Abstract
Acid Mine Drainages (AMDs) are extreme environments characterized by acidic and oligotrophic conditions and by metal contaminations. A function-based screening of an AMD-derived metagenomic library led to the discovery and partial characterization of two non-homologous endo-acting amylases sharing no sequence similarity with any known amylase nor glycosidase. None carried known amylolytic domains, nor could be assigned to any GH-family. One amylase displayed no similarity with any known protein, whereas the second one was similar to TraC proteins involved in the bacterial type IV secretion system. According to the scarce similarities with known proteins, 3D-structure modelling using I-TASSER was unsuccessful. This study underlined the utility of a function-driven metagenomic approach to obtain a clearer image of the bacterial community enzymatic landscape. More generally, this work points out that screening for microorganisms or biomolecules in a priori incongruous environments could provide unconventional and new exciting ways for bioprospecting.
Soil microorganisms constitute a reservoir of a countless quantity of genes with potential interesting properties for fundamental research, medical and industrial applications. For this reason, cultured microorganisms have been widely screened for decades for diverse enzymes useful in the degradation of pollutants1 or polymers2 as well as for antimicrobial agents production3. However, these screenings were restricted to a very small fraction of the total microorganisms i.e. those which have been successfully cultured in laboratory. This phenomenon, called « the great plate count anomaly »4, is responsible for the lack of knowledge of all recalcitrant-to-culture bacteria. With the development of novel molecular techniques two decades ago, especially metagenomic tools, new insights into this unexplored microbial majority became possible. The access to their genetic content opened new perspectives for biologists, as they contain a huge quantity of genes encoding proteins of unknown function.
Amylases were the first enzymes to be discovered and described5 and are among the most important enzymes used for biotechnological purposes, from food to paper industries6. Numerous studies were focused on the discovery of new amylases originating from cultured bacteria, and more recently from metagenomic DNA. These investigations allowed the detection of amylases with interesting characteristics, such as amylases active in various conditions such as low6 or high7 temperature, low8 or high9 pH. Amylases belong to glycoside hydrolases (GH-)10 and are clustered into several families, mainly GH-13, GH-14, GH-15 and GH-577,11, based on similarities in primary and tertiary structures together with conservation of the catalytic residues.
In previous investigations, amylases were found in many different extreme environments7,9,12. Nevertheless, no study reported the screening of metagenomic DNA derived from Acid Mine Drainages (AMDs) for any polymer degradation activity. The AMD located in Carnoulès (France) is, as most AMDs, considered to be an oligotrophic environment13. Furthermore, this site is characterized by very acidic conditions (pH 2.7–3.4) and is highly contaminated by heavy metals such as iron (up to 2,700 mg/L) and arsenic (up to 350 mg/L)14. A recent metagenomic dataset allowed the reconstruction of 7 genomes corresponding to the 7 dominant bacteria in the sediment of Carnoulès, called CARN1 to CARN7, five of which corresponded to uncultured bacteria13. The authors proposed a functioning model for the community, based principally on in silico analysis of the genomes but also on experimental data obtained by RT-PCR and proteomic. Those genomes contained between 36.3% and 53.7% of genes that could not be assigned to any function13. So, this model eluded roughly half of the information carried by the genomes.
AMDs are studied since decades, especially toward the inorganic compartment. Because metagenomic DNA from AMDs were never screened for polymer-degrading enzymes and because of the extreme conditions found in such environments –giving the possibility to discover enzymes with interesting properties-, we decided to perform a function-based screening for the well-known amylases, using standard techniques. This strategy allowed the isolation of 28 positive clones, 2 of them being subcloned, the proteins purified and characterized in vitro. In silico analyses based on the nucleotidic sequence and both the primary and the predicted tertiary structures revealed that they are completely different from other known hydrolases as both genes encode a « protein of unknown function » and display no known conserved amylolytic domain. Nevertheless, in vitro tests confirmed the amylolytic activity of these 2 enzymes. Thus, the present work showed the possibility to assign functions to genes encoding previously uncharacterized proteins. With regards to the extreme conditions found in such ecosystems, AMDs can therefore represent an unexpected reservoir for new proteins.
Results
Roughly 80,000 clones were screened on LB plates supplemented with 1% wheat starch and 20 mg/L chloramphenicol. Among them, 28 (0.035%) revealed an amylolytic activity after a 48 hours exposure with starch and both extremities of the corresponding inserts (3–5 kb) were sequenced. Nucleotidic sequences of 10 inserts corresponded to sequences of the CARNs genomes (CARN1, CARN2, CARN4 and CARN7)13. Thanks to the genome reconstructions, the complete nucleotidic sequences of the 10 inserts could be obtained. Furthermore, the coverage obtained (see Figure S1 from13) for the reconstruction of the CARN genomes allowed us to rule out any point mutations. The 18 other positive clones –not belonging to any CARN genome– were discarded.
Among the 10 sequences, none contained an ORF automatically annotated in Genoscope (http://www.genoscope.cns.fr/agc/microscope/carnoulescopes) as an “amylase”. Moreover, numerous ORFs were not automatically assigned to any known function, and were classified as “uncharacterized protein”. Inserts containing more than one unknown ORF and ORFs lacking either the 5′ or the 3′ region were discarded for further analysis. Finally, two ORFs corresponding to 2 clones (CARN4_1025 and CARN7_2759) were subcloned into the pUC19 vector to confirm the amylolytic activity conferred to the recipient DH5α cells. After 48 hours incubation on plates supplemented with 1% starch, both sub-clones expressed the ability to degrade the polysaccharide, confirming the amylolytic activities of the corresponding proteins. To further rule out any point mutation after subcloning, both ORFs were re-sequenced. The 5 kb-sequences of the 8 remaining clones are given in supplementary data S1.
The local alignment of amino acid sequences of CARN7_2759 and CARN4_1025 with the default options of the water program (EMBOSS package15) implementing the Smith & Waterman algorithm16 did not show any convincing sequence similarity between the two sequences, supporting the non-homologous character of those proteins.
The amino acid sequence of CARN4_1025 (479 amino acids, 47.45 kDa, pI 4.10) presented no sequence similarity with any entry in the nucleotide and protein databases (NCBI-nr and Uniprot), as no significant hit could be found when searching these databases with tBlastN and BlastP algorithms. It is noteworthy though that this gene has an ortholog in the genome of CARN1 (CARN1_1051, emb|CBH75884.1|). CARN1 and CARN4 represent a new species within a new uncultured phylum detected in Carnoulès and other AMDs, for which the name Candidatus Fodinabacter communificans was given. This protein was predicted by InterProScan to contain a prokaryotic membrane lipoprotein lipid attachment site but no conserved domain from the Conserved Domain Database could be identified by the NCBI web server. Furthermore, it was not possible to identify any known amylolytic domain in this protein which could therefore not be assigned to any Glycosyl Hydrolase (GH-) family.
Five 3D structure models were obtained by the I-TASSER platform. However, the C-score obtained for all those models was −2.84, which is below the −1.5 cut off value for high confidence 3D structure prediction17.
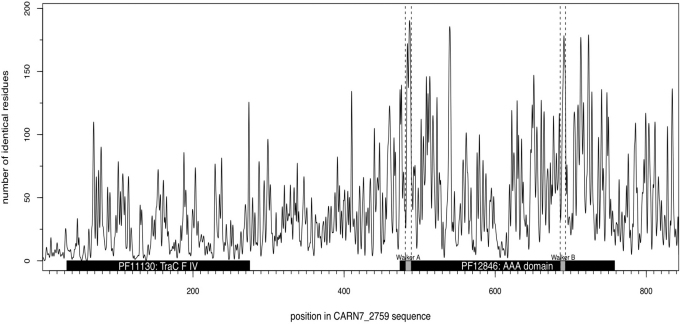
Blast database similarity searches show that the sequence of CARN7_2759 (843 amino acids, 95.18 kDa, pI 6.14) is similar to the type-IV secretion system protein TraC of various bacteria both at the protein (up to 38% identity) and nucleotide levels. However, no known amylolytic domain was found in this sequence and the protein could not be assigned to any GH- family. The predictions in CARN7_2759 of domains and signatures by InterProScan yielded hits with TraC signature (PFam: PF11130) and a AAA domain (PFam: PF12846) corresponding in TraC and other conjugative transfer proteins to the P-loop NTPase domain (Fig. 1). Interestingly, the conservation profile and the PipeAlign multiple sequence alignment showed that both the NTPase conserved motifs18 Walker A (GxxGxGK[S/T]) and Walker B (hhhhDE where h is a hydrophobic aminoacid) could be found in CARN7_2759, with the exception of the first glycine residue in Walker B which is replaced by an alanine (Supplementary data S1 online).
Figure 1. Conservation profile and Pfam domain prediction of CARN7_2759.
The graph displays the conservation profile deduced from the multiple alignment of 200 protein sequences obtained from PipeAlign using CARN7_2759 as a query. For each residue of CARN7_2759 protein sequence, this profile shows the number of identical amino-acids at the corresponding position of the multiple sequence alignment. The black boxes show the TraC signature (Pfam:PF11130 from 33 to 275) and the AAA domain (Pfam:PF12846 from 474 to 758) predicted by InterProScan. Both Walker A and Walker B motifs (grey boxes) characteristic of AAA domains could be identified in this region (respectively from 481 to 489, and from 686 to 693) and correspond to peaks of conservation.
These results are in agreement with the fact that, among the top 10 templates used by the I-TASSER platform for the 3D structure prediction, the most significant ones (Z-score above 3.8) corresponded to the structure of the ATPase subunit of TrwB conjugative transfer protein.
Out of the 5 models obtained with I-TASSER platform17 for CARN4_2759, 3 reached a C-score above −1.5, the most significant model having a C-score of −1.32. All 3 models are quite similar in shape and are characterized by numerous α-helices but lack the beta sheets that are characteristics of AAA domain. No EC-score for I-TASSER enzyme assignment reached the default 1.1 threshold. Some GO-terms were predicted with a score higher than the 0.5 default threshold: “protein transporter activity” (GO:0008565, GO-Score 0.70), “protein binding site” (GO:0005515, GO-Score 0.65), “nucleus cellular location” (GO:0005634, GO-Score 0.60) and “cytoplasm location” (GO:0005737, GO-Score 0.59). There was no prediction related to polymer degradation though.
Submitting the two potential domains (TraC and AAA) separately did not yield any significant model at all, suggesting further that I-TASSER predictions on CARN_2759 are marginal and should be taken with caution.
Both proteins were then produced in E. coli and purified. First attempts to produce soluble proteins in E. coli strain BL 21 were unsuccessful, since they were produced in inclusion bodies as insoluble proteins. To overcome this problem, the proteins were produced in E. coli strain Arctic DE3, working at lower temperature. Using this strain, the proteins could be produced in the soluble fraction. The two proteins were purified and tested for their amylolytic activity using AZCL-amylose as substrate at different pH (pH5 in acetate buffer, pH6 and pH7 with phosphate buffer, and pH8 and pH9 with Tris buffer) and temperature (30, 35, 40, 45, 50 and 60°C). For both proteins, the optimum activity was obtained at pH 7 and 45°C (0.32 +/− 0.01 UDO/h.mg protein for CARN4_1025 and 0.026 +/− 0.001 UDO/h.mg for CARN7_2759). Therefore, all subsequent experiments were performed in these conditions. For both proteins, this activity was shown to be specific to starch degradation, as no activity was observed for AZCL-xylan, AZCL-xyloglucan nor AZCL-cellulose.
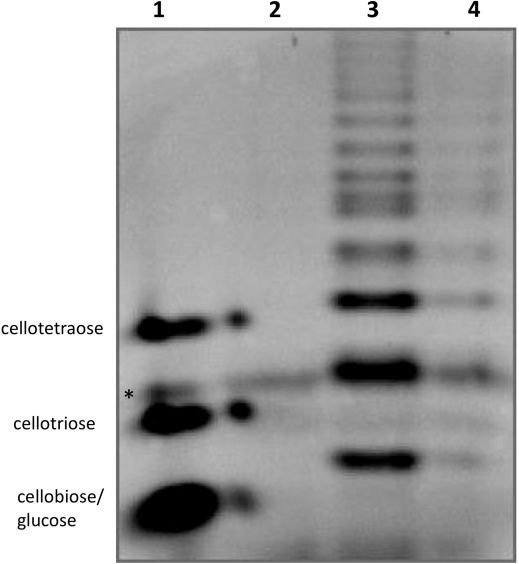
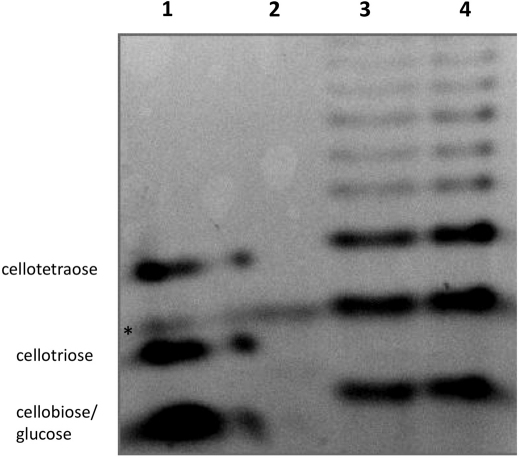
The purified amylases were tested also on natural substrates: starch from wheat and potato and on purified amylose. Despite several attempts with varying incubation time and protein concentration, no glucose could be measured after incubation of both enzymes with any polysaccharide. Two hypotheses could be made assuming this result. First, both enzymes correspond to endo-amylases and therefore do not release significant quantities of glucose from polysaccharides. Second, the activity is too low for the glucose to be detected. Objectively, the activities found with AZCL-amylose as substrate were quite low. To decide between the two hypotheses and considering these sub-detectable values, more sensitive methods are required to further characterize the enzymes. This is the reason why Polysaccharide Analysis using Carbohydrate gel Electrophoresis (PACE) experiments were performed after incubation of the enzymes with AZCL-amylose, starch and amylose as substrate. When amylose was used as substrate, degradation products could be clearly seen on the gel (Fig. 2), with numerous bands corresponding to oligosaccharides of diverse lengths. From the fact that glucose (degree of polymerisation 1, DP1) and cellobiose (DP2) comigrated in these conditions (see the standard lane) and from the absence of glucose measured, it can be concluded that the smaller product formed was cellobiose (DP2). DP 2, 3 and 4 were the major products formed but many other products with higher DP were generated. This typically corresponded to a product pattern of endo-acting enzymes after long term hydrolyses. Product pattern were very similar with both potato and wheat starch and with AZCL-amylose (Fig. 3 and supplementary Fig. S1 online).
Figure 2. PACE of the product formed by the purified amylases with amylose (1%) as substrate.
Lane 1: from top to bottom, the standards cellotetraose, cellotriose and cellobiose-glucose (DP2 and DP1 comigrate), lane 2: amylose, 3: products formed by the action of CARN4_1025 on amylose and 4: products formed by the action of CARN7_2759 on amylase. Experiments were performed at 45°C and at pH 7 in 1 ml with 269 μg and 41.5 μg of CARN4_1025 and CARN7_2759, respectively. * denotes an aspecific signal found on each lanes.
Figure 3. PACE of the product formed by the purified amylases with AZCL-amylose (0.4%) as substrate.
Lane 1: from top to bottom, the standards cellotetraose, cellotriose and cellobiose-glucose (DP2 and DP1 comigrate), lane 2: AZCL-amylose, 3: products formed by the action of CARN4_1025 on the substrate and 4: products formed by the action of CARN7_2759 on the substrate. Experiments were performed at 45°C and at pH 7 in 1 ml with 269 μg and 41.5 μg of CARN4_1025 and CARN7_2759, respectively. Loading volumes were adjusted in order to get quite similar band intensities. * denotes an aspecific signal found on each lanes.
Discussion
Since their discovery in 18335, many amylases have been characterized (for review see19). Moreover, this number has largely increased with the recent advances in molecular biology, especially with metagenomic approaches (both culture-based and sequence-based approaches) and with the accession of the genetic potentialities of yet-uncultured bacteria6,7,20,21.
In the present study, function-based screening was done with metagenomic DNA obtained from an acid mine drainage environment. AMDs are considered as extreme environments, especially with regards to pH and metals concentrations. Such sites are also considered to harbour a low bacterial diversity, and the oligotrophic conditions13,22 would not encourage the screening of amylases or other polymer-degrading enzymes. However, despite the oligotrophic conditions found in AMDs, organic matter can be produced23,24 and polymer-degrading activities should be present25. When performing this kind of experiment, 28 clones were positive for starch degradation among 80,000. Interestingly, 18 clones could not be affiliated to any of the 7 CARNs genomes previously reconstructed from the site13. In addition, among the 10 sequences left, none carried an ORF annotated as an amylase.
In order to precisely define the ORFs conferring the amylolytic activity of the CARN bacteria, two genes were chosen and subcloned and the corresponding proteins were expressed, purified and characterized. The two proteins allow the degradation of AZCL-amylose, amylase and starch, confirming the amylolytic activity. PACE experiment confirmed unambiguously the amylolytic potentialities and highlighted the endo-acting activity for both enzymes, as oligomers of sugars but not glucose were produced from amylose or starch (Fig. 2).
Notwithstanding the in vitro enzyme efficiency, biochemical experiments showed a specific amylolytic activity. The most significant advance in this study is that the nucleotidic and protein sequences of the new amylases are completely different to all the known amylases, and they share no known amylolytic domain with nor were assigned to Glycosyl Hydrolase (GH-) families. They were affiliated in previous annotations to “protein of unknown function” with few to no similarity with any other proteins. However, it should be noted that CARN7_2759 presented some similarity with TraC proteins and P-loop NTPase domains from conjugative proteins. TraC is essential for the type IV secretion system of many bacteria, but the exact role and the mechanism remain unclear26,27,28. It has been proposed that TraC functions as an adhesin that mediates host-cell targeting through binding to specific host receptors26. Here, we showed the existence of a protein displaying an amylolytic function (CARN7_2759) and sharing up to 38% identity with TraC proteins.
Amylases are enzymes studied since two centuries and many sequences, structures and hydrolysis mechanisms have already been discovered. However, when screening in an AMD, we showed the possibility to obtain new amylases without homologues. Firstly, these results imply that function-based screening allows the assignation of a function to genes encoding proteins of previously unknown function. This is crucial for every (meta)genomic approach, since the assignation of a gene to a function needs functional experiments in addition to in silico analyses. Indeed, functional screening allowed the assignation of both CARN4_1025 and CARN7_2759 to “amylases”, although the former one shows no similarity with any protein and the latter one is similar to TraC proteins, for which no hydrolytic function has been reported yet.
Secondly, it implies that sequence-based screening is not effective to discover non-homologous enzymes. This strategy is however frequently used for many enzymes such as amylases29 and arsenite oxidase30.
Thirdly, in terms of community function, the results imply that other bacteria than the ones which carry known “amylase” genes are able to degrade starch, and that those bacteria were previously missed out for this function. Here, we show that CARN1, CARN2, CARN4 and CARN7 as well as other bacteria that do not correspond to any of the CARN genomes (the 18 other clones) possess amylolytic functions that were previously not detected in the metagenomic study in this AMD13. This can be extended to any other function and (meta)genomes, as even the model species E. coli presents more than 30% of its genome encoding proteins of unknown function31. With the development of cheaper sequencing techniques, more and more raw data are injected in databases. However, without physiological experiments, no novel functional insight has to be expected from this tremendous quantity of sequences.
Lastly, the discovery of completely new amylases in AMDs should encourage researchers to screen for other interesting molecules, of biotechnological (e.g. cellulase, protease) as well as for medicinal (e.g. antibiotics) interest. Obviously, AMDs can be considered as unexpected new reservoirs for biological molecules with interesting properties especially with regards to the extreme in situ conditions.
To summarize, we reported the discovery and partial characterization of two novel amylases sharing no sequence similarity with any other amylase or glycoside hydrolase, nor possessing any known amylolytic domain. Metagenomic screening of DNA from AMDs would therefore provide other new sequences to fill the gap between genes of unknown functions and functions assignation. This work points out that screening for microorganisms or biomolecules in a priori incongruous environments, such as amylases in AMDs or thermophilic bacteria in Antarctic, could provide unconventional and new exciting ways for bioprospecting.
Methods
Bacterial strains, plasmids and growth conditions
Escherichia coli DH10β and the plasmid pCNS were previously used respectively as host cell and cloning vector for the construction of the metagenomic DNA library13. The bacteria E. coli DH5α and TOP10 (Invitrogen) and the plasmid pUC19 (Invitrogen) were used as cell host and cloning vector for the subcloning of the putative amylase genes. Proteins were expressed in E. coli Arctic DE3 (Stratagene) by using the expressing vector pET30a+.
The strain E. coli DH10β harbouring pCNS was routinely grown on LB agar (MP Biomedicals) plates supplemented with 20 mg/L chloramphenicol. The strain E. coli DH5α [pUC19] was grown on LB plates supplemented with 100 mg/L ampicillin and E. coli TOP10 [pET30a+] was maintained by addition of 50 mg/L kanamycin. Strain E. coli Arctic DE3 carrying the vector pET30a+ was grown on LB plates supplemented with 20 mg/L gentamycin and 30 mg/L kanamycin.
Screening for amylolytic activity in the metagenomic library
The previously constructed metagenomic library13 was used for amylolytic activity screening. Clones were inoculated using a 384 pin-replicator and were allowed to grow on LB plates supplemented with 20 mg/L chloramphenicol and 1% starch (Sigma) for 2 days. Plates were then overlaid with lugol solution (Sigma) and scored for clear halo zone around the colonies. Positive clones were streaked again on the same solid medium to confirm their amylolytic activities.
Sequencing and subcloning of the DNA sequences
Plasmid DNAs were extracted using the QIAprep Spin Miniprep kit (Qiagen) and the inserts were sequenced (Millegen) using the primers AHM and FM (Table 1). Subcloning was performed for two genes, CARN4_1025 and CARN7_2759. DNA was amplified for CARN4_1025 using the primers 1025-HindIII and 1025-XbaI, whereas CARN7_2759 was amplified using the primers 2759-HindIII and 2759-XbaI (Table 1). PCRs were performed using the iProof DNA polymerase (Biorad) with the following conditions: 35 amplification cycles of 98°C for 10 s, 68°C for 30 s, 72°C for 3 min, followed by a final elongation cycle (72°C for 10 min). Amplicons and purified pUC19 were digested with HindIII and XbaI primers (Fermentas), ligated and transformed into electrocompetent DH5α cells (Invitrogen). Amylolytic activities were confirmed for the subclones after inoculation on LB plates supplemented with 1% starch and 100 mg/L ampicillin.
Table 1. Primers used in this work.
| Primer | Sequence (5′→3′) | RS* |
|---|---|---|
| AHM | CAATTTCACACAGGAAACAGC | - |
| FM | TGTAAAACGACGGCCAGT | - |
| 1025-HindIII | cccaagcttgggCGCTTCCGTTCCTCAATAG | HindIII |
| 1025-XbaI | gctctagagcGCTCGACGACCCGCTTTG | XbaI |
| 2759-HindIII | cccaagcttgggCCCAATATCGAGGTCACGT | HindIII |
| 2759-XbaI | gctctagagcCGCCGCACCGATCAATAAG | XbaI |
| 1025-NcoI | catgccatggtcATGTTTACTTCCGGACGAGAATATAAAG | NcoI |
| 1025-XhoI | ccgctcgagcggTTACGGGGAGACGCCGGTAAC | XhoI |
| 2759-5 | atcgggtctcccATGTCATTTGCTTCCACGCTAAAAAAAG | BsaI |
| 2759-3 | atcgggtctcgaattTTATTTACGTTGGTTAAGCATC | BsaI |
Underlined, the added oligosequence flanking the hybridization site.
*RS, restriction site (in bold).
Full CDS were amplified using the following primers: 1025-NcoI and 1025-XhoI for CARN4_1025 (479aa) and 2759-5 and 2759-3 for CARN7_2759 (843aa) using iProof DNA polymerase (BioRad) with the following conditions: 30 amplification cycles of 98°C for 10 s, 70°C for 30 s, 72°C for 50 sec, followed by a final elongation cycle (72°C for 10 min) for CARN4_1025; 30 amplification cycles of 98°C for 10 s, 57°C for 30 s, 72°C for 2 min, followed by a final elongation cycle (72°C for 10 min) for CARN7_2759. The amplicon and the expression vector pET30a+ were digested with XhoI and NcoI for CARN4_1025 and ligated together. For CARN7_2759, the amplicon was digested by BsaI and ligated into the XhoI and NcoI sites of pET30a+. Plasmids were used to transform E. coli Top10 and inserts of several recombinants were sequenced. Plasmids with the correct sequences were introduced into the E. coli Arctic DE3 strain.
The sequences of CARN7_2759 (named amy7c) and CARN4_1025 (named amy4c) were submitted to the EMBL databases under the accession numbers HE617176 and HE617177, respectively.
Preparation of cell-free extracts and purification of the enzymes
Transformed E. coli Arctic DE3 strains were incubated in LB medium supplemented with 20 mg/L gentamycin and 30 mg/L kanamycin at 30°C until OD600 reached 0.6 to 0.7. 500 µL of cultures were then kept at 12°C for 20 min followed by induction of protein expression with 1 mM IPTG. Following a 16 h incubation time, cells were harvested by centrifugation (5,000 g, 20 min at 4°C) and lysed using the cell Lytic B solution (Sigma). Crude extracts were loaded onto Hi-Trap chelating columns to purify CARN4_1025 and CARN7_2749.
In vitro characterization of the purified enzymes
During elution, fractions of 500 μL were collected and protein concentrations were evaluated using the Bradford reagent (BioRad) with BSA as a standard. Fractions were analysed by SDS-PAGE and those corresponding to the pure proteins were pooled. Except when otherwise stated, the activities of the enzymes on polysaccharides were tested by incubating 50 μL of pure protein with a 1 mL solution of each substrate (1% potato starch, wheat starch and amylase, all from Sigma; 0.4% of AZCL-substrates from Megazyme) in 0.1 M phosphate buffer pH 7.0. Incubations were performed under agitation at 45°C. For natural substrates, 100 μL samples were taken regularly and subsequently boiled for 10 min to stop enzymatic reaction. Glucose content was estimated using the chromogen 2,2′-azino-bis(3-ethyl benzthiazoline-6-sulfonate). For AZCL-substrates, OD595 was recorded periodically after a mild centrifugation. For each determination, controls were performed without protein in the reaction mixture (blank corresponding to the substrate response) and also with the eluates of the E. coli Arctic DE3 strain bearing the empty plasmid pET30a+ (corresponding to the blank linked to the production and the purification processes).
To visualize the oligosaccharides produced during hydrolyses, Polysaccharide Analysis using Carbohydrate Electrophoresis (PACE)32 was performed. Briefly, after 48 hours of incubation in the same conditions than described below, 50 μL of the reaction mixture were dried, the products labelled with the fluorophore 8-amino-naphthalene-1,3,6-trisulfonic acid, dried again, resuspended in 6 M urea, thereafter separated by polyacrylamide electrophoresis and finally visualized under UV.
Bioinformatic analyses of the sequences
Domain and family signature prediction
Pfam33 putative domains and family signatures were predicted for both CARN4_1025 and CARN7_2759 protein sequences using the InterProScan34 on-line server.
Database similarity search
The NCBI-nr database was searched for similarity to nucleotidic sequences of CARN4_1025 and CARN7_2759 on the NCBI Blast web server35 using the BlastN (nucleotide vs nucleotide), tBlastN (protein vs translated nucleotide) and tBlastX (translated nucleotide vs translated nucleotide) algorithms. BlastP and PSI-BLAST (protein vs protein) database similarity searches were also performed with protein sequences of CARN4_1025 and CARN7_2759 against the NCBI-nr protein database and Uniprot36. The NCBI Blast server also predicted putative conserved domains by comparison with the Conserved Domain Database37.
3D Structure prediction
CARN4_1025 and CARN7_2759 protein sequences were submitted to the I-TASSER web server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/)17,38 in an attempt to predict their structural characteristics by iterative threading assembly refinement. This platform allows the prediction of structure and calculates the statistical significance of the result. It also allows the prediction of the Enzyme Commission (EC) number and GO terms (molecular function, biological process and cellular location) of the protein, based on the 3D model obtained.
Multiple sequence alignment
The protein sequence of CARN7_2759 was submitted to the PipeAlign server39 with CARN7_2759 as the reference sequence. The pipeline was configured to align a sample of 200 similar sequences identified by Blast and Ballast.
Author Contributions
F.D., V.P., M-C.L. and D.L. conceived, supervised and coordinated this study. F.D. carried out the cloning, screening and sequencing steps. V.P. and A.F. carried out all in vitro biochemical experiments. F.P. carried out all bioinformatic analyses. F.D. F.P. and V.P. performed the data interpretations. F.D. wrote the manuscript and V.P., F.P., M-C.L. and D.L. critically revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Complete sequence of the 8 other clones conferring an amylolytic activity
Acknowledgments
FD was supported by a grant from the French Ministry of Education. This work was performed as part of the program of the «Groupement de Recherche - Métabolisme de l'Arsenic chez les Microorganismes (GDR2909-CNRS)» (http://gdr2909.alsace.cnrs.fr/) in the frame of the RARE project.
References
- Jeon C. O. et al. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc Natl Acad Sci Unit States Am 100 (23), 13591 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. T. et al. Cloning and characterization of a xylanase, KRICT PX1 from the strain Paenibacillus sp. HPL-001. Biotechnol Adv 28 (5), 594 (2010). [DOI] [PubMed] [Google Scholar]
- Ballestriero F. et al. Identification of compounds with bioactivity against the nematode Caenorhabditis elegans by a screen based on the functional genomics of the marine bacterium Pseudoalteromonas tunicata D2. Appl Environ Microbiol 76 (17), 5710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. T. & Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39, 321 (1985). [DOI] [PubMed] [Google Scholar]
- Payen A. & Persoz J. F. Mémoire sur la diastase, les principaux produits de ses réactions, et leurs applications aux arts industriels. Ann. Chim. Phys. 53, 73 (1833). [Google Scholar]
- Sharma S., Khan F. G. & Qazi G. N. Molecular cloning and characterization of amylase from soil metagenomic library derived from Northwestern Himalayas. Appl Microbiol Biotechnol 86 (6), 1821 (2010). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Identification and characterization of a novel thermostable gh-57 gene from metagenomic fosmid library of the Juan De Fuca ridge hydrothermal vent. Appl Biochem Biotechnol 164 (8), 1323 (2011). [DOI] [PubMed] [Google Scholar]
- Castro G. R., Santopietro L. M. D. & Siñeriz F. Acid pullulanase from Bacillus polymyxa MIR-23. Appl Biochem Biotechnol 37 (3), 227 (1992). [Google Scholar]
- Ballschmiter M., Fütterer O. & Liebl W. Identification and characterization of a novel intracellular alkaline α-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol 72 (3), 2206 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. I. et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res 37 (SUPPL. 1), D233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijttebier A., Goesaert H. & Delcour J. A. Amylase action pattern on starch polymers. Biologia 63 (6), 989 (2008). [Google Scholar]
- Zhang J. & Zeng R., Molecular cloning and expression of an extracellular α-amylase gene from an Antarctic deep sea psychrotolerant Pseudomonas stutzeri strain 7193. World Journal of Microbiology and Biotechnology 27 (4), 841 (2011). [Google Scholar]
- Bertin P. N. et al. Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteo-genomics. ISME J 5 (11), 1735 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc M., Casiot C., Elbaz-Poulichet F. & Personnè C. Arsenic removal by oxidizing bacteria in a heavily arsenic-contaminated acid mine drainage system (Carnoulès, France).. J Geol Soc 198, 267 (2002). [Google Scholar]
- Rice P., Longden L. & Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16 (6), 276 (2000). [DOI] [PubMed] [Google Scholar]
- Smith T. F. & Waterman M. S. Identification of common molecular subsequences. J Mol Biol 147 (1), 195 (1981). [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protocol 5 (4), 725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J. & Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1 (8), 945 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M. & Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol 24 (4), 329 (1989). [DOI] [PubMed] [Google Scholar]
- Lämmle K. et al. Identification of novel enzymes with different hydrolytic activities by metagenome expression cloning. J Biotechnol 127 (4), 575 (2007). [DOI] [PubMed] [Google Scholar]
- Yun J. et al. Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl Environ Microbiol 70 (12), 7229 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C. et al. Microbial diversity in acid mine drainage of Xiang Mountain sulfide mine, Anhui Province, China. Extremophiles 14 (5), 465 (2010). [DOI] [PubMed] [Google Scholar]
- Jiao Y. et al. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl Environ Microbiol 76 (9), 2916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter D. et al. In situ proteo-metabolomics reveals metabolite secretion by the acid mine drainage bio-indicator. Euglena mutabilis. ISME J (2012) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavat F. et al. Deciphering the role of Paenibacillus strain Q8 in the organic matter recycling in the acid mine drainage of Carnoules. Microbial Cell Factories 11, art. 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Fronzes R. & Waksman G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? .Trends Microbiol 16 (9), 409 (2008). [DOI] [PubMed] [Google Scholar]
- Fronzes R., Christie P. J. & Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol 7 (10), 703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo H. J. et al. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci Unit States Am 100 (26), 15947 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amylase T. et al. A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low pH, thermostable α-amylase. J Biol Chem 277 (29), 26501 (2002). [DOI] [PubMed] [Google Scholar]
- Quéméneur M. et al. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl Environ Microbiol 74 (14), 4567 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson A. D., Pribat A. & de Crécy-Lagard V. ‘Unknown’ proteins and ‘orphans’ enzymes: The mising half of the engineering part list - And how to find it. Biochem J 425 (1), 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapito R., Carapito C., Jeltsch J. M. & Phalip V. Efficient hydrolysis of hemicellulose by a Fusarium graminearum xylanase blend produced at high levels in Escherichia coli. .Biores Technol 100 (2), 845 (2009). [DOI] [PubMed] [Google Scholar]
- Finn R. D. et al. The Pfam protein families database. Nucleic Acids Res 38 (SUPPL.1), D211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E. et al. InterProScan: Protein domains identifier. Nucleic Acids Res 33 (SUPPL. 2), W116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. et al. NCBI BLAST: a better web interface. Nucleic Acids Res 36 (Web Server issue), W5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R. et al. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res 39 (SUPPL. 1), D214 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39 (SUPPL. 1), D225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewniak F. et al. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res 31 (13), 3829 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete sequence of the 8 other clones conferring an amylolytic activity