Abstract
Genetic transformation of monocotyledonous plants still presents a challenge for plant biologists and biotechnologists because monocots are difficult to transform with Agrobacterium tumefaciens, whereas other transgenesis methods, such as gold particle-mediated transformation, result in poor transgene expression because of integration of truncated DNA molecules. We developed a method of transgene delivery into monocots. This method relies on the use of an in vitro-prepared nano-complex consisting of transferred DNA, virulence protein D2, and recombination protein A delivered to triticale microspores with the help of a Tat2 cell-penetrating peptide. We showed that this approach allowed for single transgene copy integration events and prevented degradation of delivered DNA, thus leading to the integration of intact copies of the transgene into the genome of triticale plants. This resulted in transgene expression in all transgenic plants regenerated from microspores transfected with the full transferred DNA/protein complex. This approach can easily substitute the bombardment technique currently used for monocots and will be highly valuable for plant biology and biotechnology.
Transgenesis, or genetic transformation, finds many applications in plant biology, e.g. in cell biology and gene function studies. Importantly, it also allows for the generation of plants with improved agricultural traits significantly faster than any conventional breeding practice. The technology is based on the delivery of genes of interest from a broad range of sources into a plant genome. Two major transformation techniques include Agrobacterium-mediated DNA delivery and biolistic DNA transfer (Birch, 1997; Chawla, 2002; Slater et al., 2008). Agrobacterium-mediated transformation relies on the ability of Agrobacterium tumefaciens to transfer a portion of its DNA, called transferred DNA (T-DNA), into plant cells (Tzfira and Citovsky, 2006). During its transfer from the bacterial cell to the plant nucleus, the single-stranded T-DNA is protected by a single-stranded DNA binding protein, virulence protein E2 (VirE2; Rossi et al., 1996), and guided by virulence protein D2 (VirD2). VirD2 is also important for integrating T-DNA into the plant genome (Gelvin, 2000; Citovsky et al., 2007). The integration process apparently requires broken DNA, or at least an area of active replication or transcription (Ziemienowicz et al., 2008). Agrobacterium-mediated transformation is an efficient process and typically, in dicotyledonous plants (dicots), predominantly results in integration of the transgenes at a single locus; integrated T-DNA is mostly intact and allows for normal expression of the transgene (Windels et al., 2008).
Although A. tumefaciens is not a natural pathogen of most monocotyledonous plants (Cleene, 1985; Binns and Thomashow, 1988), several monocots, including cereal crop plants such as barley (Hordeum vulgare), maize (Zea mays), rice (Oryza sativa), sorghum (Sorghum bicolor), triticale (Triticosecale spp.), and wheat (Triticum aestivum), have been transformed with this method (Grimsley et al., 1989; Gould et al., 1991; Chan et al., 1993; Cheng et al., 1997; Gurel et al., 2009; Harwood et al., 2009; Hensel et al., 2009). The main difficulty with using A. tumefaciens for transformation of monocots is that it is not very efficient, probably due to limited integration rates (Frame et al., 2002). The stable transformation of monocots requires significant modifications of the standard transformation protocol, and this has been achieved for a few monocot species (e.g. maize; Ishida et al., 2007). This creates substantial problems because a number of crops are monocots. In addition, Agrobacterium is not an effective delivery method for some cell types (e.g. microspores). An alternative method used for monocots is based on simple gold particle-mediated bombardment of naked DNA into plant tissue. Typically, such biolistic transformation generates multiple integrations of truncated, duplicated, and/or rearranged transgenes (Travella et al., 2005), resulting in a low transgene expression or no transgene expression at all. This is not surprising because the naked DNA delivered into plant cells is not protected against exo- and endonucleases and relies on various host import proteins to carry DNA inside the nucleus (Li et al., 2005). In addition, this method is not always ideal for cell biology studies. The same is also true for another method of gene transfer, polyethylene glycol-mediated transformation of protoplasts because protoplasts are usually highly stressed cells (Genschik et al., 1992); therefore, they are not suitable for some studies (e.g. detailed protein localization; Reyes et al., 2010).
The solution that is proposed in the current work is the delivery of a T-DNA molecule independent of Agrobacterium. We hypothesized that this can be achieved by using an in vitro-prepared T-DNA/protein complex. The complex was composed of single-stranded (ss) T-DNA, covered with the recombination protein A (RecA), and attached to VirD2. Escherichia coli RecA protein was used instead of Agrobacterium tumefaciens VirE2 because both proteins share the same ssDNA binding features, and RecA was shown previously to be able to substitute for VirE2 function in nuclear import of T-DNA (Ziemienowicz et al., 2001). To date, there are no reports on the generation of transgenic plants from in vitro reconstituted T-DNA/protein complex. Numerous studies, however, suggest that such a T-DNA/protein complex can reach the nucleus (Ziemienowicz et al., 1999, 2001; Pelczar et al., 2004) and integrate into the cell genome (Pelczar et al., 2004). We conjectured that this approach will result in a low copy number of transgenes in intact cells/tissues.
RESULTS AND DISCUSSION
Reconstitution of the Modified T-DNA Complex
T-DNA used to reconstruct the T-DNA/protein complex in vitro was designed to contain a GUS expression cassette consisting of the uidA (GUS) gene under the control of the rice Actin promoter and the Agrobacterium nos terminator (Fig. 1). This expression cassette, originating from an expression plasmid pACT-1D (McElroy et al., 1990), includes a region initially assigned as PAct-intron, which contains the actual Actin1 promoter-intron fusion preceded by a 0.9-kb-long rice genomic sequence (RS). The RS might be dispensable for transgene expression because no function has been assigned to this sequence thus far, and the deletion of this region has not changed the expression of the GUS transgene in rice protoplasts (McElroy et al., 1990). The actual PAct-intron fusion contains the enhancer, the promoter, the 5′ noncoding exon, and the first intron of the rice Actin1 gene. All of these elements contribute to efficient gene expression, with the intron sequence being required for efficient in vivo mRNA splicing and/or another splicing-related function, such as the nuclear export of mRNA (McElroy et al., 1990). The rice PAct-intron fusion also has been shown to function as a useful regulatory element for transgene expression in other monocots (e.g. barley; Chibbar et al., 1993).
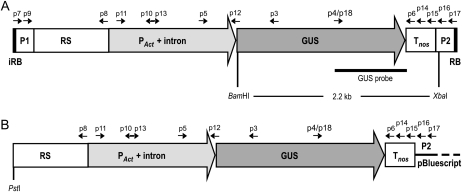
Figure 1.
Scheme of the GUS gene expression cassette in constructs used in transfection experiments: 4.8-kb-long T-DNA (A) and pACT-1D/PstI (B). RB and iRB indicate the right border and inverted right border of the Agrobacterium pTi plasmid; P1 and P2 represent 200-bp-long fragments of the pBluescript vector backbone flanking the GUS expression cassette in pACT-1D. PAct-intron, Fusion of rice actin promoter and intron (1.4 kb) preceded by a 0.9-kb-long rice genomic sequence (RS). GUS indicates 1.8-kb-long gene coding for bacterial β-glucuronidase, whereas Tnos indicates the 0.3-kb-long terminator of the nopaline synthase gene (nos). Arrows, Position and direction of primers used for the detection of the transgene and for analysis of its intactness. The BamHI and XbaI recognition sites, as well as the annealing site for the GUS probe used in Southern-blot analysis, are also indicated.
To form the T-DNA/protein complex in vitro, the GUS expression cassette was amplified by PCR and cloned into the LITMUS29 vector. PCR amplification was performed using primers annealing 200 bp upstream (forward primer) and downstream (reverse primer) of the GUS cassette in the pACT-1D plasmid. The primers were designed to contain the A. tumefaciens right border sequence (RB) in direct and inverted (iRB) orientation at the 3′ and 5′ ends of the GUS cassette, respectively (Fig. 1). This strategy resulted in a PCR product carrying the cleavage site for VirD2 on each strand of the double-stranded (ds) DNA molecule. After separation of DNA strands by heat denaturation, each strand can be processed by VirD2, thus leading to the formation of the VirD2-ssT-DNA complex, composed of ssT-DNA carrying the GUS expression cassette with the VirD2 protein covalently attached to its 5′ end. Such design allows for both DNA strands to serve as T-DNA, thus maximizing the use of each dsDNA molecule to produce the T-DNA/protein complex. In addition, the primers contained the NcoI restriction site for convenient cloning of the amplified iRB_PAct-GUS-Tnos_RB insert into the LITMUS29 vector. The recombinant vector was then used to produce dsT-DNA in high quantities; the LITMUS29_iRB_PAct-GUS-Tnos_RB plasmid DNA was digested with the NcoI enzyme, and the DNA fragment corresponding to the 4.8-kb-long iRB_PAct-GUS-Tnos_RB (dsT-DNA) was purified from an agarose gel. Then, dsT-DNA was converted into the single-stranded form (ssT-DNA) through heat denaturation by incubation for 10 min at 95°C followed by fast chilling on ice. Next, the VirD2-ssDNA complex was formed by the reaction of ssT-DNA with the purified recombinant protein VirD2. Finally, this partial complex was reacted with RecA to form the VirD2-ssT-DNA-RecA complex. The amounts of VirD2 and RecA proteins required for the formation of complexes with the DNA substrate were estimated based on the activity assays (Ziemienowicz et al., 1999). The RB cleavage activity of VirD2 was tested using fluorescently labeled 4.8-kb ssT-DNA. The cleavage efficiency of VirD2 on ssT-DNA was approximately 75% if 1 μg of protein was used per 200 ng of the DNA substrate (Fig. 2A). The application of higher amounts of VirD2 resulted in a lower cleavage efficiency (data not shown), most likely because of the inhibitory effect of salt and glycerol present in the VirD2 storage buffer. The ssDNA binding activity of RecA was assayed using ΦX174 virion ssDNA as a substrate and various amounts of protein (Fig. 2B). The binding efficiency was 100%, which means that all ssDNA molecules formed complexes with RecA, yet the use of the maximum amount of protein that did not show an adverse effect due to high glycerol concentration might not have allowed the fully complete coating of ssDNA. However, such an amount of RecA was sufficient to protect the entire ssDNA molecule from nucleolytic degradation, as shown in our recent work (S. Basu and F. Eudes, unpublished data). Based on the activity test results, the optimal (under our experimental conditions) protein-to-DNA ratio was applied to produce the VirD2-ssDNA-RecA complex (see “Materials and Methods” for details).
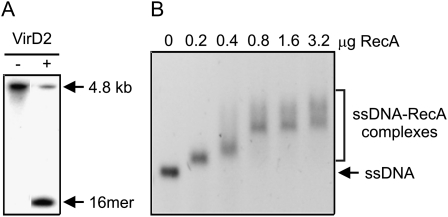
Figure 2.
Activity tests for VirD2 and RecA proteins. A, The cleavage activity of the recombinant VirD2 protein was tested using a 5′ fluorescently labeled 4.8-kb ssT-DNA substrate in the presence or absence of 1 μg of VirD2. The efficiency of the cleavage reaction was calculated as the ratio of fluorescence of the cleavage product (16mer) to the total amount of fluorescence in the reaction (4.8-kb DNA + 16 mer). B, The single-stranded DNA binding activity of the RecA protein was tested using ΦX174 ssDNA as a substrate and various amounts of the RecA protein. The binding of the RecA protein to the ssDNA substrate resulted in ssDNA-RecA complexes, and retardation of the DNA mobility due to the presence of protein was proportional to the amount of protein bound to DNA.
Transfection of Triticale Microspores
The reconstituted full T-DNA/protein complex (VirD2-ssT-DNA-RecA), partial complexes (ssT-DNA-RecA and VirD2-ssT-DNA), and naked DNA (ssT-DNA, dsT-DNA, and pACT-1D/PstI) were used as cargos in the transfection experiments with the GUS gene. Triticale var Ultima microspores were transfected with DNA or T-DNA/protein complexes in the presence or absence of the Tat2 cell-penetrating peptide (CPP) carrier, which was previously shown to be able to deliver linearized plasmid DNA and protein into the isolated microspores of triticale plants (Chugh et al., 2009). Embryos were developed from treated and nontreated microspores by the cultivation on a Ficoll-containing medium (Supplemental Fig. S1). No significant differences in the embryogenesis of microspores from various treatments were observed, except for an occasional reduction in embryogenesis of microspores that were treated with DNA/protein complexes containing RecA protein (Supplemental Fig. S1; data not shown). Interestingly, the development of embryos of various sizes was observed in all treatments (Fig. 3A). Next, a small portion of the developed embryos was used for histochemical staining to detect GUS expression. The similar GUS staining patterns were observed for embryos originating from microspores treated with the DNA and DNA/protein complexes in the presence of the Tat2 CPP (Fig. 3A). The developed embryos were then subjected to regeneration (Fig. 3B). Most embryos (>50%) were not able to undergo any type of organogenesis and died, some generated only roots or albino plantlets, and only a few regenerated into green plantlets (Table I). Efficiency of regeneration of green plantlets varied between samples from independent experiments and treatments as indicated by high sd values. The average regeneration efficiency values did not exceed 10% in most cases, with the lowest value observed for transfection with the full T-DNA/protein complex (VirD2-ssT-DNA-RecA) among all treatments in the presence of the Tat2 CPP (Table I). In total, 303 plants were regenerated in vitro and then transferred into soil. The potting survival rate was 93%, resulting in 281 plants grown in soil. Next, leaf samples were collected from green plants and used for detection of the transgene as well as for analysis of the transgene intactness and transgene expression.
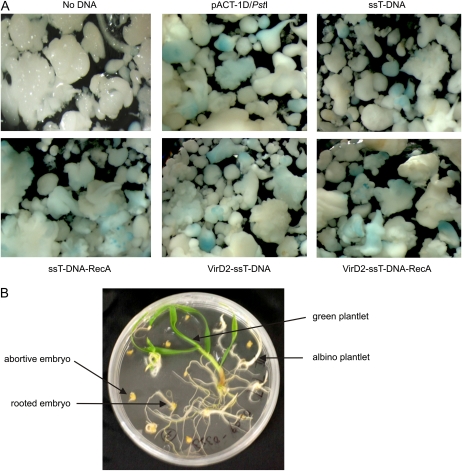
Figure 3.
GUS detection in triticale embryos and plant regeneration. A, Embryos developed from triticale microspores treated with various DNAs (e.g. linearized plasmid, ssT-DNA) or DNA/protein complexes (ssT-DNA-RecA, VirD2-ssT-DNA, and VirD2-ssT-DNA-RecA) in the presence of the Tat2 peptide were analyzed by histochemical GUS staining. B, The remaining embryos were cultured for 6 to 8 weeks on germination solid medium to regenerate into green plantlets. The examples of other regeneration effects such as albino plantlets, rooted embryos, and abortive embryos are indicated by arrows.
Table I. Plant regeneration.
Plant regeneration from embryos obtained from control (untreated) microspores and from microspores treated with various DNAs and DNA/protein complexes in the presence or absence of the Tat2 peptide. Numbers in the table represent average values (± sd) from four (treatments with Tat2) or three (treatments without Tat2) independent experiments.
| Treatment | Embryos Plated | Abortive Embryos | Rooted Embryos | Albino Plantlets | Green Plantlets | Regeneration Efficiency |
| % | ||||||
| No treatment | 90.8 ± 7.4 | 49.3 ± 14.2 | 31.0 ± 14.4 | 4.3 ± 3.0 | 6.0 ± 2.8 | 6.6 ± 3.0 |
| No DNA + Tat2 | 113.3 ± 24.8 | 73.8 ± 19.7 | 27.5 ± 16.3 | 5.3 ± 2.6 | 6.8 ± 6.1 | 6.1 ± 5.7 |
| pACT-1D/PstI + Tat2 | 140.8 ± 39.5 | 71.3 ± 17.9 | 45.5 ± 21.0 | 10.3 ± 5.6 | 16.3 ± 14.2 | 11.5 ± 10.5 |
| dsT-DNA + Tat2 | 163.0 ± 3.5 | 88.8 ± 19.3 | 46.3 ± 11.8 | 12.5 ± 4.4 | 9.5 ± 1.3 | 5.9 ± 0.9 |
| ssT-DNA + Tat2 | 85.5 ± 36 6 | 35.3 ± 26.4 | 37.3 ± 4.8 | 7.5 ± 1.9 | 7.8 ± 7.6 | 8.3 ± 6.2 |
| ssT-DNA-RecA + Tat2 | 126.8 ± 35.3 | 73.8 ± 37.8 | 31.8 ± 2.6 | 12.5 ± 4.5 | 6.3 ± 1.0 | 5.4 ± 1.9 |
| VirD2-T-DNA + Tat2 | 126.3 ± 42.6 | 82.0 ± 36.2 | 33.0 ± 6.4 | 5.0 ± 6.0 | 6.3 ± 3.0 | 4.9 ± 1.4 |
| VirD2-T-DNA-RecA + Tat2 | 173.0 ± 73.1 | 113.0 ± 40.7 | 33.0 ± 20.7 | 21.5 ± 16.7 | 5.3 ± 3.6 | 3.0 ± 1.5 |
| No DNA | 107.7 ± 42.4 | 74.3 ± 34.0 | 18.7 ± 17.7 | 9.7 ± 15.9 | 5.0 ± 6.1 | 6.6 ± 9.4 |
| pACT-1D/PstI | 109.7 ± 34.5 | 79.7 ± 14.0 | 22.0 ± 24.3 | 5.0 ± 4.6 | 3.0 ± 2.6 | 2.5 ± 2.2 |
| dsT-DNA | 74.3 ± 6.7 | 59.7 ± 11.0 | 5.3 ± 2.1 | 8.0 ± 6.2 | 1.3 ± 2.3 | 1.8 ± 3.1 |
| VirD2-ssT-DNA-RecA | 70.7 ± 20.2 | 50.0 ± 6.6 | 12.7 ± 11.0 | 4.7 ± 7.1 | 3.3 ± 5.8 | 3.5 ± 6.1 |
Transgene Detection
The presence of the GUS transgene in the genome of plants regenerated after transfection was determined by PCR analysis and confirmed by Southern-blot analysis. To detect the GUS gene by PCR, GUS-specific primers (p3 and p4) were combined with Actin intron (p5)- and nos terminator (p6)-specific primers (Fig. 1; Supplemental Table S1). PCR analysis revealed that omission of the Tat2 peptide resulted in the low (if any) efficiency of stable transfection because no, or very few, GUS-positive plants could be found (Table II). These few transformants could be attributed to the effect of the Lipofectamine transfection reagent. In contrast, DNA and DNA-protein complexes were transfected into triticale microspores via the Tat2 peptide relatively efficiently. Interestingly, the percentage of GUS-positive plants was comparable when complete T-DNA complex (VirD2-ssT-DNA-RecA), VirD2-ssT-DNA complex, or linear naked dsDNA (pACT-1D/PstI, dsTDNA) was used for transfection (Table II). Slightly lower values were observed for the ssT-DNA-RecA complex, whereas use of naked ssT-DNA generated a low number of GUS-positive plants (Table II), most likely because of the lack or incomplete protection of DNA from nucleases. Among naked DNA molecules, dsDNA was better protected from nucleolytic degradation than ssDNA. This is not surprising because many plant nucleases show higher specificity for ssDNA (Rangarajan and Shankar, 2001). All of the control untreated plants were GUS negative, and only a very few GUS-positive plants were found among those treated without DNA. The latter plants most likely represent false-positive cases.
Table II. Analysis of the intactness of the GUS expression cassette in plants regenerated from triticale microspores.
The presence and intactness of the integrated DNA carrying the GUS expression cassette were analyzed by PCR on genomic DNA isolated from triticale plants regenerated from microspores treated with DNA or DNA/protein complexes in the presence or absence of the Tat2 CPP or from untreated (control) microspores. Primers and segments of the GUS expression cassette, which contain the primer annealing sites, are indicated (Fig. 1; Supplemental Fig. S2).
| Plants Containing the Amplified Regions of the Transgene Source DNA |
||||||||||
| Treatment | P1-RS p7-p8 | P1-Act p9-p10 | Act-GUS p11-p12 | Act-GUS p13-p3 | Act-GUS p5-p3 | GUS-nos p4-p6 | GUS-nos p4-p14 | GUS-nos p4-p15 | GUS-P2 p4-p16 | GUS-P2 p4-p17 |
| No treatment | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (21 plants) | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| No DNA + Tat2 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 2 | 0 | 0 |
| (26 plants) | 0% | 0% | 0% | 0% | 3.9% | 7.7% | 7.7% | 7.7% | 0% | 0% |
| pACT-1D/PstI + Tat2 | 0 | 0 | 12 | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| (63 plants) | 0% | 0% | 19.1% | 22.2% | 22.2% | 22.2% | 22.2% | 22.2% | 22.2% | 22.2% |
| dsT-DNA + Tat2 | 5 | 7 | 9 | 10 | 13 | 13 | 13 | 11 | 6 | 4 |
| (44 plants) | 11.4% | 15.9% | 20.5% | 22.7% | 29.6% | 29.6% | 29.6% | 25.0% | 13.6% | 9.1% |
| ssT-DNA + Tat2 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 |
| (27 plants) | 3.7% | 3.7% | 3.7% | 3.7% | 7.4% | 7.4% | 7.4% | 7.4% | 3.7% | 3.7% |
| ssT-DNA-RecA + Tat2 | 2 | 2 | 3 | 3 | 4 | 4 | 4 | 4 | 2 | 2 |
| (23 plants) | 8.7% | 8.7% | 13.0% | 13.0% | 17.4% | 17.4% | 17.4% | 17.4% | 8.7% | 8.7% |
| VirD2-ssT-DNA + Tat2 | 3 | 3 | 4 | 5 | 6 | 6 | 5 | 4 | 3 | 3 |
| (23 plants) | 13.0% | 13.0% | 17.4% | 21.7% | 26.1% | 26.1% | 21.7% | 17.4% | 13.0% | 13.0% |
| VirD2-ssT-DNA-RecA + Tat2 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 |
| (20 plants) | 25.0% | 25.0% | 25.0% | 25.0% | 25.0% | 25.0% | 25.0% | 25.0% | 25.0% | 20% |
| No DNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (14 plants) | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| pACT-1D/PstI | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 |
| (Nine plants) | 0% | 0% | 0% | 0% | 11.1% | 11.1% | 11.1% | 11.1% | 0% | 0% |
| dsT-DNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (Three plants) | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| VirD2-ssT-DNA-RecA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (10 plants) | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
Next, Southern-blot analysis was performed to verify PCR results. Genomic DNA (gDNA) from GUS-positive triticale lines was digested with BamHI and XbaI and probed first with the GUS-specific probe and then with a probe specific for the wheat EF1α gene. The presence of the GUS gene was confirmed in all lines except in Ultima wild-type plants and plant lines regenerated from microspores transfected without DNA (e.g. line no. 41; Fig. 4). Because PCR analysis showed that line number 41 was GUS positive, the latter result indicated that this line represented a false-positive PCR case. The intensity of the Southern-blot signal detected by the GUS-specific probe varied between lines, whereas the intensity of the endogenous control (EF1α gene) was relatively constant in all lines (Fig. 4). This result suggested variations in the transgene copy number. Next, the GUS-positive plants were analyzed for the intactness of the integrated T-DNA.
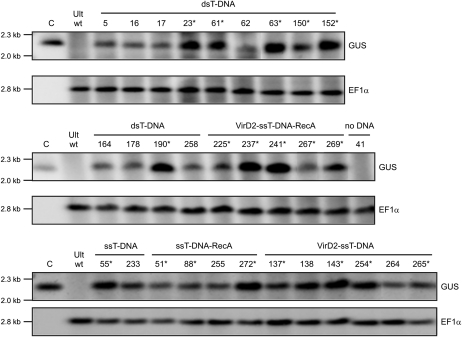
Figure 4.
Southern-blot analysis of transgenic triticale plants for the detection of the GUS transgene in gDNA of GUS-PCR-positive plants. DNA was digested with BamHI and XbaI enzymes and hybridized with the GUS-specific and EF1α-specific probes. Ult wt, Nontransgenic triticale var Ultima (wild type). C, Positive control; the 4.8-kb GUS transgene cassette digested with BamHI and XbaI. The numbers above other lanes indicate the regenerated plant lines. Plant lines that were expected to express the GUS transgene are marked by asterisks. The positions of the molecular size markers are indicated by short lines on the left.
Intactness of the Transgene
Integration of T-DNA delivered to plant cells by A. tumefaciens shows specific patterns; in most cases, the 5′ end of T-DNA is preserved because of protection by covalently attached VirD2 (precise integration), whereas the 3′ end of the integrated T-DNA is poorly conserved (truncations of 3–100 nucleotides; Tinland et al., 1995; Tinland, 1996; Windels et al., 2008). Analysis of the intactness of the integrated transgene cassette was performed by PCR using 10 sets of primers specific to various regions of the GUS cassette (Fig. 1; Table II). The analysis revealed that when naked dsT-DNA, ssT-DNA, and ssT-DNA combined with RecA or VirD2 protein, and reacted with the Tat2 peptide were used to transfect triticale microspores, some truncations of 5′, 3′, or of both ends of the transgene cassette were observed (Table II; Supplemental Fig. S2). In contrast, the use of the complete nano-complex (VirD2-ssT-DNA-RecA + Tat2) resulted in nearly 100% intact integration events, with only one case of short truncation of the 3′ end (Table II; Supplemental Fig. S2). A similar tendency was also observed in the classical Agrobacterium-mediated plant transformation; in some cases, the 3′ end of the integrating T-DNA was more prone to truncation (Tinland et al., 1995; Tinland, 1996; Brunaud et al., 2002). It seems that T-DNA delivered to triticale microspores in vitro in the form of the T-DNA/CPP complex could be better preserved than Agrobacterium-delivered T-DNA because we have not observed any truncations at the 5′ end of the GUS transgene expression cassette. According to our data, about 50% of the GUS-positive plants obtained after Tat2-mediated transfection of triticale microspores with naked dsT-DNA or ssT-DNA were expected to be expressed (Supplemental Fig. S2). In the case of plants from transfection experiments where ssT-DNA is complexed with either RecA or VirD2 protein alone (+Tat2), the percentage of GUS-positive plants that express the transgene could be higher (75% and 67%, respectively). The highest percentage (100%) was expected for GUS-positive plants regenerated from microspores transfected with the complete nano-complex (VirD2-ssT-DNA-RecA + Tat2) because they contain complete regulatory elements (promoter and terminator) required for normal transgene expression. These predictions are supported by previously reported observations showing that both VirD2 and VirE2 proteins were required for precise integration of T-DNA delivered into mammalian cells in the form of the in vitro-generated complex (Pelczar et al., 2004). However, our predictions regarding plant lines containing the PAct-GUS-Tnos cassette in their genomes may not be absolutely accurate. This is due to the fact that we were not able to detect a sequence located closer than 100 bp to the 5′ end of the PAct-intron fusion sequence because primers complementary to the most distal 5′ end of the sequence generated false-positive, unspecific PCR products. Nevertheless, analysis of the GUS-positive plants generated from triticale microspores transfected with various T-DNAs and T-DNA/protein complexes in the presence of the Tat2 peptide proved our predictions to be nearly perfect, as revealed by studying the GUS transgene expression in these plants.
Transgene Expression
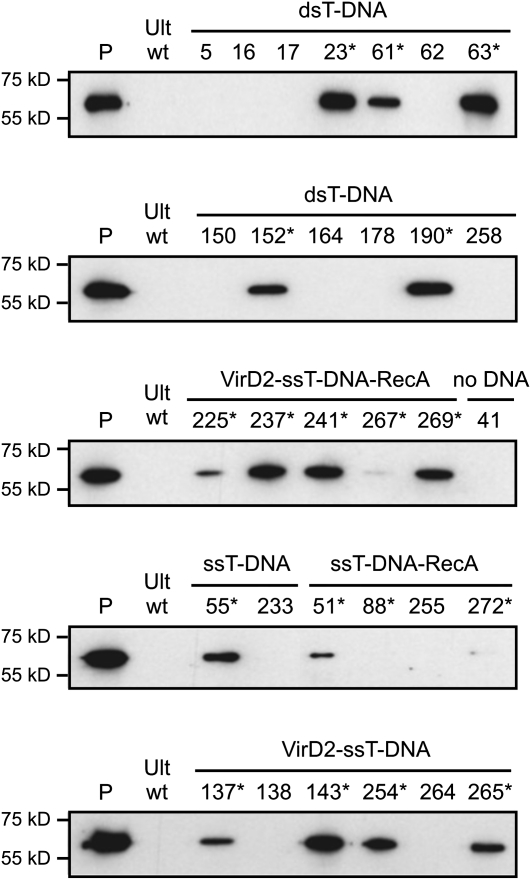
Analysis of the transgene expression was performed at the protein level using western-blotting technique on crude extracts from GUS-positive plants. The GUS transgene was expected to be expressed in lines 23, 61, 63, 152, and 190 (from treatment with dsT-DNA + Tat2); 55 (ssT-DNA + Tat2 treatment); 51, 88, and 272 (ssT-DNA-RecA + Tat2 treatment); 137, 143, 254, and 265 (VirD2-T-DNA + Tat2 treatment); and 225, 237, 241, 267, and 269 (VirD2-T-DNA-RecA + Tat2 treatment). The results of the western-blot analysis showed that the bacterial GUS protein was detected in all lines, which were predicted to express the transgene according to PCR and Southern-blot analyses (Fig. 5; Table III), with one line (no. 88) expressing the GUS transgene at an extremely low level, if at all. Line number 88 most likely represents the case of a GUS-positive plant carrying an incomplete PAct-GUS-Tnos cassette that lacks less than 100 bp of the promoter’s 5′ end. We also tested some GUS-positive plants that were predicted not to express the GUS gene due to larger truncations of the transgene cassette (line nos. 5, 16, 17, 62, 150, 164, 178, 258, 233, 252, 138, and 264) or due to the absence of the transgene (line no. 41); indeed, no GUS protein was detected in these lines (Fig. 5). Significant differences in the GUS protein level were observed in GUS-positive plants from all treatments with T-DNA and T-DNA/protein complexes (+Tat2), with some levels of the GUS protein being very high (Fig. 5). These differences may reflect variations in the transgene copy number.
Figure 5.
GUS transgene expression at the protein level. Analysis of the transgene expression was done using western-blotting technique on protein extracts isolated from GUS-positive plants that were regenerated from triticale microspores treated with various DNAs (e.g. dsT-DNA and ssT-DNA) or DNA/protein complexes (ssT-DNA-RecA, VirD2-ssT-DNA, and VirD2-ssT-DNA-RecA) in the presence of the Tat2 peptide. Lane P, Purified bacterial GUS protein (1 ng); lane Ult wt, protein extract isolated from untreated (wild type) triticale var Ultima plants. The numbers above other lanes indicate the regenerated plant lines. Plant lines that were expected to express the GUS transgene are marked by asterisks. The positions of the molecular size markers are indicated by short lines on the left.
Table III. Comparison of the transgene copy number and transgene expression.
The transgene copy number and transgene expression were compared in plants that were regenerated from microspores transfected with various types of T-DNA and T-DNA/protein complexes in the presence of the Tat2 CPP.
| Treatment/Plant Line | Protein Amounta | BamHI Fragment Sizes | Transgene Copy No. | Integration Locus No. | |||
| kb | |||||||
| dsT-DNA | |||||||
| 23 | 50.3 ± 2.5 | 5.0 | 6.4 | 7.8 | 3 | 3 | |
| 61 | 29.0 ± 3.2 | 2.8 | ∼20 | 2 | 2 | ||
| 63 | 50.7 ± 3.5 | 4.8b | 7.4 | 14.4 | 4 | 3 | |
| 152 | 30.6 ± 1.8 | 4.4 | 4.6c | 10.8 | 4 | 3 | |
| 190 | 43.5 ± 2.2 | 4.8b | 5.4 | 5.6 | 7.6 | 5 | 4 |
| 3.6 ± 1.1 | 3.0 ± 0.7 | ||||||
| VirD2-ssT-DNA-RecA | |||||||
| 225 | 12.3 ± 1.2 | 8.8 | 1 | 1 | |||
| 237 | 45.7 ± 3.4 | 4.8b | 19.2 | 3 | 2 | ||
| 241 | 43.5 ± 2.2 | 4.4 | 4.6c | 7.4 | 12.0 | 5 | 4 |
| 267 | 2.5 ± 0.5 | 5.8 | 1 | 1 | |||
| 269 | 36.6 ± 2.1 | 2.6 | 15.0 | 2 | 2 | ||
| 2.4 ± 1.7 | 2.0 ± 1.2 | ||||||
| ssT-DNA | |||||||
| 55 | 28.3 ± 2.6 | 5.3 | 9.4 | 2 | 2 | ||
| ssT-DNA-RecA | |||||||
| 51 | 11.1 ± 0.8 | 11.0 | 1 | 1 | |||
| 88 | 0.8 ± 0.2 | 2.8 | 1 | 1 | |||
| 272 | 1.3 ± 1.9 | 4.2 | 4.8b | 7.2 | 4 | 3 | |
| 2.0 ± 1.7 | 1.7 ± 1.2 | ||||||
| VirD2-ssT-DNA | |||||||
| 137 | 17.1 ± 1.5 | 9.6 | 1 | 1 | |||
| 143 | 40.6 ± 3.5 | 5.0 | 10.8 | 2 | 2 | ||
| 254 | 27.5 ± 1.8 | 2.7 | 6.0 | 14.8 | 3 | 3 | |
| 265 | 22.3 ± 2.1 | 5.8 | 8.8 | 2 | 2 | ||
| 2.0 ± 0.8 | 2.0 ± 0.8 | ||||||
The protein amount is expressed in ng of the GUS protein per 1 g of fresh leaf weight; the data represent mean values (± sd) from three independent experiments.
Head-to-tail or tail-to-head integration (4.8 kb).
Head-to-head or tail-to-tail integration (4.6 kb).
Transgene Integration Pattern
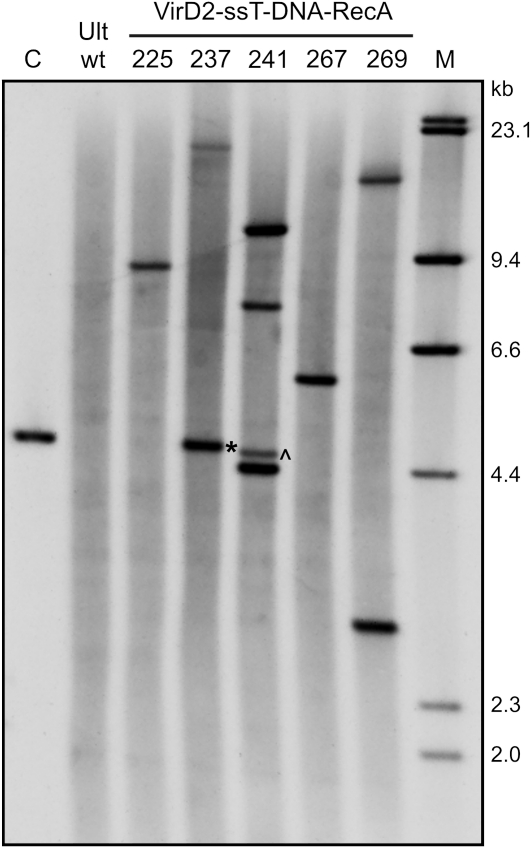
Standard procedures of plant transformation using Agrobacterium often result in clear integration patterns, such as a low number of copies of integrated transgenes (Windels et al., 2008), whereas most other methods used in plant biotechnology (direct gene transfer, bombardment, etc.) result in the integration of multiple DNA molecules and, as a consequence, multiple copy or multiple loci insertion patterns that may lead to variations in the transgene expression (Latham et al., 2006). The transgene copy number and integration pattern were analyzed by Southern blotting, using gDNA of transgenic plants expressing the transgene at the detectable level. gDNA was digested with BamHI and hybridized with the GUS-specific probe. The number of integration loci was concluded from the number of bands detected by Southern blotting, whereas the presence of bands corresponding to DNA fragments of 4.6 and 4.8 kb in length indicated the insertion of at least two transgene copies into a single locus in the head-to-head or tail-to-tail and head-to-tail or tail-to-head arrangements (Supplemental Fig. S3). The results of the Southern-blot analysis of plants from transfection experiments using the DNA (dsT-DNA and ssT-DNA) and DNA/protein complexes (ssT-DNA-RecA, VirD2-ssT-DNA, and VirD2-ssT-DNA-RecA) with the Tat2 CPP are presented in Table III, and an example image of Southern blot performed on gDNA from plants transfected with the full T-DNA complex (VirD2-ssT-DNA-RecA) is shown in Figure 6. The application of the CPP-mediated transgene delivery resulted in a low copy number (≤5) and relatively simple patterns of transgene integration (Fig. 6; Table III). However, transgenic plants regenerated from microspores transfected with dsT-DNA + Tat2 were found to contain the transgene copy number and the integration locus number that were 1.5-fold higher than those numbers in plants obtained by transfection with the VirD2-ssT-DNA-RecA + Tat2 nano-complex. These values are actually higher, as the 2-fold difference in the number of DNA molecules in transfection samples should also be taken into consideration. More importantly, 40% of the nano-complex-transfected plants showed a single-copy single locus integration pattern (Fig. 6; Table III), which was not observed in plants transfected with dsT-DNA (Table III). In addition, the latter plants showed more frequent events of integration of at least two DNA molecules into a single locus, such as head-to-head or tail-to-tail and head-to-tail or tail-to-head integrations (Table III).
Figure 6.
Southern-blot analysis of the transgene copy number and integration patterns in transgenic triticale plants that were regenerated from microspores transfected with the nano-complex (VirD2-ssT-DNA-RecA + Tat2). gDNA was digested with BamHI and hybridized with the GUS-specific probe. Lane Ult wt, Nontransgenic triticale plant var Ultima (wild type); lane C, positive control (the 4.8-kb GUS transgene cassette); lane M, DIG-labeled DNA size markers. The numbers above other lanes indicate the regenerated plant lines. Asterisk, Head-to-tail or tail-to-head integration event (4.8 kb); ^, Head-to-head or tail-to-tail integration event (4.6 kb).
Finally, the transgene copy number and the expression level were compared. In most cases, the comparison revealed a clear correlation between these two factors (Table III). Two lines containing a single copy of the transgene (line nos. 267 and 88) showed very low protein levels. This may result from transgene silencing due to the position effect (Filipecki and Malepszy, 2006) or from the incomplete promoter sequence in the case of line number 88. In the instance of another line (no. 272), the low level of GUS expression was most likely caused by gene silencing induced by additional copies of the transgene. Variations in the transgene protein level were also noted in plants regenerated from microspores transfected with the complete nano-complex (VirD2-ssT-DNA-RecA + Tat2; Fig. 5; Table III). Such variations in transgene expression are sometimes observed in plants generated by the classical Agrobacterium-mediated transformation, with most transformants expressing transgenes at relatively low levels (Peach and Velten, 1991; Filipecki and Malepszy, 2006). The improvement of the expression pattern of transgenes should be possible in the future by changing the ratio of Tat2 to T-DNA, which would allow for the formation of smaller DNA-CPP complexes and result in the delivery of fewer T-DNA molecules into plant cells.
CONCLUSION
We developed a method for plant transformation and showed that this method can be used for plant species and cell types that are difficult to transform by the classical Agrobacterium-mediated technique. Moreover, we have shown that CPP-mediated delivery of the T-DNA complex results in an Agrobacterium-like type of transgene integration pattern as far as transgene intactness, copy number, and expression are concerned. In particular, the nano-complex strategy yields the more frequent integration of intact transgene molecules into a single locus of the triticale genome, which leads to the efficient expression of the transgene in transgenic plants. Fewer copies of transgenes were also observed in maize callus lines resulting from “agrolistic” transformation (Hansen and Chilton, 1996; Hansen et al., 1997) as compared with the classical “biolistic” transformation. In the agrolistic approach, T-DNA complexes were produced in planta by expressing A. tumefaciens virulence genes (virD1 and virD2, with or without virE2) that were codelivered into maize protoplasts together with the target plasmid carrying T-DNA. Both tactics, agrolistics and nano-complex, proved to combine the advantages of Agrobacterium T-DNA-type transgene integration with high efficiency of either biolistic or CPP-mediated DNA delivery.
Our nano-complex-mediated method of transgene delivery was shown to work in germ line cells (microspores) of triticale plants. Both the plant species and cell type are difficult to transform by other methods. We speculate that our approach might find applications in gene transfer to other plant cells and species, including perhaps somatic cells and dicot plants. Our supposition is supported by previous findings related to the ability of CPP peptides to transfer their cargos (e.g. DNA, proteins) to various cells of monocot and dicot plants (Chen et al., 2007; Chugh and Eudes, 2007, 2008a, 2008b, Chugh et al., 2010). Further studies need to be performed to verify our assumption.
Additionally, the fact that the nano-complex approach (the delivery of the in vitro-generated VirD2-ssT-DNA-RecA + Tat2 nano-complex into plant microspores) resulted in successful regeneration of transgenic plants provides new information about the role of the single-stranded DNA binding proteins in T-DNA delivery into the plant cell genome. Previous studies showed that RecA, but not the SSB protein of E. coli, was able to substitute for VirE2 function in nuclear targeting of the T-DNA complex in vitro (Ziemienowicz et al., 2001). On the other hand, RecA produced in transgenic tobacco (Nicotiana tabacum) plants was not able to complement the virE2-deficient A. tumefaciens strain (Ziemienowicz et al., 2001), thus indicating that VirE2 may play a role not only in the nuclear import but also in other intracellular steps of T-DNA transfer. The ability of the VirD2-ssT-DNA-RecA complex to transform triticale microspores suggests species- and/or cell type-dependent differences in the T-DNA complex delivery into plant cells. Alternatively, the Tat2 CPP may substitute for VirE2 function in cellular entry and/or intracellular trafficking of the T-DNA complex containing RecA.
MATERIALS AND METHODS
Overexpression and Purification of VirD2
Production of VirD2 from pET3aVirD2 (pFSVirD2) was performed as previously described (Ziemienowicz et al., 1999). The recombinant protein was produced in Escherichia coli as 6× His fusion and purified using affinity and ion-exchange chromatography (Pelczar et al., 2004).
Activity Tests for VirD2 and RecA Proteins
The cleavage activity of the recombinant VirD2 protein was tested as previously described (Ziemienowicz et al., 1999). The reaction was performed for 1 h at 37°C in TKM buffer (50 mm Tris-Cl, pH 8.0, 150 mm KCl, and 1 mm MgCl2) using 200 ng of the 5′ 6-carboxy-fluorescin-labeled heat-denatured 4.8-kb ssT-DNA substrate in the presence or absence of 1 μg of VirD2.
The single-stranded DNA binding activity of the RecA protein was tested as previously described for its human homolog (Yokoyama et al., 2003), using 200 ng of ΦX174 ssDNA as substrate. The reaction was performed at 37°C for 30 min in the reaction mixture containing TKM buffer and various amounts of the RecA protein.
Formation of the ssT-DNA-VirD2-RecA Complex
The DNA insert was produced by PCR using primers p1 and p2 containing the RB sequence and NcoI site (Supplemental Table S1) and pACT-1D plasmid as a template. The PCR reaction mixture (25 μL) contained GC buffer, 0.2 mm deoxyribonucleoside triphosphates, 0.5 μm of each primer, 5 ng of DNA template, and 0.5 units of Phusion polymerase (Fermentas). The amplification reactions consisted of a preliminary denaturation step at 98°C for 30 s, followed by 10 cycles of 98°C for 10 s, 59°C for 30 s, and 72°C for 75 s, 20 cycles of 98°C for 10 s, 67°C for 30 s, and 72°C for 75 s, followed by incubation at 72°C for 10 min. The PCR product was extracted from agarose gel, purified using a QiaQuick gel extraction kit (Qiagen), and cloned into the NcoI site of the LITMUS29 plasmid vector, resulting in recombinant vector LITMUS29_iRB_PAct-GUS-Tnos_RB. T-DNA (4.8-kb-long iRB_PAct-GUS-Tnos_RB cassette) was released from the recombinant vector by digestion with NcoI restrictase and extracted from agarose gel using a QiaQuick gel extraction kit (Qiagen). This dsT-DNA was converted into the ssDNA form by heat denaturation for 10 min at 95°C and immediate cooling on ice. The VirD2-ssT-DNA complex was formed by reacting 2.0 μg of ssT-DNA with 10 μg of the VirD2 protein in TKM buffer for 1 h at 37°C. Next, the VirD2-ssT-DNA complex was reacted with 16 μg of E. coli RecA (New England Biolabs) during a 30-min incubation at 37°C to form the VirD2-ssT-DNA-RecA complex, as previously described (Ziemienowicz et al., 1999, 2001).
Transfection of Triticale Microspores
Microspores were isolated from triticale (Triticosecale spp.) var Ultima plants and subjected to transfection procedure as previously described (Chugh et al., 2009), using the Tat2 peptide (RKKRRQRRRRKKRRQRRR; synthesized by CanPeptide Inc.). For transfection experiments, DNA was used in the form of dsT-DNA, ssT-DNA, ssT-DNA-RecA complex, VirD2-ss-T-DNA complex, or VirD2-ssT-DNA-RecA complex. Plasmid pACT-1D (pAct-1GUS), linearized with the PstI restrictase, was used as a positive control. The Tat2 peptide-to-DNA ratio in all experiments was 4:1 (w/w). An additional step of treatment of the DNA-Tat2 complexes with 5 μg of lipofectamin for 15 min was introduced prior to the transfection step. Four and three independent experiments were performed for treatments with and without the Tat2 peptide, respectively.
Embryogenesis and Plant Regeneration
Transfected microspores were cultured in 30-mm petri dishes containing liquid NPB-99 medium supplemented with 10% (v/v) Ficoll in the presence of four ovaries per plate. Plates were incubated for 4 to 6 weeks at 28.5°C in the dark. Then, 1- to 2-mm-long embryos were transferred germ-side-up from liquid medium to standard petri dishes containing solid germination of embryo of monocot medium (Eudes et al., 2003) and incubated in a growth chamber with a 16-/8-h photoperiod at 80 μmol m−2 s−1 and 16°C. Green plantlets were then transferred to root trainers containing Cornell mix soil and cultivated in a greenhouse.
Histochemical GUS Assay
Histochemical GUS analysis was performed according to the previously described protocol (Kosugi et al., 1990), with some modifications. Embryos were incubated overnight in GUS buffer (50 mm NaH2PO4, 10 mm EDTA, 0.3 m mannitol, 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 20% methanol; pH 7.0) at 37°C in the dark.
PCR on Genomic DNA
gDNA was isolated from 100-mg leaf samples using the cetyltrimethylammonium bromide method (Doyle and Dickson, 1987) which was modified according to the DArT protocol (http://www.diversityarrays.com/sites/default/files/pub/DArT_DNA_isolation.pdf). PCR reactions were performed in 25-μL reaction mixtures containing 1× Phusion CoralLoad PCR buffer, 0.2 mm deoxyribonucleoside triphosphates, 0.1 μm of each primer (Supplemental Table S1), 0.025 units of Taq polymerase (Qiagen), and 100 ng of genomic DNA. The amplification reactions consisted of a preliminary denaturation step at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1 to 1.5 min, and incubation at 72°C for 10 min. PCR products were analyzed by electrophoresis in 0.8% (w/v) agarose/1× Tris-acetate-EDTA gel containing ethidium bromide at the concentration of 0.5 μg mL−1.
Southern-Blot Analysis
Transgenic and nontransformed triticale genomic DNA was isolated as described above and treated with RNaseA (the final concentration: 80 μg mL−1) for 10 min at 65°C, followed by purification using the phenol-chloroform method and precipitation with ethanol (Sambrook and Russell, 2001). To test for the presence of the transgene, gDNA was digested using BamHI and XbaI restriction enzymes (New England Biolabs) in a 1-mL reaction mixture containing New England Biolabs buffer no. 3 (NEBuffer 3), 1 mg mL−1 of bovine serum albumin, 80 μg of gDNA, and 1,000 units of each restrictase. To test for the transgene copy number, gDNA was digested using BamHI alone in a 500-μL reaction mixture containing New England Biolabs buffer no. 3 (NEBuffer 3), 1 mg mL−1 bovine serum albumin, 30 μg of gDNA, and 400 units of the restrictase. The reactions were incubated at 37°C overnight. The digested DNA was purified using the phenol/chloroform method and concentrated by precipitation with ethanol (Sambrook and Russel, 2001). Southern-blot analysis was performed according to a standard protocol (Sambrook and Russel, 2001), with some modifications. Sixty micrograms of gDNA digested with BamHI and XbaI or 20 μg of gDNA digested with BamHI were separated on a 0.8% (w/v) agarose gel at 35 V for 16 h in 1× TAE buffer. The gel was rinsed in distilled water (dH2O), depurinated for 15 min in 0.25 n HCl, rinsed in dH2O, denatured for 30 min in 0.4 n NaOH, rinsed again in dH2O, neutralized for 15 min in 0.5 m Tris-HCl (pH 7.5) containing 3 m NaCl, and soaked in transfer buffer (10× SSC; 1.5 m NaCl and 0.15 m sodium citrate) for 10 min. DNA transfer onto a positively charged nylon membrane (Roche) was performed for 2.5 h using a vacuum blotter (Appligene). DNA was then cross linked to the membrane at 120 mJ cm−2 in Spectrolinker (Spectromics Corp.). The probes were prepared with the PCR DIG Probe Synthesis kit (Roche) according to the protocol provided by the supplier, using primers p18 and p6 for the GUS-specific probe and primers p19 and p20 for the EF1α-specific probe. Hybridization with the GUS-specific probe was carried out at 42°C using DIG Easy Hyb solution (Roche). The detection was performed using AP-conjugated anti-DIG antibodies (Roche; diluted 1:2,500) in blocking solution containing 1% (w/v) Blocking Reagent (Roche) in maleic acid buffer (0.1 m maleic acid, 0.5 m NaCl, pH 7.5) and CPD Star (Roche) as a substrate. Images of the membrane were taken with FluorChem HD2 (Alpha Innotech).
For the transgene detection analysis, the membrane was rehybridized with the EF1α-specific probe. Particularly, the membrane was rinsed in dH2O, washed twice in stripping buffer (0.2 n NaOH, 0.1% [w/v] SDS) at 37°C for 15 min, equilibrated in 2× SSC solution, and hybridized with the EF1α probe at 65°C, followed by signal detection performed as described above.
Protein Extraction and Western-Blotting Analysis
Crude protein extracts were prepared from leaf tissue (100 mg) according to a previously published protocol (Stoger et al., 1999), using extraction buffer supplemented with Complete Protease Inhibitor Cocktail (Roche). Aliquots of 10 μg of total protein were analyzed by western blotting in accordance with a standard protocol (Towbin et al., 1979; Sambrook and Russel, 2001), using polyclonal rabbit antibodies raised against the N-terminal peptide of bacterial GUS protein (Abcam; 1:2,000; primary antibody) and donkey antibody to rabbit IgG (horseradish peroxidase conjugate; Abcam, 1:10,000; secondary antibody). Detection was carried out with ECL Plus according to the manufacturer’s recommendations (GE Healthcare). The membranes were then exposed to x-ray films, and the intensity of signals was quantified using the ImageJ program.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Triticale embryogenesis.
Supplemental Figure S2. Summary of the transgene intactness analysis.
Supplemental Figure S3. Scheme for the interpretation of transgene integration patterns.
Supplemental Table S1. Sequences of primers used in PCR reactions.
Supplementary Material
Acknowledgments
Dr. Ray Wu (Cornell University) is kindly acknowledged for the pACT-1D construct. We thank Dr. Valentina Titova for English revision of the manuscript.
References
- Binns AN, Thomashow MF. (1988) Cell biology of Agrobacterium infection and transformation of plants. Annu Rev Microbiol 42: 575–606 [Google Scholar]
- Birch RG. (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Physiol Plant Mol Biol 48: 297–326 [DOI] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al. (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MT, Chang HH, Ho SL, Tong WF, Yu SM. (1993) Agrobacterium-mediated production of transgenic rice plants expressing a chimeric alpha-amylase promoter/beta-glucuronidase gene. Plant Mol Biol 22: 491–506 [DOI] [PubMed] [Google Scholar]
- Chawla HS. (2002) Gene transfer in plants. In Introduction to Plant Biotechnology, Ed 2. Science Publishers Inc., Enfield, NH, pp 359–395 [Google Scholar]
- Chen C-P, Chou J-C, Liu BR, Chang M, Lee H-J. (2007) Transfection and expression of plasmid DNA in plant cells by an arginine-rich intracellular delivery peptide without protoplast preparation. FEBS Lett 581: 1891–1897 [DOI] [PubMed] [Google Scholar]
- Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y. (1997) Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol 115: 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibbar RN, Kartha KK, Datla RJJ, Leung N, Caswell K, Mallard CS, Steinhauer L. (1993) The effect of different promoter-sequences on transient expression of gus reporter gene in cultured barley (Hordeum vulgare L.) cells. Plant Cell Rep 12: 506–509 [DOI] [PubMed] [Google Scholar]
- Chugh A, Amundsen E, Eudes F. (2009) Translocation of cell-penetrating peptides and delivery of their cargoes in triticale microspores. Plant Cell Rep 28: 801–810 [DOI] [PubMed] [Google Scholar]
- Chugh A, Eudes F. (2007) Translocation and nuclear accumulation of monomer and dimer of HIV-1 Tat basic domain in triticale mesophyll protoplasts. Biochim Biophys Acta 1768: 419–426 [DOI] [PubMed] [Google Scholar]
- Chugh A, Eudes F. (2008a) Cellular uptake of cell-penetrating peptides pVEC and transportan in plants. J Pept Sci 14: 477–481 [DOI] [PubMed] [Google Scholar]
- Chugh A, Eudes F. (2008b) Study of uptake of cell penetrating peptides and their cargoes in permeabilized wheat immature embryos. FEBS J 275: 2403–2414 [DOI] [PubMed] [Google Scholar]
- Chugh A, Eudes F, Shim YS. (2010) Cell-penetrating peptides: nanocarrier for macromolecule delivery in living cells. IUBMB Life 62: 183–193 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, Vyas S, Tovkach A, Tzfira T. (2007) Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol 9: 9–20 [DOI] [PubMed] [Google Scholar]
- Cleene M. (1985) The susceptibility of monocotyledons to Agrobacterium tumefaciens. J Phytopathol 113: 81–89 [Google Scholar]
- Doyle JJ, Dickson EE. (1987) Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 36: 715–772 [Google Scholar]
- Eudes F, Acharya S, Laroche A, Selinger LB, Cheng KJ. (2003) A novel method to induce direct somatic embryogenesis, secondary embryogenesis and regeneration of fertile green cereal plants. Plant Cell Tissue Organ Cult 73: 147–157 [Google Scholar]
- Filipecki M, Malepszy S. (2006) Unintended consequences of plant transformation: a molecular insight. J Appl Genet 47: 277–286 [DOI] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol 51: 223–256 [DOI] [PubMed] [Google Scholar]
- Genschik P, Parmentier Y, Durr A, Marbach J, Criqui M-C, Jamet E, Fleck J. (1992) Ubiquitin genes are differentially regulated in protoplast-derived cultures of Nicotiana sylvestris and in response to various stresses. Plant Mol Biol 20: 897–910 [DOI] [PubMed] [Google Scholar]
- Gould J, Devey M, Hasegawa O, Ulian EC, Peterson G, Smith RH. (1991) Transformation of Zea mays L. using Agrobacterium tumefaciens and the shoot apex. Plant Physiol 95: 426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley N, Hohn B, Ramos C, Kado C, Rogowsky P. (1989) DNA transfer from Agrobacterium to Zea mays or Brassica by agroinfection is dependent on bacterial virulence functions. Mol Gen Genet 217: 309–316 [DOI] [PubMed] [Google Scholar]
- Gurel S, Gurel E, Kaur R, Wong J, Meng L, Tan HQ, Lemaux PG. (2009) Efficient, reproducible Agrobacterium-mediated transformation of sorghum using heat treatment of immature embryos. Plant Cell Rep 28: 429–444 [DOI] [PubMed] [Google Scholar]
- Hansen G, Chilton MD. (1996) “Agrolistic” transformation of plant cells: integration of T-strands generated in planta. Proc Natl Acad Sci USA 93: 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen G, Shillito RD, Chilton MD. (1997) T-strand integration in maize protoplasts after codelivery of a T-DNA substrate and virulence genes. Proc Natl Acad Sci USA 94: 11726–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood WA, Bartlett JG, Alves SC, Perry M, Smedley MA, Leyland N, Snape JW. (2009) Barley transformation using Agrobacterium-mediated techniques. Methods Mol Biol 478: 137–147 [DOI] [PubMed] [Google Scholar]
- Hensel G, Kastner C, Oleszczuk S, Riechen J, Kumlehn J. (2009) Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int J Plant Genomics 2009: 835608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Hiei Y, Komari T. (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2: 1614–1621 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. (1990) An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci 70: 133–140 [Google Scholar]
- Latham JR, Wilson AK, Steinbrecher RA. (2006) The mutational consequences of plant transformation. J Biomed Biotechnol 2006: 25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. (2005) Involvement of KU80 in T-DNA integration in plant cells. Proc Natl Acad Sci USA 102: 19231–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach C, Velten J. (1991) Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol 17: 49–60 [DOI] [PubMed] [Google Scholar]
- Pelczar P, Kalck V, Gomez D, Hohn B. (2004) Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic T-DNA complexes in mammalian cells. EMBO Rep 5: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Shankar V. (2001) Sugar non-specific endonucleases. FEMS Microbiol Rev 25: 583–613 [DOI] [PubMed] [Google Scholar]
- Reyes FC, Sun B, Guo H, Gruis DF, Otegui MS. (2010) Agrobacterium tumefaciens-mediated transformation of maize endosperm as a tool to study endosperm cell biology. Plant Physiol 153: 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B. (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 93: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel D. (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Slater A, Scott NW, Fowler MR. (2008) Techniques for plant transformation. In Plant Biotechnology: The Genetic Manipulation of Plants. Oxford University Press, Oxford, pp 54–76 [Google Scholar]
- Stoger E, Williams S, Christou P, Down RE, Gatehouse JA. (1999) Expression of the insecticidal lectin from snowdrop (Galanthus nivalis agglutinin; GNA) in transgenic wheat plants: effects on predation by the grain aphid Sitobion avenae. Mol Breed 5: 65–73 [Google Scholar]
- Tinland B. (1996) The integration of T-DNA into plant genomes. Trends Plant Sci 1: 178–184 [Google Scholar]
- Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel AM, Hohn B. (1995) The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J 14: 3585–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travella S, Ross SM, Harden J, Everett C, Snape JW, Harwood WA. (2005) A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep 23: 780–789 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V. (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17: 147–154 [DOI] [PubMed] [Google Scholar]
- Windels P, De Buck S, Depicker A. (2008) Agrobacterium tumefaciens-mediated transformation: patterns of T-DNA integration into the host genome. In Agrobacterium: from Biology to Biotechnology. Springer, New York, pp 442–483 [Google Scholar]
- Yokoyama H, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. (2003) Holliday junction binding activity of the human Rad51B protein. J Biol Chem 278: 2767–2772 [DOI] [PubMed] [Google Scholar]
- Ziemienowicz A, Görlich D, Lanka E, Hohn B, Rossi L. (1999) Import of DNA into mammalian nuclei by proteins originating from a plant pathogenic bacterium. Proc Natl Acad Sci USA 96: 3729–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Merkle T, Schoumacher F, Hohn B, Rossi L. (2001) Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Tzfira T, Hohn B. (2008) Mechanisms of T-DNA integration. In: Agrobacterium: from Biology to Biotechnology. Springer, New York, pp 396–441 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.