Abstract
Background
We hypothesized that drug resistance mutations would impact clinical outcomes associated with HIV-1 infection.
Methods
A matched case-control study of participants in AIDS Clinical Trials Group Longitudinal Linked Randomized Trials (ALLRT). Cases experienced an AIDS-defining event (ADE) or mortality, and controls did not. One hundred thirty four cases were identified and matched to a total of 266 controls by age, sex, treatment regimen, and length of follow-up. Both cases and controls had HIV RNA levels of ≥ 500 copies/mL within 24 weeks of an event. Population-based genotyping at or near the time of the event was used to evaluate the impact of resistance mutations on incidence of ADE and/or death using conditional logistic regression models.
Results
One hundred and four cases and 183 controls were analyzed. Median time to event was 99 weeks; six cases were deaths. At baseline, cases had lower CD4 (median 117 vs. 235 cells/mm3, p<0.0001) and higher HIV RNA levels (median 205,000 vs. 57,000 copies/mL, p=0.03). No significant differences in resistance were seen between cases and controls.
Conclusions
In this rigorously designed case-control study, lower CD4 cell counts and higher virus loads, not antiretroviral drug resistance, were strongly associated ADE and mortality.
Keywords: Human Immunodeficiency Virus, Acquired Immunodeficiency Syndrome, Virologic Resistance, Opportunistic Infection, Mortality
INTRODUCTION
The selection of antiretroviral drug resistance in the treatment of HIV-1 infection has been shown to be a major risk factor for treatment failure.1-4 The presence of specific drug resistance mutations is strongly associated with decreased virologic response to therapy.5,6 Prospective studies have also demonstrated that using resistance testing in designing antiretroviral regimens after treatment failure may enhance virologic response to treatment7-9 and based on this, resistance testing has been incorporated into standards of care for both antiretroviral-experienced, and naïve patients.10,11 Recent guidelines in the US and Europe now recommend resistance testing before initiating ART, and upon the detection of viremia on treatment.
The important clinical question of whether, and if so, how viral resistance impacts risk of clinical disease progression has not been satisfactorily answered. Drug resistance is well documented in association with virologic failure, but the relationship with disease progression and mortality is less clear. It has been postulated that attenuation of viral fitness as drug resistance is selected may have a beneficial effect on the course of disease through a decrease in replication competence, reduced virus load and maintenance of CD4 cell numbers.12-15 We therefore implemented a targeted investigation to evaluate whether individuals with a broader spectrum of drug-resistant mutations had reduced risk of clinical progression. By comparing the drug resistance mutations among matched viremic patients on similar randomized drug regimens, with and without subsequent adverse clinical outcomes, we endeavored to test the hypothesis that drug resistant HIV-1 impacts pathogenicity as indicated by disease progression and mortality.
METHODS
This study used a nested case-control sampling scheme with, on average, two planned controls per “case” matched by parent study, study regimen/treatment history, observational time, sex, age and viral load status. All cases and controls were identified from the ACTG Longitudinal Linked Randomized Trials (ALLRT) database. Selection criteria for this observational study included parent trials that randomly assigned subjects to anti-HIV therapies or strategies for management of anti-HIV interventions. Clinical trials for both treatment naïve and experienced were included, but the treatment-experienced studies did not incorporate baseline resistance testing in selection of antiretroviral regimens. AIDS-defining events were reviewed by the ALLRT protocol chairs. Approval to use data from the following ALLRT parent studies was obtained: 347, 373, 364, 372 (Group A only), 384, 388, 398, 400, A5014, A5025, A5095 and A5143. Figure 1 illustrates the study schema. The titles and brief descriptions of studies with at least one identified case (defined below) are listed in Table 1. The time period from development of a final version of the protocol through completion of all protocol activities is also included.
Figure 1.
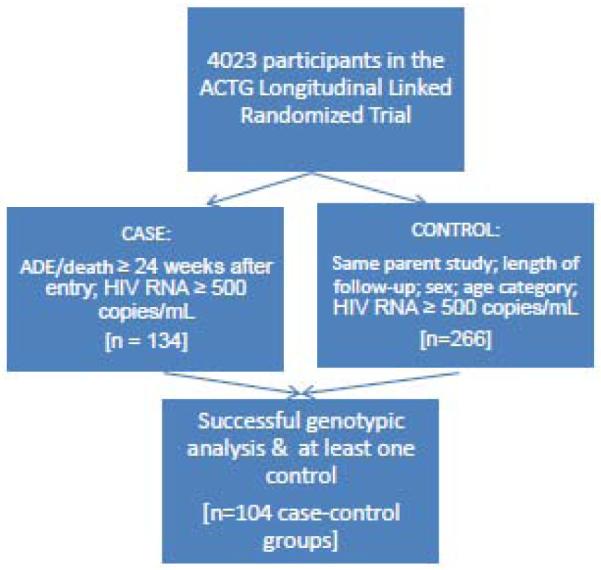
Table 1.
Description of Relevant Parent Studies
| Protocol | Protocol title | Study population |
Study regimen | Total Sampl e size |
# of cases (controls ) used in analysis |
Time Period |
|---|---|---|---|---|---|---|
| 34725 | A Randomized, Double-Blind Phase 2 Study of APV Monotherapy vs APV plus ZDV plus 3TC in HIV-Infected Individuals |
PI and 3TC naïve HIVinfected subjects |
Arm 1: APV + ZDV + 3TC Arm 2: APV + ZDV placebo + 3TC placebo |
92 | 1 (1) | 12/13/1996 - 05/01/2000 |
| 36426 | Comparison of the Virologic Efficacy of Nelfinavir and/or Efavirenz in Combination with Two Nucleoside Analogs in Nucleoside Experienced Subjects: A Rollover Study of ACTG 302/303 |
NRTI therapy experienced, but HAART naïve HIVinfected subjects |
Arm I: NFV + EFV placebo Arm II: NFV placebo + EFV Arm III: NFV + EFV Plus 2 NRTI |
237 | 7 (9) | 05/15/1997 - 12/01/2006 |
| 372A27 | A Phase II Study of the Prolongation of Virologic Success & Options for Virologic Failure in HIVInfected Subjects Receiving IDV in Combination with Nucleoside Analogs: A Rollover Study to ACTG 320 |
HIV-infected subjects who were virologically suppressed on IDV+ZDV (or d4T) + 3TC regimen |
Arm 1: IDV + ZDV + 3TC + ABC Arm 2: IDV + ZDV + 3TC + ABC placebo |
299 | 6 (11) | 07/22/1997 - 06/28/2003 |
| 38428 | Study of Protease Inhibitor and/or Non- Nucleoside Reverse Transcriptase Inhibitor with Dual Nucleosides in Initial Therapy of HIV Infection |
HIV-infected antiretroviralnaive subjects (defined as <7 days of prior antiretroviral therapy) with plasma HIV-1 RNA levels ≥ 500 copies/mL |
Arm A: ddI + d4T + EFV + NFV placebo Arm B: ddI + d4T + EFV placebo + NFV Arm C: 3TC/ZDV + EFV + NFV placebo Arm D: 3TC/ZDV + EFV placebo + NFV Arm E: ddI + d4T + EFV + NFV Arm F: 3TC/ZDV + EFV + NFV |
987 | 18 (32) | 08/24/1998 - 11/03/2002 |
| 38829 | A Phase III Randomized, Controlled Trial of Efavirenz (EFV) or Nelfinavir (NFV) in Combination with Fixed-Dose Combination Lamivudine/Zidovudi ne (3TC/ZDV) and Indinavir (IDV) in HIV-infected Subjects with <200 CD4 cell/mm3 or > 80,000 HIV RNA copies/mL in Plasma |
HIV-infected subjects with limited (no prior 3TC, NNRTI, or protease inhibitor) or no prior antiretroviral treatment and ≤ 200 CD4 cells/mm3 or 80,000 HIV RNA copies/mL of plasma |
Arm 1: 3TC/ZDV + IDV Arm 2: 3TC/ZDV + IDV + EFV Arm 3: 3TC/ZDV + IDV + NFV |
517 | 16 (31) | 02/16/1999 - 10/15/2005 |
| 39830 | A Phase II, Randomized Trial of APV as Part of Dual PI Regimens (Placebo Controlled) in Combination with ABC, EFV & ADV vs. APV Alone in HIV- Infected Subjects with Prior Exposure. To Approved PIs and Loss of Virologic Suppression. as Reflected By A Plasma HIV-1 RNA of ≥ 1000 copies/ml |
Subjects failing IDV, NFV, SQV, or RTV as part of a single- or dual-PI regimen |
Arm 1: APV + SQV + ABC + EFV +ADV Arm 2: APV + IDV + ABC + EFV + ADV Arm 3: APV + NFV + ABC + EFV + ADV Arm 4: APV + PI placebo + ABC + EFV +ADV |
481 | 41 (74) | 07/31/1998 - 05/01/2000 |
| A502531 | A Phase II Randomized Study of the Safety and Efficacy of Hydroxyurea in Patients on Potent Antiretroviral Therapy with Less than 200 copies/mL of HIV RNA in the Plasma |
HIV-infected subjects with suppressed viral load after having received at least six (6) months of IDV, ZDV (or d4T) and 3TC |
Arm A: IDV + ddI + d4T + HU placebo Arm B: IDV + ddI + d4T + HU Am C: IDV + 3TC/ZDV (or d4T + 3TC) |
207 | 1 (1) | 08/31/1998 - 08/01/2004 |
| A509532 | Phase III, Randomized, Double- Blind Comparison of Three Protease Inhibitor-Sparing Regimens for the Initial Treatment of HIV Infection |
Antiretroviral -naive subjects aged 16 and over with HIV-1 RNA ≥ 400 copies/mL |
Arm A: ABC/3TC/ZD V + 3TC/ZDV placebo + EFV Arm B: ABC/3TC/ZD V Placebo + 3TC/ZDV + EFV Arm C: ABC/3TC/ZD V + 3TC/ZDV placebo +EFV placebo |
1147 | 14 (24) | 12/22/2000 - 06/29/2005 |
KEY: 3TC = lamivudine ABC = abacavir ADV = adefovir APV = amprenavir d4T = stavudine ddI = didanosine EFV = efavirenz HAART = highly active antiretroviral therapy HU = hydroxyurea IDV = indinavir NFV = nelfinavir NRTI = nucleoside reverse transcriptase inhibitor NNRTI = nonnucleoside reverse transcriptase inhibitor PI = protease inhibitor RTV = ritonavir SQV = saquinavir ZDV = zidovudine
A case was defined as a subject from ALLRT study who met the following two criteria:
Had an AIDS-defining opportunistic infection (OI) or malignancy event (confirmed by A5001 chairs/clinicians) or non-accidental death on or after 24 weeks after parent study entry. If a subject had multiple events, only the first event was considered. The time period from the parent study entry to the date the first OI/Malignancy/Non-accidental-death is referred to as the “case follow-up period”.
Had at least one HIV RNA level ≥ 500 copies/mL within 24 weeks of the event as defined in (a).
For each case, a control had to satisfy the following conditions:
Be enrolled in the same parent study as the case.
Been followed from study entry for at least the length of the “case follow-up period”, or no more than 12 weeks shorter than the “case follow-up period”.
Have at least one HIV RNA level ≥ 500 copies/mL within 24 weeks of the end of “case follow-up period”.
Be matched by study regimen/treatment history within each study. Study treatments were categorized as “protease inhibitor (PI) containing”; “non-nucleoside reverse transcriptase inhibitor (NNRTI) containing”; “PI and NNRTI containing”; and “nucleoside reverse transcriptase inhibitor (NRTI) only”. Except for study 398, all cases and controls were matched by the category of their study regimens. For study 398, cases and controls were matched by their prior exposure status to NNRTI.
Same sex as the case.
Same age category as the case, where the age categories are defined as ≤ 40 and > 40 years of age.
For each case with only one control available, another case with at least 3 potential controls was randomly selected to be matched to 3 controls. For all cases and controls, specimens on the date of RNA evaluation that was closest to and within 6 months of the event date were used. To increase the chance of having stored specimens available, up to two alternative dates with RNA results within 6 months of the event date were identified for each subject. All specimens were sent to the Stanford ACTG Virology Support Laboratory for genotyping.
Genotyping Methods
Population-based genotyping was performed using standard RT-PCR and dideoxyterminator sequencing procedures. Two hundred to 500 microliters of plasma was centrifuged for 30 min at >20,000 x g to pellet virus particles. The supernatant was removed and discarded, and the pellet extracted with Qiagen Viral RNA Prep Kits (Qiagen, Chatsworth, CA), according to manufacturer’s instructions. The resulting RNA was added to a one-step RT-PCR reaction, where the RNA was reverse-transcribed into DNA, and a 2 kb fragment containing the entire protease gene and the first 300 bases of the RT gene amplified. A portion of this amplicon was amplified again using nested PCR primers, generating a 1.8 kb PCR amplicon. This PCR product was sequenced using at least 6 overlapping sequencing reactions with BigDye dideoxyterminators, which were analyzed on an ABI377 instrument.
The resulting raw sequencing data were assembled and edited using sequence analysis software into a contiguous nucleotide sequence. Quality of the sequence data was controlled by running each sequence through the Stanford CFAR Sequence QC database, which compares each sequence with all other sequences ever generated in the laboratory. Only those sequences whose analysis shows that they are unique or that match other sequences from the same patient qualified for export.
STATISTICAL ANALYSIS
The primary endpoint was genotypic resistance scores as measured by the average Stanford resistance scores [version 4.3.6 (May 2008)] and the Future Drug Options (FDO) Measure.16 The Stanford University HIV Resistance Database [available at http://hivdb.stanford.edu/index.html] is a publicly available integrated genotypic resistance interpretation system, which includes the HIVdb system. The drug resistance estimate for an ARV is obtained by adding together the scores of each for the mutations associated with resistance to that drug. The scores are titrated to fall within the following ranges: (i) 0 to 9: Susceptible, no evidence of reduced susceptibility compared with wildtype; (ii) 10 to 14: Potential low-level resistance. The virus is likely to be fully susceptible yet it contains mutations that may be indicative of previous exposure to the ARV class of the drug; (iii) 15 to 29: Low-level resistance. Virus isolates of this type have reduced in-vitro drug-susceptibility and/or patients with viruses of this genotype may have a suboptimal virologic response to treatment compared with the treatment of a wildtype virus; (iv) 30 to 59: The genotype suggests a degree of drug resistance greater than low-level resistance but lower than high-level resistance; (v) >=60: the genotype is similar to that of isolates with the highest levels of in vitro drug resistance and/or patients infected with isolates having similar genotypes generally have little or no virologic response to treatment with the drug.
The FDO measure quantifies the effect of any given therapeutic strategy on drug resistance. Specifically, the metric quantifies the number of antiretroviral drugs that are likely to be effective, on the basis of genotypic and/or phenotypic resistance measurements. Other measures of resistance, such as the genotype summary score, make use of genotype-interpretation systems to count the number of drugs in a regimen to which a given virus is expected to be sensitive. The FDO extends this concept to consider sensitivity to all possible future drug regimens. Using the Stanford resistance scores and a cutpoint of 15 to evaluate the number of drug classes to which a virus strain is sensitive (if resistance score <15), the FDO measure assigns extra credit to full-class susceptibility to the NRTI or PI classes. The FDO measure can therefore be used to quantify the number of drug classes that remain useful for specific patients. Larger values of FDO correspond to greater drug susceptibility and more drug/regimen options in the future. For example, an FDO score of ≥ 3 means that the genotype is susceptible to at least one drug in all three main classes (NRTI, NNRTI and PI).
Conditional logistic regression models were used to evaluate the effect of each unit increase of the resistance measure on the risk of ADE or death, after adjusting for CD4 cell count and log10 HIV RNA levels. With 115 anticipated evaluable cases, we had estimated >80% statistical power to detect a difference of 0.3 FDO units between cases and controls (assuming a standard deviation for FDO=1 for both cases and controls).
RESULTS
One hundred thirty four cases were identified from 8 ALLRT parent trials and matched to a total of 266 controls. The final sample size included 104 case-control groups with sequence data available for the case and at least one control subject (Table 1). Among them, 34 cases (33%) were matched with only 1 control, 61 cases (59%) were matched with 2 controls, and 9 cases (9%) were matched with 3 controls. 46% were treatment-naïve at entry to their parent study. The median time to event was 99 weeks; six cases were deaths. Of the causes of the OIs reported, 6 were parasitic; 47 were fungal; 6 bacterial/mycobacterial; and 34 viral in origin. Five subjects developed AIDS-related malignancies.
Table 2 presents a summary of the patient characteristics of cases and controls, ignoring the matching. As guaranteed by the matching algorithm, the cases and controls were well balanced by age and sex. The race/ethnicity was also comparable between the cases and controls. However, the cases had higher HIV-1 RNA levels at entry to the parent study (median value 204,913 copies/mL for the cases and 56,885 copies/mL for the controls, p=0.03) and lower CD4 counts at entry to the parent study (median 117 cells/mm3 for cases and 235 cells/mm3 for controls, p<0.0001). Compared to controls, case patients also had higher HIV-1 RNA levels at the end of the follow-up period (median level 45,022 copies/mL for cases and 8,785 copies/mL for controls, p=0.009). The controls had significantly greater viral load reduction from study entry to the end of the follow-up period under their parent study regimens, as compared to the cases. The median viral load reduction was −0.30 log10 for cases and −0.67 log10 for controls (p = 0.003).
Table 2.
Subject Characteristics
| Total (n=287) |
Cases (n=104) |
Controls (n=183) |
P value* | ||
|---|---|---|---|---|---|
| Sex | M | 250 (87%) | 90 (87%) | 160 (87%) | N/A |
| Race/ethnicity | W | 144 (50%) | 54 (52%) | 90 (49%) | 0.56 |
| B | 73 (25%) | 30 (29%) | 43 (23%) | ||
| H | 60 (21%) | 18 (17%) | 42 (23%) | ||
| Age in years (median) (Q1, Q3) |
39 (33, 44) |
39 (33, 44) |
39 (33, 44) |
0.48 | |
| Entry HIV-1 RNA (median) (Q1, Q3) |
83,653 copies/mL (17662, 377637) |
204,913 copies/mL (21294, 454654) |
56,885 copies/mL (14622, 247378) |
0.03 | |
| Entry CD4 (median) (Q1, Q3) |
194 cells/mm3 (67, 332) |
117 cells/mm3 (27, 242) |
235 cells/mm3 (98, 362) |
<0.0001 | |
| Proximal HIV-1 RNA (median) (Q1, Q3) |
17,270 copies/mL (2881, 68409) |
45,022 copies/mL (8514, 308449) |
8,785 copies/mL (1726, 44041) |
0.009 | |
| Proximal CD4 (median) (Q1, Q3) |
291 cells/mm3 (121, 465) |
119 cells/mm3 (46, 300) |
333 cells/mm3 (251, 580) |
<0.0001 |
P-values were derived from the conditional logistic regression model. N/A = Not Applicable as sex was a matching factor.
Table 3 presents the resistance scores for cases and controls to each class of antiretroviral drug and effect on risk of ADE or death in terms of odds ratio. Overall, there were no significant differences in resistance scores between cases and controls. The overall mean FDO was 2.53 for the cases and 2.69 for controls (p=0.3). There were no differences in the proportion of patients with high level resistance (defined as a Stanford resistance score of >59.5) between cases and controls. The mean number of classes with high level resistance was 0.58 for cases, and 0.48 for controls (p=0.5).
Table 3.
Resistance scores for cases and controls
| Variables | Cases (n=104) Percentages or Mean (Standard Deviation) |
Controls (n=183) Percentages or Mean (Standard Deviation) |
Conditional logistic results unadjusted OR (95% CI), p-value |
Conditional logistic results adjusted for baseline CD4 and HIV-1 RNA OR (95% CI), p-value |
|---|---|---|---|---|
| FDO Mean (standard deviation) |
2.53 (1.25) |
2.69 (1.14) |
0.89 (0.70, 1.13), 0.3 |
0.89 (0.68, 1.17), 0.4 |
| NRTI high level resistance (HLR), defined as average Stanford resistance score > 59.5 |
17.3% | 11.5% | 1.49 (0.76, 2.91), 0.2 |
1.19 (0.57, 2.46), 0.6 |
| NNRTI HLR | 23% | 22% | 0.91 (0.48,1.75), 0.8 |
0.92 (0.46, 1.86), 0.8 |
| PI HLR | 17% | 14% | 1.31 (0.63, 2.73), 0.5 |
1.13 (0.49, 2.64), 0.8 |
| Number of classes with HLR Mean (Standard Deviation) |
0.58 (0.91) | 0.48 (0.82) | 1.38 (0.55, 3.46), 0.5 |
1.12 (0.40, 3.12), 0.8 |
The median (IQR) of average NRTI Stanford resistance scores were 12.9 (0 - 49.7) and 16.6 (0 - 40.1) respectively for cases and controls, and were not significantly different from each other (p=0.7).
The median (IQR) number of NRTI resistance mutations was 1 (0-5) and 1 (0-4) for cases and controls, respectively. Univariate conditional logistic regression analyses showed that each additional NRTI mutation was associated with an odds ratio of 1.04 and 95% confidence interval of (0.94, 1.15), p=0.4. For NNRTI, median (IQR) Stanford resistance scores were 5 (0 - 58.8) and 0 (0 - 50), respectively for cases and controls, and again were not significantly different from each other (p=0.14). The median (IQR) number of NNRTI resistance mutations was 1 (0-2) and 0 (0-2) for cases and controls, respectively. Each additional NNRTI mutation was associated with an odds ratio of 1.20 and 95% confidence interval of (0.95, 1.51), p=0.14. For PI, the median (IQR) Stanford resistance scores were 1 (0 – 40.4) and 0 (0 – 32.8), respectively for cases and controls, and also not significantly different from each other (p=0.28). The median (IQR) number of PI resistance mutations was 1 (0-5) and 0 (0-4) for cases and controls, respectively. Each additional PI mutation was associated with an odds ratio of 1.03 and 95% confidence interval of (0.93, 1.15), p=0.5.
A conditional logistic regression model indicated that for any two subjects whose average NRTI scores differed by 10 units, the odds ratio of developing an ADE or experiencing non-accidental death was 1.02 with a 95% confidence interval, for an odds ratio ranging from 0.92 to 1.14 (p=0.66). There was no evidence that subjects who experienced ADE or death carried more (or less) resistant virus at the time of the events compared to control subjects who were matched by parent studies, type of antiretroviral treatments received, age, sex and follow-up period.
Similar conditional logistic regression models were used to compare the average NNRTI scores, average PI scores, and the number of drug class specific mutations. Each 10 unit increase in the average NNRTI score was estimated to be associated with about a 5% increase in the odds of experiencing ADE or non-accidental death (the point estimate of odds ratio for two subjects with the average NNRTI scores differing by 10 was 1.05 and the 95% confidence interval for OR ranged from 0.98 to 1.13). The effect was not statistically significant (p=0.14).
Similarly, the average PI score was not found to be associated with increased risk of ADE or non-accidental death. For two subjects whose average PI scores differed by 10, the odds ratio of developing ADE or non-accidental death was estimated to be 1.06 and 95% confidence interval was 0.95-1.18 (p=0.3).
Univariate conditional logistical regression modeling showed that higher baseline HIV RNA levels and lower baseline CD4 cell counts were associated with increased risk of developing OI/death over the same follow-up period. One log10 increase in entry HIV RNA level was associated with a 76% increase in the odds of developing ADE or death (95% confidence interval for OR was 1.21-2.55, p = 0.003). A 10 cell increase in CD4 cells was associated with a 4% decrease in the odds of developing ADE or death (95% confidence interval for OR was 0.94-0.98, p<0.0001). After adjusting for the baseline HIV RNA level and/or baseline CD4, multiple conditional logistical regression showed that none of the average Stanford or FDO resistance scores was significantly associated with the risk of ADE or death (Table 3).
DISCUSSION
In this retrospective study of virologic failure, drawn from a large antiretroviral clinical trial database, we did not find evidence that the presence of drug resistance mutations or summary measures of drug resistance were associated with serious clinical outcomes. Although genotyping and the interpretation of sequence data to identify drug options is recommended in the management of antiretroviral therapy (ART), viral load and CD4 cell numbers have been shown to contribute to risk of developing an OI.17 In the era of effective ART, diagnostic tools and drug options enable detection of viremia, identification of drug susceptibility and suppression of HIV RNA by providing appropriate drugs. Thus, AIDS-defining opportunistic infections and HIV-associated malignancies have become less frequent. Here we found that among 4023 patients enrolled in the ALLRT longitudinal follow-up study, only 134 cases of ADE were identified despite a larger number of participants with virologic failure who continued treatment on the “failing” regimen. The cases of ADE and death were matched to subjects participating in the same study, receiving the same regimen who, despite viremia, did not progress to an ADE or death. Examining the drug resistance mutations in RT and protease, calculating the class specific drug resistance mutation rates, and summing the future drug options (remaining viral susceptibility) each indicated that there was no evidence that drug resistance increased, or decreased the risk of ADE and clinical outcomes.
Previous attempts to identify the relationship between drug resistance and clinical outcome have been less than satisfactory. Recsky et al. investigated the prevalence of antiretroviral resistance among persons enrolled in the British Columbia HIV/AIDS Treatment Program who died between July 1997 and December 2001.18 The investigators found relatively low prevalence of multidrug resistance in this cohort, suggesting that drug resistance is not a significant contributor to mortality. However, 37% of the “cases” used in the analysis had undetectable viral load and were assumed to carry no resistance mutation. Lucas et al performed an observational cohort study to evaluate the association between antiretroviral resistance and disease progression or death.19 The resistance testing data used were only from patients whose providers ordered the tests, leading to potential bias. However, the authors concluded that patients with drug resistance were not at higher risk of disease progression or death. Similarly, a larger European cohort study demonstrated an association between the detection of resistance at failure and risk of clinical progression.20 As noted in the present study, in all of these analyses patients on treatment with events of interest had significantly lower CD4 cell counts. However, unlike the present study, resistance was not examined at or near the time of an event.
Some studies have explored the association of drug resistance and disease progression for different drug classes and in different populations, for example treatment-naïve or treatment-experienced patients. In treatment-naïve patients from British Columbia, emergence of resistance to non-nucleoside reverse transcriptase inhibitors was associated with a greater risk of death than was emergence of protease inhibitor resistance.5 Similarly, Kozal et al. found that in treatment-naïve participants, the risk of AIDS or death was increased for those who failed virologically with solitary NNRTI resistance and those who failed with no known drug resistance compared to those with no virologic failure.21
Limitations of our study include the retrospective nature of the analyses, the overall low prevalence of resistance-associated mutations, lack of data about possible interruptions of antiretroviral therapy as a cause of plasma viremia, and the fact that participants in clinical trials often undergo more close monitoring of plasma viremia which may lead to earlier detection of treatment failure and decrease risk of clinical events. In addition, resistance testing prior to initiation of antiretroviral therapy was not performed for all study participants, therefore baseline genotype data are not available.
These results need to be considered in the context of continued activity of ART among patients on treatment, despite viremia, compared to those who abandon therapy. A large meta-analysis of survival in EuroSIDA by Miller and colleagues provides evidence that the continued provision of ART provided a significant survival advantage even from sub-optimal (non suppressive) therapy compared to those who had stopped taking treatment.22 This analysis, as well as studies of dual therapy in the 1990’s23 provided strong evidence for the activity of ART despite viremia and resistance, and confirm that ART continues to provide clinical benefit despite viremia and possible drug resistance.
This has led to speculation that certain drug resistance mutations impair viral fitness, and thus the maintenance of specific resistance mutations may be associated with decreased replication capacity and the maintenance of CD4 cell numbers and immunocompetence. Class-specific drug resistance mutations that impair viral fitness and replicative capacity are associated with adherence and drug resistance.24-26 These measures of drug resistance and fitness suggest that drug resistance mutations may contribute to discordant or paradoxical CD4+ T cell responses due, at least in part, to reduced viral fitness. Theoretically, this decreased replication capacity should lead to decreased risk for clinical events, although this has not been examined systematically.
CONCLUSION
In this rigorously designed case-control study, compared to matched controls, patients who developed an AIDS-defining event or who died demonstrated significantly lower baseline CD4 counts and higher virus loads, as well as lower CD4 counts and higher viral loads proximal to a new ADE or death. Viral resistance at or near the time of an event was not associated with increased risk of ADE or death.
ACKNOWLEDGMENTS
Robert Zackin, ScD originally designed the study. Xiao Ding implemented the nested random matching algorithm. We thank the protocol teams for ACTG 347, 373, 364, 372, 384, 388, 398, 400, 5001, 5014, 5025 and 5095; the participating site personnel; and especially the patients.
Financial support: This work was supported by NIH grants AI 68636, AI 68634, AI 38858, AI 38855. This includes a Virology Support Laboratory subcontract to Stanford University. Roche Laboratories provided financial support for the virologic assays.
Footnotes
Potential conflicts of interest: S.S. reports receiving research grants or contracts from or was a consultant for Abbott Pharmaceuticals, Bavarian Nordic, Novartis Pharmaceuticals Corporation, Pfizer, and Tibotec Therapeutics. D.K. is a consultant for Bristol-Myers Squibb and his laboratory research receives research support from Pfizer, GSK and Gilead.
Presented in part: 16th Conference on Retroviruses and Opportunistic Infections, Montreal, Canada, February 8-11, 2009 (abstract M-169).
REFERENCES
- 1.Hammer SM, Eron JJ, Reiss P, et al. Antiretroviral treatment of adult HIV infection – 2008 recommendations of the International AIDS Society-USA Panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG. Determinants of virological response to antiretroviral therapy: implications for long-term strategies. Clin Infect Dis. 2000;30:S177–184. doi: 10.1086/313855. [DOI] [PubMed] [Google Scholar]
- 4.Ledergerber B, Egger M, Opravil M, et al. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Lancet. 1999;353:863–868. doi: 10.1016/s0140-6736(99)01122-8. [DOI] [PubMed] [Google Scholar]
- 5.Hogg RS, Bangsberg DR, Lima VD, et al. Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med. 2006;3:e356, 1570–1578. doi: 10.1371/journal.pmed.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen ML, van der Laan MJ, Napravnik S, Eron JJ, Moore RD, Deeks SG. Long-term consequences of the delay between virologic failure of highly active antiretroviral therapy and regimen modification. AIDS. 2008;22:2097–2106. doi: 10.1097/QAD.0b013e32830f97e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eron JJ, Jr, Park JG, Haubrich R, et al. Predictive value of pharmacokinetics-adjusted phenotypic susceptibility on response to ritonavir-enhanced protease inhibitors (PIs) in human immunodeficiency virus-infected subjects failing prior PI therapy. Antimicrob Agents Chemother. 2009;53:2335–2341. doi: 10.1128/AAC.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JA, Jiang H, Ding X, et al. Genotypic susceptibility scores and HIV type 1 RNA responses in treatment-experienced subjects with HIV type 1 infection. AIDS Res Hum Retroviruses. 2008;24:685–694. doi: 10.1089/aid.2007.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vray M, Meynard JL, Dalban C, et al. Predictors of the virological response to a change in the antiretroviral treatment regimen in HIV-1-infected patients enrolled in a randomized trial comparing genotyping, phenotyping and standards of care (Narval trial, ANRS 088) Antivir Ther. 2003;8:427–434. doi: 10.1177/135965350300800510. [DOI] [PubMed] [Google Scholar]
- 10.Department of Health & Human Services Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents – December 1. 2009 Available at: http://aidsinfo.nih.gov.
- 11.Aberg JA, Kaplan JE, Libman H, et al. Primary care guidelines for the management of persons infected with human immunodeficiency virus: 2009 Update by the HIV Medicine Association of the Infectious Diseases Society of America. Clin Infect Dis. 2009;49:651–681. doi: 10.1086/605292. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Picado J, Martinez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 2008;134:104–123. doi: 10.1016/j.virusres.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Dykes C, Demeter LM. Clinical significance of HIV Type 1 replication fitness. Clin Micro Reviews. 2007;20:550–578. doi: 10.1128/CMR.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin BS, Choi J, Nam JG, et al. Inverse relationship between viral load and genotypic resistance mutations in Korean patients with primary HIV type 1 infections. AIDS Res Hum Retroviruses. 2006;22:1142–1147. doi: 10.1089/aid.2006.22.1142. [DOI] [PubMed] [Google Scholar]
- 15.Castagna A, Danise A, Menzo S, Galli L, Gianotti N, Carini E, Boeri E, Galli A, Cernuschi M, Hasson H, Clementi M, Lazzarin A. Lamivudine monotherapy in HIV-1-infected patients harbouring a lamivudine-resistant virus: a randomized pilot study (E-184V study) AIDS. 2006 Apr 4;20(6):795–803. doi: 10.1097/01.aids.0000218542.08845.b2. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Deeks SG, Kuritzkes DR, Lallemant M, Katzenstein D, Albrecht M, DeGruttola V. Assessing Resistance costs of antiretroviral therapies via measures of future drug options. J Infect Dis. 2003;188:1001–1008. doi: 10.1086/378355. [DOI] [PubMed] [Google Scholar]
- 17.Swindells S, Evans S, Zackin R, et al. Predictive value of HIV-1 viral load on risk for opportunistic infection. J Acquir Immune Defic Syndr. 2002;30:154–158. doi: 10.1097/00042560-200206010-00003. [DOI] [PubMed] [Google Scholar]
- 18.Recsky MA, Brumme ZL, Chan KJ, et al. Antiretroviral resistance among HIV-infected persons who have died in British Columbia, in the era of modern antiretroviral therapy. J Infect Dis. 2004;190:285–292. doi: 10.1086/422007. Epub 2004 Jun 11. [DOI] [PubMed] [Google Scholar]
- 19.Lucas GM, Gallant JE, Moore RD. Relationship between drug resistance and HIV-1 disease progression or death in patients undergoing resistance testing. AIDS. 2004;18:1539–1548. doi: 10.1097/01.aids.0000131339.68666.1a. [DOI] [PubMed] [Google Scholar]
- 20.Cozzi-Lepri A, Phillips AN, Clotet B, Mocroft A, Ruiz L, Kirk O, Lazzarin A, Wiercinska-Drapalo A, Karlsson A, Lundgren JD, EuroSIDA Study Group Detection of HIV drug resistance during antiretroviral treatment and clinical progression in a large European cohort study. AIDS. 2008 Oct 18;22(16):2187–98. doi: 10.1097/QAD.0b013e328310e04f. [DOI] [PubMed] [Google Scholar]
- 21.Kozal MJ, Hullsiek KH, Macarthur RD, et al. The incidence of HIV drug resistance and its impact on progression of HIV disease among antiretroviral naïve participants started on 3 different antiretroviral therapy strategies. HIV Clin Trials. 2007;8:357–370. doi: 10.1310/hct0806-357. [DOI] [PubMed] [Google Scholar]
- 22.Miller V, Mocroft A, Reiss P, et al. Relations among CD4 lymphocyte count nadir, antiretroviral therapy, and HIV-1 disease progression: results from the EuroSIDA study. Ann Intern Med. 1999;130:570–577. doi: 10.7326/0003-4819-130-7-199904060-00005. [DOI] [PubMed] [Google Scholar]
- 23.Katzenstein D, Hughes M, Albrecht M, et al. Virologic and CD4+ cell responses to new nucleoside regimens: switching to stavudine or adding lamivudine after prolonged zidovudine treatment of human immunodeficiency virus infection. AIDS Res Hum Retroviruses. 2001;16:1031–1037. doi: 10.1089/08892220050075282. [DOI] [PubMed] [Google Scholar]
- 24.Bangsberg DR, Acosta EP, Gupta R, et al. Adherence-resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS. 2006;20:223–231. doi: 10.1097/01.aids.0000199825.34241.49. [DOI] [PubMed] [Google Scholar]
- 25.Ho SK, Coman RM, Bunger JC, et al. Drug-associated changes in amino acid residues in Gag p2, p7(NC), and pg(Gag)/p6(Pol) in human immunodeficiency virus type 1 (HIV-1) display a dominant effect on replicative fitness and drug response. Virology. 2008;378:272–281. doi: 10.1016/j.virol.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross L, Parkin N, Lanier R. Short communication: The number of HIV major NRTI mutations correlates directly with other antiretroviral-associated mutations and indirectly with replicative capacity and reduced drug susceptibility. AIDS Res Hum Retroviruses. 2008;24:617–620. doi: 10.1089/aid.2007.0188. [DOI] [PubMed] [Google Scholar]
- 27.Murphy RL, Gulick RM, DeGruttola V, et al. Treatment with amprenavir alone or amprenavir with zidovudine and lamivudine in adults with human immunodeficiency virus infection. AIDS Clinical Trials Group 347 Study Team. J Infect Dis. 1999;179:808–816. doi: 10.1086/314668. [DOI] [PubMed] [Google Scholar]
- 28.Albrecht M, Bosch R, Hammer S, et al. Nelfinavir, efavirenz, or both after the failure of nucleoside treatment of HIV infection. N Engl J Med. 2001;345:398–407. doi: 10.1056/NEJM200108093450602. [DOI] [PubMed] [Google Scholar]
- 29.Hammer SM, Bassett R, Squires KE, et al. A randomized trial of nelfinavir and abacavir in combination with efavirenz and adefovir dipivoxil in HIV-1-infected persons with virological failure receiving indinavir. Antivir Ther. 2003;8:507–518. [PubMed] [Google Scholar]
- 30.Robbins GK, DeGruttola V, Shafer RW, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2293–2303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischl MA, Ribaudo HJ, Collier AC, et al. A randomized trial of 2 different 4-drug antiretroviral regimens versus a 3-drug regimen, in advanced human immunodeficiency virus disease. J Infect Dis. 2003;188:625–634. doi: 10.1086/377311. [DOI] [PubMed] [Google Scholar]
- 32.Hammer SM, Vaida F, Bennett KK, et al. Dual vs single protease inhibitor therapy following antiretroviral treatment failure: a randomized trial. JAMA. 2002;288:169–180. doi: 10.1001/jama.288.2.169. [DOI] [PubMed] [Google Scholar]
- 33.Havlir DV, Gilbert PB, Bennett K, et al. Effects of treatment intensification with hydroxyurea in HIV-infected patients with virologic suppression. AIDS. 2001;15:1379–1388. doi: 10.1097/00002030-200107270-00007. [DOI] [PubMed] [Google Scholar]
- 34.Gulick RM, Ribaudo HJ, Shikuma CM, et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med. 2004;350:1850–1861. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]


