Preface
Conformational changes involving coding and non-coding RNAs form the basis for genetic regulatory elements and provide an important source of complexity for driving many fundamental processes of life. While RNA is highly flexible, the underlying dynamics are robust and limited to transitions between the few conformations that preserve favorable base-pairing and stacking interactions. The mechanisms by which cellular processes harness RNA’s intrinsic dynamic behavior and steer it towards functionally productive pathways are complex. Versatile functions and ease of integration into a wide variety of genetic circuits and biochemical pathways suggests a general and fundamental role for RNA dynamics in cellular processes.
Introduction
Peering into the first protein X-ray structure of myoglobin1 begged the question, how do ligands reach the deeply buried heme iron center? This simple but powerful observation inspired decades of investigation into the dynamic behavior of proteins and today, it is well established that protein structures are in constant motion, and that these fluctuations are critical to, and sometimes drive function. Early X-ray structures of RNA also foresaw the importance of conformational dynamics; large changes in tRNA’s helical arms were observed on binding tRNA synthetase2, and changes in the conformation of ribozymes needed to be invoked to envision catalytically active states3–5. However, no one could have anticipated the existence of new genetic circuits that are based on RNA conformational switches, or that the acrobatic nature of a biopolymer consisting of only four chemically similar nucleotides would be at the heart of the inner-workings of a complex macromolecular machine such as the ribosome.
Dynamic changes in RNA structure serve a dazzling range of ever-increasing functions using a common two-step process; a cellular signal triggers RNA dynamics, and this in turn is transduced into a specific biological output. This review provides a critical account of RNA dynamics as a regulatory mechanism and source of functional complexity. We first review what is known about the dynamic properties of RNA structure and emphasize unique properties that enable large changes in structure to take place in a biologically specific and robust manner. We then examine the wide range of cellular inputs used to interface with RNA dynamics and the various mechanisms that are used to steer the dynamics to achieve a broad spectrum of functional outputs.
RNA free energy landscape
It is important to distinguish between two types of dynamics; ‘equilibrium fluctuations’ and ‘conformational transitions’. Equilibrium fluctuations correspond to thermally activated motions that occur in all RNAs. Conformational transitions arise when cellular cues create a non-equilibrium state that relaxes back to equilibrium. While this review is principally focused on conformational transitions given their dominant role in regulatory mechanisms, the two motions are intricately related, as highlighted by numerous studies of RNA and also protein dynamics6,7. This and other aspects of RNA dynamic behavior that are relevant to function are best understood by examining RNA’s free energy landscape8,9.
The free energy landscape specifies the free energy of every possible RNA conformation (Fig 1A). Here, equilibrium fluctuations correspond to spontaneous jumps between various conformers along the free energy landscape. The population of a given conformer depends on its free energy whereas the transition rate between conformers depends on the free energy barrier of separation (Fig 1A). Conformational transitions arise when cellular cues perturb the energy landscape, leading to a conformational redistribution (Fig 1A). Studies show that the RNA free energy landscape is punctuated by deep local minima corresponding to narrow sets of very different conformations, and that these conformations are the ones that are significantly sampled by equilibrium motions and that are stabilized by cellular cues to effect conformational transitions10–12 (Fig 1A).
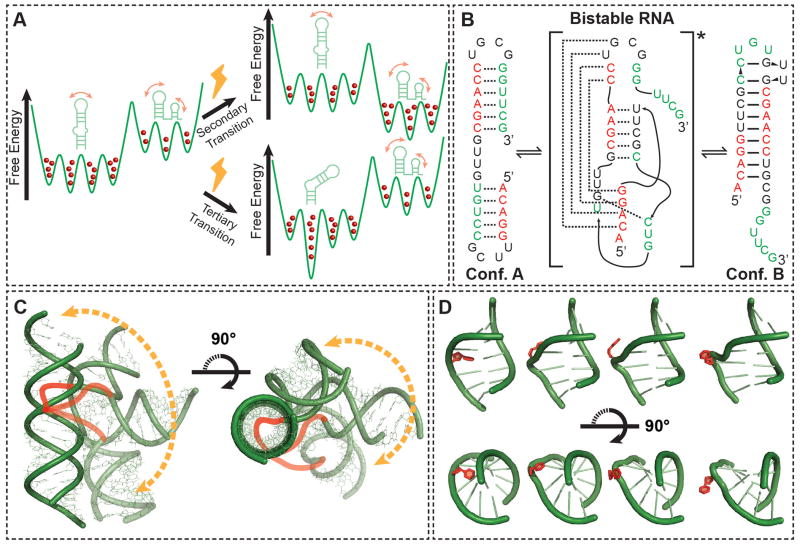
Figure 1.
Shape and form of RNA dynamics. (A) A RNA free energy landscape (green). Shown are secondary and tertiary RNA conformations of low-lying energy. The relative population of each conformation is indicated using red balls. Cellular effectors (bolts) can modify the energy landscape to favor an alternative secondary structure (top), or preferentially stabilize an alternate tertiary conformation (bottom). (B) Exchange between alternative, isoenergetic secondary structures that are separated by large energetic barriers due to disruption of base-pairs in the transition state13. Note that RNA helices tend to be shorter than ~15 base pairs. (C) The accessible range of inter-helical conformations for an RNA two-way junction consisting of a trinucleotide bulge, with the possible paths of the bulge, which were excluded during the modeling, illustrated as cartoons (red)14,15. The allowed range of conformations is restricted towards a specific and directed conformational pathway by steric and stereochemical forces. (D) Flipping out of a non-canonical base pair with an RNA internal loop (red) from an intra-helical stacked to extra-helical unstacked conformation. The motion occurs without perturbing flanking Watson-Crick pairs (green).
For example, the degeneracy of base-pairing and stacking interactions, together with the high stability of RNA duplexes, gives rise to deep local minima corresponding to different yet isoenergetic secondary structures that are separated by large kinetic barriers13 (Fig 1B). As few as two secondary structures may dominate the RNA dynamic landscape because the loss of energy accompanying disruption of even one base-pair markedly destabilizes alternative conformations. A given secondary structure can in turn undergo more facile dynamic excursions in tertiary structure involving smaller energetic barriers. These dynamics are commonly dominated by large changes in the relative orientation of helical domains, which carry motifs involved in tertiary contacts, and occur about flexible pivot points consisting of bulges, internal loops, and higher-order junctions (Fig 1C). Although these excursions can lead to very large changes in tertiary structure, they are limited to a narrow set of conformations. For example, calculations of the set of conformations accessible to two helices connected by a three-residue bulge reveals that the inter-helical bend angle, when combined with inter-helical twisting, can range between 0 and 180°. Yet, despite this large range, the connectivity constraints imposed by the bulge junction, as well as steric forces, direct changes between inter-helical orientations along a highly directional pathway and restricts the conformational space to <20% of that which is theoretically possible. 10,14–16 (Fig 1C). Likewise, due to the high stability of duplexes, non-canonical residues can loop out from intra- to extra-helical conformations without significantly perturbing the structure of flanking helices17,18 (Fig 1D). Precise control over the dynamics is encoded within the sequence, and small sequence variations can greatly alter the relative populations of different RNA secondary structures and their rates of inter-conversion11,19. For example, distinct inter-helical orientations can be sampled by changing the length and asymmetry of junctions10,14,15, and the tendency of residues to loop out can be modulated based on sequence-specific stacking interactions20,21.
The above features help explain three remarkable aspects of RNA conformational transitions that are of fundamental importance for regulatory functions. First, the landscape is hierarchical; due to the height of energy barriers separating alternative secondary structures, changes in tertiary contacts rarely entail changes in secondary structure, and the two types of conformational changes can be used to serve different functions. Throughout this review, we will use “secondary” and “tertiary” conformational changes to distinguish between these two types of dynamics. Second, the limited landscape of energetically favorable conformations allows RNA to undergo very large changes in structure yet be directed towards a very specific set of conformations from an astronomical number of possibilities. Finally, there is growing evidence that RNA dynamics are strongly determined by the underlying RNA free energy landscape, and to lesser extent by cellular cues7,22,23. Thus, conformational transitions can be considered perturbations that steer preexisting equilibrium fluctuations towards specific functionally productive pathways. In this manner, even an imperfect force or cellular signal will drive changes in RNA structure along a predetermined pathway, making the transitions highly robust.
Triggers of RNA conformational transitions
RNA dynamics can be triggered by a remarkably diverse set of molecular effectors and environmental cues through a number of different mechanisms. This provides many different points of entry for integrating RNA conformational transitions into biological circuits and biochemical pathways.
Specific protein binders
The most common effectors are proteins that bind their target RNA specifically through well-defined structural features, thereby stabilizing one or a subset of conformations from the preexisting energy landscape. For example, the mitochondrial tyrosyl-tRNA synthetase CYT-18 from Neurospora crassa binds specifically to group I introns, a class of large self-splicing ribozymes that catalyze their own excision from mRNA, tRNA and rRNA precursors, and stabilizes the conformation required for catalytic activity24. Protein binding often leads to large changes in the overall orientation of RNA helices about junctions such as bulges25, three-way junctions26, and other motifs such as the K-turn27. For example, the spliceosomal U4 snRNA undergoes a sharp transition in inter-helical bend angle, from ~69° to ~25° about a K-turn motif on binding its cognate protein target28 (Fig 2A). These changes in inter-helical conformation are driven in part by non-specific electrostatic interactions between basic amino acids and high negative charge density that builds up at inter-helical junctions and are often observed as equilibrium dynamics in the absence of effector29–31. For example, unbound HIV-1 TAR RNA dynamically samples the many different inter-helical orientations that are observed upon binding to seven distinct ligands, including peptide mimics of its cognate protein Tat31 (Fig 2B).
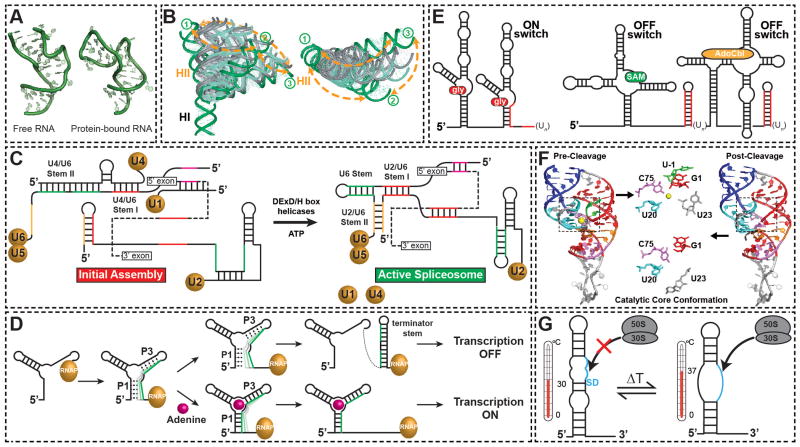
Figure 2.
Triggering RNA conformational transitions. (A) Conformational changes in the spliceosomal U4 snRNA K-turn motif (2KR8) triggered upon binding to the protein hPrp31-15.5K (2OZB)28. (B) Similarity between the TAR RNA inter-helical conformational conformations that are triggered by binding to small molecules (in grey) and that are sampled by equilibrium dynamics (in green) in the unbound state. Adapted from31. (C) RNA conformational transitions during spliceosome assembly on pre-mRNA (dashed line). (D) Modulating RNA structure by steering the co-transcriptional folding pathway, with the adenine transcription terminating riboswitch as a prototypical example. Shown is the progression of co-transcriptional folding with and without the ligand. The RNA polymerase is shown in gold. (E) Examples of tandem riboswitch architectures. Cooperative binding of glycine by the gly riboswtich using tandem aptamer domains and one expression platform (left panel). Tandem SAM and AdoCbl riboswitches in which either of two ligands yields an output of gene repression (right panel). Transcription terminator stems are shown in red. (F) Conformations of HDV ribozyme pre-cleavage with Mg2+ (1VC7) and post-cleavage (1DRZ). The catalytic core is highlighted in the two states, with the substrate and Mg2+ ion shown in green and yellow, respectively. (G) Melting of secondary structure around the ribosome binding site of virulence genes in the pathogen triggered by an increase in temperature makes the Shine-Dalgarno sequence available for ribosome binding and translation initiation.
In a growing number of cases protein binding does not entail stabilization of a specific conformational well of the RNA energy landscape, but rather, binding selectively lowers surrounding energy barriers to accentuate or alter the equilibrium dynamics of the RNA. For example, binding of the U1A protein to its cognate RNA target does not lead to the arrest of pre-existing equilibrium inter-helical motions, but rather induces mobility in regions of the RNA that are in direct contact with the protein32. The CBP2 protein from yeast mitochondria binds specifically to the bI5 group I intron and activates large scale RNA equilibrium motions33. Even simple small molecule ligands lead to reorganization of the TAR RNA equilibrium dynamics34. These observations underscore the importance of embracing a broader view of trigger factors as elements that perturb the entire energy landscape and thereby steer RNA dynamics rather than simply stabilize a single conformation from a dynamic range.
RNA chaperones and helicases
As is often the case in RNAs that possess alternative secondary structures, the large energy barriers associated with base pair melting can limit dynamics between RNA conformational wells. In this scenario and RNA become kinetically trapped in a metastable conformation, unable to reach thermodynamic equilibrium. In response, cells have evolved a variety of proteins that possess the RNA chaperone activity needed to efficiently drive RNA secondary structural transitions over large energetic barriers35,36. One strategy, taken by the HIV nucleocapsid (NC) protein, involves using nonspecific interactions between the RNA and protein to destabilize RNA helices37. This has the effect of lowering the energetic barrier to conformational exchange, accelerating relaxation to equilibrium and converting metastable RNAs into their thermodynamically more favorable conformations.
Other chaperones, such as RNA helicases, use energy to traverse the high barriers, unwinding helices and disrupting RNA structure in addition to promoting formation of RNA duplexes in a nonprocessive way, thus accelerating conformational transitions in RNAs and ribonucleoprotein (RNP) complexes38. These proteins play important roles in remodeling of RNA/RNP structures by virtue of having the capacity to anneal and unwind RNA strands and choose one process over the other in a manner dependent on environmental cues39. For example, helicases play a critical role in the assembly of the spliceosome, which is a complex ribonucleoprotein (RNP) consisting of 5 RNAs and dozens of proteins that catalyzes excision of introns from a nuclear pre-mRNA40,41. Assembly proceeds via a series of transitions that entail the melting and annealing of RNA duplexes that are catalyzed by DExD/H box ATPase helicases (Fig 2C). For example, the U4 RNA escorts the U4/U6/U5 triple small nuclear ribonucleoprotein complex (tri-snRNP) to the pre-mRNA, but is subsequently released by DEXD-box helicase Brr2 catalyzed melting of two stems within U4 and U6. This frees up the U6 stem to base-pair with U2 snRNA and leads to a new RNA structure that is required for the first transesterification reaction42 (Fig 2C). DExD/H box proteins also play a role in the release of product mRNA in pre-mRNA splicing reactions. For example, DEAH box splicing factor Prp22 gets deposited on mRNA downstream of the exon-exon junction and catalyzes the disruption of contacts with U5 snRNP, thereby liberating the spliced mRNA from the U5/U6/U2 spliceosomal assembly43. In another example showing disparity in the roles played by RNA chaperones, DExD/H box protein CYT-19 carries out ATP-dependent unfolding of native and misfolded conformations of a group I catalytic RNA. A large free energy gap between the native and the misfolded conformers directs CYT-19 to act more frequently on misfolded conformers and also to redistribute the populations of the two, allowing native RNA to populate a wider range of conformations than would otherwise be possible44.
Metabolites and physiochemical conditions
Another ingenious strategy modulates RNA secondary structure in response to a strikingly wide range of metabolite-based effectors, including small molecules such as amino-acids, coenzymes, and nucleotides23,45 and changes in physiochemical conditions, such as Mg2+ concentration46 and pH47. Such effectors and cellular cues would be difficult, if not impossible to endow with the chaperone activity needed to efficiently drive secondary structural transitions. Instead, the strategy acts on the initial RNA folding process itself, taking advantage of the unidirectional and comparatively slow rate (relative to RNA folding and effector binding) with which RNA is transcribed from the 5′ to the 3′ direction to alternate between two folding pathways that favor either of two distinct secondary structures (Fig 2D). This trigger mechanism is implicated in a growing list of other RNA switches, though it has been best described for metabolite sensing riboswitches23,45.
Riboswitches are RNA-based genetic elements typically embedded in the 5′ untranslated region (5′ UTR) of bacterial genes that regulate expression of metabolic genes in response to changes in cellular metabolite concentration23,45. In a prototypical metabolite riboswitch, a given metabolite, such as adenine, binds the aptamer domain with exceptional affinity and selectivity. This stabilizes an otherwise shallow energy well, inducing a redistribution of aptamer conformational states towards one that, in most riboswitches, sequesters an RNA element into a helix of the aptamer domain48 (Fig 2D). The unavailability of this element in turn changes the folding pathway of a downstream decision-making expression platform, directing it towards structures that turn off (and in some cases, on) gene expression, either by forming a transcription terminating helix (Fig 2D) or sequestering the Shine-Dalgarno sequence where the ribosome binds, thereby inhibiting translation. This system also minimizes spontaneous conformational transitions, or premature switching in the absence of ligands, because very large barriers separate the alternative secondary structural forms of the expression platform.
More complex functions can also be achieved by tandem architectures. For example, the glycine riboswitch uses two aptamer domains in tandem to cooperatively bind glycine, thereby providing greater responsiveness to changing ligand concentrations49 (Fig 2E). Tandem arrangement of two entire riboswitches that respond to two distinct ligands allows for the construction of more sophisticated genetic circuits such as two-input Boolean NOR logic gates, in which either of two ligands can trigger the conformational switch and yield an output of gene repression50 (Fig 2E). In another example, the c-di-GMP-sensing riboswitch and a GTP-dependent self-splicing group I ribozyme in the 5′UTR of Clostridium difficile work in tandem to regulate translation51. In the presence of c-di-GMP and GTP, a structure that stabilizes a 5′-splice site is formed and the ribozyme self-splices to yield an RNA transcript with a perfect ribosome binding site (RBS) located upstream of the start codon. Conversely, in the presence of GTP alone, the UTR forms a structure that promotes splicing at an alternative site, resulting in a splicing product that lacks a RBS, and thus downregulates translation. This RNA arrangement represents the first natural example of an allosteric ribozyme.
Chemical reactions
Chemical reactions, such as cleavage of the RNA phosphodiester backbone, can also reshape the underlying RNA energy landscape. Thus, a previously equilibrium state becomes a non-equilibrium one, triggering changes in RNA secondary and tertiary structure. For example, X-ray structures of precursor and product states of the hepatitis delta virus (HDV) ribozyme, which catalyzes site-specific self-cleavage of the viral RNA phosphodiester backbone, reveal changes in the local arrangement of catalytic groups along with the ejection of a catalytically important metal ion52. These conformational changes may help accelerate product release53,54 (Fig 2F). Cleavage can also trigger changes in RNA secondary structure. This is observed in the RNA secondary structural switch that is triggered by a cleavage of the 3′ end of the pre-18S rRNA during eukaryotic ribosome maturation and which is used to enforce a sequential order to the maturation process55.
Thermal and mechanical triggers
Other energy dependent processes can induce the complete melting of RNA hairpins. RNA thermosensors alter expression of genes during heat-shock response and pathogenic invasion in response to increases in temperature56 (Fig 2G). For example, during invasion of Listeria monocytogenes into an animal host, the pathogen encounters a warmer environment thereby activating a thermosensor located at the 5′ UTR of the prfa mRNA57. The higher host temperature shifts the energy landscape from one that favors the formation of the thermosensor hairpin to one where the melted, single strand conformation dominates. This melting transition exposes ribosome-binding sites (RBSs) required for ribosome binding and translation. Other triggers are mechanical, such as the translation induced melting of mRNA hairpins, which is thought to slow down the rate of ribosome elongation to allow for the proper folding of autonomously folding proteins and protein domains58.
Functions of secondary structural changes
Secondary structural transitions are widely used as binary switches that can be activated by cellular cues. The switch can be transduced into a wide range of outputs by simply sequestering or exposing key RNA regulatory elements.
Transcription
Many RNA switches regulate gene expression at the transcriptional level by producing transcription-terminating helices. In addition to metabolite sensing riboswitches, other RNA switches use this strategy to regulate gene expression in response to more complex molecules23,45. For example, non-aminoacylated tRNAs can activate transcription of their cognate aminoacyl-tRNA synthetase gene once they reach a specific concentration through specific interactions between the anti-codon and acceptor stem within the T-box region in the 5′ UTR of the mRNA (Fig. 3A). This interaction disrupts formation of a terminator hairpin during co-transcriptional folding that would otherwise abort transcription. However, the interaction with the acceptor stem is blocked upon aminoacylation, resulting in formation of the terminator stem that aborts transcription59 (Fig. 3A). Few proteins have been identified that modulate transcription by influencing folding of transcription-terminating helices. A rare example is the tryptophan-activated RNA-binding attenuation protein (TRAP), which binds trp mRNA to regulate gene expression at both the transcriptional and translational level by, as an example, promoting the formation of a terminator hairpin that terminates transcription60.
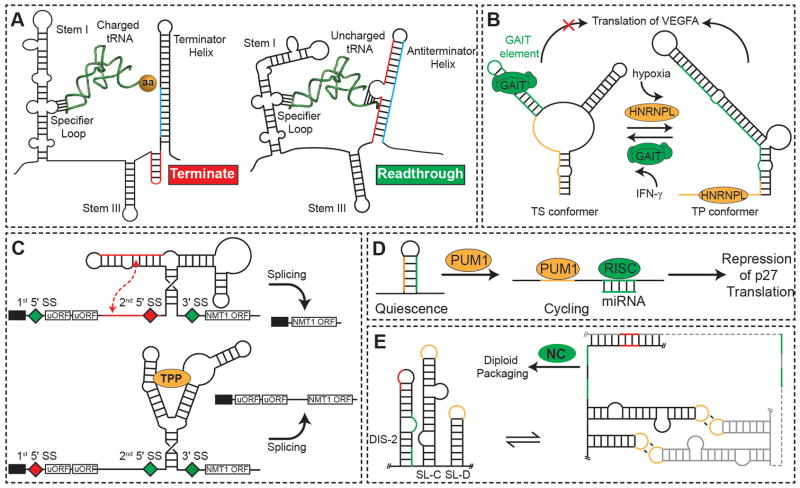
Figure 3.
Functional outputs of secondary structural changes. (A) Transcriptional activation of the aminoacyl-tRNA synthetase gene by uncharged tRNA by steering co-transcriptional folding away from a transcription terminating helix99. (B) Translation control of VEGFA expression via a dual protein-dependent RNA secondary structural switch that responds to IFN-γ (left panel) and hypoxic stress (right panel). (C) TPP-riboswitch-regulated alternative splicing and gene expression of NMT1. On binding of TPP, the aptamer domain undergoes a conformational change, which exposes a proximal splice site (diamond). Spliced mRNAs now contain uORFs, thus reducing expression of the NMT1 ORF. (D) Pumilio protein-mediated mRNA secondary structural switch controls accessibility of microRNA binding sites and regulates expression of p27 protein. Binding of PUM1 induces a conformational change to expose the miR-211/miR-222 binding site to allow for p27 silencing. (E) Secondary structural switch couples dimerization and diploid genome packaging of the Moloney murine leukemia virus. Dimerization leads to a coupled frame shift that exposes NC protein binding sites (green) required for genome packaging.
Translation
There is a growing list of protein and RNA triggered61 RNA switches that regulate translation by sequestering or exposing ribosomal binding sites or by affecting the structure of ribosomal RNA, thereby blocking translation. For example, a protein-dependent RNA switch has recently been identified in the 3′ UTR of human vascular endothelial growth factor-A (VEGFA) mRNA in myeloid cells that regulates translation of VEGFA in response to proteins associated with two disparate stress stimuli (Fig. 3B). The interferon (IFN)-gamma-activated GAIT complex binds a structural GAIT element within a family of inflammatory mRNAs to silence their translation by promoting formation of a translational-silencing (TS) conformer62. During oxidative stress, the heterogeneous nuclear ribonucleoprotein L (HNRNPL) overrides GAIT silencing by triggering a secondary structural RNA switch to a translation-permissive (TP) conformer, in which the GAIT element is occluded. In this way, the RNA alternates between two mutually exclusive conformers in response to binding of the GAIT complex or HNRNPL, thereby functioning as an ‘AND NOT’ Boolean logic gate switch in which the presence of one protein but not the other yields an output of gene repression (Fig. 3B).
Post-transcriptional processing
A growing number of RNA switches are also implicated in regulating post-transcriptional processing, including splicing, gene silencing by microRNA (miRNA), and RNA editing. Though detailed mechanistic insights are still lacking for many of these systems, in all cases the RNA switch serves to expose, occlude, or modulate the structure of processing sites, thus regulating the degree of processing and post-transcriptional regulation. For example, one of the thiamine pyrophosphate (TPP) riboswitches discovered in eukaryotes regulates alternative splicing63 (Fig. 3C). Here, changes in the secondary structure serve to sequester or expose splice sites (Fig. 3C).
An RNA switch has recently been identified in the 3′ UTR of p27 mRNA that simultaneously sequesters miRNA target sites from cleavage by the RNA-induced silencing complex (RISC) and a Pumilio-recognition element (PRE), which binds the Pumilio RNA-binding protein (PUM1)64. Binding of PUM1 to the PRE region induces a secondary structural switch that exposes the miRNA target site leading to miRNA silencing (Fig. 3D). There is also evidence that a pre-existing equilibrium between two RNA secondary structures involving a kinetically trapped conformation and thermodynamically more favorable state determines type 3 RNA editing levels in HDV65. These initial discoveries suggest a wide role for RNA switches in post-transcriptional processing.
Viral replication
RNA secondary structural switches are widely used by the RNA genomes of retroviruses to transition between different roles required by various steps of the viral lifecycle. For example, there is evidence that the HIV-1 5′ UTR genome can adopt two mutually exclusive secondary structures; a meta-stable branched multiple hairpins (BMH) conformation which plays roles in dimerization and packaging, and an energetically more favorable long-distance interaction (LDI) conformation which plays roles in transcription and translation. The transition from the LDI to BMH conformation is catalyzed by the RNA chaperone NC66.
RNA switches are also used to couple distinct processes within a given step. For example, an RNA switch is used to couple dimerization and selective encapsidation of two copies of the Moloney murine leukemia (MML) virus RNA genome. Here, dimerization of the RNA genome induces a register shift in base pairing within the ψ-RNA packaging signal, which serves to expose conserved UCUG elements that bind NC with high affinity, thereby promoting genome packaging67 (Fig. 3E). These elements are base-paired and bind NC weakly in the monomeric RNA (Fig 3E).
Functions of tertiary conformational changes
RNA tertiary conformational changes can range from large global changes in the orientation of helices to more subtle local changes in the structure of motifs involved in tertiary interactions. These conformational transitions allow RNA molecules to adaptively bind a wide range of molecular partners and can help direct the assembly of RNPs.
Polyvalent binding
Early structures of RNA-protein complexes revealed a remarkable ability of RNA to undergo adaptive changes in conformation2,25 that could potentially allow optimization of intermolecular interactions with disparate targets. Indeed, such conformational changes allow tRNA to interact with many diverse partners, including RNase P, various nucleotide modifying enzymes, tRNA synthetase, EF-Tu, the ribosome, and other RNA elements. High-resolution structures of tRNA, tRNA-protein, and tRNA-RNP complexes reveal that binding is often accompanied by significant conformational changes, which range from reorientation of helical domains to finer changes in local structure, all of which serve to optimize intermolecular interactions68 (Fig 4A).
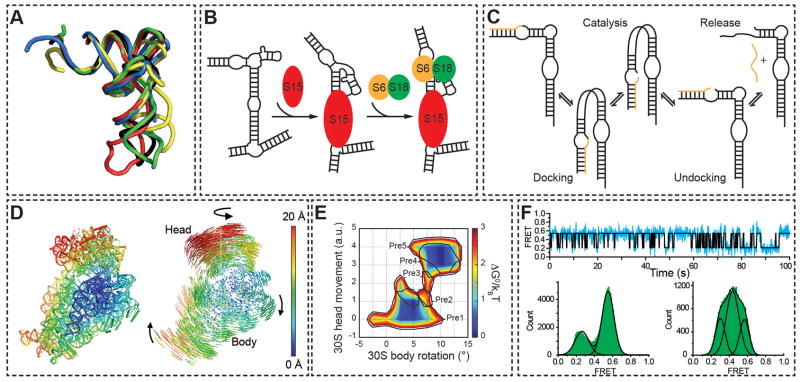
Figure 4.
Functional outputs of tertiary conformational changes. (A) X-ray structures of tRNAPhe in the unbound state (black, 1EHZ), in complex with RNaseP (blue, engineered anticodon stem removed, 3Q1Q), the ribosome in the P/E state (green, 3R8N), isopentenyl-tRNA transferase (red, 3FOZ), and phenyalanyl-tRNA synthetase (yellow, 1EIY). The structures are superimposed by the acceptor stem. (B) Hierarchical assembly of the central domain of the 30S ribosomal subunit by successive protein-induced changes in the conformation of 16S rRNA. (C) Enzymatic cycle of the hairpin ribozyme. (D) Ratcheting motions of the ribosome as observed by X-ray crystallography. The degree of 30S subunit atomic displacement between the unratcheted and R2 ratcheted states with the 50S subunit as a reference (not shown) are color-coded by Å (left). Atomic displacement vectors and arrows indicate the directionality of the change (right). From reference82. Reprinted with permission from AAAS. (E) Free energy landscape of ribosomal ratcheting, as calculated from sub-classification of cryo-EM particles. Movements of the 30S subunit body and head domains in relation to the 50S subunit are shown in units of degrees and arbitrary units, respectively, with corresponding tRNA translocation intermediates outlined in black. Reprinted by permission from Macmillan Publishers Ltd: Nature84, copyright (2009). (F) Dynamics of the 50S ribosomal L1 stalk monitored by single molecule FRET. Representative smFRET trace (top) and histogram (bottom left) of the L1 stalk dynamically sampling open and closed conformations in A and P-site tRNA-bound ribosome complexes. Upon translocation by EF-G • GTP and tRNA occupation of the E and P-sites the L1 stalk conformation shifts dramatically (bottom right). Adapted with permission from the National Academy of Sciences, USA100.
Ordering RNP assembly
RNA tertiary conformational changes induced by successive protein binding are thought to help direct the order of assembly of complex RNP machines, including the 30S ribosome69,70, the signal recognition particle (SRP)71, and telomerase72. For example, the binding of ribosomal protein S15 to 16S rRNA initiates the ordered assembly of the central domain in the 30S ribosomal subunit73 and leads to a change in the orientation of helical domains that favors binding of ribosomal proteins S6 and S1874 (Fig. 4B). Premature binding of S6 and S18 to the unbound 16S rRNA may be disfavored in part by an entropic penalty associated with the partial freezing out of inter-helical motions. Even in telomerase, which consists of one RNA and two protein components, binding of the first protein, p65, induces a conformational change in the RNA that facilitates binding of telomerase reverse transcriptase, thus ordering the assembly72.
Assembly can also involve coupled protein binding that leads to coupled changes in secondary and tertiary structure. For example, coupled binding of the maturase and Mrs1 protein cofactors to the RNA of the bI3 group I intron RNP stabilizes both native tertiary contacts and promotes a reorganization of a non-native intermediate secondary structure75. While both Mrs1 dimers and maturase can independently bind and stabilize portions of the bI3 tertiary structure, binding by both proteins is required to induce a partial secondary structure rearrangement and assembly to the native, active state.
Ribozyme catalysis
Tertiary conformational transitions are frequently observed in small ribozymes such as the hairpin and HDV that are thought to be important for transitioning between the various steps of catalytic cycles. These transitions also involve large changes in the orientation of helical arms. Typically, an undocked conformation binds substrate, which in turn promotes docking into a conformation required for catalysis. Following catalysis, another undocking transition allows product release (Fig. 4C). The importance of these motions is underscored by the strong impact they have on the overall catalytic rate constant76. Large hinge-like motions of the J2a/b bulge in the human telomerase have also been proposed to facilitate dynamic telomere repeat synthesis77. As an extreme example, the Tetrahymena group I ribozyme has recently been shown to inter-convert between alternative tertiary conformations that have a range of substrate binding affinities but similar enzymatic activities78. The rates of inter-conversion between these states is slower than the rate of catalysis, implying the existence of multiple native states. Such long-lived heterogeneities have been observed in the tertiary folds of many other RNAs, though some of these may be the result of RNA purification side-products79. The structural differences between these various conformations and the source of the severe heterogeneity remains unknown at the atomic level but may constitute yet another mechanism for allowing a narrow set of differentiated RNA conformations to be sampled and should serve as an exciting topic for future research.
Protein Synthesis
Perhaps the ultimate example of the cell manipulating the intrinsic dynamic landscape of RNA to achieve a desired biological outcome is ribosome catalysis. Translation requires large scale ratcheting motions where the small and large subunits reorient with respect to one another through numerous intermediates that are driven by conformational change in both the ribosomal RNAs and proteins80–84 (Fig. 4D). Recent data strongly indicate that all of these intermediates are relatively low-lying energy states that readily interconvert, as highlighted by the ability of the ribosome to spontaneously undergo full tRNA retrotranslocation84,85 (Fig. 4E). This has led to the model of the ribosome known as the “Brownian machine”, whose function derives from thermally driven equilibrium fluctuations that are innately biased to promote the translation process22 (Fig. 4F).
The cell couples these intrinsic ribosome dynamics with numerous effectors to achieve tight control over the complex transactions required by translation. One such transaction is the selection and proof-reading of incoming tRNAs that underlies the ribosome’s remarkable fidelity in discriminating between cognate versus near- or non-cognate tRNAs, where small differences between the mini-helices of incorrect and correct anticodon-codon pairs lead to tRNA accommodation or rejection. Here, formation of a cognate mini-helix leads to a kinked tRNA structure that triggers a 30S ‘domain closure’ motion86–88. This stabilizes tRNA-ribosome interactions and in turn promotes conformational rearrangements in the EF-Tu protein that delivers the tRNA to the ribosome, resulting in EF-Tu • GTP hydrolysis, release of tRNA from EF-Tu, and initial tRNA selection89,90. The second proofreading step that follows EF-Tu dissociation is thought to be driven by relaxation of the kinked tRNA. In cognate tRNAs the strong interactions between the codon and anticodon bias this relaxation towards a fully accommodated state within the A-site, whereas for near-cognate tRNAs with weak codon-anticodon interactions the relaxation can also occur through other pathways, leading to rejection91,92. Following tRNA accommodation, other effectors including EF-G93, other initiation factors94, recycling factors95, release factors96, and even the identity and deacylation/acylation state of the P-site tRNA come into play97, manipulating the ribosome’s dynamic landscape so as to efficiently drive the desired translation outcome.
Although the relative roles of the RNA and protein components in driving ribosome dynamics is unclear, there is little doubt that the RNA components help confer dynamic specificity and robustness to ribosome dynamics98. This is another exciting area of future inquiry.
Looking ahead
The conventional view that one sequence codes for one structure and one function is giving way to a dynamic view of RNA as a pre-existing superposition of conformational states that can be resolved into a directed and synchronized motion by dedicated cellular machinery, leading to a broad range of functional outcomes. This makes it all the more important to study RNA dynamics within the complex in vivo environment of living cells, an important goal for the future. At the same time, there is a need to deepen our basic understanding of RNA dynamic behavior, even within the simpler in vitro environment. It is remarkable that even for an iconic molecule such as tRNA that has been studied for more than half a century, very little experimental data is available regarding its equilibrium fluctuations at the atomic level; the same is true for catalytically important motions in ribozymes. Similarly, little is known about the structure and dynamics of large RNAs, such as eukaryotic mRNAs. This will require hand-in-hand developments in computational and experimental tools. Slowly, we will inch closer towards atomic movies of RNA in dynamic action within living cells and a predictive understanding of RNA dynamic behavior. In the meantime, great advances will come by simply embracing this new dynamic view of RNA, and always being on the lookout for another myoglobin.
Supplementary Material
Acknowledgments
We thank Catherine Eichhorn and Dr. Qi Zhang for input and assistance with preparation of figures and Dr. Sam Butcher and Dr. Sasha Serganov for critically reading the manuscript and providing comments. A.M.M. acknowledges support by a NSF graduate research fellowship. The authors gratefully acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for the support of the purchase of 600 MHz spectrometer. This work was supported by the US National Institutes of Health (R01 AI066975 and R01 GM089846) and the US National Science Foundation (NSF Career Award CHE-0918817).
Footnotes
Author contributions
All authors contributed to the writing and synthesis of all aspects of the review. E.A.D. performed a broad literature survey of RNA dynamics, primarily wrote sections related to RNA switches, and contributed to preparation of Figures 2 and 3. J.C. performed literature review on mechanism of RNA dynamics, primarily wrote sections related to tertiary dynamics, and prepared Figures 1–3. A.M.M. performed the literature review on translation, primarily wrote the section on ribosome dynamics and prepared Figure 4. H.M.A. oversaw the writing of the review with assistance from the co-authors.
Links to movies and animations of experimentally determined RNA dynamics are included in supplemental information. Also included are additional references to reviews of RNA and RNA dynamics.
References
- 1.Kendrew JC, et al. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 2.Rould MA, Perona JJ, Soll D, Steitz TA. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 3.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 4.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 6.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hashimi HM, Walter NG. RNA Dynamics: It is about time. Curr Opin Struct Biol. 2008;18:321–9. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 9.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Bailor MH, Mustoe AM, Brooks CL, 3rd, Al-Hashimi HM. Topological constraints: using RNA secondary structure to model 3D conformation, folding pathways, and dynamic adaptation. Curr Opin Struct Biol. 2011;21:296–305. doi: 10.1016/j.sbi.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat Struct Mol Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- 12.Schultes EA, Hraber PT, LaBean TH. Estimating the contributions of selection and self-organization in RNA secondary structure. J Mol Evol. 1999;49:76–83. doi: 10.1007/pl00006536. [DOI] [PubMed] [Google Scholar]
- 13.Fürtig B, Wenter P, Pitsch S, Schwalbe H. Probing mechanism and transition state of RNA refolding. ACS Chem Biol. 2010;5:753–765. doi: 10.1021/cb100025a. [DOI] [PubMed] [Google Scholar]
- 14.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. Simple topological constraints induced by steric and stereochemical forces severely restrict the allowed orientation of helices across two-way junctions. [DOI] [PubMed] [Google Scholar]
- 15.Mustoe AM, Bailor MH, Teixeira RM, Brooks CLI, Al-Hashimi HM. New Insights Into the Fundamental Role of Topological Constraints as a Determinant of Two-Way Junction Conformation. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu VB, et al. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venditti V, Clos L, 2nd, Niccolai N, Butcher SE. Minimum-energy path for a U6 RNA conformational change involving protonation, base-pair rearrangement and base flipping. J Mol Biol. 2009;391:894–905. doi: 10.1016/j.jmb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fourmy D, Yoshizawa S, Puglisi JD. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J Mol Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- 19.Le SY, Zhang K, Maizel JV., Jr RNA molecules with structure dependent functions are uniquely folded. Nucleic Acids Res. 2002;30:3574–3582. doi: 10.1093/nar/gkf473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew Chem Int Ed Engl. 2010;49:5731–3. doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shankar N, et al. NMR reveals the absence of hydrogen bonding in adjacent UU and AG mismatches in an isolated internal loop from ribosomal RNA. Biochemistry. 2007;46:12665–12678. doi: 10.1021/bi700802s. [DOI] [PubMed] [Google Scholar]
- 22.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res. 2011 doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 24.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 25.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 26.Orr JW, Hagerman PJ, Williamson JR. Protein and Mg2+-induced conformational changes in the S15 binding site of 16 S ribosomal RNA. J Mol Biol. 1998;275:453–464. doi: 10.1006/jmbi.1997.1489. [DOI] [PubMed] [Google Scholar]
- 27.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DM. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falb M, Amata I, Gabel F, Simon B, Carlomagno T. Structure of the K-turn U4 RNA: a combined NMR and SANS study. Nucleic Acids Res. 38:6274–6285. doi: 10.1093/nar/gkq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HD, et al. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zacharias M, Hagerman PJ. The influence of symmetric internal loops on the flexibility of RNA. J Mol Biol. 1996;257:276–289. doi: 10.1006/jmbi.1996.0162. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 32.Shajani Z, Drobny G, Varani G. Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry. 2007;46:5875–5883. doi: 10.1021/bi602658x. [DOI] [PubMed] [Google Scholar]
- 33.Bokinsky G, et al. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardaro MF, Jr, Shajani Z, Patora-Komisarska K, Robinson JA, Varani G. How binding of small molecule and peptide ligands to HIV-1 TAR alters the RNA motional landscape. Nucleic Acids Res. 2009;37:1529–1540. doi: 10.1093/nar/gkn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyle AM, Green JB. RNA folding. Curr Opin Struc Biol. 1995;5:303–310. doi: 10.1016/0959-440x(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 37.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struc Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 38.Hirling H, Scheffner M, Restle T, Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 40.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosowski TR, Keys HR, Quan TK, Ruby SW. DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle. RNA. 2009;15:1345. doi: 10.1261/rna.1065209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struc Mol Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. RNA switch discovered in the 5' UTR of bacterial mRNA regulates gene expression in response to ligands without assistance from proteins. [DOI] [PubMed] [Google Scholar]
- 46.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 47.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 50.Sudarsan N, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 51.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferre-D'Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 53.Ke A, Zhou K, Ding F, Cate JHD, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. A significant local conformational change in the active site of the HDV ribozyme is observed post-cleavage which is associated with ejection of the substrate and a catalytically critical divalent metal ion. [DOI] [PubMed] [Google Scholar]
- 54.Harris DA, Rueda D, Walter NG. Local conformational changes in the catalytic core of the transacting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 55.Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J Mol Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nocker A, et al. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 2001;29:4800–4807. doi: 10.1093/nar/29.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansson J, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 58.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diaz-Toledano R, Ariza-Mateos A, Birk A, Martinez-Garcia B, Gomez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5' region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498–5510. doi: 10.1093/nar/gkp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ray PS, et al. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. The 3' UTR of human VEGFA mRNA undergoes a binary conformational switch in response to inflammatory and hypoxic protein stress signals to regulate VEGFA expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. A secondary structural change in a eukaryotic TPP riboswitch regulates gene expression through control of alternative splicing. [DOI] [PubMed] [Google Scholar]
- 64.Kedde M, et al. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 65.Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 66.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 67.Miyazaki Y, et al. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J Mol Biol. 2010;396:141–152. doi: 10.1016/j.jmb.2009.11.033. Dimerization of the 5' UTR of the Moloney murine leukemia virus results in a secondary structural change that promotes genome packaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giege R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- 69.Mulder AM, et al. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Menichelli E, Isel C, Oubridge C, Nagai K. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J Mol Biol. 2007;367:187–203. doi: 10.1016/j.jmb.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 72.Stone MD, et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 74.Agalarov SC, Prasad GS, Funke PM, Stout CD, Williamson JR. Structure of the S15,S18-rRNA complex: Assembly of the 30S ribosome central domain. Science. 2000;288:107–112. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 75.Duncan CD, Weeks KM. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry. 2010;49:5418–5425. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson TJ, Nahas M, Ha T, Lilley DM. Folding and catalysis of the hairpin ribozyme. Biochem Soc Trans. 2005;33:461–465. doi: 10.1042/BST0330461. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. Observation of slowly inter-converting catalytically active states in a ribozyme establishes the coexistence of multiple native states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenfeld M, Solomatin SV, Herschlag D. Removal of covalent heterogeneity reveals simple folding behavior for P4-P6 RNA. J Biol Chem. 2011;286:19872–19879. doi: 10.1074/jbc.M111.235465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 81.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 82.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. Cryo-EM observation of thermally driven tRNA retro-translocation on the ribosome. [DOI] [PubMed] [Google Scholar]
- 85.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 87.Valle M, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 88.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struc Mol Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 92.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struc Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. This important single molecule FRET study directly observes the dynamics of tRNA initial selection and proofreading by the ribosome. [DOI] [PubMed] [Google Scholar]
- 93.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 97.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tama F, Valle M, Frank J, Brooks CL., 3rd Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci U S A. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.