Abstract
After the discovery of the complete repertoire of D. melanogaster Olfactory Receptors (ORs), candidate ORs have been identified from at least 12 insect species from four orders (Coleoptera, Lepidoptera, Diptera, and Hymenoptera), including species of economic or medical importance. Although all ORs share the same G-protein coupled receptor structure with seven transmembrane domains, they share poor sequence identity within and between species, and have been identified mainly through genomic data analyses. To date, D. melanogaster remains the only insect species where ORs have been extensively studied, from expression pattern establishment to functional investigations. These studies have confirmed several observations made in vertebrates: one OR type is selectively expressed in a subtype of olfactory receptor neurons, and one olfactory neuron expresses only one type of OR. The olfactory mechanism, further, appears to be conserved between insects and vertebrates. Understanding the function of insect ORs will greatly contribute to the understanding of insect chemical communication mechanisms, particularly with agricultural pests and disease vectors, and could result in future strategies to reduce their negative effects. In this study, we propose molecular models for insect olfactory receptor co-receptor OR83b and its possible functional oligomeric states. The functional similarity of OR83b to GPCRs and ion channels has been exploited for understanding the structure of OR83b. We could observe that C-terminal region (TM4-7) of OR83b is involved in homodimer amd heterodimer formation (with OR22a) which suggests why C-terminus of insect ORs are highly conserved across different species. We also propose two possible ion channel pathways in OR83b: one formed by TM4-5 region with intracellular pore-forming domain and the other formed by TM5-6 with extracellular pore forming domain using analysis of the electrostatics distribution of the pore forming domain.
Keywords: olfaction, homology modeling, ion channels, heterodimers, distant relationships
Introduction
Chemoreception in insects has long been a major focus of insect ecology. Candidate receptor proteins mediating olfaction were identified from the genome of D. melanogaster in 1999 using bioinformatics approaches.1 The functional organization of olfactory system is remarkably similar in organisms ranging from insects to mammals. Thus, principles elucidated in one experimental organism often apply to many others.2
The evolutionary dynamics of odour receptors show a great deal of fluidity. The olfactory receptor families, in particular, show great diversity. Within a species, many pairs of receptors show little sequence identity and between species of same order, the number of orthologs is few.2
D. melanogaster Olfactory Sensory Neurons (OSNs) express two olfactory receptors (ORs)—a divergent member of the OR family and a highly conserved, broadly expressed olfactory receptor co-receptor OR83b (Dme Orco).3 OR83b is essential in vivo for OR localization and function (signal transduction). OR83b is selectively expressed only in OSNs throughout all four stages of D. melanogaster development. OR/OR83b complex is necessary and sufficient to promote odor evoked signalling.4 Membrane topology of OR83b has been determined by analysing engineered glycosylation sites which are used as topological markers.5 D. melanogaster ORs have a novel membrane topology with their N-terminus and most conserved loops in the cytoplasm, which mediate direct association of OR with OR83b.6 This makes OR/OR83b complex an attractive target for developing insect repellents. OR/OR83b heteromeric complex7 is present in sensory cilia which are concentrated at the site of odor detection. Dendritic localization of conventional ORs is abolished in the OR83b mutants.
Conventional ORs fail to associate directly as homomeric complexes without OR83b reinforcing the specificity of formation of OR83b homomers and OR/OR83b heteromers. Yeast two hybrid assay demonstrates the interaction between intracellular loop 3 (IC3) of OR43a and IC3 of OR83b but not any other combination.7,8 Recent electrophysiological analysis provided strong evidence that insect ORs are in fact ligand-gated non-specific cation channels. The different subunits of OR/OR83b complex are also able to shift ion selectivity of measured amount, a property directly related to ion channels. This suggests that ORs themselves are necessary and sufficient to produce odor-evoked response.9
Another group showed ORs are able to induce opening of a c-AMP dependent CNG channel, suggesting the involvement of G-proteins directly in intracellular signalling.10 Finally, a dual functional nature of ORs as both functional GPCR and non-selective cation channels could rise interesting questions as to how substantially different functions developed within the same protein family.11,12
Despite a weak similarity to known potassium channel pores,13 there is not a clear consensus on where the pore of the channel is located in insect OR83b and to what extent different subunits in OR complex contribute to pore formation. Or83b is thought to function as a non-selective cation channel. Ruthenium red, a cation channel blocker is shown to specifically inhibit the conductance of OR/OR83b complex.9
Structure and function prediction of Inositol trisphosphate receptor (Ip3R) channels using potassium channel as template proved to be successful. Ip3R is a membrane glycoprotein complex acting as Ca2+ channel activated by inositol trisphosphate and is necessary for the control of cellular and physiological processes including cell division, cell proliferation, apoptosis, fertilization, development, behaviour, learning and memory. Inositol triphosphate receptor represents a dominant second messenger leading to the release of Ca2+ from intracellular store sites. The Ip3R complex is formed of four subunits which can form homo—or hetero-oligomers. The study was able to predict calcium ion binding regions and selectivity filter region using the homology model structure of Ip3R channels.14 The fold prediction server PHYRE15 predicts a part of OR83b to have paddle chimera voltage gated potassium channel like structure with 90% precision (albeit at 4% sequence identity) at an E-value of 0.21 (Supplementary Fig. S1). Thus, a similar approach as mentioned above, could be used to identify channel properties of OR83b using potassium channel as template. This will help us to understand how ORs bind chemicals with different structure and how conformational changes within proteins play an important role in olfactory signalling.
The potassium channel16 is an integral membrane protein with four identical subunits that create an inverted tepee (cone) containing the selectivity filter of the pore in its outer end. A bacterial potassium channel structure containing tetrameric unit was obtained in 1998 (PDB ID: 1BL8).16 The structure revealed the presence of two transmembrane helices that are connected by a short ‘pore’ helix and a glycine-rich loop which performs the functional role of a ‘selectivity filter’. Four such similar units combine to form an ion channel.
Methodology
Transmembrane domain prediction of OR83b
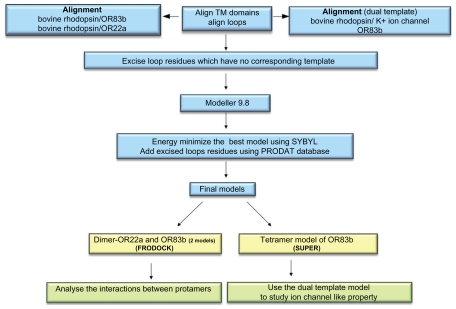
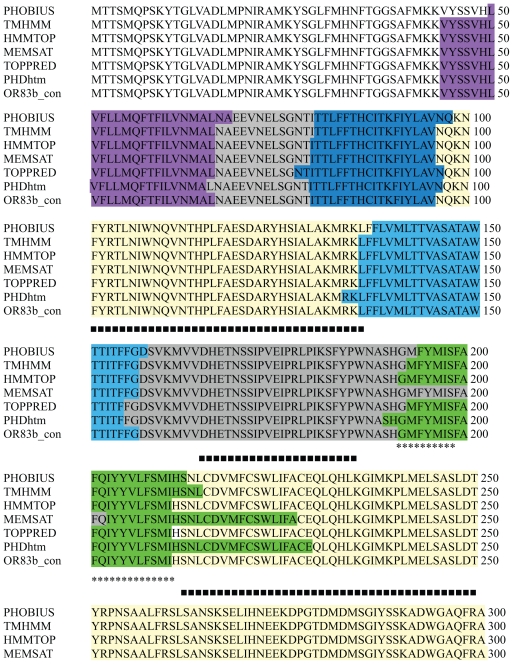
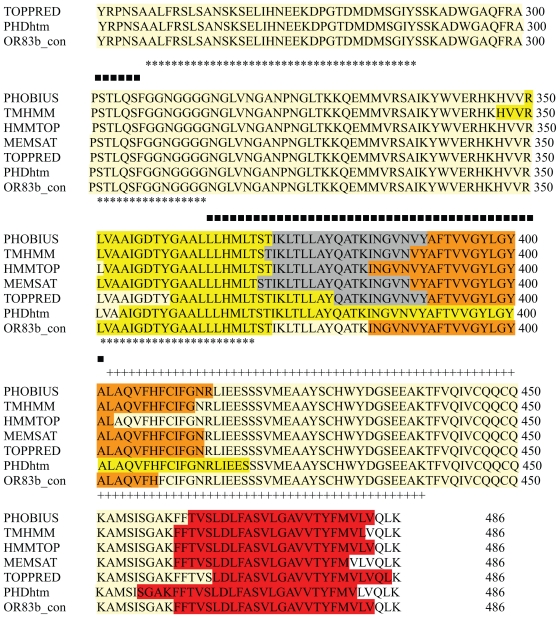
Detailed steps for the modelling of OR83b were as described in Figure 1. The membrane topology of OR83b was determined using different transmembrane prediction servers and consensus was obtained. The average length of helices in the seven GPCR proteins (X-ray crystallographic structures determined till date) is about 29 residues. The length of helices varies from 21–35 residues and the exact boundaries might differ based on the method used to determine them. Consensus Transmembrane (TM) helices boundaries for OR83b were predicted (Fig. 2). Three intracellular and three extracellular loops were labelled thereafter.
Figure 1.
Molecular modelling of D. melanogaster olfactory receptors.
Figure 2.
Transmembrane domain prediction of OR83b sequence using different servers. PHOBIUS,30 TMHMM,31 HMMTOP,32 MEMSAT,33 TOPPRED34 and PHDhtm.35
Notes: Helices marked with VIBGYOR colour. EC loops marked with GREY. IC loops marked with pale yellow. ‘*’ shows the IC2 region modelled using ion channel template. ‘+’ shows the IC3 region modelled using ion channel template. ‘■’
Sequence analysis of OR83b
Sequences of all the GPCRs with known structural information and homologous sequences of OR83b were used to generate a multiple sequence alignment. Sequences of the transmembrane domains of ORs were aligned using ClustalW17 (Gap opening penality 10.00, Gap extension penality 0.20, Blossum 62 matrix, Toggle residue specific penalities ON and Hydrophillic residues GPSNDQEKR) and manually edited using JALVIEW18 to retain high structural equivalence (81%, 149 out of 184 alignment positions had matching transmembrane regions predicted as in the template) in the conserved regions. The loop regions were aligned to available loop regions of template.
Molecular modelling of OR83b
The fold prediction server PHYRE15 was used to determine the fold of OR83b transmembrane domains. The server predicts OR83b to have a fold similar to potassium channels (Supplementary Figure S1) and rhodopsin but not any other GPCR. Although there is strong remote homology with any of the GPCRs, we found best correspondence with rhodopsin in the position and size of transmembrane and loop regions (Supplementary Table S1). The presence of similar overall secondary structural topology (namely seven transmembrane helices and connecting loops) and overall biological function (namely signal transduction in response to ligand binding) are compelling evidences that the two families share similar structures. The model was built using the software MODELLER19 based on the manually edited alignment of OR83b and bovine rhodopsin20 (1F88: PDB ID) sequence (Supplementary Fig. S2A) giving rise to Model 1. OR22a sequence and bovine rhodopsin (1F88: PDB ID) sequence were aligned (Fig. S2D) in a method similar to that mentioned for OR83b (Fig. 1) and model of OR22a was built using MODELLER.
Since OR83b is predicted to have ion channel like property, IC2 loop (connecting TM4 and TM5) and EC3 loop (connecting TM5 and TM6) were aligned independently to the pore forming domain of shows the residues deleted from the sequence due to lack of template. potassium ion channel monomer (1BL8). OR83b is known to cluster with ion channels in phylogenetic analysis21 (MEGA 5,22 Neighbour joining method using JTT matrix). On further analysis of the phylogenetic tree, we found IC2 region of OR83b is aligned to the ion channel sequence. Thus, this study guided us to obtain an alignment for generating Model 2 (using alignment in Fig S2B). Since EC3 loop contains a motif similar to ion channel selectivity filter, we generated an alignment guided by presence of similar motifs to generate Model 3 (using alignment in Fig. S2C). Experimental evidence shows the mutation of the selectivity filter motif in EC3 to ion channel motif increased the conductivity of OR83b to calcium ions.12 Thus the region TM5-6 and EC3 should form the probable ion channel tetramer pore.
Finally, three alignments were generated between query (OR83b) and template (PDB Structure). One using bovine rhodopsin (1F88) as template and the other two using both bovine rhodopsin and potassium ion channel as template corresponding to two different regions of the query. We refer this as dual-template approach to modelling OR83b. Previous analysis has suggested that the architecture of the channels is similar, irrespective of the direction of ion transfer.16 Thus, one would not expect large structural differences in the ion channel with flip in orientation of N-terminal domain.
Energy minimization of generated models
The initial five low-energy models obtained from MODELLER were validated using Ramachandran plot analysis from PROCHECK23 server. The best model (the model with maximum number of residues in allowed and partially allowed regions of Ramachandran plot) was then chosen for energy minimization. The structure was energy minimized using Tripos force field (SYBYL7.1, Tripos Inc) using 200 iterations of Powell’s gradient with a distance dependent dielectric constant equal to 1 and non-bonded interactions cut-off value of 8 and was terminated at a convergence of 0.05 Kcal mol Å−1.
The energy minimized structures were further used to build excised loop regions using PRODAT DB (inbuilt loop DB) from the SYBYL software package (version7.1) (Tripos associate Inc). The final structures were further energy minimized for 500 iterations of Powell’s gradient and 200 iterations of conjugate gradient, while the other parameters remained same as above. The models were energy minimized until the short contacts between neighbouring residues were removed and the final energy of the macromolecule was negative. The energy values did not change much between two succesive steps. The final structures were validated based on Ramachandran plot values.
Generating homodimers and heterodimers of ORs
Homodimers of OR83b (For both GPCR and Dual template model) were generated using protein-protein docking tool, FRODOCK,24 where two copies of the same molecular model was provided as inputs. 100 outputs were generated and the dimer models, with correct membrane orientation and low energy, were chosen for analysis of regions that form the interface. WHATIF server25 was used to find regions that interact (contact distances 1 Å) in the dimers. Heterodimer models were generated for OR83b (bovine rhodopsin and Dual template model) and OR22a using the same method as mentioned above (Fig. 1). The best model was then chosen for energy minimization. The structure was energy minimized using Tripos force field (SYBYL7.1, Tripos Inc) using 500 iterations of Powell’s gradient with a distance dependent dielectric constant equal to 1 and non-bonded interactions cut-off value of 8 and was terminated at a convergence of 0.05 Kcal mol Å−1.
Generating tetramer model of OR83b from the initial model
Using the dual template models of OR83b, a tetramer model of OR83b was generated. SUPER26 (B.S. Neela, personal communications) was used where in the ion channel tetramer template was kept rigid and the model of OR83b was superposed on each protomer of 1BL8 independently to get a final tetramer model of OR83b. The final model was energy minimized using SYBYL software package (version 7.2) (Tripos Associates Inc.). Tripos force field, using 100 iterations of Powell’s gradient with a distance dependent dielectric constant of 1 and a non-bonded interaction cut off value of 8 and was terminated at a convergence of 0.05 kcal/mol. The protomers showed severe short contacts after superposition as tetramer. Therefore, each protomer of the tetramer was moved 2 Å away14 from the pore axis symmetrically (calculated by SCHELAX)27 to obtain a final tetramer structure which is energetically favourable, without short contacts at the inter-protomer interfaces.28 Since no experimental data are available for the pore dimensions of OR83b, the pore size was not altered further.
Electrostatic charge distribution
The Adaptive Poisson-Boltzmann Solver (APBS) in PYMOL visualization tool was employed to calculate the electrostatic charge distribution on the tetramer models. The consideration of a membrane environment has been indirectly included by reducing the dielectric constant to 70.29 The electrostatics was mapped on the model using the molecular surface representation and the charges in intracellular and extracellular pore forming faces for Model 2 and Model 3, respectively, were observed.
Results
Transmembrane domain prediction of OR83b
The membrane topology of OR83b was determined using different transmembrane prediction servers (Fig. 2). The consensus from the results of all methods was chosen to define the boundaries of the transmembrane domain regions. The alignment of the query and the template used to generate models has been given in Supplementary Figure S1. OR83b contains unusually long loops between the TM domains and thus these loops were excised from the alignment and would have to be added later during homology modelling using ab initio or loop modelling methods.
Molecular modelling
The alignments (in Supplementary Fig. S1) were used as an input for MODELLER. The final structure after loop building and energy minimization through SYBYL was validated using PROCHECK. The PROCHECK results for the Olfactory Receptor protein models, excluding the loop regions, shows more than 95% of residues in allowed regions (including strictly allowed and partially allowed) of the Ramachandran plot. The full-length structure for the models, Model 1, shows that more than 90% in the allowed regions (including strictly allowed and partially allowed) of the Ramachandran plot. The residues that were found in the disallowed regions were mainly in the loop regions which are highly variable in length and low in sequence identity.
Analysis of OR83b molecular models
The regions of the protomer at the putative dimer interface, as identified by WHAT IF server, for all the above mentioned models is given in Table 1. The pictorial representation of the same has been shown in Supplementary Figure S3. The interacting regions were analysed only for dimer models with correct orientation in the membrane (N-in and C-out for both protomers).
Table 1.
The regions of the protomers found to be interacting in the dimer models.
| Model name | Regions in the interface for protomer 1 (OR83b) | Regions in the interface for protomer 2 |
|---|---|---|
| OR83b (rhodopsin template)-homodimer (Model 1) | TM1, TM3, TM4, TM5, TM6, TM7, IC3 | (Or83b) TM1, TM4, TM5, TM6, TM7, IC3, IC2 |
| OR83b (dual template)-homodimer (Model 2) | TM4, TM5, IC2 | (Or83b) TM4, TM5, IC2 |
| OR83b (dual template)-homodimer (Model 3) | TM5, TM6, EC3 | (Or83b) TM5, TM6, EC3 |
| OR83b (rhodopsin template)-OR22a heterodimer | TM4, TM5, TM6, IC3, IC2 | (OR22a) TM1, TM7, IC3 C-terminal |
| OR83b (dual-template)-OR22a heterodimer | TM4, TM5, TM6, IC3, IC2 | (OR22a) TM1, TM7, TM6, IC3 |
We could observe that C-terminal region (TM4-7) of OR83b is involved in homodimer formation which explains why the C-terminus of Dme ORs and insect ORs are highly conserved.8 Orthologs of Dme Orco are found in many other insect species and show high homology in sequence and function. Thus, all insect olfactory systems might function by a similar mechanism of oligomerization. OR83b-Or22a complex is also showed to interact through C-terminal ends in both the models (transmembrane region and dual template dimer model), indicating C-terminus plays important role in oligomerization. In the dual template dimer model of OR83b, we found interaction between regions which are thought to form ion channels (TM4-5), indicating OR83b forms an ion channel like pore in the oligomeric state in order to increase the response to odors and acting as ligand gated ion channels. The interactions of residues of TM1 in OR83b and OR22a might be due to its structural proximity to TM7.
Analysis of tetramer model of OR83b
A tetramer of OR83b model was generated using the SUPER program26 (B.S. Neela, personal communications). PROCHECK output of Ramachandran plot showed 95.9% residues in the allowed confirmation (87.8% residues in allowed regions and 8.1% residues in partially allowed regions). Each protomer of the tetramer was moved 2 Å away from pore axis symmetrically to get a final tetramer structure (picture as shown in Supplementary Fig. S4).
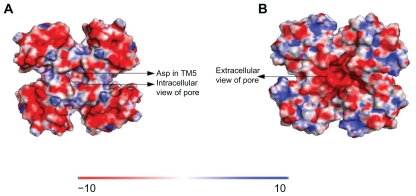
The fact that OR83b functions as a non-selective channel has been proved by several groups. Since the ion-selectivity filter in OR83b was not obvious by mere sequence alignment, we had built two models (Models 2 and 3 involving IC2 and EC3 as the possible selectivity filter region, respectively) and analysed the electrostatic distribution of the protein around these two loops to recognise the likely pore region. The pore entry along the intracellular region (IC2) shows a strong negatively charged patch (Fig. 3A) (including aspartate residue in TM5) which could suggest a pore region for cations to move along the pore. For Model 3, likewise, a negatively charged patch was found in the extracellular end of the pore along the EC3 loop (Fig. 3B) which resembles potassium channel where there is cation influx. The motif TVVGYLG in TM6 lines the ‘pore’ in Model 3. There is a very high negative charge along the pore which would also help in movement of cations across the channel. Our study, therefore, predicts two possible ion channel pathways in OR83b: one formed by TM4-5 region with intracellular pore-forming domain and the other formed by TM5-6 with extracellular pore forming domain.
Figure 3.
Surface representation of intracellular view of Model 2 (Fig. 3A) and extracellular view of Model 3 (Fig. 3B). Electrostatics is represented by calculated charge from red (acidic residues; À10 kbT/ec) to blue (basic residues; +10 kbT/ec) as in Adaptive Poisson–Boltzmann Solver (APBS) program in PYMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.).
Discussion
It is known that most D. melanogaster olfactory neurons express two types of odorant receptors, OR83b, a broadly expressed receptor of unknown function and one or more members of 61 selectively expressed olfactory receptors. OR83b is remarkably conserved between the insect species. Experimental studies show that OR83b has to be expressed for the OSN to be functional. Without OR83b, the receptors do not even reach the dendritic membrane of OSN. Also known is that expression of OR83b alone leads to functional ion channels not directly responding to odorants, but activated by cAMP or cGMP. It is postulated that there might be two systems, like OR83b and functional ORs where one is energy independent and ionotropic and the second is energy dependent and metabotropic. Thus to analyse how structure of OR83b would be to carry out these two functions we went on with molecular modelling of OR83b using different templates and different approaches.
Since OR83b activates a G-protein receptor coupled pathway and is predicted to have seven transmembrane domains, we used bovine rhodopsin as the template to homology model the receptor. Further, since it has an ion channel like property we analysed which regions of OR83b sequence were similar to ion channel (potassium channel). Firm evidence on which loop region of OR83b serves as an ion channel selectivity filter is not available from the literature and which helices participate in the ion channel pore remained dubious. We found some regions of the intracellular loop 2 (IC2) and extracellular loop 3 residues are quite similar to selectivity filter region of ion channels like potassium channel. IC2 is predicted to have 140 residues and therefore it could form an individual domain. Thus, we modelled the region around IC2 and EC3 using an ion channel template and further modelled a tetramer as potassium channel functional as tetramer. Though the two transmembrane regions with intracellular loop were modelled as a tetramer, a full-length model of OR83b as tetramer consisting of 28 helices was difficult to accomplish. Loop regions shown in alignment which had no suitable template and which could not find a loop template from PRODAT DB in SYBYL was not incorporated in the model. It will require regress energy minimization and MD analysis to get a full-length ion channel like model in the absence of available crystal structure template.
Olfactory receptors in D. melanogaster are known to form heterodimers and OR83b is specifically known to form homomer (dimer or tetramer). Yeast two hybrid assays have shown that olfactory receptors do not form homodimers in the absence of OR83b; which means OR83b forms heterodimer with olfactory receptors and then oligomerizes to give larger functional units. The structural basis of this data was analysed by generating oligomers of OR83b and OR83b/OR22a heterodimers. The possibility that there could be C-terminal domain-swap oligomers cannot be ruled out as there is high sequence similarity between C-terminal regions of insect ORs. The regions which could possibly interact were reported after choosing the best dimer model.
The tetramer model of OR83b shows pore lined with negatively charged patch which would allow cations to pass through. Since Or83b is known to be non-selective cation channel without any known ion channel motifs, it will be difficult to predict which residues bind to the cation from the model. The aspartate residue in Model 2 and residues in TVVGYLY motif of TM6 in Model 3 might guide the movement as they contribute to the electronegative charge along the pore. From our homology modelling studies and the analysis of electrostatics charge distribution in Model 2 and Model 3, both possibilities seem to be valid ie, regions around IC2 and EC3 could serve to be in the pore region suggesting that OR83b could be an inward or an outward rectifier. However, there is a higher extent of negatively charged residues at the mouth of EC3 (as in Model 3). Further analysis using known ligands for D. melanogaster olfactory receptors and mutation data would help us to understand which model is functional and also to uncover whether olfactory receptor OR83b/OR22a complex is functional or a higher order oligomer is required for receptor activation.
Supplementary Data
The fold prediction server PHYRE aligns TM4 and TM5 of OR83b sequence to pore forming transmembrane helices of paddle chimera voltage gated potassium channel.
Alignments between query and template used as an input for MODELLER. (A) The alignment of OR83b and bovine rhodopsin sequence giving rise to Model 1. (B) The alignment of OR83b (Model 2), bovine rhodopsin and potassium channel sequence giving rise to Model 2. Possible selectivity filter region is shown between green and yellow colour in the alignment. (C) Alignment of OR83b (Model 3), bovine rhodopsin and potassium channel giving rise to Model 3. Possible selectivity filter region is shown between yellow and orange colour. (D) The alignment of OR22a and bovine rhodopsin.
Dimer models to show regions predicted at the interface. (A) Homodimer model built using dual template model of OR83b. The figure shows interactions in the region of transmembrane helices 4 and 5. (B) Heterodimer model of OR83b and OR22a. The 7 TM helices are marked in VIBGYOR colour. The TM helices that are predicted to be at the interface are labelled as TM (int). Figures generated using PYMOL.
Tetramer model of OR83b (Model 2) superposed on K+ channel (PDB ID:1BL8). Figures generated using PYMOL.
Table S1.
The number of residues in TM helices and loop regions of OR83b (query) and two possible templates (bovine rhodopsin and beta-2 adrenergic receptors).
| Name | TM 1 | TM 2 | TM 3 | TM 4 | TM 5 | TM 6 | TM 7 |
|---|---|---|---|---|---|---|---|
| 1f88 (Bovine rhodopsin) | 30 | 30 | 33 | 23 | 26 | 31 | 21 |
| 2rh1 (Beta-2 AR) | 32 | 30 | 34 | 25 | 33 | 32 | 24 |
| OR83b | 23 | 18 | 23 | 20 | 20 | 19 | 24 |
| Loop 1 | Loop 2 | Loop 3 | Loop 4 | Loop 5 | Loop 6 | ||
| 1f88 (Bovine rhodopsin) | 12 | 17 | 20 | 30 | 30 | 8 | |
| 2rh1 (Beta-2 AR) | 6 | 8 | 10 + 160 | 38 | 27 | 7 | |
| OR83b | 12 | 38 | 34 | 141 | 12 | 57 |
Notes: We do not find much variation in the length of TM helices between two templates, while bovine rhodopsin has loop lengths closer to the query compared to beta-2 adrenergic receptors. One of the loops in beta-2 adrenergic receptors (marked in red) is T4 bacteriophage insert. Similarly, loop 4 and loop 6 have large inserts in OR83b.
Acknowledgments
We thank NCBS for infrastructural facilities.
Footnotes
Author Contributions
Conceived and designed the experiments: RS. Analysed the data: KH. Wrote the first draft of the manuscript: KH. Contributed to the writing of the manuscript: RS. Agree with manuscript results and conclusions: RS, KH. Jointly developed the structure and arguments for the paper: RS, KH. Made critical revisions and approved final version: RS. All authors reviewed and approved of the final manuscript.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
K.H. and the work are supported by Indo-Japan grant funded by Department of Science and Technology (DBT), India and AIST (Japan).
References
- 1.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron. 1999;22(2):327–8. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 2.Su Chih-Ying, Menuz Karen, Carlson John R. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139(1):45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vosshall Leslie B, Hansson Bill S. A unified nomenclature system for the insect olfactory coreceptor. Chem Senses. 2011;36(6):497–8. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- 4.Larsson Mattias C, Domingos Ana I, Jones Walton D, Chiappe M Eugenia, Amrein Hubert, Vosshall Leslie B. Or83b encodes a broadly expressed odorant receptor essential for drosophila olfaction. Neuron. 2004;43:703–14. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Lundina Carolina, Kallb Lukas, Kreherc Scott A, Kappd Katja, Sonnhammerb Erik L. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Letters. 2007;581:5601–4. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benton R. On the origin of smell: Odorant receptors in insects. Cell Mol Life Sci. 2006;63(14):1579–85. doi: 10.1007/s00018-006-6130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuhaus Eva M, Gisselmann Gunter, Zhang Weiyi, Dooley Ruth, Stortkuhl Klemens, Hatt Hanns. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nature Neuroscience. 2004;8(1):15–7. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- 8.Miller Raymond, Tu Zhijian. Odorant receptor c-terminal motifs in divergent insect species. Journal of Insect Science. 2008;8:53. [Google Scholar]
- 9.Nichols Andrew S, Chen Sisi, Luetje Charles W. Subunit contributions to insect olfactory receptor function: Channel block and odorant recognition. Chem Senses. 2011;36(9):781–90. doi: 10.1093/chemse/bjr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richard Benton. Molecular basis of odor detection in insects. Annals of the New York Academy of Sciences. 2009;1170:478–81. doi: 10.1111/j.1749-6632.2009.03880.x. [DOI] [PubMed] [Google Scholar]
- 11.Benton Richard, Sachse Silke, Michnick Stephen W, Vosshall Leslie B. Atypical membrane topology and heteromeric function of drosophila odorant receptors in vivo. PLoS Biology. 2006;4(2) doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wicher Dieter, Schafer Ronny, Bauernfeind Ren, et al. Hanssona, dOr83b— Receptor or ion channel? Annals of the New York Academy of Sciences. 2009;1170:164–7. doi: 10.1111/j.1749-6632.2009.04101.x. [DOI] [PubMed] [Google Scholar]
- 13.Metpally RR, Sowdhamini R. Cross genome phylogenetic analysis of human and Drosophila G protein-coupled receptors: Application to functional annotation of orphan receptors. BMC Genomics. 2005;6:106. doi: 10.1186/1471-2164-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah PK, Sowdhamini R. Structural understanding of the transmembrane domains of inositol triphosphate receptors and ryanodine receptors towards calcium channeling. Prot Engng. 2001;14:867–74. doi: 10.1093/protein/14.11.867. [DOI] [PubMed] [Google Scholar]
- 15.Kelley LA, Sternberg MJE. Protein structure prediction on the web: A case study using the Phyre server. Nature Protocols. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 16.Doyle DA, Morais Cabral J, Pfuetzner RA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2: A multiple sequence alignment and analysis workbench. Bioinformatics. 2009;25(9):1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: A G-protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 21.Prasad Raghu, Metpally Rao. PhD Thesis. [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laskowski RA, Mac Arthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereo chemical quality of protein structures. J Appl Crystallogr. 1993;26:283–91. [Google Scholar]
- 24.Garzon Jose Ignacio, Lopez-Blanco Jose Ramon, Pons Carles, et al. FRODOCK: A new approach for fast rotational protein-protein docking. Bioinformatics. 2009;25(19):2544–51. doi: 10.1093/bioinformatics/btp447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vriend G. WHAT IF: A molecular modelling and drug design program. J Mol Graph. 1990;8:52–6. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 26.Kearsley SK. On the orthogonal transformation used for structural comparisons. Acta Cryst. 1989;A45:208. [Google Scholar]
- 27.Chou KC, Nemethy G, Scheraga HA. Energetic approach to the packing of α-helices. 2. General treatment of non-equivalent and nonregular helices. J Am Chem Soc. 1984;106:3161–70. [Google Scholar]
- 28.Srinivasan N, White HE, Emsley J, Wood SP, Pepys MB, Blundell TL. Comparative analyses of pentraxins: Implications for protomer assembly and ligand binding. Structure. 1984;2:1017–27. doi: 10.1016/S0969-2126(94)00105-7. [DOI] [PubMed] [Google Scholar]
- 29.Kanagarajadurai Karuppiah, Malini Manoharan, Bhattacharya Aditi, Panicker Mitradas M, Sowdhamini Ramanathan. Molecular modeling and docking studies of human 5-hydroxytryptamine A (5-HT2 A) receptor for the identification of hotspots for ligand binding. Molecular Bio Systems. 2009;5:1877–88. doi: 10.1039/b906391a. [DOI] [PubMed] [Google Scholar]
- 30.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 32.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–50. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 33.Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–49. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 34.von Heijne G. Membrane protein structure prediction hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–94. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 35.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–18. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The fold prediction server PHYRE aligns TM4 and TM5 of OR83b sequence to pore forming transmembrane helices of paddle chimera voltage gated potassium channel.
Alignments between query and template used as an input for MODELLER. (A) The alignment of OR83b and bovine rhodopsin sequence giving rise to Model 1. (B) The alignment of OR83b (Model 2), bovine rhodopsin and potassium channel sequence giving rise to Model 2. Possible selectivity filter region is shown between green and yellow colour in the alignment. (C) Alignment of OR83b (Model 3), bovine rhodopsin and potassium channel giving rise to Model 3. Possible selectivity filter region is shown between yellow and orange colour. (D) The alignment of OR22a and bovine rhodopsin.
Dimer models to show regions predicted at the interface. (A) Homodimer model built using dual template model of OR83b. The figure shows interactions in the region of transmembrane helices 4 and 5. (B) Heterodimer model of OR83b and OR22a. The 7 TM helices are marked in VIBGYOR colour. The TM helices that are predicted to be at the interface are labelled as TM (int). Figures generated using PYMOL.
Tetramer model of OR83b (Model 2) superposed on K+ channel (PDB ID:1BL8). Figures generated using PYMOL.
Table S1.
The number of residues in TM helices and loop regions of OR83b (query) and two possible templates (bovine rhodopsin and beta-2 adrenergic receptors).
| Name | TM 1 | TM 2 | TM 3 | TM 4 | TM 5 | TM 6 | TM 7 |
|---|---|---|---|---|---|---|---|
| 1f88 (Bovine rhodopsin) | 30 | 30 | 33 | 23 | 26 | 31 | 21 |
| 2rh1 (Beta-2 AR) | 32 | 30 | 34 | 25 | 33 | 32 | 24 |
| OR83b | 23 | 18 | 23 | 20 | 20 | 19 | 24 |
| Loop 1 | Loop 2 | Loop 3 | Loop 4 | Loop 5 | Loop 6 | ||
| 1f88 (Bovine rhodopsin) | 12 | 17 | 20 | 30 | 30 | 8 | |
| 2rh1 (Beta-2 AR) | 6 | 8 | 10 + 160 | 38 | 27 | 7 | |
| OR83b | 12 | 38 | 34 | 141 | 12 | 57 |
Notes: We do not find much variation in the length of TM helices between two templates, while bovine rhodopsin has loop lengths closer to the query compared to beta-2 adrenergic receptors. One of the loops in beta-2 adrenergic receptors (marked in red) is T4 bacteriophage insert. Similarly, loop 4 and loop 6 have large inserts in OR83b.