ongoing infection needing relaparotomy and/or with in-hospital death. ‘Ongoing infection needing relaparotomy’ points to residual abdominal infection or a new infectious focus in the abdominal cavity. If patients did not receive a relaparotomy but died within 14 days following initial emergency laparotomy, patients were also determined as having ‘ongoing infection needing relaparotomy’. ‘No ongoing infection, not needing relaparotomy’ was defined as patients who either underwent a relaparotomy for secondary peritonitis yielding no residual infection or new pathology, or patients who did not receive a relaparotomy and survived the acute phase of the disease (at least 14 days). In-hospital death was defined as patients who died during hospitalization, when the initial operation for abdominal sepsis due to secondary peritonitis was performed.
Data collection
Data were prospectively collected. Data on cultures of abdominal fluid obtained at index laparotomy were evaluated. Abdominal fluid cultures were not obtained systematically at index laparotomy. Other assessed data included: patient characteristics, disease and operation related information, and postoperative variables including the development of major peritonitis-related complications for 12 months following index operation.
Microbial profiles
Patients were divided over five different microbial profiles based on the spectrum reported in the culture results: (1) no microbial growth, (2) coliform species (gram negative microorganisms) in absence of gram positive species, (3) gram positive cocci in absence of gram negative species, (4) coliforms and positive cocci, and (5) only anaerobes. Furthermore, patients with yeast (mono- or polymicrobial) or with pseudomonas (mono- or polymicrobial) infections were also identified. The latter two categories were not mutually exclusive from the five main groups. These main groups are considered relevant as literature describes possible differences in outcome (in-hospital death) for abdominal sepsis patients [18, 21–30].
Antibiotic regimen at the initial laparotomy
All patients received empirical antibiotic treatment at the emergency laparotomy, covering gram positive cocci (amoxicillin), gram negative rods (gentamicin) and anaerobes (metronidazole). This broad spectrum abdominal flora coverage is considered to be adequate in light of resistance patterns in more than 95% of cases in a study of microbial resistance patterns of our country [30]. Noteworthy, resistance of gram negative microorganisms to amoxicillin is only seen occasionally in our country, certainly in community-acquired peritonits [30]. Gentamicin dosage (4 mg/kg) was systematically monitored by obtaining “peak” and “trough” levels when administrated more than 3 days. Dosing was adjusted according to levels measured and in case of (preexistent) renal dysfunction.
Antibiotic susceptibility tests were performed on all obtained isolates, in order to identify the efficiency of the empiric antibiotic regime.
Statistical analysis
Demographic data and baseline characteristics were compared for patients with ‘ongoing infection needing relaparotomy’ and ‘no ongoing infection, relaparotomy not needed’ as well as for patients that died in-hospital and those who survived. Culture results were evaluated for the outcomes of ‘ongoing infection needing relaparotomy’ and of in-hospital death. The culture results were also compared between nosocomial (post-operative intra-abdominal infection) and community-acquired peritonitis (intra-abdominal infection at presentation on the Emergency Department). Continuous variables were expressed as medians with associated 25–75% interquartile ranges and compared using Mann-Whitney U tests. Categorical numbers were reported as absolute numbers (frequencies with percentages) and compared using a χ2 test. In hypothesis testing, p-values of <0.05 were considered significant.
Whether or not a certain microbial profile was predictive for either ongoing infection needing relaparotomy or in-hospital death was evaluated by univariate analyses. A P-value <0.10 was considered to depict association with the evaluated outcome variable. In case of a univariate association, a multivariate regression analysis was performed to evaluate if there was an independent association.
All statistic analyses were performed using SPSS for Windows version 18 (SPSS® Inc, Chicago, Illinois).
Results
Patient inclusion and demographic data
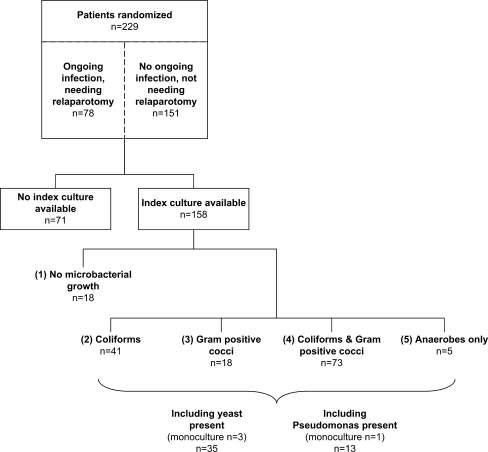
In total 229 of the 510 eligible secondary peritonitis patients were included and randomized in the RELAP trial, to either an on-demand strategy (n = 114) or a ‘planned strategy' (n = 115; Fig. 1). Tables 1 and 2 present the demographic and baseline characteristics for both all included patients (n = 229) and patients with culture results available (n = 158). Patients with ‘ongoing infection needing a relaparotomy’ (overall n = 78, cultures available n = 49) and patients with ‘no ongoing infection, not needing relaparotomy’ were compared in Table 1. Patients that died in-hospital (overall n = 50, cultures available n = 31) were compared to those who survived the initial hospital admission (overall n = 179, cultures available n = 127) in Table 2.
Fig. 1.
Flow chart of patient inclusion and overall culture results per microbial profile
Table 1.
Demographic and clinical characteristics compared for patients with ‘ongoing infection needing relaparotomy’ and patients with ‘no ongoing infection, not needing relaparotomy’
| Variables | Ongoing infection needing relaparotomy | No ongoing infection, not needing relaparotomy | P values for available cultures | ||
|---|---|---|---|---|---|
| All (N = 78) | Cultures available (n = 49) | All (N = 151) | Cultures available (n = 109) | ||
| Age (years), median (IQR) | 67 (56–73) | 65 (55–73) | 70 (57–76) | 69 (57–76) | 0.182 |
| Male | 43 (55%) | 28 (43%) | 66 (44%) | 49 (45%) | 0.156 |
| Major comorbidity present, no. (%) | 48 (62%) | 29 (59%) | 88 (58%) | 63 (58%) | 0.870 |
| Malignancy | 22 (28%) | 14 (29%) | 35 (23%) | 25 (23%) | 0.447 |
| Cardiovascular disease | 19 (24%) | 15 (31%) | 35 (23%) | 27 (25%) | 0.442 |
| Respiratory disease (COPD) | 9 (12%) | 3 (6%) | 22 (15%) | 14 (13%) | 0.207 |
| Renal disease | 8 (10%) | 7 (14%) | 9 (6%) | 4 (4%) | 0.044 |
| Diabetes | 6 (8%) | 2 (4%) | 14 (9%) | 12 (11%) | 0.156 |
| Severity of disease | |||||
| APACHE II score at study entry, median (IQR) | 15 (13–18) | 15 (12–19) | 15 (13–18) | 15 (13–18) | 0.812 |
| Etiology of peritonitis, no. (%) | 0.048 | ||||
| Perforation | 40 (51%) | 27 (55%) | 93 (62%) | 67 (62%) | |
| Anastomotic leakage | 23 (30%) | 13 (27%) | 40 (27%) | 29 (27%) | |
| Ischemia | 3 (4%) | 1 (2%) | 11 (7%) | 9 (8%) | |
| Inflammation | 5 (6%) | 3 (6%) | 4 (3%) | 2 (2%) | |
| Othera | 7 (9%) | 5 (10%) | 3 (2%) | 2 (2%) | |
| Nosocomial (postoperative) peritonitis | 37 (47%) | 22 (45%) | 71 (47%) | 48 (44%) | 0.920 |
| Localization | 0.972 | ||||
| Upper GI tract (incl. small bowel) | 19 (24%) | 10 (20%) | 39 (26%) | 28 (26%) | |
| Lower GI tract | 51 (65%) | 33 (67%) | 100 (66%) | 71 (65%) | |
| Biliary tract | 6 (8%) | 4 (8%) | 8 (5%) | 6 (6%) | |
| Otherb | 2 (3%) | 2 (4%) | 4 (3%) | 4 (4%) | |
| In-hospital mortality | 27 (35%) | 13 (27%) | 23 (15%) | 18 (17%) | 0.143 |
| Type of contamination | 0.300 | ||||
| Clear | 4 (5%) | 0 (0%) | 10 (7%) | 5 (5%) | |
| Turbid/cloudy | 19 (24%) | 11 (22%) | 28 (19%) | 23 (21%) | |
| Purulent | 24 (31%) | 14 (29%) | 51 (34%) | 39 (36%) | |
| Fecal | 30 (38%) | 23 (47%) | 56 (37%) | 37 (34%) | |
| Bile | 1 (1%) | 1 (2%) | 6 (4%) | 5 (5%) | |
a‘Other’ consisted of 8x no evident infectious focus in contaminated abdomen, 1x bile leakage, 1x infected haematoma
b‘Other’ consisted of either an infectious focus localized at the upper as well as the lower GI tract or at a gynecologic site.
All continuous data are analyzed with the Mann Whitney-U test
All categorical data are analyzed with the Chi square test
IQR interquartile range
Table 2.
Demographic and clinical characteristics compared for patients with in-hospital death and patients who survived the first hospital admission
| Variables | In-hospital death | Survival | P values for available cultures | ||
|---|---|---|---|---|---|
| All (N = 50) | Cultures available (n = 31) | All (N = 179) | Cultures available (n = 127) | ||
| Age (years), median (IQR) | 73 (68–77) | 74 (67–78) | 66 (55–73) | 64 (55–75) | 0.004 |
| Male | 21 (42%) | 15 (48%) | 88 (49%) | 62 (49%) | 0.966 |
| Major comorbidity present, no. (%) | 31 (62%) | 17 (55%) | 105 (59%) | 75 (59%) | 0.670 |
| Malignancy | 11 (22%) | 6 (19%) | 46 (26%) | 33 (26%) | 0.443 |
| Cardiovascular disease | 9 (18%) | 7 (23%) | 45 (25%) | 35 (28%) | 0.574 |
| Respiratory disease (COPD) | 14 (28%) | 7 (23%) | 17 (9%) | 10 (8%) | 0.018 |
| Renal disease | 8 (16%) | 0 (0%) | 9 (5%) | 1 (1%) | 0.009 |
| Diabetes | 2 (4%) | 1 (3%) | 18 (10%) | 13 (10%) | 0.218 |
| Severity of disease | |||||
| APACHE II score at study entry, median (IQR) | 19 (16–24) | 20 (16–24) | 14 (12–17) | 14 (12–17) | <0.001 |
| Etiology of peritonitis, no. (%) | 0.029 | ||||
| Perforation | 34 (68%) | 25 (81%) | 99 (55%) | 69 (54%) | |
| Anastomotic leakage | 7 (14%) | 2 (6%) | 56 (31%) | 40 (31%) | |
| Ischemia | 6 (12%) | 3 (10%) | 8 (4%) | 7 (6%) | |
| Inflammation | 1 (2%) | 0 (0%) | 8 (4%) | 5 (4%) | |
| Othera | 2 (4%) | 1 (3%) | 8 (4%) | 6 (5%) | |
| Nosocomial (postoperative) peritonitis | 14 (28%) | 4 (13%) | 94 (53%) | 66 (52%) | <0.001 |
| Localization | 0.500 | ||||
| Upper GI tract (incl small bowel) | 15 (30%) | 10 (32%) | 43 (24%) | 28 (22%) | |
| Lower GI tract | 34 (68%) | 20 (65%) | 117 (65%) | 81 (64%) | |
| Biliary tract | 1 (2%) | 1 (3%) | 13 (7%) | 9 (7%) | |
| Otherb | 0 (0%) | 0 (0%) | 6 (3%) | 6 (5%) | |
| Type of contamination | 0.697 | ||||
| Clear | 5 (10%) | 1 (3%) | 9 (5%) | 4 (3%) | |
| Turbid/cloudy | 9 (18%) | 7 (23%) | 38 (21%) | 27 (21%) | |
| Purulent | 17 (34%) | 10 (32%) | 58 (32%) | 43 (34%) | |
| Fecal | 17 (34%) | 11 (35%) | 69 (39%) | 49 (39%) | |
| Bile | 2 (4%) | 2 (6%) | 5 (3%) | 4 (3%) | |
a‘Other’ consisted of 8x no evident infectious focus in contaminated abdomen, 1x bile leakage, 1x infected haematoma
b‘Other’ consisted of either an infectious focus localized at the upper as well as the lower GI tract or at a gynecologic site
All continuous data are analyzed with the Mann Whitney-U test
All categorical data are analyzed with the Chi square test
IQR interquartile range
Culture results
Abdominal fluid from the index laparotomy was cultured in 69% of patients (n = 158, Fig. 1). In 110 out of 158 (70%) patients more than one species were cultured (polymicrobial) at initial emergency laparotomy, while in 30 out of 158 (19%) patients one single strain of microorganisms (monomicrobial) was cultured (Table 3). Only 18 out of 158 cultures showed no microbial growth (11%).
Table 3.
Distribution of microbes in the 140 patients with positive cultures, compared for monomicrobial versus polymicrobial cultures and for nosocomial peritonitis versus community-acquired peritonitis
| Microbes | Mono-microbiala | Poly-microbialb | Nosocomialc | Community-acquiredd | Total number of isolates |
|---|---|---|---|---|---|
| Gram (−) rods (coliforms) | |||||
| E. coli | 10 | 75 | 42 | 43 | 85 |
| Klebsiella pneumoniae | 1 | 16 | 8 | 9 | 17 |
| Enterobacter cloacae | 1 | 12 | 9 | 4 | 13 |
| Proteus mirabilis | 0 | 10 | 4 | 6 | 10 |
| Gram (−) rods not specified | 3 | 6 | 5 | 4 | 9 |
| Klebsiella oxytoca | 1 | 8 | 4 | 5 | 9 |
| Citrobacter freundii | 0 | 7 | 2 | 5 | 7 |
| Morganella (proteus) morganii | 0 | 4 | 2 | 2 | 4 |
| Serratia marcescens | 0 | 2 | 2 | 0 | 2 |
| Acinetobacter, not specified | 0 | 1 | 1 | 0 | 1 |
| Citrobacter amalonaticus | 0 | 1 | 1 | 0 | 1 |
| Hafnia alfeii | 0 | 1 | 1 | 0 | 1 |
| Proteus vulgaris | 0 | 1 | 1 | 0 | 1 |
| Subtotal | 16 | 144 | 82 | 78 | 160 |
| Gram (+) cocci | |||||
| Enterococci, not specified | 6 | 59 | 39 | 26 | 65 |
| Enterococcus faecalis | 2 | 11 | 7 | 6 | 13 |
| Streptococci viridans | 0 | 12 | 4 | 8 | 12 |
| Streptococcus milleri | 0 | 1 | 3 | 4 | 7 |
| Coagulase negative Staphylococci | 0 | 7 | 3 | 4 | 7 |
| Streptococci, not specified | 0 | 3 | 1 | 2 | 3 |
| Staphylococcus hemolyticus | 0 | 2 | 2 | 0 | 2 |
| Staphylococci not specified | 0 | 2 | 0 | 2 | 2 |
| Stapgylococcus aureus | 0 | 1 | 0 | 1 | 1 |
| Streptococci (Group B) | 0 | 1 | 0 | 1 | 1 |
| Streptococcus constellatus | 0 | 1 | 0 | 1 | 1 |
| Subtotal | 8 | 106 | 59 | 55 | 114 |
| Anaerobes | |||||
| Anaerobes not further specified | 2 | 46 | 25 | 23 | 48 |
| Bacteroides fragilis | 1 | 6 | 1 | 6 | 7 |
| Bacillus not specified | 1 | 3 | 3 | 1 | 4 |
| Bacteroides vulgaris | 0 | 2 | 1 | 1 | 2 |
| Bacteroides ovatus | 1 | 0 | 1 | 0 | 1 |
| Bacteroides difteroides | 0 | 1 | 1 | 0 | 1 |
| Clostridium septicum | 0 | 1 | 0 | 1 | 1 |
| Subtotal | 5 | 59 | 32 | 32 | 64 |
| Yeast | |||||
| Yeast not further specified | 2 | 16 | 9 | 9 | 18 |
| Candida albicans | 2 | 11 | 5 | 8 | 13 |
| Candida glabrata | 0 | 3 | 1 | 2 | 3 |
| Candida tropicalis | 0 | 1 | 0 | 1 | 1 |
| Subtotal | 4 | 31 | 15 | 20 | 35 |
| Pseudomonas | |||||
| Pseudomonas aeruginosa | 1 | 13 | 5 | 9 | 14 |
| Total | 34 | 353 | 193 | 194 | 387 |
aIn 36 patients
bIn 104 patients
cIn 70 patients
dIn 88 patients
Patients were divided over the five clinically relevant microbial categories according to culture results, including the group with negative culture results. Forty-one patients (25%) had coliforms without gram positive species, 18 patients (11%) had gram positive cocci without gram negative species, 73 patients (46%) had both coliforms and gram positive cocci and five patients (3%) had anaerobes only (Fig. 1). In addition, 35 patients (22%) had yeast-positive cultures (3 patients monomicrobial yeast), while 13 patients (8%) were positive for Pseudomonas aeruginosa (1 patient monomicrobial; Fig. 1). Overall Escherichia coli was most frequently cultured (85 strains), followed by Enterococcus species (65 strains). A complete overview of cultured micro-organisms is presented in Table 3.
Susceptibility results of cultures obtained at the initial laparotomy were available for 116 patients (83%). Overall resistance against antibiotics used as empiric regimen is depicted in Table 4. Resistance of Enterococcus species against amoxicillin specifically was 11% (11/99), whereas as much as 82% (61/99) of strains showed multidrug resistance. Regarding the ‘positive cocci only’ group 35% (6/17) of patients had strains resistant against amoxicillin. In the group where positive cocci and coliforms were cultured this resistance was only present in 8% (5/62) of patients. Gentamicin resistance of coliform strains was 3% (4/133) whereas 76% (101/133) of strains were multidrug resistant. Three of 14 (21%) strains with Pseudomonas showed multidrug resistance.
Table 4.
Overall resistance for microbial subgroups specified for empiric regimen and multidrug resistance where antibiotic susceptibility is known (284 strains in 116 patients)
| Microbial group | Amoxicillin | Gentamicin | Metronidazol | Multidrug resistancea |
|---|---|---|---|---|
| Positive cocci | 11% (11/99) | n.a. | n.a. | 62% (61/99) |
| Coliforms | n.a. | 3% (4/133) | n.a | 76% (101/133) |
| Anaerobes | n.a. | n.a. | 0% (0/52) | 21% (11/52) |
a Resistance for more then one antibiotic
n.a. not applicable
Outcome
In total, 78 out of 229 (34%) patients had ‘ongoing infection needing a relaparotomy’ and 50 out of 229 (22%) patients died in-hospital. When patients with cultures available were compared to those without cultures, comparable proportions of patients with ‘ongoing infection needing relaparotomy’ (with culture 49/158, 31% vs. without culture 29/71, 41%, P = 0.13) and comparable proportions of patients who died in-hospital (with culture 31/158, 20% vs. without culture 18/71, 26%, P = 0.32) were seen. As can be expected, mortality was higher in the group with ongoing infection (35% vs. 15% for all patients and 27% vs. 17% for those with available culture results; Table 1). Poly- and monomicrobial growth was seen in comparable proportions of patients with or without ongoing infection and of patients who did or did not survive (Tables 5 and 6).
Table 5.
Predictive value of microbial profile for ‘ongoing infection needing relaparotomy’ in patients with culture available from index laparotomy (n = 158)
| Analysis | Ongoing infection needing relaparotomy (N = 49) | No ongoing infection, not needing relaparotomy (N = 109) | OR | 95% CI | P | ||
|---|---|---|---|---|---|---|---|
| Univariate analysis | |||||||
| Monomicrobiala | 11 | (23%) | 19 | (20%) | 0.84 | 0.36–1.5 | 0.686 |
| Microbial profile | |||||||
| Negative culture result | 2 | (4%) | 16 | (15%) | 0.25 | 0.06–1.12 | 0.070 |
| Coliform | 12 | (25%) | 29 | (27%) | 0.90 | 0.41–1.95 | 0.779 |
| Gram (+) cocci | 6 | (12%) | 12 | (11%) | 1.13 | 0.40–3.20 | 0.821 |
| Coliforms and gram (+) cocci | 26 | (53%) | 47 | (43%) | 1.49 | 0.76–2.94 | 0.247 |
| Only anaerobes | 2 | (4%) | 3 | (3%) | 1.50 | 0.24–9.30 | 0.661 |
| Yeast present | 10 | (20%) | 25 | (23%) | 0.86 | 0.38–1.97 | 0.724 |
| Pseudomonas present | 2 | (4%) | 11 | (11%) | 0.38 | 0.08–1.78 | 0.219 |
| Amoxicilline resistant gram (+) coccib | 6 | (15%) | 5 | (5%) | 3.27 | 0.94–11.45 | 0.063 |
| Gentamicin resistant coliformsb | 3 | (8%) | 1 | (1%) | 7.83 | 0.79–77.78 | 0.079 |
| Multi drug resistanceb | 31 | (79%) | 63 | (66%) | 1.36 | 0.60–3.06 | 0.465 |
| Multivariate analysisb | |||||||
| Negative culture result | 2 | (5%) | 16 | (17%) | 0.29 | 0.06–1.35 | 0.189 |
| Amoxicilline resistant gram (+) cocci | 6 | (15%) | 5 | (5%) | 3.43 | 0.95–12.38 | 0.060 |
| Gentamicin resistant coliforms | 3 | (8%) | 1 | (1%) | 7.61 | 0.75–76.94 | 0.085 |
| Perforation or ischemia | 23 | (59%) | 65 | (68%) | 0.67 | 0.30–1.52 | 0.342 |
OR odds ratio, CI confidence interval
a Regression analysis in 140 patients with positive culture result (47 ongoing infection, 93 no ongoing infection), since patients without growth are in neither of the categories.
b Regression analysis in 134 patients with known susceptibility or negative culture result (39 ongoing infection, 95 no ongoing infection).
Table 6.
Predictive value of microbial profile for in-hospital death in patients with culture available from index laparotomy (n = 158)
| Analysis | In-hospital death (N = 31) | Survival (N = 127) | OR | 95% CI | P | |||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | ||||||||
| Monomicrobiala | 7 | (23%) | 23 | (18%) | 1.29 | 0.49–3.40 | 0.607 | |
| Microbial profile | ||||||||
| Negative culture result | 3 | (10%) | 15 | (12%) | 0.80 | 0.22–2.96 | 0.738 | |
| Coliforms | 6 | (19%) | 35 | (28%) | 0.60 | 0.22–1.61 | 0.310 | |
| Gram (+) cocci | 8 | (26%) | 10 | (8%) | 4.08 | 1.43–11.61 | 0.008 | |
| Coliforms and gram (+) cocci | 12 | (39%) | 61 | (48%) | 0.63 | 0.27–1.45 | 0.274 | |
| Only anaerobes | 1 | (3%) | 4 | (3%) | 1.00 | 0.1–9.3 | 1.000 | |
| yeast present | 8 | (26%) | 27 | (21%) | 1.26 | 0.50–3.18 | 0.626 | |
| Pseudomonas present | 2 | (6%) | 11 | (9%) | 0.71 | 0.15–3.39 | 0.664 | |
| Amoxicilline resistant gram (+) coccib | 3 | (12%) | 8 | (7%) | 1.63 | 0.40–6.26 | 0.494 | |
| Gentamicin resistant coliformsb | 1 | (4%) | 3 | (3%) | 1.40 | 0.14–14.03 | 0.775 | |
| Multi-drug resistanceb | 18 | (69%) | 76 | (70%) | 0.91 | 0.35–2.23 | 0.830 | |
| Multivariate analysis | ||||||||
| Gram (+) cocci | 8 | (26%) | 10 | (8%) | 3.69 | 0.99–13.80 | 0.052 | |
| APACHE II score, median (IQR) | 20 | (16–24) | 14 | (12–17) | 1.22 | 1.10–1.36 | <0.001 | |
| Community-acquired | 27 | (87%) | 61 | (48%) | 5.57 | 1.68–18.47 | 0.005 | |
| Upper GI perforation | 8 | (26%) | 16 | (13%) | 2.06 | 0.60–7.06 | 0.253 | |
| Age, median (IQR) | 74 | (67–78) | 64 | (55–75) | 1.04 | 1.00–1.0 | 0.069 | |
OR odds ratio, CI confidence interval
a Regression analysis in 140 patients with positive culture result (28 in-hospital death, 112 survival), since patients without growth are in neither of the categories
b Regression analysis in 134 patients with known susceptibility or negative culture result (26 in-hospital death, 108 survival)
Predictive value of microbial profile
‘Ongoing infection needing relaparotomy’
No microbial profile cultured from abdominal fluid samples at the initial emergency laparotomy was associated with ongoing infection (Table 5). Only the absence of microbial growth was associated with a lower rate of ongoing abdominal infection needing relaparotomy, as can be expected (OR 0.25, 95%CI 0.06–1.12, P = 0.070). Furthermore, resistance to the empiric therapy of gram positive cocci and coliforms was associated with ongoing infection (OR 3.27, 95%CI 0.94–11.45, P = 0.063 and OR 7.83, 95%CI 0.79–77.78, P = 0.079). These variables were entered in a multivariate regression model along with perforation or ischemia as the cause of abdominal sepsis because this was less frequent in patients with ongoing infection. The multivariate analysis showed that although not statistically significant, there was a strong independent association of amoxicillin resistant gram positive cocci (OR 3.43, 95%CI 0.95–12.38, P = 0.060) and gentamicin resistant coliforms (OR 7.61, 95%CI 0.75–76.94, P = 0.085) with ongoing infection.
In-hospital death
Monoculture of gram positive cocci in the abdominal fluid at the initial emergency laparotomy was highly associated with in-hospital death (OR 4.08, 95%CI 1.5–11, P = .008). Other microbial profiles were not clearly associated with in-hospital death (Table 6). Looking closer at patients with only gram positive cocci, no major differences were seen in severity of disease and baseline peritonitis characteristics compared to the other patients. We did find, however, that a large proportion (72%) underwent surgery for peritonitis due to a perforation (Table 7). Moreover, more patients with only gram positive bacteria had an upper gastrointestinal (GI) source of peritonitis compared to patients with other bacterial profiles (44% versus 21%). Although patients with gram positive bacteria only showed high rates of resistance to empiric therapy, this was not associated with death.
Table 7.
Disease characteristics comparing patients with only gram positive cocci (n = 18) to those with other cultured micro-organisms (n = 140) from index laparotomy (n = 158)
| Variables | Only gram (+) cocci | Others | P value | ||
|---|---|---|---|---|---|
| N = 18 | N = 140 | ||||
| Severity of disease | |||||
| APACHE II score at study entry, median (IQR) | 17 | (13–20) | 15 | (12–17) | 0.178 |
| Etiology of peritonitis, no. (%) | 0.485 | ||||
| Perforation | 14 | (78%) | 80 | (57%) | |
| Anastomotic leakage | 3 | (17%) | 39 | (28%) | |
| Ischemia | 1 | (6%) | 9 | (6%) | |
| Inflammation | 0 | (0%) | 5 | (4%) | |
| Othera | 0 | (0%) | 7 | (5%) | |
| Nosocomial (postoperative) peritonitis | 6 | (33%) | 64 | (46%) | 0.320 |
| Localization | 0.454 | ||||
| Upper GI tract (incl. small bowel) | 8 | (44%) | 30 | (21%) | |
| Lower GI tract | 9 | (50%) | 95 | (68%) | |
| Biliary tract | 1 | (6%) | 9 | (6%) | |
| Otherb | 0 | (0%) | 6 | (4%) | |
| Amoxicilline resistancec | 6 | (35%) | 5 | (4%) | <0.001 |
a‘Other’ consisted of 8x no evident infectious focus in contaminated abdomen, 1x bile leakage, 1x infected haematoma
b‘Other’ consisted of either an infectious focus localized at the upper as well as the lower GI tract or at a gynecologic site.
c In 134 patients with known susceptibility or negative culture result (n = 17 only gram positive cocci profile, n = 117 other profiles)
Continuous data were analyzed with the Mann Whitney U-test. All categorical data were analyzed with the chi-square test
IQR interquartile range
Multivariate analysis was performed to determine whether gram positive cocci were independently associated with mortality. The variables ‘gram positive cocci only’ (yes/no), severity of disease (APACHE-II score), upper GI perforation (yes/no), community-acquired (yes/no) and age were entered into the model. Community-acquired peritonitis (OR 5.57, 95%CI 1.68–18.47, P = .0.005) and APACHE-II score (OR 1.22 per point increase, 95%CI 1.10–1.36, P < .0.001) were independently associated with in-hospital death. Monoculture of gram positive cocci (OR 3.7, 95%CI 0.99–13.8, P = .0.021) was borderline associated with in-hospital death.
Univariate analysis showed there was no association of amoxicillin resistant gram positive cocci and in-hospital death (OR 1.63, 95%CI 0.40–6.26, P = 0.494) despite the independent association with ongoing infection.
Nosocomial versus community-acquired peritonitis
The similar distribution of micro-organisms between community-acquired and nosocomial peritonitis (Table 3) is also reflected by comparable microbial profiles (Table 8).
Table 8.
Outcomes comparing patients with nosocomial (n = 108) peritonitis versus community-acquired (n = 121) peritonitis
| Variables | Nosocomial peritonitis | Community-acquired peritonitis | P values for available cultures | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All (N = 108) | Cultures available (N = 70) | All (N = 121) | Cultures available (N = 88) | ||||||
| Severity of disease | |||||||||
| APACHE II score at study entry, median (IQR) | 14 | (13–16) | 14 | (12–16) | 16 | (13–20) | 16 | (12–20) | 0.075 |
| Primary outcomes | |||||||||
| Ongoing infection needing relaparotomy | 37 | (34%) | 22 | (31%) | 41 | (34%) | 27 | (31%) | 0.920 |
| In-hospital death | 14 | (13%)a | 4 | (6%) | 36 | (30%)* | 27 | (31%) | <0.001 |
| Cultures | |||||||||
| Monomicrobial | 12 | (17%) | 18 | (20%) | 0.478 | ||||
| Microbial profile | |||||||||
| Negative culture results | 6 | (9%) | 12 | (14%) | 0.320 | ||||
| Coliforms | 18 | (26%) | 23 | (26%) | 0.952 | ||||
| Gram (+) cocci | 6 | (9%) | 12 | (14%) | 0.320 | ||||
| Coliforms and gram (+) cocci | 39 | (56%) | 34 | (39%) | 0.032 | ||||
| Only anaerobes | 1 | (1%) | 4 | (5%) | 0.266 | ||||
| Yeast present | 15 | (21%) | 20 | (23%) | 0.845 | ||||
| Pseudomonas present | 5 | (7%) | 8 | (9%) | 0.658 | ||||
| Amoxicilline resistant gram (+) coccib | 5 | (9%) | 6 | (10%) | 0.891 | ||||
| Gentamicin resistant coliformsb | 2 | (4%) | 2 | (3%) | 0.916 | ||||
| Multi-drug resistanceb | 50 | (91%) | 44 | (72%) | 0.001 | ||||
a P = 0.002 (all patients)
b In 134 patients with known susceptibility or negative culture result (n = 61 nosocomial peritonitis, n = 73 community-acquired peritonitis)
Continuous data were analyzed with the Mann Whitney U-test. All categorical data were analyzed with the chi-square test
IQR interquartile range
The finding that community-acquired peritonitis is an independent predictor of death is reflected in significantly more patients with community-acquired peritonitis who died in-hospital (31% vs. 6%, P < .0.001). However, since there is a similar distribution of the microbial profiles, other factors in community-acquired peritonitis were examined. Initial severity of disease was somewhat more profound for the community-acquired peritonitis group (APACHE-II score 16 [IQR 12–20] vs. nosocomial 14 [IQR 12–16], P = .0.075). On the other hand, just the same proportion of patients with community-acquired peritonitis and nosocomial peritonitis needed a relaparotomy for ongoing infection (31%; Table 8).
Discussion
In this study culture results from the initial emergency laparotomy of secondary peritonitis patients were studied for their microbial patterns as well as for their effect on ongoing abdominal infection and on in-hospital death. Patients participated in a randomized trial to evaluate effectiveness of two surgical treatment strategies. In a large proportion of patients (31%) no abdominal cultures were drawn at initial surgery. Surgeons were blinded for the allocated surgical treatment strategy at the time of the initial laparotomy [12]. This would eliminate selection bias in retrieval of fluid samples for culture merely related to whether or not a relaparotomy would follow. Nevertheless, macroscopic appearance of the abdominal contamination or source of infection might have been reason for selection of patients needing cultures. This bias can work in either direction: some surgeons might feel that cultures might be superfluous in clear-cut fecal contamination due to anastomotic leakage, whereas others might anticipate different microorganisms in nosocomial peritonitis resulting in a higher tendency to culture. No differences were found in demographic and clinical characteristics, showing that potential selection bias did not affect distribution of patients in the cultured group versus the total trial cohort.
Culture results from the initial emergency laparotomy revealed a large contribution of gram positive cocci as infectious agent. In more than half of the patients (58%) gram positive species were retrieved, with or without presence of other microorganisms. As can be expected, the absence of microbial growth was associated with a lower rate of ongoing infection. Nevertheless, these patients did have peritonitis at index operation, predominantly caused by perforation or anastomotic leakage (14 out of 18 patients without microbial growth). Furthermore, a larger proportion of patients with ongoing infection needing relaparotomy exhibited amoxicillin resistant positive cocci and gentamicin resistant coliforms. Although a significant association could not be determined, it can be argued that these results are indeed clinically relevant. On the other hand, results are based on sub analyses and therefore study numbers were small and probably under powered.
Secondary peritonitis caused by gram positive cocci, predominantly Entercoccus spp., in the absence of gram negative microorganisms was associated with in-hospital death. This association appears to be very relevant for clinical practice. Resistance to empiric therapy was associated with ongoing infection, but this resistance was not associated with increased risk of in-hospital death. Possibly the current sample size limits the capacity to determine this association. There is evidence that suggests that complicated intra-abdominal infections involving mixed flora can be treated with surgery and non-enterococcal antibiotic coverage [22]. However, in settings without routine empiric coverage of enterococci, enterococcal infections are associated with a higher mortality [22]. Patients in our study all received empiric coverage of Enterococcus spp. by amoxicillin. The high prevalence of Enterococcus spp. and the reported higher mortality due to Enterococcus spp. suggests benefit from empiric coverage. The regimen used should take into account regional resistance patterns, including resistance to amoxicillin of gram negative microorganisms.
In our hospital and for this study suitability of empirical antibacterial treatment was based on national resistance surveillance data [30]. In the Nethmap database nationwide microbial resistance patterns are evaluated, a surveillance that is performed yearly. Based on these data coverage of empiric therapy consisting of amoxicillin, gentamicin and metronidazole should have been appropriate in the vast majority of cases. Resistance of Enterococcus spp. against amoxillin in this study, however, surprisingly exceeded regional prevalence.
We can conclude, based only on present results, that amoxicillin coverage may be insufficient in these severely ill patients. Importantly, in our setting vancomycin resistant Enterococcus spp. are rare (sporadic encounter, 0% www.swab.nl) and vancomycin is not part of empiric therapy.
Yeast strains are common in early severe secondary peritonitis (22%), but not clearly related to mortality in this cohort of patients. Patients did not receive prophylactic or preemptive antifungal therapy, but were treated in case of clinically suspected or confirmed invasive candidiasis. The absence of any relation between yeast in polymicrobial infection and death is not likely caused by a lack of prophylaxis. A recent systematic review of prospective trials evaluating single drug antifungal prophylaxis, demonstrates that prophylaxis is not as effective for surgical patients as opposed to medical patients [31]. These results and presented data do not support the view to broaden the empiric regimen with coverage of yeast in patients with peritonitis. Also, empiric coverage of Pseudomonas aeruginosa can not be propagated based on present results. Pseudomonas is a notorious hospital microbe, opportunistic and resistant to many antibiotics [32]. Here, prevalence of Pseudomonas was similar among survivors and nonsurvivors, and surprisingly not more frequent in nosocomial peritonitis than in community-acquired peritonitis for early disease.
Furthermore, nosocomial peritonitis was associated with similar proportions of patients with yeast or Pseudomonas spp. as was community-acquired peritonitis. Nosocomial infection is often associated with other types of pathogens than community-acquired infection. This is well-known for pneumonia [33]. It is likely that during initial disease, be it an anastomotic leakage (nosocomial) or perforated diverticulitis (community-acquired), the abdominal invasion of abundantly available enteric bacteria is quite similar. Therefore, when empiric antibiotic coverage is adequate and frequently evaluated by local resistance surveillance, cultures of abdominal fluid during initial laparotomy may not contribute to clinical management decisions in the individual patient.
In conclusion, in this study on secondary peritonitis no microbial profile was associated with ongoing infection needing a relaparotomy except for a negative culture result. Although not statistically significant, the association between resistance to empiric therapy and ongoing infection compels more attention to be paid to resistance in the selection of empiric antibiotic coverage. Present data do not support empiric coverage of yeast and Pseudomonas spp. in abdominal sepsis. Gram positive cocci, in particular Enterococcus spp., appeared to be a larger threat to peritonitis patients than previously assumed.
Acknowledgments
Dutch Peritonitis Study Group
RELAP trial clinical centers and investigators of the Dutch Peritonitis Study Group are listed herein. All investigators are from the Departments of Surgery unless specified as Clinical Epidemiology and Biostatistics (E), Intensive Care (I) or Medical Psychology (MP).
O van Ruler MD; KR Boer MSc (E); JB Reitsma MD, PhD (E); J.J.S. Kiewiet MD; CW Mahler MD; EA Reuland MSc; JWO van Till MD; BC Opmeer PhD (E); PMM Bossuyt PhD (E); MJ Schultz MD, PhD (I); MA Sprangers MD, PhD (MP); DJ Gouma MD, PhD; H Obertop MD, PhD; AJM de Borgie MD, PhD (E); MA Boermeester MD, PhD, Academic Medical Center, Amsterdam; EPh Steller MD, PhD; P. Tanis MD, PhD; H Hart MD (I), St Lucas Andreas Hospital, Amsterdam; MF Gerhards MD, PhD; M Guijt MD; HM Oudemans MD, PhD (I), Onze Lieve Vrouwe Gasthuis, Amsterdam; K. Bosscha MD, PhD; E Ritchie MD; M Vermeer, Bosch Medical Centre, Den Bosch; PW de Graaf MD, PhD; B van Etten MD, PhD; C Haazer (I); E Salm MD, PhD (I); Reinier de Graaf Hospital, Delft; B Lamme MD, PhD; EJ Hesselink MD, PhD; H Rommes MD, PhD (I), Gelre Hospitals, Lucas Hospital Apeldoorn; RJ Oostenbroek MD, PhD; L te Velde MD; G Govaert MD; HH Ponssen MD (I), Albert Schweitzer Hospital, Dordrecht; HG Gooszen MD, PhD; MK Dinkelman MD; LPH Leenen MD, PhD (I), University Medical Centre Utrecht; EGJM Pierik MD, PhD; KWW Lansink MD; J Bakker MD, PhD (I), Isala Clinics, Zwolle.
Conflicts of interests
There were no authors nor members of the Dutch Peritonitis Study Group with conflicts of interests
Financial support
This study was supported by the Netherlands Organization for Health Research and Development (grant no. 945-02-028).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Lamme B, Mahler CW, van Till JW, van Ruler O, Gouma DJ, Boermeester MA. Relaparotomy in secondary peritonitis. Planned relaparotomy or relaparotomy on demand? Chirurg. 2005;76:856–867. doi: 10.1007/s00104-005-1086-y. [DOI] [PubMed] [Google Scholar]
- 2.Dellinger RP, Carlet JM, Masur H, Gerlach H, Calandra T, Cohen J, Gea-Banacloche J, Keh D, Marshall JC, Parker MM, Ramsay G, Zimmerman JL, Vincent JL, Levy MM. Surviving Sepsis Campaign guidelines for management of severe sepsis and septic shock. Crit Care Med. 2004;32:858–873. doi: 10.1097/01.CCM.0000117317.18092.E4. [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest. 2000;118:146–155. doi: 10.1378/chest.118.1.146. [DOI] [PubMed] [Google Scholar]
- 4.Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115:462–474. doi: 10.1378/chest.115.2.462. [DOI] [PubMed] [Google Scholar]
- 5.Anaya DA, Nathens AB. Risk factors for severe sepsis in secondary peritonitis. Surg Infect (Larchmt) 2003;4:355–362. doi: 10.1089/109629603322761418. [DOI] [PubMed] [Google Scholar]
- 6.Bohnen J, Boulanger M, Meakins JL, McLean AP. Prognosis in generalized peritonitis. Relation to cause and risk factors. Arch Surg. 1983;118:285–290. doi: 10.1001/archsurg.1983.01390030017003. [DOI] [PubMed] [Google Scholar]
- 7.Fry DE, Garrison RN, Heitsch RC, Calhoun K, Polk HC., Jr Determinants of death in patients with intraabdominal abscess. Surgery. 1980;88:517–523. [PubMed] [Google Scholar]
- 8.Grunau G, Heemken R, Hau T. Predictors of outcome in patients with postoperative intra-abdominal infection. Eur J Surg. 1996;162:619–625. [PubMed] [Google Scholar]
- 9.Holzheimer RG, Gathof B. Re-operation for complicated secondary peritonitis—how to identify patients at risk for persistent sepsis. Eur J Med Res. 2003;8:125–134. [PubMed] [Google Scholar]
- 10.Lamme B, Mahler CW, van Ruler O, Gouma DJ, Reitsma JB, Boermeester MA (2006) Clinical predictors of ongoing infection in secondary peritonitis: systematic review. World J Surg 30:2170–2181 [DOI] [PubMed]
- 11.Pusajo JF, Bumaschny E, Doglio GR, Cherjovsky MR, Lipinszki AI, Hernandez MS, Egurrola MA. Postoperative intra-abdominal sepsis requiring reoperation. Value of a predictive index. Arch Surg. 1993;128:218–222. doi: 10.1001/archsurg.1993.01420140095015. [DOI] [PubMed] [Google Scholar]
- 12.van Ruler O, Mahler CW, Boer KR, Reuland EA, Gooszen HG, Opmeer BC, de Graaf PW, Lamme B, Gerhards MF, Steller EP, van Till JW, de Borgie CJ, Gouma DJ, Reitsma JB, Boermeester MA, Dutch Peritonitis Study Group (2007) Comparison of on-demand versus planned relaparotomy strategy in patients with severe peritonitis. A randomized trial. JAMA 298(8):865–872 [DOI] [PubMed]
- 13.van Goor H. Interventional management of abdominal sepsis: when and how. Langenbecks Arch Surg. 2002;387:191–200. doi: 10.1007/s00423-002-0309-7. [DOI] [PubMed] [Google Scholar]
- 14.van Ruler O, Lamme B, Gouma DJ, Reitsma JB, Boermeester MA. Variables associated with positive findings at relaparotomy in patients with secondary peritonitis. Crit Care Med. 2007;35:468–476. doi: 10.1097/01.CCM.0000253399.03545.2D. [DOI] [PubMed] [Google Scholar]
- 15.Wacha H, Hau T, Dittmer R, Ohmann C. Risk factors associated with intraabdominal infections: a prospective multicenter study. Peritonitis Study Group. Langenbecks Arch Surg. 1999;384:24–32. doi: 10.1007/s004230050169. [DOI] [PubMed] [Google Scholar]
- 16.Wacha H, Linder MM, Feldmann U, Wesch G, Gundlach E, Steifensand RA (1987) Mannheim Peritonitis Index—prediction of risk of death from peritonitis: construction of a statistical and validation of an emperically based index. Theor Surg 169–177
- 17.Wittmann DH, Muller M. Patient stratification in intra-abdominal infection. Chemioterapia. 1987;6:460–462. [PubMed] [Google Scholar]
- 18.Solomkin JS, Mazuski JE, Baron EJ, Sawyer RG, Nathens AB, DiPiro JT, Buchman T, Dellinger EP, Jernigan J, Gorbach S, Chow AW, Bartlett J. Guidelines for the selection of anti-infective agents for complicated intra-abdominal infections. Clin Infect Dis. 2003;37:997–1005. doi: 10.1086/378702. [DOI] [PubMed] [Google Scholar]
- 19.Wong PF, Gilliam AD, Kumar S, Shenfine J, O'Dair GN, Leaper DJ (2005) Antibiotic regimens for secondary peritonitis of gastrointestinal origin in adults. Cochrane Database Syst Rev CD004539 [DOI] [PMC free article] [PubMed]
- 20.Barie PS, Hydo LJ, Shou J, Larone DH, Eachempati SR. Influence of antibiotic therapy on mortality of critical surgical illness caused or complicated by infection. Surg Infect. (Larchmt.) 2005;6:41–54. doi: 10.1089/sur.2005.6.41. [DOI] [PubMed] [Google Scholar]
- 21.Barie PS, Christou NV, Dellinger EP, Rout WR, Stone HH, Waymack JP. Pathogenicity of the enterococcus in surgical infections. Ann Surg. 1990;212:155–159. doi: 10.1097/00000658-199008000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee I, Iredell JR, Woods M, Lipman J. The implications of enterococci for the intensive care unit. Crit Care Resusc. 2007;9:69–75. [PubMed] [Google Scholar]
- 23.Christou NV, Turgeon P, Wassef R, Rotstein O, Bohnen J, Potvin M. Management of intra-abdominal infections. The case for intraoperative cultures and comprehensive broad-spectrum antibiotic coverage. The Canadian Intra-abdominal Infection Study Group. Arch Surg. 1996;131:1193–1201. doi: 10.1001/archsurg.1996.01430230075014. [DOI] [PubMed] [Google Scholar]
- 24.Garbino J, Villiger P, Caviezel A, Matulionyte R, Uckay I, Morel P, Lew D. A randomized prospective study of cefepime plus metronidazole with imipenem-cilastatin in the treatment of intra-abdominal infections. Infection. 2007;35:161–166. doi: 10.1007/s15010-007-6237-2. [DOI] [PubMed] [Google Scholar]
- 25.Harbarth S, Uckay I. Are there patients with peritonitis who require empiric therapy for enterococcus? Eur J Clin Microbiol Infect Dis. 2004;23:73–77. doi: 10.1007/s10096-003-1078-0. [DOI] [PubMed] [Google Scholar]
- 26.Krobot K, Yin D, Zhang Q, Sen S, Altendorf-Hofmann A, Scheele J, Sendt W (2004) Effect of inappropriate initial empiric antibiotic therapy on outcome of patients with community-acquired intra-abdominal infections requiring surgery. Eur J Clin Microbiol Infect Dis 23(9):682–687 [DOI] [PubMed]
- 27.Solomkin JS, Wilson SE, Christou NV, Rotstein OD, Dellinger EP, Bennion RS, Pak R, Tack K. Results of a clinical trial of clinafloxacin versus imipenem/cilastatin for intraabdominal infections. Ann Surg. 2001;233:79–87. doi: 10.1097/00000658-200101000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Top J, Willems R, Blok H, de Regt M, Jalink K, Troelstra A, Goorhuis B, Bonten M. Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin Microbiol Infect. 2007;13:316–319. doi: 10.1111/j.1469-0691.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 29.Wacha H, Warren B, Bassaris H, Nikolaidis P. Comparison of sequential intravenous/oral ciprofloxacin plus metronidazole with intravenous ceftriaxone plus metronidazole for treatment of complicated intra-abdominal infections Surg. Infect (Larchmt) 2006;7:341–354. doi: 10.1089/sur.2006.7.341. [DOI] [PubMed] [Google Scholar]
- 30.Nethmap (2009) Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands. Available at www.swab.nl. Accessed 05 January 2011
- 31.van Till JWO, van Ruler O, Lamme B, Weber RJP, Reitsma JB, Boermeester MA (2007) Single-drug therapy or selective decontamination of the digestive tract as antifungal prophylaxis in critically ill patients: a systematic review. Crit Care 11 [DOI] [PMC free article] [PubMed]
- 32.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 33.Herold CJ, Sailer JG. Community-acquired and nosocomial pneumonia. Eur Radiol. 2004;14(Suppl 3):E2–E20. doi: 10.1007/s00330-003-2162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]