Abstract
The coat proteins in tailed, dsDNA phage and in herpesviruses include a conserved core, similar to the bacteriophage HK97 subunit. Additions to the HK97 core often include Ig-like domains, such as the telokin-domain in P22. Eighty-six P22-like phages and prophages share a similar set of virion assembly genes and, based on comparisons of the thirteen viral assembly proteins, these phages are classified into three groups (P22-like, Sf6-like, and CUS-3-like). We used cryo-electron microscopy and 3D image reconstruction methods to determine the icosahedrally-averaged structures of Sf6 procapsids and virions at subnanometer resolutions, and to determine the structure of the entire, asymmetric Sf6 virion at 16-Å resolution. Compared to P22, the Sf6 coat protein is similar yet shows key differences in the telokin-domain and in the overall quaternary organization of the capsid. Furthermore, thermal stability and agarose gel experiments show that Sf6 particles are slightly less stable than P22. Finally, bacterial outermembrane proteins A and C were identified in lipid vesicles that co-purify with Sf6 particles, but these proteins are not components of the capsid.
Keywords: cryoEM, image reconstruction, bacteriophage Sf6, virus assembly, procapsid, virion
Introduction
The polypeptide backbones of the major coat proteins (CPs) of eukaryotic viruses and bacteriophages adopt a small number of unique folds, which is noteworthy since viral pathogens infect hosts in all kingdoms of life. Conservation of CP fold suggests that the assembly of viable virus shells is tightly controlled by a small set of specific conformations and intersubunit interactions. The CPs of the icosahedral shells of tailed, dsDNA-containing bacteriophage and members of the eukaryotic Herpesviridae family such as herpes simplex virus 1 (HSV-1) share the “HK97-core” fold. This refers to the structure adopted by the ~280 residue CP of bacteriophage HK97 (Figure 1A), whose capsid structure was the first of any bacteriophage to be solved by X-ray crystallography (Wikoff et al., 2000). CPs with this fold often share 10–15% or less sequence identity, yet they exhibit several common features. These include a long (~20–30 a.a.) “spine helix”; an α-helical N-arm domain; a long, two-stranded, β-sheet P-domain; and an A-domain with 4–6 loops and 2–4 helices and sometimes with 1-3β-strands. Some HK97-like CPs have evolved an additional, surface-exposed protrusion that often consists of an Ig-like domain (Bamford, Grimes, and Stuart, 2005; Morais et al., 2005; Parent et al., 2010a). The major capsid protein of HSV-1 (1374 a.a.) displays even more elaborate additions to it’s HK97-core (Baker et al., 2005; Bamford, Grimes, and Stuart, 2005), and this indicates that extensive surface modifications can be tolerated without compromising the assembly of these proteins into functional virions.
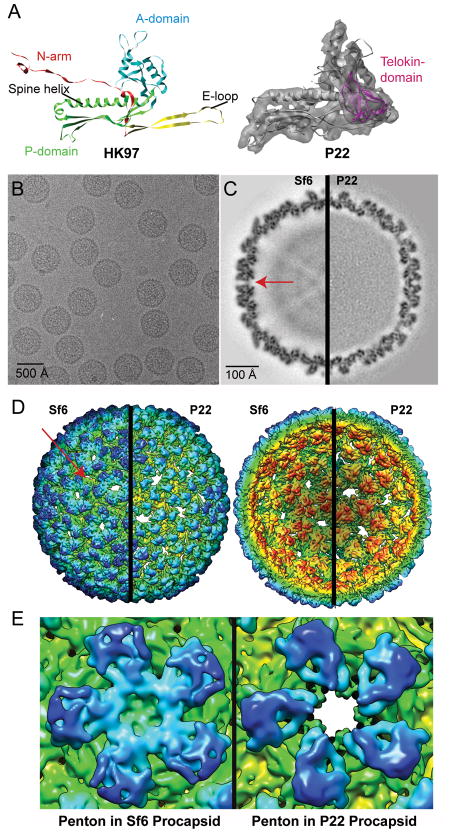
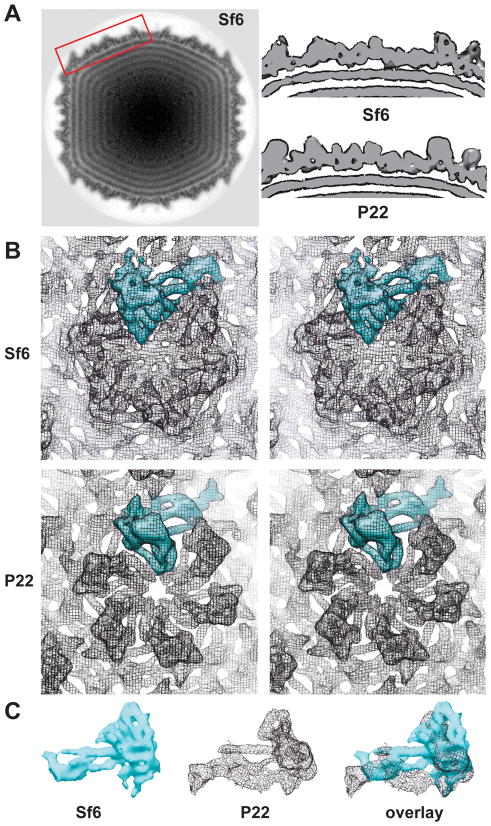
Figure 1. 3D cryo-reconstructions of Sf6 and P22 procapsids.
A. (Left) Ribbon diagram of HK97 CP (PDB ID 1OHG) color-coded to highlight domain organization. (Right) Segmented density for one CP subunit from the P22 procapsid cryo-reconstruction (transparent grey density, EMDB ID 5150) with a fitted ribbon diagram of the pseudo-atomic model of the P22 CP (PDB ID 3IYH) (Parent et al., 2010a). The telokin domain (absent in HK97) is highlighted in magenta. The HK97 and P22 subunit models are aligned with respect to their spine helices. B. Micrograph of an unstained, vitrified sample of Sf6 procapsids. C. Equatorial slabs (1 pixel, or ~1-Å thick) through the Sf6 (left half) and P22 (right half) procapsid density maps (features of highest and lowest density are depicted in black and white, respectively). Red arrow points to a solvent accessible channel in the Sf6 procapsid. D. Radially color-cued, surface representations of the Sf6 and P22 procapsid structures. For clarity, only the front (left pair) and back (right pair) quadrants of each structure are shown. The view direction in each pair is along an icosahedral two-fold axis of symmetry. Scale bar in panel (C) also refers to panel (D). E. Close-ups of the penton in Sf6 (left) and in P22 (right), each viewed along an icosahedral five-fold axis. The density maps (also in panels C and D) were scaled and rendered at the same threshold, which highlights the closed verses open axial channels in Sf6 and P22, respectively.
The propagation of lytic viruses occurs in a well-orchestrated series of steps. These include recognition and attachment of the virus and delivery of its genome to the host, replication, transcription, and translation of the viral genes, virion assembly, and release. All viruses with HK97-like CPs assemble into intermediate, precursor structures called “procapsids”. The genome is packaged into these procapsids, which then mature and, in the case of tailed phages, this is followed by the addition of the tail machinery.
P22, a dsDNA bacteriophage (family Podoviridae), has long served as an informative model system for exploring how phages and viruses utilize this basic assembly strategy (Casjens and Weigele, 2005; Prevelige, 2006; Teschke and Parent, 2010). There are currently 86 members of the P22-like phage group that have syntenic and largely homologous sets of twelve essential, virion assembly genes (Casjens and Thuman-Commike, 2011). The CPs of these “closely related” phages can be classified into three distinct types as represented by P22 (Zinder and Lederberg, 1952), Sf6 (Gemski, Koeltzow, and Formal, 1975), and CUS-3 (Casjens and Thuman-Commike; King et al., 2007). However, the assembly pathway and virion structure have only been studied in detail for P22 (Casjens and Thuman-Commike, 2011; Prevelige, 2006). In this study, we have determined the structures of phage Sf6 procapsids and virions. Most of the assembly proteins of Sf6 have amino acid sequences that are quite distinct and divergent from those of P22 (Table 1) (Casjens et al., 2004). For example, the large terminase subunits, portal proteins, and CPs of P22 and Sf6 share only about 15%, 30%, and 14% identity, respectively. The overall mechanisms of virion morphogenesis are likely conserved for these phages although they may differ in detail, and comparison of close relatives is often more informative than comparison of very distant relatives. High-resolution, X-ray crystallographic structures have been determined for three of the 13 Sf6 virion assembly proteins. These include the receptor-binding domain of the tailspike protein (gp14) (Muller et al., 2008), the small terminase (gp1) (Zhao et al.), and the knob domain of the tail needle (gp9) (Bhardwaj et al., 2011).
Table 1.
Sf6 and P22 virion assembly proteins
| Gene Product | Common name | Residues | % Identity* | ||
|---|---|---|---|---|---|
| Sf6 | P22 | Sf6 | P22 | ||
| 1 | 3 | Small terminase subunit | 140 | 162 | 12 |
| 2 | 2 | Large terminase subunit | 470 | 499 | 15 |
| 3 | 1 | Portal protein | 708 | 725 | 30 |
| 4 | 8 | Scaffolding protein | 294 | 303 | 15 |
| 5 | 5 | Coat protein | 423 | 430 | 14 |
| 7 | 4 | Tail plug protein | 160 | 166 | 35 |
| 8 | 10 | Tail plug protein | 472 | 472 | 93 |
| 9 | 26 | Tail needle | 282 | 233 | 35 |
| 11 | 7 | Ejection protein | 230 | 229 | 82 |
| 12 | 20 | Ejection protein | 431 | 471 | 33 |
| 13 | 16 | Ejection protein | 665 | 609 | 32 |
| 14 | 9 | Tailspike-endorhamnosidase | 623 | 667 | 25 |
Some values from Casjens and Thuman-Commike, 2011 (Casjens and Thuman-Commike, 2011).
Many temperate phage, including Sf6, encode virulence factors that are expressed from the prophage in bacteria (Boyd and Brussow, 2002; Canchaya, Fournous, and Brussow, 2004; Casjens and Hendrix, 2005; Cheetham and Katz, 1995). These factors can increase the fitness of the host by changing its surface characteristics, such as, for example, by affecting the composition of the lipopolysaccharide layer (Allison and Verma, 2000; Banks, Beres, and Musser, 2002; Broadbent, Davies, and van der Woude). In turn, this can protect the bacterium from attack by other bacteriophages or the eukaryotic host’s immune system. Sf6 infects the human pathogen, Shigella flexneri, and, in the prophage state, Sf6 can affect the host pathogenecity by encoding an acetylase that alters the external, O-antigen polysaccharide (Clark, Beltrame, and Manning, 1991; Verma et al., 1991). Shigella cause bacillary dysentery (“shigellosis”), and some strains cause 10–15% fatality. Shigella includes four subgroups (Liu et al., 2008), and the most frequent isolate, S. flexneri, is present in ~60 % of shigellosis cases (Yang et al., 2005). Transmission of S. flexneri occurs via the fecal-oral route and ~165 million cases occur annually, predominantly in developing countries (~18,000 in the U.S) (Kotloff et al., 1999). Bacillary dysentery typically affects children (1.1 million deaths in children under the age of 5), but individuals with compromised immune systems such as AIDS patients are highly susceptible to S. flexneri infection (Kotloff et al., 1999). Shigella is quite virulent, as evidenced by the striking statistic that ingestion of as few as 10 bacteria is sufficient to induce shigellosis. Given that Sf6 interaction with S. flexneri could result in a more dangerous human pathogen, this has acted as a stimulus to investigate Sf6 structure and function in more detail.
Here, we have employed electron cryo-microscopy (cryoEM) and three-dimensional (3D) image reconstruction methods to begin to explore Sf6 structure and assembly. We have computed icosahedrally-symmetrized, cryo-reconstructions of Sf6 procapsids and mature particles at subnanometer resolutions. In addition, we have determined the structure of the entire, asymmetric Sf6 virion at moderate resolution (~16 Å). The unsymmetrized Sf6 cryo-reconstruction shows that Sf6 has a “tail machine” and extended portal “barrel” architecture quite similar to that seen in P22 (Olia et al., 2011; Tang et al., 2011) even though the homologous proteins in the two phage share little sequence identity (Table 1). Furthermore, the Sf6 CP has the conserved, HK97-like core and a surface-exposed, Ig-like “telokin” domain, similar to that in P22 (Figure 1A) (Chen et al., 2011; Parent et al., 2010a). Despite showing overall morphological similarity, the CPs of Sf6 and P22 exhibit distinct differences in tertiary structure and inter-subunit contacts, and the Sf6 capsid is slightly less stable than that of P22.
Results
Sf6 procapsid structure
Sf6 procapsids harvested from phage-infected cells (see Materials and Methods) were vitrified and imaged under standard cryoEM conditions (Figure 1B) (Baker, Olson, and Fuller, 1999). Image preprocessing and icosahedral reconstruction methods were performed as described (Parent et al., 2010a). In brief, 139 transmission electron micrographs were recorded, and 6,851 particle images were included in the final 3D reconstruction at an estimated resolution of 7.8 Å (Table 2). Density for procapsid components that do not strictly follow the icosahedral symmetry of the CP, such as the portal protein complex, the ejection proteins, and all or most of the scaffolding proteins, was reduced to insignificant levels in the cryo-reconstruction as a result of the icosahedral averaging imposed during the image processing procedures. The gross morphology of the Sf6 procapsid appears quite similar to that of the P22 procapsid shell (EMDB ID 5149) (Parent et al., 2010a) and both exhibit T=7 laevo symmetry. Sf6 and P22 shells are similar in diameter and are built of CPs that all have a distinct protrusion on the outer surface. The CPs in both shells are organized as oligomers (“capsomers”), 12 of which are pentamers (“pentons”) and 60 of which are hexamers (“hexons”) (Figure 1D). Hexons of the Sf6 procapsid are skewed significantly and exhibit two-fold, but not six-fold symmetry. This hexon skewing is similar to that seen in P22 (Thuman-Commike et al., 1996), T7 (Agirrezabala et al., 2005), λ (Dokland and Murialdo, 1993), HK97 (Conway et al., 1995), 80α (Spilman et al., 2011), and herpes simplex 1 (Trus et al., 1996). Also, the inner surfaces of Sf6 and P22 procapsids show evidence that the capsomers cluster as trimers (Figure 1D). Though the Sf6 and P22 procapsids share many common structural features, close inspection of finer details in the respective cryo-reconstructions reveals numerous, striking differences.
Table 2.
Reconstruction statistics
| Symmetry applied | Sample: | Micrographs | Images† | Defocus (μm)†† | Pixel size (Å) | Resolution (Å)*† |
|---|---|---|---|---|---|---|
| 532 | Procapsids | 139 | 6,851 | 0.58–4.61 | 1.07 | 7.8 |
| 532 | Virions | 469 | 15,483 | 0.53–3.95 | 1.07 | 7.0 |
| 1 | Virions | 611 | 16,253 | 0.10–4.61 | 2.14 | 16.0 |
Number of boxed particles used in image reconstruction
Range of objective lens underfocus settings for micrographs
Estimation based on FSC0.5 criterion (van Heel and Schatz, 2005)
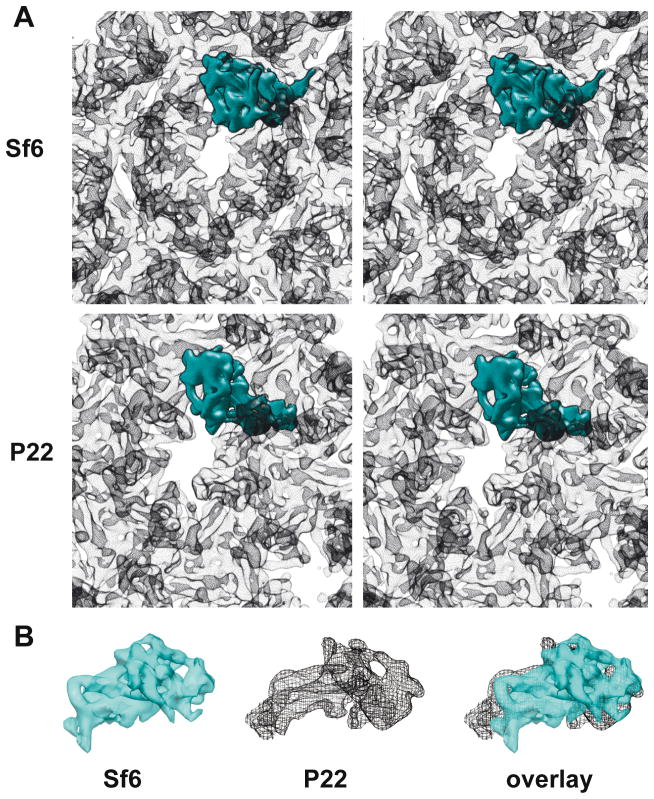
The Sf6 procapsid cryo-reconstruction contains many tube-shaped density features, consistent with the presence of α-helical secondary structural elements. These and other clearly defined features indicate that the CP subunits of Sf6 have an HK97-like core, but the overall shape and tertiary structure of the subunit differs from that seen in the P22 procapsid (Figure 2A & B). For example, when the Sf6 and P22 procapsid CP structures were aligned with respect to the spine helices, two distinct differences became obvious. First, changes in the Sf6 P-domain lead to inter-capsomer contacts at the two-fold symmetry axes that are far less extensive than those observed in P22. Second, the surface-exposed, telokin domain in Sf6 adopts a completely different orientation relative to that seen in P22 and this results in quite extensive, intra-capsomer subunit contacts that are absent in P22.
Figure 2. Folds of Sf6 and P22 CPs are similar.
A. Stereo views of hexons in Sf6 and P22 procapsids with the density envelope of a single subunit in each highlighted in cyan. B. Segmented density envelopes of individual CPs from Sf6 (cyan, solid density) and P22 (black mesh), and the two aligned with respect to the spine helices and superimposed using Chimera (Goddard, Huang, and Ferrin, 2007).
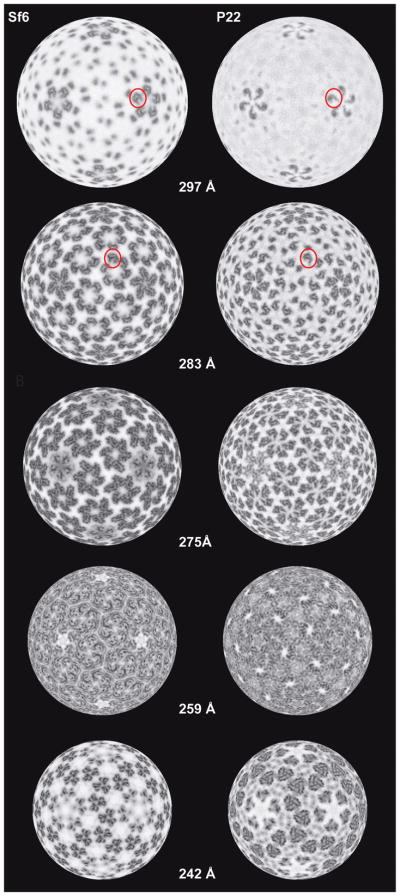
Large differences in the quaternary structures of Sf6 and P22 procapsids are evident in radial density projections (Figure 3). For example, these show the disposition of the high radius, CP protrusions in pentons (r = 297 Å) and hexons (r = 283 Å). In Sf6, the telokin domains adopt a different rotational orientation compared to P22, which results in dramatic changes in intra-capsomer contacts (r = 275 Å). As a consequence, the axial hole in Sf6 hexons is slightly smaller than that in P22 (Figure 2). The penton is completely closed in Sf6 but remains open in P22. This difference, however, is more likely attributed to changes in the A-domain than to changes in telokin orientations (Figures 1E and 2). Substantial differences in inter-capsomer contacts between Sf6 and P22 create a markedly more porous shell in Sf6, as is especially evident at the interfaces between hexons (Figure 1D and 2). As reported previously for lower resolution density maps of mature Sf6 virions (Casjens and Thuman-Commike, 2011; Zhao et al., 2011)(EMDB-5201), distinct gaps in the procapsid shell are located between Sf6 capsomers at both the local and strict two-fold axes of symmetry (Figure 1C & D, arrows). No such gaps occur in P22. Furthermore, these solvent-accessible regions are considerably larger in Sf6 procapsids compared to mature virions (see below). Contacts between Sf6 subunits are most extensive at both the quasi- and strict three-fold axes of symmetry. This is similar to what was observed in P22 and was hypothesized to contribute to overall capsid stability (Parent et al., 2010a). Lastly, the putative N-terminal arm domain and spine helix in Sf6, when compared to P22, lie at a steeper angle at the inner surface of the capsid shell and protrude further towards the particle center (Figure 1E, and Figure 2, r = 242 Å).
Figure 3. Radial density projections of Sf6 and P22 procapsids.
Projected density distributions at different radii illustrate similarities and differences in the tertiary and quaternary structures of the two procapsids. Each projection shows the density contained within a spherical, ~1-Å thick annulus. Individual telokin domains are highlighted (red circles) in Sf6 and P22 in a penton (r = 297 Å) and in a hexon (r = 283 Å).
Little if any density is present on the inside of the Sf6 procapsid that we can assign with confidence to the scaffolding protein (gp4; 33.5 kDa). This likely indicates that gp4 is not organized with the icosahedral symmetry that was imposed during the reconstruction process. This is also true of P22 procapsids; however, a recent P22 procapsid density map at 3.8-Å resolution suggested that a “V”-shaped volume of density represents the C-terminal 65 residues of the P22 scaffolding protein (gp8; 33.6 kDa), (Chen et al., 2011; Sun et al., 2000). Thorough biochemical analysis is needed to confirm the precise identity of this density in Sf6.
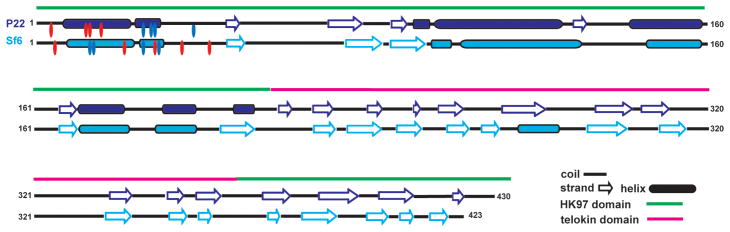
Secondary structure predictions for the CPs of Sf6 and P22, as determined with PSIPRED (McGuffin, Bryson, and Jones, 2000) (GenBank accession Nos. Sf6, AF547987; P22, BK000583), support the notion that the two subunits are remarkably similar despite being only ~14% identical in sequence (Casjens and Thuman-Commike, 2011) (Figure 4). We also analyzed the sequence of the Sf6 CP using FFAS03, a fold and function assignment system used to identify and quantify homology between proteins with ~30% or less sequence identity (Jaroszewski et al., 2005). The top match for the Sf6 CP was a pseudo-atomic model of the P22 CP (PDB ID 3IYH) that was derived from subnanometer resolution cryoEM data (Parent et al., 2010a) (Table 3). Additional hits with lower, albeit reliable scores, included the CPs of other dsDNA bacteriophage that adopt the HK97 fold and also the human FC gamma receptor, which has an Ig-like fold similar to telokin (Table 3). Despite the compelling, predicted similarities between Sf6 and P22, detailed homology modeling of the Sf6 CP proved to be problematic. First, two models of the P22 CP have been reported that differ primarily in how the C-terminal ~130 residues were interpreted (Chen et al., 2011; Parent et al., 2010a). Second, there is no reliable way to constrain or confirm the modeling process given the current paucity of biochemical and biophysical data for the Sf6 CP.
Figure 4. Secondary structure predictions for the CPs of Sf6 and P22.
The PSIPRED server (McGuffin, Bryson, and Jones, 2000) was used to predict the secondary structures of the CPs of Sf6 and P22. Regions of the CPs that correspond to predicted helices and β-strands are depicted as cylinders and arrows, respectively. Regions that correspond to the HK97-like fold and the telokin domain, as defined by a pseudo-atomic model of P22 (Parent et al., 2010a), are shown as green and magnetic lines, respectively. Charged residues in the N-arm are marked by ovals (red for acidic and blue for basic).
Table 3.
FFAS03 results
| Score | PDB ID | Protein | # Residues |
|---|---|---|---|
| −101 | 3IYH (Parent et al., 2010a) | P22 CP | 430 |
| −45 | 2XVR (Ionel et al.) | Phage T7 CP | 345 |
| −42 | 2XD8 (Liu et al., 2010) | Cyanophage P-SSP7 CP | 375 |
| −6 | 1OHG (Wikoff et al., 1999) | HK97 CP (Mature state) | 282 |
| −6 | 3E8K (Gertsman et al., 2009) | HK97 CP (Prohead II state) | 273 |
| −4 | 1E4J (Sondermann et al., 2000) | Human FC gamma receptor | 176 |
Despite the high conservation of secondary structural elements in the P22 and Sf6 CPs, the two procapsids exhibit clear differences in tertiary and quaternary structures, which must be dictated by the large differences in the amino acid sequences. Indeed, the two proteins have very different isoelectric points (pI) of 6.09 and 4.97 for Sf6 and P22, respectively, as predicted by the ProtParam server (Gasteiger et al., 2005). Such differences are consistent with the slower mobilities of Sf6 procapsids and virions compared to P22 in agarose gels, which reflect surface charge changes between particles with very similar sizes and masses (described below).
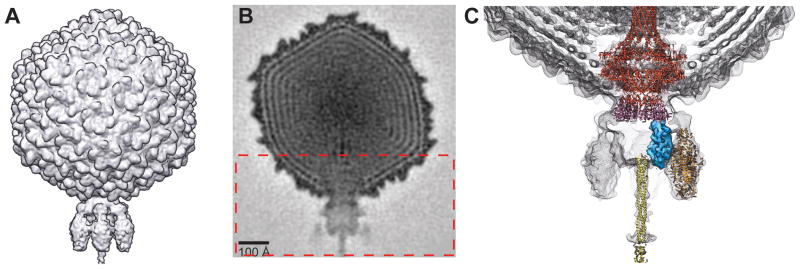
Icosahedral 3D reconstruction of Sf6 virion
An icosahedrally-averaged, 3D cryo-reconstruction of the Sf6 virion was computed from 15,483 particle images, yielding a density map of the mature Sf6 head structure at an estimated resolution of 7.0 Å (Table 2; Materials and Methods). As was also true for the Sf6 procapsid, the overall morphologies of the Sf6 and P22 heads are similar but they differ in many of the finer details. The dsDNA genome is tightly packed inside the Sf6 capsid within a series of discreet, concentric shells (at least six are visible in Figure 5A). Dense packing of the genome is found in P22 and many other tailed dsDNA phages and herpesviruses (Johnson and Chiu, 2007). The Sf6 CP appears to contact the outermost shell of the genome (Figure 5A) in a manner similar to that seen in bacteriophage φ29 (Tang et al., 2008) but, in P22, the CP does not appear to associate directly with the genome. Though CP-DNA interactions in Sf6 and φ29 may help stabilize the highly condensed genome, additional experimental evidence is needed to verify this hypothesis and to explain why P22 apparently lacks such interactions.
Figure 5. Comparisons of icosahedrally-averaged capsids of Sf6 and P22 virions.
A. Equatorial density section from the Sf6 virion cryo-reconstruction, with features of highest and lowest density depicted in black and white, respectively. Rectangular box outlines the region of Sf6 shown as a surface-rendered slab at top right. The corresponding region of P22 is shown at the bottom right. B. Stereo views of hexons with a CP monomer in each view shown in cyan, highlight marked differences in the tertiary structures of the CPs in Sf6 and P22. C. Segmented density envelopes of individual CPs from Sf6 (cyan, solid density) and P22 (black mesh), and the two aligned with respect to the spine helices and superimposed using Chimera (Goddard, Huang, and Ferrin, 2007).
Differences in the structures of procapsid and phage heads provide one means to explore the process by which the CP shell matures (expands) when procapsids convert into virions. Maturation in the well-studied HK97 system involves dramatic movements of the spine helix and N-arm, which lead to considerable thinning and increased stability of the capsid shell (Gertsman et al., 2009). P22 maturation includes similar conformational changes and also movements of the A-domain that close off the axial holes of the capsomers (Teschke and Parent, 2010). In Sf6, maturation also involves capsid expansion (~90 Å larger maximum diameter of virions versus procapsids) and wall thinning. The axial holes of the hexons become occluded in virions (Figure 5B), but the pentons are occluded prior to maturation (Figure 1E). The gaps that occur between capsomers in the Sf6 procapsid shell remain in the mature shell, although they are less pronounced and are only recognized when the cryo-reconstruction is contoured at a high density threshold. The Sf6 CP spine helix and putative N-arm rearrange during maturation (data not shown). Movement of these structural elements is consistent with observations of P22 maturation (Parent et al., 2010a). In P22, the N-arm domain of CP has been suggested to provide a binding site for scaffolding protein (Chen et al., 2011; Parent et al., 2010a). The conformation of the N-arm switches during dsDNA packaging, and this likely triggers release of the scaffolding proteins (Chen et al.; Parent et al., 2010a). In spite of large differences in sequence, the highly charged CP N-termini and scaffolding protein C-termini in P22 and Sf6 suggest that the N-arm in the Sf6 CP functions similarly in assembly and maturation.
Asymmetric cryo-reconstruction of Sf6 virion
The structure of the Sf6 virion was also reconstructed without imposing any symmetry during the processing procedures (Figure 6A). This asymmetric reconstruction, computed from 16,253 particle images and reaching an estimated 16-Å resolution (Table 2), includes reliable structural details for the dodecameric portal complex (gp3) and the tail machine, which contains the tail body (twelve gp7 and six gp8 subunits), six tailspikes (each a trimer of gp14), and the tail needle (trimer of gp9). A central density section of the Sf6 virion shows the organization of many virion components (Figure 6B). This includes the outer capsid (415 copies of gp5), the concentric layers of densely packed dsDNA genome, the portal complex at a unique vertex in the head at the junction between capsid and tail, and the tail machine. The portal complex includes an extended barrel domain that surrounds a linear stretch of DNA. With the aid of Chimera (Goddard, Huang, and Ferrin, 2007), we docked the crystal structure of the receptor-binding domain of one Sf6 tailspike trimer (PDB ID 2VBK (Muller et al., 2008)) as a rigid body into the density map. In addition, we also fit other known structures of P22 homologues (Figure 6B). These included the crystal structures of the complete, gp1, dodecameric portal complex (PDB ID 3LJ4 (Olia et al.)), the trimeric, gp26 tail needle (PDB ID 3C9I (Olia, Casjens, and Cingolani, 2007)), the gp4 tail protein (PDB ID 1VT0 (Olia et al.)), and the segmented density of the gp10 tail plug protein (EMDB ID 5051) extracted from a cryo-reconstruction of the isolated P22 tail machine (Lander et al., 2009). All of these virion components, which are similar in size in Sf6 and P22 (Table 1), fit nicely without remodeling into the Sf6 density map. Hence, the Sf6 and P22 virions have quite similar structures. Of note, the Sf6 portal structure closely mimics that in P22, and includes the long barrel domain that was hypothesized to be present in many other tailed phage (Olia et al.; Tang et al.). In Sf6, the barrel is at least 80% the height of the P22 barrel, but, at the current resolution of our virion cryo-reconstruction, we cannot unambiguously determine the full extent of the Sf6 barrel because this portion is masked by disordered genome density near the center of the phage head. Finally, as remains true for P22, we were unable to locate density in the virion reconstruction that could be assigned unambiguously to any of the three ejection proteins.
Figure 6. Sf6 virion structure.
A. Shaded-surface representation of the Sf6 virion, reconstructed by asymmetric methods to an estimated resolution of 16 Å. B. Equatorial, cross-section (1 pixel, or ~2.1 Å thick) from the Sf6 virion density map shown in A, with highest and lowest density features depicted in black and white, respectively. Rectangular box outlines the region of the virion map enlarged in C. C. Sf6 virion density map (grey mesh) plus the fitted crystal structures of the Sf6 tailspike (orange; PDB ID 2VBK), the P22 gp1 portal (red; PDB ID 3LJ4), the P22 gp4 tail plug (pink; PDB ID 1VT0), and the P22 gp26 tail needle (yellow; PDB ID 3C91), and the segmented density for the P22 gp10 tail plug (blue; EMDB ID 505) extracted from the tail machine cryo-reconstruction (Lander et al., 2009).
OmpA and OmpC are not components of Sf6 procapsids or virions
Recently, a cryo-reconstruction of the Sf6 virion head structure was determined at ~20 Å resolution, and contained density features adjacent to the inner surface of the CP shell that were interpreted as arising from about 36 and 32 copies each of the S. flexneri major outer membrane proteins, OmpA and OmpC, respectively (Zhao et al.). This interpretation was based on two observations: First, OmpA and OmpC were identified by SDS-PAGE and mass spectrometry in preparations of phage particles that had been purified through CsCl gradients. Second, an Sf6 minus P22 virion difference density map computed from icosahedrally-averaged cryo-reconstructions showed the presence of positive difference density. OmpA and OmpC were also present in our preparations of virions from Sf6-infected Shigella, and N-terminal sequencing and mass spectrometric analyses of these SDS-PAGE protein bands from Sf6 virions purified on a CsCl, step-gradient (the two SDS-PAGE bands labeled “?” in part A of Figure 2 of Casjens et al., 2004) showed the presence of both proteins. However, we found no evidence for Omp proteins in any of the three current Sf6 cryo-reconstructions (icosahedrally-symmetrized procapsids and virions and the unsymmetrized virion). We don’t, for example, see any features in the 3D density maps that correspond to a β–barrel in the positions reported by Zhao et al. (2011) and which is a structural element common to OmpA and OmpC (PDB IDs 1QJP and 2J1N; (Basle et al., 2006; Pautsch and Schulz, 2000)). When we calculate a difference density map similarly to Zhao et al, 2011 (Sf6 minus P22), but using our 7Å-resolution data, we also see positive difference density in the same places. However, the higher-resolution achieved in our structures combined with prior knowledge of the P22 CP fold (Parent et al., 2010a) allows us to trace the Sf6 CP backbone, and it appears that the observed differences result from quaternary structural changes between Sf6 and P22 (as discussed above).
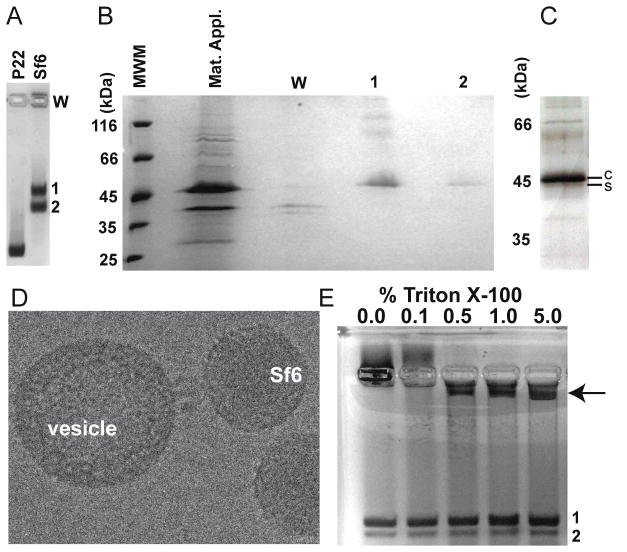
To unequivocally determine if OmpA or OmpC are components of Sf6, we used native, agarose gel electrophoresis to separate the Sf6 procapsid and virion products from phage infected cells (see Materials and Methods). The results showed two distinct bands and a significant amount of material that remained in the well and did not enter the gel (Figure 7A). As determined by ethidium bromide staining, the slower migrating band (band 1) contained dsDNA but the faster migrating band (band 2) did not (data not shown). We used SDS-PAGE and mass spectrometry to identify proteins in each band and in the well (Materials and Methods). In short, bands 1 and 2 and the well were excised from the native agarose gel (Fig. 7A), and the proteins in each extracted slice were TCA-precipitated and visualized by SDS-PAGE (Figure 7B). The major components in the wells corresponded to OmpA and OmpC and Sf6 phage proteins appeared as minor components. Band 1 contained all of the expected Sf6 virion proteins. Although Coomassie staining revealed CP as the sole component of band 2 (Figure 7B), a silver-stained SDS gel revealed the presence of all expected procapsid proteins (Figure 7C). The silver staining showed that the Sf6 scaffolding protein migrates very near to CP in SDS-PAGE (Figure 7C) despite substantial differences in their predicted molecular masses (Table 1). Slower than expected mobility has been reported for scaffolding proteins in phages T7 (Cerritelli and Studier, 1996) and cyanophage Syn5 (Raytcheva et al.) because these proteins contain a large number of charged residues, extended conformations, and low theoretical pIs (4.3 and 4.1, respectively for T7 and Syn5). The theoretical pI of the Sf6 scaffolding protein is 4.6, which may explain its altered SDS-PAGE mobility. Several different types of purification and separation procedures were performed (CsCl-purified, CHCl3-extracted, SEC-purified +/− detergent, and sucrose gradient-purified), and in all instances OmpA and OmpC were not found as components of Sf6 procapsids or virions, and were identified only in the material that remained in the wells. This therefore raised questions about what the source of the OmpA and OmpC proteins was and why they co-purified with Sf6 particles.
Figure 7. Omp A & C are not associated with assembled particles.
A. Coomassie-stained agarose gel showing P22 procapsids as a control (left lane) and assembly products from an Sf6 infection (right lane). B. SDS gel of the Sf6 sample shown in panel A, stained with Coomassie (W=well). C. Silver-stained SDS gel of material extracted from band 2 (from gel shown in A), enlarged to emphasize the separation between the scaffolding protein (“s”) and CP bands (“c”). D. Cryo-electron micrograph of a representative Sf6 virion that appears to be docked to a lipid vesicle that co-purified with virions in a CsCl gradient. E. Coomassie-stained agarose gel of Sf6 virions from a Triton X-100 titration. Increasing amounts of the material associated with the well in the first lane (marked “0%”) enter the gel as the detergent concentration increases (arrow points to this material in lane 5).
Sf6 virions from Shigella lysates show, even after CsCl, step-gradient purification, a few percent of particles with their tails apparently attached to host-derived membrane vesicles (Figure 7D). P22 particles prepared from Salmonella lysates (as well as other P22-like phages purified from their respective hosts) do not show this type of behavior (data not shown). We hypothesized that OmpA and OmpC found in the vesicles in the Shigella lysate could account for their presence as a contaminant in the purified phage samples. To test this, we treated the Sf6 samples with increasing amounts of Triton X-100 and examined the products by agarose gel electrophoresis and mass spectrometry. At levels of Triton X-100 exceeding 0.1%, conditions under which Sf6 virions remain intact, material retained in the agarose gel wells decreased, and a new band appeared (Figure 7E, arrow). Mass spectrometry analysis of this band revealed that it contained several S. flexneri, membrane-associated proteins, with the major components being OmpA and OmpC. We conclude that increasing concentrations of detergent likely liberate OmpA and OmpC from the lipid vesicles that co-purify with Sf6 virions. Furthermore, the material trapped in the wells of the agarose gel (Figures 7A and 7E) represents the virion-bound vesicles seen in the electron cryo-micrographs (a representative particle is shown in Figure 7D).
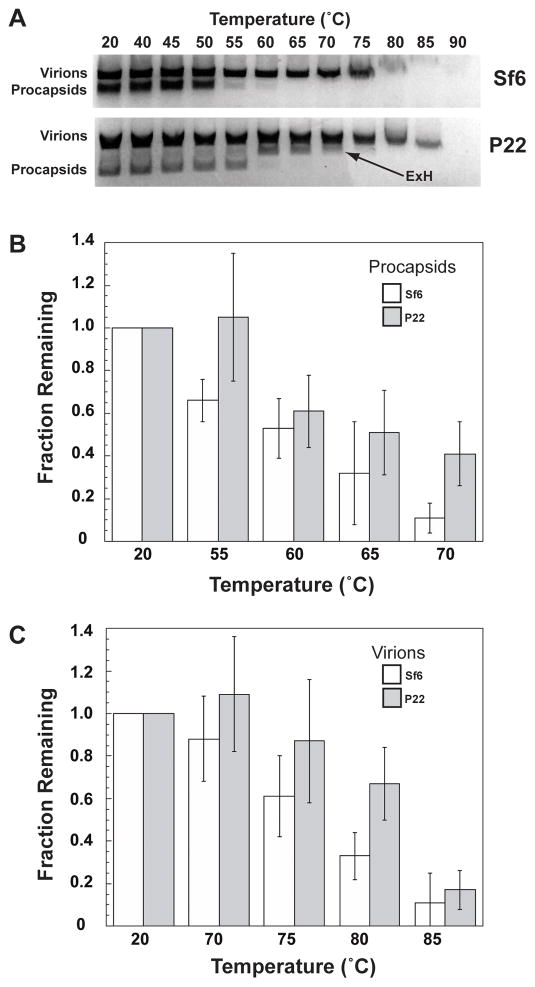
Sf6 is slightly less stable than P22
Products obtained after heat-treatment of P22 samples have been examined with agarose gel electrophoresis to monitor the relative stabilities of wild-type and variant CP procapsids (Parent et al., 2010b). If maturation or disruption of a capsid shell is induced at a lower than normal temperature, this generally indicates that intersubunit interactions between CP subunits are less stable than in wild-type particles. We performed heating and electrophoresis experiments with Sf6 procapsids and virions and also repeated them with P22 samples to compare the stabilities of the various particles. The results showed that Sf6 procapsids and virions are slightly less stable than their P22 counterparts (Figure 8A). As previously reported and reproduced again here, heat treatment caused P22 procapsids to swell and transform into “expanded heads” (ExH), which are particles that lack pentons but otherwise have matured (Parent et al., 2010a; Teschke, McGough, and Thuman-Commike, 2003). No such band corresponding to expanded heads was found in heat-treated Sf6 samples, and negative stain, transmission electron microscopy confirmed that heated P22 samples contained ExH particles but heated Sf6 samples did not (data not shown). The lack of ExH in combination with significant differences in the procapsid structures at the base of the CP shell (Figure 3, r= 259) supports the idea that the inter-capsomeric interactions, and thus assembly pathways, differ between Sf6 and P22.
Figure 8. Relative stabilities of Sf6 and P22.
A. Agarose gel of phage assembly products treated at different temperatures for 15 minutes. The expanded, penton-less heads (“ExH”) in P22 (arrow) appear as a major component of the heat-treated sample. B and C. Quantification of the fraction of intact proheads (B) and virions (C) remaining after heat treatment (data shown only for select temperatures). These data are an average from three independent experiments (error bars denote the standard deviation of the measurements).
Discussion
Sf6 CP structure consists of an HK97-like core and a telokin domain
The CPs of all tailed bacteriophages (Myoviridae, Siphoviridae, Podoviridae) and some large dsDNA eukaryotic viruses (Herpesviridae) share a common fold (Bamford, Grimes, and Stuart, 2005). It is not surprising therefore, that our cryo-reconstructions of Sf6 reveal a CP protein with an “HK97-like” core. However, like P22 but unlike other known virion structures, the Sf6 CP includes a surface-exposed, Ig-like, “telokin” domain (Parent et al., 2010a). The presence of this domain suggests that it is likely a conserved feature specific to the P22-like phages.
Most of the thirteen essential genes that encode the P22-like virion proteins show extreme variations within this phage group, and P22 and Sf6 represent two quite different members of this group (Casjens and Thuman-Commike, 2011). The respective virion assembly proteins range from having 93% identity in a.a. sequence (tail protein gp10) to having no recognizable similarity (e.g., the small terminase subunit, the scaffolding protein, and the receptor-binding domain of the tailspike). Very distantly-related species of tailed phages are proposed to have undergone “rampant” horizontal transfer of virion assembly genes during their evolution (Casjens, 2005; Hendrix, 2002). Thus, the large differences between most of the orthologous P22 and Sf6 virion assembly gene pairs raise questions about whether they have diverged in the context of the P22-like phage group or whether their differences have arisen by acceptance of genes of similar function that diverged within other phage groups and were then horizontally transferred into the P22-like group. The Sf6 and P22 CPs (only 14% identical) are among those viral proteins whose evolutionary history is uncertain. Our current results show that, unlike the CPs of other tailed phage, the Sf6 CP contains a telokin-like domain that is very similar to that of P22, and this supports the ideas that the coat protein has diverged within the P22-like group and differences among the P22-like phage CPs did not arise from horizontal exchanges with other phages. This, however, does not mean that horizontal exchange can not occur between the P22-like and other phage groups. Indeed, evidence suggests that the tailspike and small terminase subunit genes have exchanged with other phage types (Casjens and Thuman-Commike, 2011). Nonetheless, the P22-like phage group appears to be very ancient, with CP variants (and probably most other virion assembly proteins) having diverged within this group to the point of losing or nearly losing recognizable sequence homology. The P22-like portal proteins, scaffolding proteins, CPs, and the gp4 tail adaptor proteins all have very similar phylogenetic trees, which suggests that these procapsid assembly proteins have co-evolved without exchange with other phage groups, and thus that they all diverged within the P22-like phage group (Casjens and Thuman-Commike, 2011).
Sf6 capsids are likely stabilized by CP contacts at three-fold sites
Capsid assembly and subsequent stabilization that accompanies maturation in dsDNA-containing phages and in HSV-1 can occur by several different mechanisms. For example, HK97 remains unique in that it utilizes a covalent crosslinking mechanism (Wikoff et al., 2000) and forms “molecular staples” via P-loops at all three-fold sites during maturation (Gertsman et al., 2009). HK97 also includes a key, intercapsomeric salt bridge at this position, where residue substitutions reduce particle stability or eliminate capsomer assembly (Gertsman et al., 2010). The presence of auxiliary “decoration” or “cement” proteins in other phage and viruses provide a common mechanism for stabilizing virions at three-fold and quasi-three-fold positions. Examples include gpD of λ (Lander et al., 2008; Sternberg and Weisberg, 1977; Yang, Maluf, and Catalano, 2008), Dec of phage L (Gilcrease et al., 2005; Tang et al., 2006), gpE of ε15 (Jiang et al., 2008), and the triplexes of HSV-1 (Brown and Newcomb, 2011). Recently, a pseudo-atomic model of the P22 CP indicated that capsid stabilization in P22, which has no auxiliary protein and does not employ cross-links, may also occur through strong interactions between neighboring capsomers at strict and quasi three-fold symmetry axes via the P-loops (Parent et al., 2010a). The recent cryo-reconstructions of phage 80α procapsids and virions, which show CPs form “trefoil” structures at both types of three-fold sites (Spilman et al., 2011), further support the notion that stabilizing interactions commonly occur at these locations. Like P22, Sf6 CP does not form chemical crosslinks and the Sf6 genome does not encode any auxiliary decoration proteins (Casjens et al., 2004). In addition, our cryo-reconstruction of Sf6 procapsids reveals that the most extensive CP:CP contacts reside at the three-fold sites, which suggests that this feature is conserved among dsDNA-containing phages.
Capsomer maturation and A-domain flexibility
Another obvious distinction between the Sf6 and P22 structures is the presence or absence of structural elements near the centers of the capsomers. In the Sf6 procapsid, hexons have small central holes but the pentons are closed. This contrasts with the P22 procapsid, where the hexons and pentons both contain holes that are larger than that in the Sf6 hexon. Variations in the P22 CP at position 170 that increase rigidity of the A-domain result in procapsids with closed pentons (Suhanovsky et al., 2010). CP variants that resulted in increased A-domain rigidity also led to a lower energy barrier to procapsid maturation and an increase in the appearance of aberrant assembly products such as polyheads (Parent et al., 2010b). It may be that the A-domain in Sf6 is inherently more rigid than in other P22-like phages, and this favors the closed state of capsomers in procapsids. However, there does not seem to be a corresponding change in thermal stability between Sf6 and P22 procapsids, nor did we observe any aberrant assembly products in our samples. Perhaps the flexibility of the A-domain does not play the same role in regulating maturation of the Sf6-like subgroup of phages.
Cell recognition and attachment in Podoviridae
For members of the family Podoviridae, the tailspike proteins are the primary viral components that function in cell-recognition and attachment. Crystal structures have been determined for the P22 and Sf6 tailspike proteins, and the endorhamnosidase activity of both of these proteins has been thoroughly documented (Muller et al., 2008; Steinbacher et al., 1994). A secondary receptor has been postulated to be required to trigger DNA release from the virion (Casjens and Molineux, 2012; Chang et al., 2010). However, for P22, the lipopolysaccharide on the host is sufficient to allow genome release in vitro (Andres et al., 2010), and this indicates that a secondary receptor may not be needed for P22 to infect Salmonella.
The observation of host membrane vesicles stably bound to Sf6 virion tails is nonetheless provocative, since the tailspike endorhamnosidase might remove all of the O-antigen polysaccharide, thereby leaving nothing to which the tailspike could bind. This raises the question of whether the Omp proteins are involved in binding, or perhaps a different secondary receptor. OmpA is a major component of the outer membranes of enterobacteria, with ~100,000 copies present on the surface of each bacterium (Koebnik, Locher, and Van Gelder, 2000). In E.coli, OmpA is a transmembrane protein, and surface-exposed loops that connect the transmembrane domains act as the site for host recognition and attachment for many coliphage (Morona, Klose, and Henning, 1984). The ompA2 allele has ~99.6% identity between E.coli and S. flexneri (Power et al., 2006) and could be a universally present, cell recognition site for P22-like phage. OmpC has also been implicated as a cell surface receptor for bacteriophage (Ho and Slauch, 2001; Tanji et al., 2008; Yu and Mizushima, 1982). Our cryoEM results reveal that neither Omp A nor Omp C is associated with the Sf6 capsid, but instead both associate with the lipid vesicles that co-purify with virions. This phenomenon has yet to be reported for any other P22-like phage. The recently determined crystal structure of the Sf6 tail needle-knob shows significant conservation with the structures of the cell receptor binding motifs in PRD-1 phage and adenovirus (Bhardwaj et al., 2011). This suggests that the knob of the tail needle may promote lipid vesicle co-purification with Sf6 and could be driven in part owing to the high concentrations of Omp A and Omp C in our samples. Alternatively, Sf6 may not bind to S. flexneri via a secondary receptor, but rather through the tailspikes or even aided by non-specific hydrophobic interactions with the lipid vesicles, and could mean that Omp A and C are simply contaminant “cargo”.
Roles of the D-loops and telokin domains in P22-like phages
The CPs of Sf6 and P22 have a strikingly similar pattern of secondary structure elements (Figure 4) but yet very different amino acid sequences. The folds of these CPs are similar overall, but differ in a few, key sites, which tends to suggest that control of assembly and maturation may also differ in Sf6 and P22. A recent, higher-resolution cryo-reconstruction study of P22 procapsids indicates that capsid stability in P22 arises not only from interactions among P-loops at the three-fold sites, but also from intersubunit D-loop interactions (Chen et al., 2011). The Sf6 procapsid has no obvious D-loop structure and neighboring capsomers only minimally contact each other, indicating that Sf6 lacks a corresponding, P22-like, D-loop function. Whether P22 gained or Sf6 lost D-loop function during evolution remains an open question. However, Sf6 procapsids function without the D-loop, and they are only slightly less stabile than P22 when both are heated.
In P22, the telokin domains do not contact one another and each domain is only associated with a single CP monomer (Parent et al., 2010a). Hence, these domains are not responsible for stabilizing particles by bridging and tethering adjacent subunits. Instead, the domain appears to function primarily as a folding nucleus for the HK97-like core structure (Teschke and Parent, 2010). However, in striking contrast, the telokin-like domain in Sf6 makes extensive contacts between hexon subunits (Fig. 2, r = 275 Å), and these may enhance capsomer stability or they may confer some advantage during assembly of CPs that lack D-loop structures, or they may do both.
Materials and Methods
Preparation and purification of Sf6 virions and procapsids
Luria Broth (LB) was used to support bacterial growth for the preparation of phage and procapsids. S. flexneri strain PE577 (Morona et al., 1994) was grown in LB at 37 °C to 1×108 cells/mL. Phage Sf6 (clear plaque mutant (Casjens et al., 2004)) infection was initiated at a multiplicity of infection of 0.1. The culture was shaken until near-complete lysis occurred (~3.5 hours at 37 °C). Chloroform was added to ensure complete lysis, and cell debris was removed by centrifugation (Sorvall SS-34 rotor, for 10 minutes at 10,000 rpm, 4 °C). Phage and procapsids were then concentrated at 18,000 rpm for 90 minutes at 4 °C. The resulting pellet was resuspended by nutation at 4 °C, overnight, in M9 salts supplemented with 2 mM MgSO4. Aggregated material was removed by centrifugation and the phage/procapsid preparation was stored at 4 °C. Further purification using a CsCl step gradient was performed as described (Casjens et al., 2004).
Cryo-transmission electron microscopy
Small (3.5 μL) aliquots of purified phage and procapsids (at ~1×1014 phage/mL or ~10 mg/mL) were vitrified and examined using established procedures (Baker, Olson, and Fuller, 1999). Samples were applied to holey Quantifoil grids that had been glow-discharged for ~15 s in an Emitech K350 evaporation unit. Grids were then blotted with Whatman filter paper for ~5 s, plunged into liquid ethane, and transferred into a precooled, FEI Polara, multi-specimen holder, which maintained the specimen at liquid nitrogen temperature. Micrographs were recorded on Kodak SO-163 electron-image film in an FEI Polara microscope operated at 200 keV and under minimal-dose conditions (~22 e/Å2) at a nominal magnification of 59,000X. Additional data collection statistics, including the range of objective lens defocus settings used to record each set of micrographs, are listed in Table 2. The programs RobEM (http://cryoEM.ucsd.edu/programs.shtm) and ctffind3 (Mindell and Grigorieff, 2003) were used to extract individual particles and estimate the level of defocus and astigmatism for each micrograph.
Icosahedral cryo-reconstructions of Sf6 procapsid and virion
Image processing for each of the two separate data sets (procapsids and virions) was performed in the same manner as follows. Micrographs that exhibited minimal astigmatism and specimen drift were selected for further processing and digitized at 6.35 μm intervals (represents 1.07 Å per pixel) on a Nikon Supercool 8000, and image preprocessing was performed as described (Baker, Olson, and Fuller, 1999). A subset of 150 particle images was used as input to the random-model computation procedure to generate an initial 3D density map at ~25 Å resolution (Yan et al., 2007). This map was then used to initiate determination and refinement of particle orientations and origins for the complete set of images using version 4.01.07 of AUTO3DEM (Yan, Sinkovits, and Baker, 2007). Phases and amplitudes of the particle structure factor data were corrected to compensate for the effects caused by the microscope contrast-transfer function (Bowman et al., 2002). The Fourier Shell Correlation criterion (FSC0.5) was used to estimate the resolution of each reconstruction (Table 2) (van Heel and Schatz, 2005). Graphical representations were generated with the RobEM and Chimera (Goddard, Huang, and Ferrin, 2007) visualization software packages.
Asymmetric cryo-reconstruction of Sf6 virion
The general strategy we used to process the Sf6 virion images and compute an asymmetric reconstruction was similar to that used to derive an asymmetric reconstruction of the P22 virion (Lander et al., 2006). First, all digitized micrographs of the Sf6 virion samples were binned 2X, which generated a pixel size of 2.14 Å. Then, we re-boxed all virion images within a box window that was large enough to assure that the tail of each particle was not excluded as was the case during the icosahedral processing strategy described above. Also, the origin and orientation parameters of each virion image were set to match those determined during the icosahedral processing. Next, we used the “ticos_equiv” option in AUTO3DEM (v4.01.07) (Yan, Sinkovits, and Baker, 2007) to determine an initial estimate of the orientation that properly aligns the tail in each virion image to a common reference, which was the 17-Å resolution 3D cryo-reconstruction of P22 (EMDB ID 1220 (Lander et al., 2006)). Next, iterative refinement of the origin and orientation of each virion image was performed in AUTO3DEM set to operate in “asymmetric” mode, in which no symmetry is assumed or imposed on any of the data. It is important to note that the P22 reconstruction did not introduce any model bias into the Sf6 asymmetric reconstruction process. This was clearly evident because the final Sf6 reconstruction showed a head structure with all of the features that distinguish Sf6 from P22. In addition, the final Sf6 reconstruction showed a tail structure that was similar overall, yet distinct in detail from the P22 tail. Finally, the tip of the tail needle is partially missing in the Sf6 cryo-reconstruction (Figure 5) because the size of box window was reduced slightly to keep computations efficient and within memory limits, and secondarily to reduce unwanted background noise.
Agarose and polyacrylamide electrophoresis gels
Samples (~1 mg/mL) were diluted with ice-cold agarose sample buffer (0.25% bromophenol blue and 10% glycerol in TAE buffer: 40 mM Tris acetate and 1 mM EDTA). Approximately 10 μg of protein was loaded in each lane of a 1.0% Fisher agarose gel (modified from Serwer and Pichler, 1978) (Serwer and Pichler, 1978). The gel was run at 110 V for 120 min at room temperature and stained with Coomassie blue. Bands were excised with a razor blade, and the agarose was solubilized in 500 μL QB buffer from a Qiagen gel-extraction kit at 50 °C for 10 min. The total protein in each sample was either 1) TCA precipitated, visualized by SDS-PAGE (10% gel), and either Coomassie stained (wells and virion band) or silver-stained (procapsid band) using a SilverQuest staining kit (INVITROGEN) or 2) analyzed by mass spectrometry as described below.
In-gel digest for mass spectrometry
Samples were dried in a speedvac, reduced with 200 μl of 100 mM ammonium bicarbonate-10mM DTT and incubated at 56°C for 30 min. The liquid was removed and 200 ml of 100 mM ammonium bicarbonate-55mM iodoacetamide was added to the gel pieces and incubated at room temperature in the dark for 20 min. After the removal of the supernatant and one wash with 100 mM ammonium bicarbonate for 15 min, the same volume of ACN was added to dehydrate the gel pieces. The solution was then removed and samples were dried in a speedvac. For digestion, enough solution of ice-cold trypsin (0.01 ug/ml) in 50 mM ammonium bicarbonate was added to cover the gel pieces and set on ice for 30 min. After complete rehydration of the pieces, excess trypsin solution was removed and replaced with fresh 50 mM ammoniumbicarbonate and then incubated overnight at 37°C. The peptideswere extracted twice by the addition of 50 μl of 0.2% formic acid and 5% ACN and vortexed at room temperature for 30 min. The supernatant was removed and saved. A total of 50 μl of 50% ACN-0.2% formic acid was added to the sample, which was vortexed again at room temperature for 30 min. The supernatant was removed and combined with the supernatant from the first extraction. The combined extractions were analyzeddirectly by liquid chromatography (LC) in combination with tandem mass spectroscopy (MS/MS) using electrospray ionization.
LC-MS/MS analysis
Trypsin-digested peptides extracted from SDS-PAGE as describedabove were analyzed by liquid chromatography (LC)-MS/MS with electrospray ionization. All nanospray ionization experiments were performed with a QSTAR-Elite hybrid mass spectrometer (AB/MDS Sciex) that was interfaced to a nanoscale, reverse-phase, high-pressure, liquid chromatograph (Tempo) using a 10 cm-180 ID glass capillary packed with 5-μm C18 ZorbaxTM beads (Agilent). Buffer compositions were as follows: Buffer A - 98% H2O, 2% ACN, 0.2% formic acid, and 0.005% TFA; Buffer B - 100% ACN, 0.2% formic acid, and 0.005% TFA. Peptides were eluted from the C-18 column into the mass spectrometer using a linear gradient of 5–60% Buffer B over 60 min at 400 ml/min. LC-MS/MS data were acquired in a data-dependent fashion by selecting the four most intense peaks with charge states of 2 to 4 that exceed 20 counts, with exclusion of former target ions set to “360 seconds” and the mass tolerance for exclusion set to 100 ppm. Time-of-flight MS were acquired at m/z 400 to 1600 Da for 1 s with summation of 12 time bins. MS/MS data were acquired from m/z 50 to 2,000 Da by using “enhance all” (24 time bins summed), dynamic background subtract, automatic collision energy, and automatic MS/MS accumulation with the fragment intensity multiplier set to 6 and maximum accumulation set to 2 s before returning to the survey scan. Peptide identifications were made using the paragon algorithm executed in Protein Pilot 2.0 (Life Technologies).
Heat titration of assembly products
Samples containing procapsids and phage were incubated for 15 min in an Applied Biosystems GeneAmp thermalcycler at temperatures ranging from 20 to 90 °C, after which they were placed on ice and diluted with ice-cold agarose sample buffer. Electrophoresis was performed in TAE buffer. Gels were run at 110V at room temperature for 90–120 min.
Acknowledgments
We thank N. Olson for expert guidance in cryoEM methods, K. and J. Pogliano (UCSD) for access to equipment and lab space, R. Khayat and J.E. Johnson (Scripps Research Institute) for helpful discussions, and G. Lander (UC Berkeley) for a segmented gp10 cryoEM density map. We also thank C. Teschke (UConn) for helpful suggestions and use of lab space and A. Yashchenko for help with image digitization and preliminary data processing. This work was supported in part by NIH grants R37 GM-033050 and 1S10 RR-020016 to TSB, AI074825 to SRC, NIH fellowship F32A1078624 to KNP, and support from UCSD and the Agouron Foundation to TSB to establish and support the UCSD cryoEM facilities.
Abbreviations
- a.a
amino acid
- cryoEM
cryo electron microscopy
- CP
coat protein
- 3D
three-dimensional
- gp
gene product
- omp
outer membrane protein
References
- Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24(21):3820–9. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8(1):17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- Andres D, Hanke C, Baxa U, Seul A, Barbirz S, Seckler R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem. 2010;285(47):36768–75. doi: 10.1074/jbc.M110.169003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. [erratum appears in Microbiol Mol Biol Rev 2000 Mar;64(1):237.] Microbio & Mol Biol Rev. 1999;63(4):862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–63. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10(11):515–21. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006;362(5):933–42. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Molineux IJ, Casjens SR, Cingolani G. Atomic structure of bacteriophage Sf6 tail needle knob. J Biol Chem. 2011;286:30867–77. doi: 10.1074/jbc.M111.260877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol. 2002;76(23):12250–8. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10(11):521–9. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010;77(2):337–53. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Newcomb WW. Herpesvirus Capsid Assembly: Insights from Structural Analysis. Curr Opin Virol. 2011;1(2):142–9. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53(1):9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hendrix R. Bacteriophages and the Bacterial Genome. In: Higgins NP, editor. The Bacterial Chromosome. ASM Press; Washington D.C: 2005. [Google Scholar]
- Casjens S, Molineux I, Rossmann M, Rao V, editors. Viral Molecular Machines. New York: Springer; 2012. Adsorption and DNA delivery by podoviruses. Vol. In Press. [Google Scholar]
- Casjens S, Weigele P, Catalano C, editors. Viral Genome Packaging Machines. Georgetown, TX: Landes Bioscience; 2005. Headful DNA packaging by bacteriophage P22. [Google Scholar]
- Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kuhlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol. 2004;339(2):379–94. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8(4):451–8. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411(2):393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996;258(2):286–98. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Haase-Pettingell C, Weigele PR, King JA, Chiu W. Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J Mol Biol. 2010;402(4):731–40. doi: 10.1016/j.jmb.2010.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18(2):201–8. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108(4):1355–60. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CA, Beltrame J, Manning PA. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene. 1991;107(1):43–52. doi: 10.1016/0378-1119(91)90295-m. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of a virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233(4):682–94. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gemski P, Jr, Koeltzow DE, Formal SB. Phage conversion of Shigella flexneri group antigens. Infect Immun. 1975;11(4):685–91. doi: 10.1128/iai.11.4.685-691.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsman I, Fu CY, Huang R, Komives EA, Johnson JE. Critical salt bridges guide capsid assembly, stability, and maturation behavior in bacteriophage HK97. Mol Cell Proteomics. 2010;9(8):1752–63. doi: 10.1074/mcp.M000039-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458(7238):646–50. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, Casjens SR. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J Bacteriol. 2005;187(6):2050–7. doi: 10.1128/JB.187.6.2050-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157(1):281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61(4):471–80. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Ho TD, Slauch JM. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J Bacteriol. 2001;183(4):1495–8. doi: 10.1128/JB.183.4.1495-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionel A, Velazquez-Muriel JA, Luque D, Cuervo A, Caston JR, Valpuesta JM, Martin-Benito J, Carrascosa JL. Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J Biol Chem. 286(1):234–42. doi: 10.1074/jbc.M110.187211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile-profile sequence alignments. Nucl Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. backbone structure of the infectious e15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–34. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol. 2007;17:237–43. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- King MR, Vimr RP, Steenbergen SM, Spanjaard L, Plunkett G, 3rd, Blattner FR, Vimr ER. Escherichia coli K1-specific bacteriophage CUS-3 distribution and function in phase-variable capsular polysialic acid O acetylation. J Bacteriol. 2007;189(17):6447–56. doi: 10.1128/JB.00657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37(2):239–53. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–66. [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeemaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Khayat R, Li R, Prevelige PJ, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architechture of an infectious conduit. Structure. 2009;17:789–99. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige PJ, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, Reeves PR, Wang L. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008;32(4):627–53. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nature Structural & Molecular Biology. 2010;17(7):830–37. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in the tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18(2):149–59. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Morona R, Klose M, Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol. 1984;159(2):570–8. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R, Mavris M, Fallarino A, Manning PA. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176(3):733–47. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JJ, Barbirz S, Heinle K, Freiberg A, Seckler R, Heinemann U. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6. Structure. 2008;16(5):766–75. doi: 10.1016/j.str.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18(5):597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical crosslinks. Structure. 2010a;18:390–410. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Sinkovits RS, Suhanovsky MM, Teschke CM, Egelman EH, Baker TS. Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. Phys Biol. 2010b;7(4):045004. doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J Mol Biol. 2000;298(2):273–82. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- Power ML, Ferrari BC, Littlefield-Wyer J, Gordon DM, Slade MB, Veal DA. A naturally occurring novel allele of Escherichia coli outer membrane protein A reduces sensitivity to bacteriophage. Appl Environ Microbiol. 2006;72(12):7930–2. doi: 10.1128/AEM.01040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PJ. Bacteriophage P22. In: Calendar R, editor. The Bacteriophages. 2. Oxford University Press; New York City, NY: 2006. [Google Scholar]
- Raytcheva DA, Haase-Pettingell C, Piret JM, King JA. Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. J Virol. 85(5):2406–15. doi: 10.1128/JVI.01601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Pichler ME. Electrophoresis of bacteriophage T7 and T7 capsids in agarose gels. J Virol. 1978;28(3):917–928. doi: 10.1128/jvi.28.3.917-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406(6793):267–73. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- Spilman MS, Dearborn AD, Chang JR, Damle PK, Christie GE, Dokland T. A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80α capsids. Journal of Molecular Biology. 2011;405:863–76. doi: 10.1016/j.jmb.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal Structure of P22 Tailspike Protein: Interdigitated Subunits in a Thermostable Trimer. Science. 1994;265:383–86. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J Mol Biol. 1977;117:733–59. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibilty in conformational switching. Mol Micro. 2010;77(6):1568–82. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PEJ, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297(5):1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- Tang J, Lander GC, Olia A, Li R, Casjens S, Prevelige P, Jr, Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19(4):496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16:935–43. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid cementing protiens in bacteriophage L. Structure. 2006;14:837–45. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tanji Y, Hattori K, Suzuki K, Miyanaga K. Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl Environ Microbiol. 2008;74(14):4256–63. doi: 10.1128/AEM.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM, McGough A, Thuman-Commike PA. Penton release from P22 heat-expanded capsids suggests importance of stabilizing penton-hexon interactions during capsid maturation. Biophysical Journal. 2003;84:2585–92. doi: 10.1016/S0006-3495(03)75063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM, Parent KN. ‘Let the phage do the work’: Using the phage P22 coat protein structure as a framework to understand its folding and assembly mutants. Virology. 2010;401(2):119–30. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jokana J, Prasad BVV, King J, Prevelige PE, Jr, Chiu W. Three-dimensional structure of scaffolding-containing phage P22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383. [DOI] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263(3):447–62. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151(3):250–62. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Verma NK, Brandt JM, Verma DJ, Lindberg AA. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol Microbiol. 1991;5(1):71–5. doi: 10.1111/j.1365-2958.1991.tb01827.x. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Duda RL, Hendrix RW, Johnson JE. Crystallographic analysis of the dsDNA bacteriophage HK97 mature empty capsid. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):763–71. doi: 10.1107/s0907444998017661. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289(5487):2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–25. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33(19):6445–58. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Maluf NK, Catalano CE. Packaging of a unit-length viral genome: the role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J Mol Biol. 2008;383(5):1037–48. doi: 10.1016/j.jmb.2008.08.063. [DOI] [PubMed] [Google Scholar]
- Yu F, Mizushima S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J Bacteriol. 1982;151(2):718–22. doi: 10.1128/jb.151.2.718-722.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A. 107(5):1971–6. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sequeira RD, Galeva NA, Tang L. The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion. Virology. 2011;409(2):319–27. doi: 10.1016/j.virol.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64(5):679–99. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]