Abstract
The systematic translation of cancer genomic data into knowledge of tumor biology and therapeutic avenues remains challenging. Such efforts should be greatly aided by robust preclinical model systems that reflect the genomic diversity of human cancers and for which detailed genetic and pharmacologic annotation is available1. Here we describe the Cancer Cell Line Encyclopedia (CCLE): a compilation of gene expression, chromosomal copy number, and massively parallel sequencing data from 947 human cancer cell lines. When coupled with pharmacologic profiles for 24 anticancer drugs across 479 of the lines, this collection allowed identification of genetic, lineage, and gene expression-based predictors of drug sensitivity. In addition to known predictors, we found that plasma cell lineage correlated with sensitivity to IGF1 receptor inhibitors; AHR expression was associated with MEK inhibitor efficacy in NRAS-mutant lines; and SLFN11 expression predicted sensitivity to topoisomerase inhibitors. Altogether, our results suggest that large, annotated cell line collections may help to enable preclinical stratification schemata for anticancer agents. The generation of genetic predictions of drug response in the preclinical setting and their incorporation into cancer clinical trial design could speed the emergence of “personalized” therapeutic regimens2.
Human cancer cell lines represent a mainstay of tumor biology and drug discovery through facile experimental manipulation, global and detailed mechanistic studies, and various high-throughput applications. Numerous studies have employed cell line panels annotated with both genetic and pharmacologic data, either within a tumor lineage3–5 or across multiple cancer types6–12. While affirming the promise of systematic cell line studies, many prior efforts were limited in their depth of genetic characterization and pharmacologic interrogation.
To address these challenges, we generated a large-scale genomic dataset for 947 human cancer cell lines, together with pharmacologic profiling of 24 compounds across ~500 of these lines. The resulting collection, which we termed the Cancer Cell Line Encyclopedia (CCLE), encompasses 36 tumor types (Fig. 1a, Supplementary Table 1 and www.broadinstitute.org/ccle). All cell lines were characterized by several genomic technology platforms. The mutational status of >1,600 genes was determined by targeted massively parallel sequencing, followed by removal of variants likely to be germline events (Supplementary Methods). Moreover, 392 recurrent mutations affecting 33 known cancer genes were assessed by mass spectrometric genotyping13 (Supplementary Table 2 and Supplementary Fig. 1). DNA copy number was measured using high-density single nucleotide polymorphism arrays (Affymetrix SNP 6.0; Supplementary Methods). Finally, mRNA expression levels were obtained for each of the lines using Affymetrix U133 plus 2.0 arrays. These data were also used to confirm cell line identities (Supplementary Methods, Supplementary Figs. 2–4).
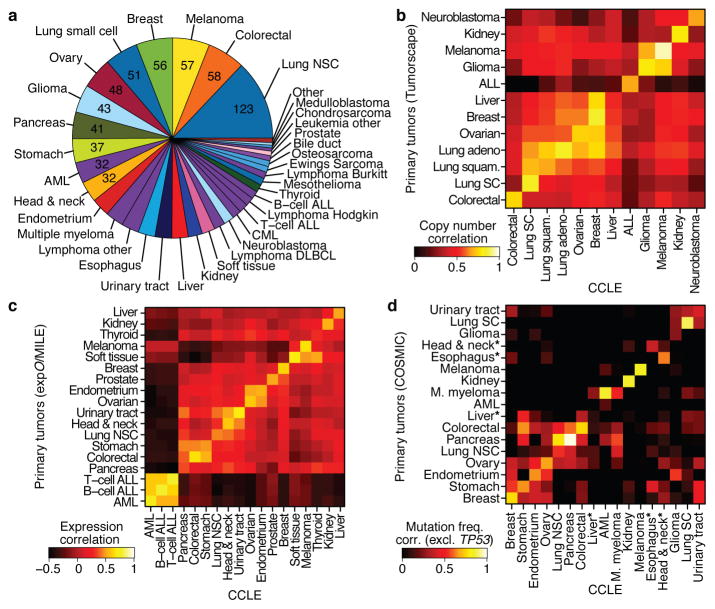
Figure 1. The Cancer Cell Line Encyclopedia (CCLE).
a. Distribution of cancer types in the CCLE by lineage. b. Comparison of DNA copy-number profiles (GISTIC G-scores) between cell lines and primary tumors. The diagonal of the heatmap shows the Pearson correlation between corresponding sample types. Because cell lines and tumors are separate datasets, the correlation matrix is asymmetric: the top left showing how well the tumor features correlate with the average of the cell lines in a lineage, and the bottom right showing the converse. c. Comparison of mRNA expression profiles between cell lines and primary tumors. For each tumor type, the log-fold-change of the 5,000 most variable genes is calculated between that tumor type and all others. Pearson correlations between tumor type fold-changes from primary tumors and cell lines are shown as a heatmap. d. Comparison of point mutation frequencies between cell lines and primary tumors in COSMIC (v56), restricted to genes that are well represented in both sample sets but excluding TP53 which is highly prevalent in most tumor types. Pairwise Pearson correlations are shown as a heatmap. *The correlations of esophageal, liver, and head and neck cancer mutation frequencies are restored when including TP53.
We next measured the genomic similarities by lineage between CCLE lines and primary tumors from Tumorscape14, expO, MILE and COSMIC datasets (Fig. 1b–d, see Supplementary Methods). For most lineages, a strong positive correlation was observed in both chromosomal copy number and gene expression patterns (median correlation coefficients of 0.77, range = 0.52–0.94, p < 10−15, for copy number and 0.60, range = 0.29–0.77, p < 10−15, for expression, respectively; Fig. 1b–c, Supplementary Table 3 and 4), as has been described previously3–5,15. A positive correlation was also observed for point mutation frequencies (median correlation coefficient = 0.71, range = −0.06–0.97, p < 10−2 for all but 3 lineages, Supplementary Fig. 5), even when TP53 was removed from the dataset (median correlation coefficient = 0.64, range = −0.31–0.97, p < 10−2 for all but 3 lineages; Fig. 1d, Supplementary Table 5). Thus, with relatively few exceptions (Supplementary Information), the CCLE may provide representative genetic proxies for primary tumors in many cancer types.
Given the pressing clinical need for robust molecular correlates of anticancer drug response, we incorporated a systematic framework to ascertain molecular correlates of pharmacologic sensitivity in vitro. First, 8-point dose response curves for 24 compounds (targeted and cytotoxic agents) across 481 cell lines were generated (Supplementary Tables 1 and 6, and Supplementary Methods). These curves were represented by a logistic sigmoidal function with a maximal effect level (Amax), the concentration at half- maximal activity of the compound (EC50), a Hill coefficient representing the sigmoidal transition, and the concentration at which the drug response reached an absolute inhibition of 50% (IC50).
Broadly active compounds, exemplified by the HDAC inhibitor panobinostat, showed a roughly even distribution of Amax and EC50 values across most cell lines (Fig. 2a). In contrast, the RAF inhibitor PLX4720 displayed a more selective profile: Amax or EC50 values for most cell lines could be categorized as “sensitive” or “insensitive” to PLX4720, with sensitive lines enriched for the BRAFV600E mutation (Fig. 2a). To capture simultaneously the efficacy and potency of a drug, we designated an “activity area” (Fig. 2b and Supplementary Fig. 6). The 24 compounds profiled showed wide variations in activity area, and those with similar mechanisms of action clustered together (Supplementary Fig. 7).
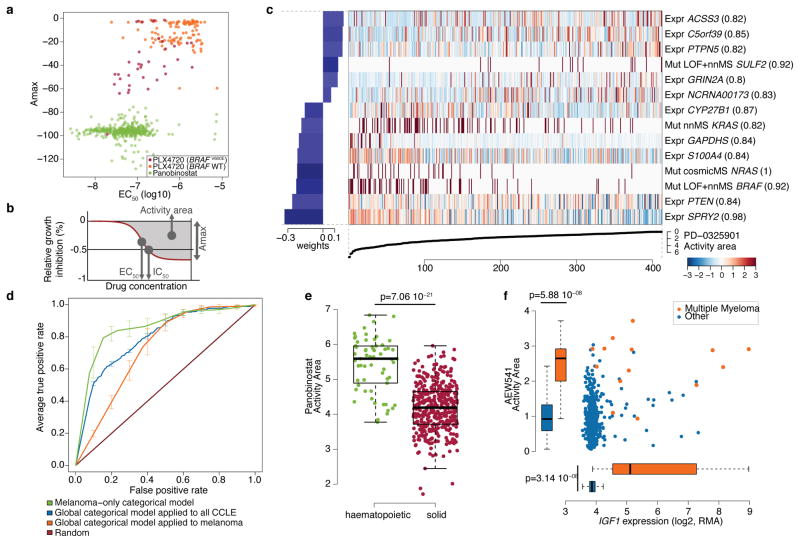
Figure 2. Predictive modeling of pharmacologic sensitivity using CCLE genomic data.
a. Drug responses for Panobinostat (green) and PLX4720 (orange/purple) represented by the high-concentration effect level (Amax) and transitional concentration (EC50) for a sigmoidal fit to the response curve (b). c. Elastic net regression modeling of genomic features that predict sensitivity to PD-0325901. The bottom curve indicates drug response, measured as the area over the dose-response curve (activity area), for each cell line. The central heatmap shows the CCLE features in the model (continuous z-score for expression and copy-number, dark red for discrete mutation calls), across all cell lines (x-axis). Bar plot (left): weight of the top predictive features for sensitivity (bottom) or insensitivity (top). Parenthesis indicate features present in >80% of models after bootstrapping. d. Specificity and sensitivity (ROC curves) of cross-validated categorical models predicting the response to a MEK inhibitor, PD-0325901 (activity area). Mean true positive rate and standard deviation (n=5) are shown when models are built using all lines (“Global categorical model” in blue and orange), or within only melanoma lines (green). e. Activity area values for LBH589 (panobinostat) between cell lines derived from hematopoietic (n=61) and solid tumors (n=387). The middle bar = median, box = inter-quartile range, and bars extend to 1.5x the inter-quartile range. f. Distribution of activity area values for AEW541 relative to IGF1 mRNA expression. Orange dots: multiple myeloma cell lines (n=14); blue dots: cell lines from other tumor types (n=434). Box-and-whisker plots show the activity area or mRNA expression distributions relative to each cell line type (line = median and box = inter-quartile range), with bars extending to 1.5x the inter-quartile range.
Genomic correlates of drug sensitivity may be extracted by predictive models using machine learning techniques6,10. We therefore assembled all CCLE genomic data types into a matrix wherein each feature was converted to a z-score across all lines (Supplementary Methods). Next, we adapted a categorical modeling approach that utilized a naive Bayes classification and discrete sensitivity calls, or an elastic net regression analysis16 for continuous sensitivity measurements. Both approaches were applied to all compounds with or without gene expression data (Supplementary Methods). Prediction performance was determined using ten-fold cross-validation, and the elastic net features were bootstrapped to retain only those that were consistent across runs (Supplementary Methods).
Out of >50,000 input features, the regression-based analysis identified multiple known features as top predictors of sensitivity to several agents (Supplementary Table 7 and Supplementary Fig. 8 and 9), with robust cross-validated performance (Supplementary Fig. 10 and 11). For example, activating mutations in BRAF and NRAS were among the top four predictors of sensitivity in models generated for the MEK inhibitor PD-032590110 (Fig. 2c). Additional predictive features for MEK inhibition included expression of PTEN, PTPN5, and SPRY2, which encodes a regulator of MAPK output. KRAS mutations were also identified, albeit with a lower predictive value (Fig. 2c, Supplementary Tables 8–9 and Supplementary Fig. 8).
Additional top predictors included EGFR mutations and ERBB2 amplification/over- expression for Erlotinib8 and Lapatinib17, respectively; BRAFV600E for RAF inhibitors (PLX472018 and RAF265); HGF expression and MET amplification for the MET/ALK inhibitor PF-234106619; and MDM2 over-expression for Nutlin-320 sensitivity. Variants affecting the EXT2 gene, which encodes a glycosyltransferase involved in heparin sulfate biosynthesis, were significantly correlated with Erlotinib sensitivity (Supplementary Fig. 12). This observation is intriguing in light of a report linking heparin sulfate with erlotinib sensitivity21. In addition, NQO1 expression was identified as the top predictive feature for sensitivity to the Hsp90 inhibitor 17-AAG, a quinone moiety metabolized by NAD(P)H:quinone oxidoreductase (NQO1). NQO1 produces a high-potency intermediate (17-AAGH2)22, and has previously been identified as a potential biomarker for Hsp90 inhibitors23.
Since some genetic/molecular alterations occur commonly in specific tumor types, lineage may become a confounding factor in predictive analyses. Indeed, a classifier built using the entire cell line dataset performed suboptimally when applied exclusively to melanoma derived-cell lines (Fig. 2d), whereas a model built with only melanoma cell lines performed better (Fig. 2d). Predictive features in the melanoma-only model showed a strong over-expression of genes regulated by the transcription factors MITF and SOX10 (Supplementary Table 10), recently identified as predictive of RAF inhibitor drug sensitivity within a melanoma-dominated cell line collection.
On the other hand, lineage emerged as the predominant predictive feature for several compounds. For example, elastic net studies of the HDAC inhibitor LBH589 (panobinostat) identified hematologic lineages as predictors of sensitivity (Fig. 2e and Supplementary Fig. 9). Interestingly, most clinical responses to panobinostat and related compounds (e.g., vorinostat and romidepsin) have been observed in hematological cancers. Similarly, most multiple myeloma cell lines (12 of 14 lines tested) exhibited enhanced sensitivity to the IGF-1 receptor inhibitor AEW541 (Fig. 2f and Supplementary Fig. 8 and 9) and showed high IGF1 expression (Fig. 2f). Interestingly, elevated IGF1R expression also correlated with AEW541 sensitivity (Supplementary Fig. 9). The CCLE results suggest that multiple myeloma may be a promising indication for clinical trials of IGF-1 receptor inhibitors24 and that these drugs may have enhanced efficacy in cancers with high IGF1 or IGF1R expression.
While BRAF and NRAS mutations are known single-gene predictors of sensitivity to MEK inhibitors, several “sensitive” cell lines lacked mutations in these genes, whereas other lines harboring these mutations were nonetheless “insensitive” (Fig. 2c). The elastic net regression model derived from the subset of cell lines with validated NRAS mutations identified elevated expression of the AHR gene (which encodes the aryl hydrocarbon receptor) as strongly correlated with sensitivity to the MEK inhibitor PD-0325901 (Fig. 3a). This finding was intriguing in light of prior studies suggesting that a related MEK inhibitor (PD-98059) may also function as a direct AHR antagonist25. We therefore hypothesized that the enhanced sensitivity of some NRAS-mutant cell lines to MEK inhibitors might relate to a coexistent dependence on AHR function.
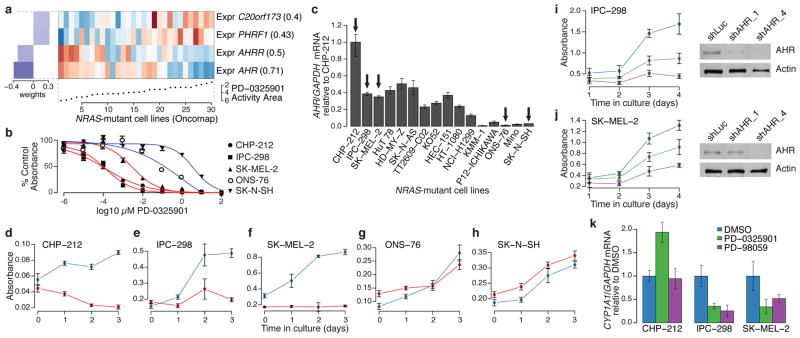
Figure 3. AHR expression may denote a tumor dependency targeted by MEK inhibitors in NRAS-mutant cell lines.
a. Predictive features for PD-0325901 sensitivity (varying baseline activity area) in validated NRAS-mutant cell lines. b. Growth inhibition curves for NRAS-mutant cell lines expressing high (red) or low (blue) levels of AHR mRNA in the presence of the MEK inhibitor PD-0325901. c. Relative AHR mRNA expression across a panel of NRAS-mutant cell lines (arrows indicate cell lines where AHR dependency was analyzed). d–h. Proliferation of NRAS-mutant cell lines displaying high (d–f) and low (g–h) AHR mRNA expression, after introduction of shRNAs against AHR (red lines) or luciferase (blue lines). i. (left) Proliferation of IPC-298 cells (high AHR) after introduction of additional shRNAs against AHR (shAHR_1 and shAHR_4; green and purple lines, respectively) or luciferase (control shLuc; blue line); (right) corresponding immunoblot analysis of AHR protein. j. Equivalent studies as in (i) with using SK-MEL-2 cells (high AHR). k. Endogenous CYP1A1 mRNA expression in the neuroblastoma line CHP-212 or the melanoma lines IPC-298 and SK-MEL-2 after exposure to vehicle (blue) or MEK inhibitors (PD-0325901, green or PD-98059, purple). Error bars: standard deviation between replicates, with n=12 (b), n=3 (c), n=6 (d–k).
To test this hypothesis, we first confirmed the correlation between AHR expression and sensitivity to MEK inhibitors in a subset of NRAS-mutant cell lines (Fig. 3b and Supplementary Fig. 13). Next, we performed shRNA knockdown of AHR in cell lines with high or low AHR expression (Fig. 3c). Silencing of AHR suppressed the growth of three NRAS-mutant cell lines with elevated AHR expression (Figs. 3d–f), but had no effect on the growth of two lines with low AHR expression (Figs. 3g–h). The growth inhibitory effect was confirmed with two additional shRNAs, where evidence for a dose-dependent knockdown effect was also apparent (Figs. 3i–j). We also tested the hypothesis that allosteric MEK inhibitors may function as AHR antagonists by measuring the effect of PD-0325901 and PD-98059 on endogenous CYP1A1 mRNA, a transcriptional target of AHR in some contexts. Both compounds reduced CYP1A1 levels in NRAS-mutant melanoma cells (IPC-298 and SK-MEL-2; Fig. 3k) but not in neuroblastoma cells (CHP-212, Fig. 3k), suggesting that other factors may govern CYP1A1 expression in the latter lineage. Together, these results suggest that AHR dependency may co-occur with MAP kinase activation in some NRAS-mutant cancer cells, and that elevated AHR may serve as a mechanistic biomarker for enhanced MEK inhibitor sensitivity in this setting.
We also looked for markers predictive of response to several conventional chemotherapeutic agents (Supplementary Fig. 7 and Supplementary Table 6) and identified SLFN11 expression as the top correlate of sensitivity to irinotecan (Fig. 4a), a camptothecin analog that inhibits the topoisomerase I (TOP1) enzyme. SLFN11 expression also emerged as the top predictor of topotecan sensitivity (another TOP1 inhibitor; Supplementary Figs. 8 and 14). Overall, 12 of 16 lineages showed significant SLFN11 associations for topotecan or irinotecan sensitivity (Pearson’s r ≥ 0.2, Supplementary Fig. 14b). This finding was independently validated using data from the NCI-60 collection (Supplementary Fig. 15). SLFN11 knockdown did not affect steady-state growth sensitivity profiles (Supplementary Fig. 14d–f).
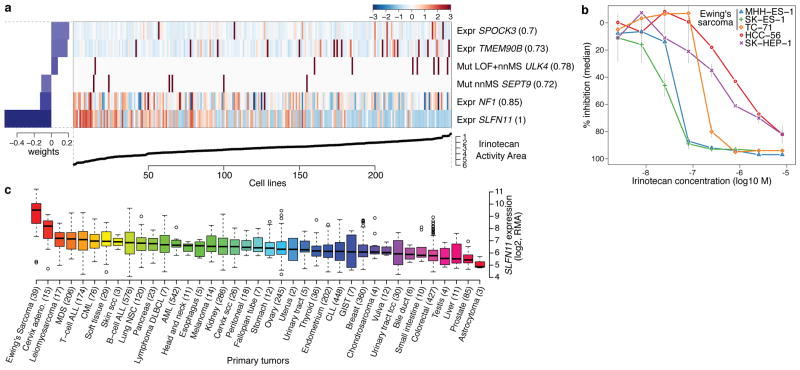
Figure 4. Predicting sensitivity to topoisomerase I inhibitors.
a. Elastic net regression analysis of genomic correlates of irinotecan sensitivity is shown for 250 cell lines. b. Dose-response curves for three Ewing’s sarcoma cell lines (MSS-ES-1, SK-ES-1, and TC-71) and two control cell lines with low SLFN11 expression (HCC-56, and SK-HEP-1). Grey vertical bars: standard deviation of the mean growth inhibition (n=2). c. SLFN11 expression across 4103 primary tumors. Box-and-whisker plots show the distribution of mRNA expression for each subtype, ordered by the median SLFN11 expression level (line), the inter-quartile range (box) and up to 1.5x the inter-quartile range (bars). Sample numbers (n) are indicated in parentheses.
All three Ewing’s sarcoma cell lines screened showed both high SLFN11 expression and sensitivity to irinotecan (Fig. 4b, Supplementary Fig. 14). Ewing’s sarcomas also exhibited the highest SLFN11 expression among 4,103 primary tumor samples spanning 39 lineages (Fig. 4c), suggesting that TOP1 inhibitors might offer an effective treatment option for this cancer type. Toward this end, several ongoing trials in Ewing’s sarcoma are examining irinotecan-based combinations, or the addition of topotecan to standard regimens26. For some lineages with high SLFN11 expression, (e.g. cervical adenocarcinoma) topoisomerase inhibitors already comprise a standard chemotherapy regimen. In other tumors where topoisomerase inhibitors are commonly used (e.g., colorectal and ovarian cancers), a range of SLFN11 expression was observed, raising the possibility that high SLFN11 expression might enrich for tumors more likely to respond. If confirmed in correlative clinical studies, SLFN11 expression may offer a means to stratify patients for topoisomerase inhibitor treatment.
By assembling the Cancer Cell Line Encyclopedia (CCLE), we have expanded the process of detailed annotation of preclinical human cancer models (www.broadinstitute.org/ccle). Genomic predictors of drug sensitivity revealed both known and novel candidate biomarkers of response. Even within genetically defined sub-populations—or when agents were broadly active without clear genetic targets—predictive modeling studies identified key predictors or mechanistic effectors of drug response. Future efforts that increase the scale and add additional types of information (e.g., whole genome/transcriptome sequencing, epigenetic studies, metabolic profiling or proteomic/phosphoproteomic analysis) should enable additional insights. In the future, comprehensive and tractable cell line systems provided through this and other efforts27 may facilitate numerous advances in cancer biology and drug discovery.
Methods Summary
A total of 947 independent cancer cell lines were profiled at the genomic level (data available at www.broadinstitute.org/ccle and Gene Expression Omnibus (GEO) using accession numbers GSE36139) and compound sensitivity data was obtained for 479 lines (Supplementary Table 11). Mutation information was obtained both by using massively parallel sequencing of >1,600 genes (Supplementary Table 12) and by mass spectrometric genotyping (OncoMap), which interrogated 492 mutations in 33 known oncogenes and tumor suppressors. Genotyping/copy number analysis was performed using Affymetrix Genome-Wide Human SNP Array 6.0 and expression analysis using the GeneChip Human Genome U133 Plus 2.0 Array. 8-point dose response curves were generated for 24 anticancer drugs using an automated compound-screening platform. Compound sensitivity data were used for two types of predictive models that utilized the naive Bayes classifier or the elastic net regression algorithm. The effects of AHR expression silencing on cell viability were assessed by stable expression of shRNA lentiviral vectors targeting either this gene or luciferase as control. The effect of compound treatment on AHR target gene expression was assessed by quantitative RT-PCR. A full description of the Methods is included in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank the staff of the Biological Samples Platform, the Genetic Analysis Platform and the Sequencing Platform at the Broad Institute. We thank S. Banerji, J. Che, C.M. Johannessen, A. Su and N. Wagle, for advice and discussion. We are grateful for the technical assistance and support of G. Bonamy, R. Brusch III, E. Gelfand, K. Gravelin, T. Huynh, S. Kehoe, K. Matthews, J. Nedzel, L. Niu, R. Pinchback, D. Roby, J. Slind, T.R. Smith, L. Tan, V. Trinh, C. Vickers, G. Yang, Y. Yao and X. Zhang. The Cancer Cell Line Encyclopedia project was enabled by a grant from the Novartis Institutes for Biomedical Research. Additional funding support was provided by the National Cancer Institute (M.M., L.A.G.), the Starr Cancer Consortium (M.F.B., L.A.G.), and the NIH Director’s New Innovator Award (L.A.G.). This resource, the Cancer Cell Line Encyclopedia (CCLE), is made available online at www.broadinstitute.org/ccle.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
For the work described herein, J.B. and G.C. were the lead research scientists; N.S., K.V., and A.M. were the lead computational biologists; M.M., W.R.S., R.S., and L.A.G. were the senior authors. J.B, G.C., S.K., P.M., J.M., J.T., A.S., N.L., and K.A., performed cell line procural and processing; P.M., and K.A., performed or directed nucleic acid extraction and quality control; S.G., W.W., and S.B.G., performed or directed genomic data generation; C.J.W., F.A.M., E.B-F., I.E., P.A., M.dS., K.J., and V.E.M., performed pharmacologic data generation; N.S., K.V., G.V.K., A.R., M.F.B., J.C., G.K.Y., M.D.J., T.L., M.R., and G.G., contributed to software development; N.S., K.V., A.A.M., J.L., G.V.K., D.S., A.R., M.L., M.F.B., A.K., P.R., J.C., G.K.Y., J.Y., M.D.J., C.H., E.P., J.P.M., V.C. and M.P.M., performed computational biology and bioinformatics analysis; J.B., G.C., N.S., L.M., J.E.M., J.J-V., M.P.M., W.R.S., R.S., and L.A.G. performed biological analysis and interpretation; N.S., K.V., A.A.M., J.L., A.R., M.L., L.M., A.K., J.J-V., J.C., G.K.Y and J.Y., prepared figures and tables for the main text and supplementary information; J.B., G.C., N.S., K.V., A.A.M., J.L., G.V.K., J.J-V., M.P.M., and L.A.G. wrote and edited the main text and supplementary information; J.B., G.C., N.S., K.V., S.K., C.J.W., J.L., S.M., C.S., R.O., T.L., L.McC., W.W., M.R., N.L., S.B.G., K.A., and V.C., performed project management; J.P.M., V.E.M., B.L.W., J.P., M.W., P.F., J.H., M.M., and T.R.G., contributed project oversight and advisory roles; and M.P.M., W.R.S., R.S., and L.A.G. provided overall project leadership.
Competing financial interests
Multiple authors are employees of Novartis, Inc., as noted in the affiliations. T.R.G., M.M., and L.A.G. are consultants for and equity holders in Foundation Medicine, Inc. M.M. and L.A.G. are consultants for and receive sponsored research from Novartis, Inc.
References
- 1.Caponigro G, Sellers WR. Advances in the preclinical testing of cancer therapeutic hypotheses. Nat Rev Drug Discov. 2011;10:179–187. doi: 10.1038/nrd3385. [DOI] [PubMed] [Google Scholar]
- 2.Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin WM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sos ML, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dry JR, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 8.Greshock J, et al. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70:3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 9.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staunton JE, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 13.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 14.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 16.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Roy Stat Soc B. 2005;67:301–320. [Google Scholar]
- 17.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 18.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou HY, et al. An Orally Available Small-Molecule Inhibitor of c-Met, PF-2341066, Exhibits Cytoreductive Antitumor Efficacy through Antiproliferative and Antiangiogenic Mechanisms. Cancer Research. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 20.Muller CR, et al. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. Int J Cancer. 2007;121:199–205. doi: 10.1002/ijc.22643. [DOI] [PubMed] [Google Scholar]
- 21.Nishio M, et al. Serum heparan sulfate concentration is correlated with the failure of epidermal growth factor receptor tyrosine kinase inhibitor treatment in patients with lung adenocarcinoma. J Thorac Oncol. 2011;6:1889–1894. doi: 10.1097/JTO.0b013e3182286d41. [DOI] [PubMed] [Google Scholar]
- 22.Guo W, et al. Formation of 17-allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: role of 17-AAG hydroquinone in heat shock protein 90 inhibition. Cancer Res. 2005;65:10006–10015. doi: 10.1158/0008-5472.CAN-05-2029. [DOI] [PubMed] [Google Scholar]
- 23.Kelland LR, Sharp SY, Rogers PM, Myers TG, Workman P. DT-Diaphorase expression and tumor cell sensitivity to 17-allylamino, 17-demethoxygeldanamycin, an inhibitor of heat shock protein 90. J Natl Cancer Inst. 1999;91:1940–1949. doi: 10.1093/jnci/91.22.1940. [DOI] [PubMed] [Google Scholar]
- 24.Moreau P, et al. Phase I study of the anti insulin-like growth factor 1 receptor (IGF-1R) monoclonal antibody, AVE1642, as single agent and in combination with bortezomib in patients with relapsed multiple myeloma. Leukemia. 2011;25:872–874. doi: 10.1038/leu.2011.4. [DOI] [PubMed] [Google Scholar]
- 25.Reiners JJ, Jr, Lee JY, Clift RE, Dudley DT, Myrand SP. PD98059 is an equipotent antagonist of the aryl hydrocarbon receptor inhibitor of mitogen-activated protein kinase kinase. Mol Pharmacol. 1998;53:438–445. doi: 10.1124/mol.53.3.438. [DOI] [PubMed] [Google Scholar]
- 26.Wagner LM, et al. Temozolomide and intravenous irinotecan for treatment of advanced Ewing sarcoma. Pediatr Blood Cancer. 2007;48:132–139. doi: 10.1002/pbc.20697. [DOI] [PubMed] [Google Scholar]
- 27.Garnett MJ, et al. A systematic screen for genomic markers of drug sensitivity in cancer cells. Nature. 2012;XXX:XXX–XXX. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.