Abstract
Protein interactions are fundamental to the molecular processes occurring within an organism and can be utilized in network biology to help organize, simplify, and understand biological complexity. Currently, there are more than 10 publicly available Arabidopsis (Arabidopsis thaliana) protein interaction databases. However, there are limitations with these databases, including different types of interaction evidence, a lack of defined standards for protein identifiers, differing levels of information, and, critically, a lack of integration between them. In this paper, we present an interactive bioinformatics Web tool, ANAP (Arabidopsis Network Analysis Pipeline), which serves to effectively integrate the different data sets and maximize access to available data. ANAP has been developed for Arabidopsis protein interaction integration and network-based study to facilitate functional protein network analysis. ANAP integrates 11 Arabidopsis protein interaction databases, comprising 201,699 unique protein interaction pairs, 15,208 identifiers (including 11,931 The Arabidopsis Information Resource Arabidopsis Genome Initiative codes), 89 interaction detection methods, 73 species that interact with Arabidopsis, and 6,161 references. ANAP can be used as a knowledge base for constructing protein interaction networks based on user input and supports both direct and indirect interaction analysis. It has an intuitive graphical interface allowing easy network visualization and provides extensive detailed evidence for each interaction. In addition, ANAP displays the gene and protein annotation in the generated interactive network with links to The Arabidopsis Information Resource, the AtGenExpress Visualization Tool, the Arabidopsis 1,001 Genomes GBrowse, the Protein Knowledgebase, the Kyoto Encyclopedia of Genes and Genomes, and the Ensembl Genome Browser to significantly aid functional network analysis. The tool is available open access at http://gmdd.shgmo.org/Computational-Biology/ANAP.
Protein interaction networks can provide a global view of cellular processes, thus facilitating the study of complex, dynamic biological systems (Jansen et al., 2003). Interactions between proteins can be direct physical interactions and also indirect, which may involve intermediate molecules to facilitate interactions. For example, an indirect interaction means that if proteins A and B, and also B and C, have direct interactions, then A and C indirectly interact. These interactions are key to cellular events associated with protein localization, translation rates, gene regulation, and posttranslational modifications (Bork et al., 2004). The development of full-genome- and proteomics-based technologies, such as next-generation sequencing, transcriptomics, and high-throughput yeast two-hybrid screening, has generated huge amounts of biological data. To capitalize upon these data for functional biological studies, this information needs to be analyzed, effectively integrated, and stored to facilitate rapid searching and in-depth analysis.
There have been a number of model organisms for which large-scale protein interaction data sets have been generated, which include Saccharomyces cerevisiae (Schwikowski et al., 2000; Uetz et al., 2000), Drosophilia melanogaster (Giot et al., 2003), Caenorhabditis elegans (Li et al., 2004), and the human protein interactome (Rual et al., 2005). These data sets, and many others, have increased the amount of available protein interaction data hugely over the past 10 years, but currently, they are all collated into different protein interaction databases (Arabidopsis Interactome Mapping Consortium, 2011). To date, a significant amount of protein interaction data have been generated for Arabidopsis (Arabidopsis thaliana); however, this has been produced using a range of methods. These data sets are stored in a variety of databases, including Agile Protein Interaction DataAnalyzer (APID; Prieto and De Las Rivas, 2006), Arabidopsis thaliana Protein Interactome Database (AtPID; Cui et al., 2008), Arabidopsis thaliana Protein Interaction Network (AtPIN; Brandão et al., 2009), the Biomolecular Interaction Network Database (BIND; Bader et al., 2003), Biological General Repository for Interaction Datasets (BioGRID; Stark et al., 2006, 2011), ChEMBL (Overington, 2009), The Database of Interacting Proteins (DIP; Xenarios et al., 2000, 2001, 2002), IntAct (Aranda et al., 2010), InteroPORC (Michaut et al., 2008), iRefIndex (Razick et al., 2008), The Molecular INTeraction database (MINT; Ceol et al., 2010), MolCon (http://www.ebi.ac.uk/Tools/webservices/psicquic/view/main.xhtml), and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; Jensen et al., 2009; Szklarczyk et al., 2011).
Currently, it is not easy to directly access these data to integrate information from different sources and methodologies to provide biological network information. Fortunately, an excellent recent resource, PSICQUIC (for Protemics Standard Initiative Common QUery InterfaCe; Aranda et al., 2010), has provided an interface for protein interaction databases to allow easy access to these data. The main goal of the PSICQUIC project is to provide a common query interface and implement data quality assessment from these disparate databases; this is now being successfully used for many projects, including Cytoscape, IntAct, and Reactome (http://code.google.com/p/psicquic/wiki/WhoUsesPsicquic).
There are a number of bioinformatics tools, such as ATTED (Obayashi et al., 2007), that utilize coexpression data for network analysis; however, these have their limitations, since they are based upon transcript levels and do not utilize protein data. One of the initial network analysis tools for visualizing the Arabidopsis interactome was the Arabidopsis Interaction Viewer (Geisler-Lee et al., 2007). The Arabidopsis Interaction Viewer currently contains nearly 99,466 Arabidopsis interacting proteins, which were collected from BIND, MINT, literature sources such as Arabidopsis Interactome Mapping (Arabidopsis Interactome Mapping Consortium, 2011), and some predictions generated by the authors. The Arabidopsis thaliana Protein Interaction Network also offers an online tool that integrates some of the available Arabidopsis protein interaction databases, including the Predicted Interactome for Arabidopsis (Geisler-Lee et al., 2007), Arabidopsis protein-protein interaction data curated from The Arabidopsis Information Resource (TAIR) curators (http://www.arabidopsis.org/index.jsp), BioGRID (Stark et al., 2006, 2011), and IntAct (Aranda et al., 2010).
There are many variables that have to be addressed to facilitate data integration between the large numbers of available protein interaction data sets. These include data standards, the use of single types of protein identifiers, and well-defined ontology terms. A large amount of these data are generated from different sources with no shared database design, many with no clearly defined standards and the use of different identifiers. Therefore, it is vital to develop a set of definitive standards for the collection, integration, and analysis of protein interaction data to enable the establishment of networks that utilize data from both small-scale experiments and high-throughput approaches. This is particularly important since, if interactions have been demonstrated by multiple approaches, it provides a greater validity and robustness to the network.
To address these issues and to facilitate effective protein interaction network construction, we have developed an interactive bioinformatics Web tool entitled the Arabidopsis Network Analysis Pipeline (ANAP) for Arabidopsis network analysis. The main aims of ANAP are to integrate the currently available Arabidopsis protein interaction data sets and to provide biologists with a novel, easy-to-use, and intuitive interface that enables researchers to carry out high-throughput detailed network analysis with limited bioinformatics experience. Protein interaction data sets were integrated and formatted from 11 public Arabidopsis protein interaction databases. At publication, ANAP contained 201,699 unique protein interaction pairs, comprising 15,208 identifiers (include 11,931 TAIR Arabidopsis Genome Initiative [AGI] codes) with 89 interaction detection methods, 73 proteins from different species that interact with Arabidopsis proteins, and 6,161 references (Table I). This provides an extensive and valuable knowledge base for generating protein interaction networks from the integrated data sets, thus producing a far more detailed and reliable network than if produced from any single protein interaction database.
Table I. Statistics of the integrated ANAP protein interaction source data.
| Category | No. |
| Protein interaction databases | 11 |
| Species interacting with Arabidopsis | 73 |
| Interaction detection methods | 89 |
| References | 6,161 |
| Unique TAIR AGI codes | 11,931 |
| Unique molecules | 15,208 |
| Protein interaction pairs | 201,699 |
ANAP allows for either single or multiple protein searches to be conducted for each query protein. The networks generated display the various interaction detection methods and data sources in unique colors to enable effective network viewing. There are additional functions available to conduct “in-depth” protein searches, which identify the indirect interactions from the original input source protein. This is very important, as a network, or a protein interaction complex, may include indirect interactions to many other proteins. This type of approach has previously been shown to be a very useful way to recognize new interactions within a complex (Jensen et al., 2009). Each protein in the network is described using its TAIR AGI code, UniProt identifier (ID), and a short description; additionally, the full TAIR locus details can be viewed by double clicking on the protein. Direct links to five popular Arabidopsis resources (AtGenExpress Visualization Tool, Arabidopsis 1,001 Genomes GBrowse, Protein Knowledgebase, Kyoto Encyclopedia of Genes and Genomes, and Ensembl Genome Browser) are also provided. The detailed evidence of the network and each interaction can be saved in various file formats, including PNG, PDF, SVG, SIF, GRAPHML, and XGMML. The file formats SIF, GRAPHML, and XGMML are particularly useful for large networks where the user wishes to import the resulting ANAP network into Cytoscape, which is a well-established network analysis tool (Shannon et al., 2003; Kohl et al., 2011; Smoot et al., 2011). ANAP also supports the import of the resulting network into other network analysis tools, such as Network Workbench (GRAPHML; NWB Team, 2006). ANAP is a fully functional integration and analysis pipeline that will serve as an extremely valuable resource for biologists. It will enable them to capitalize upon the currently available Arabidopsis protein interaction data for effective network-based analysis, enabling greater predictions of function and selection of targets for further biological analysis.
RESULTS
ANAP Framework and Searches
The ANAP tool has been developed to integrate the available Arabidopsis protein interaction data that have been generated from different sources by a variety of approaches. These data are then used to generate accurate protein interaction networks, which will facilitate greater understanding of biological processes. ANAP can be used as a platform to construct protein interaction networks based on both direct and indirect interaction analysis.
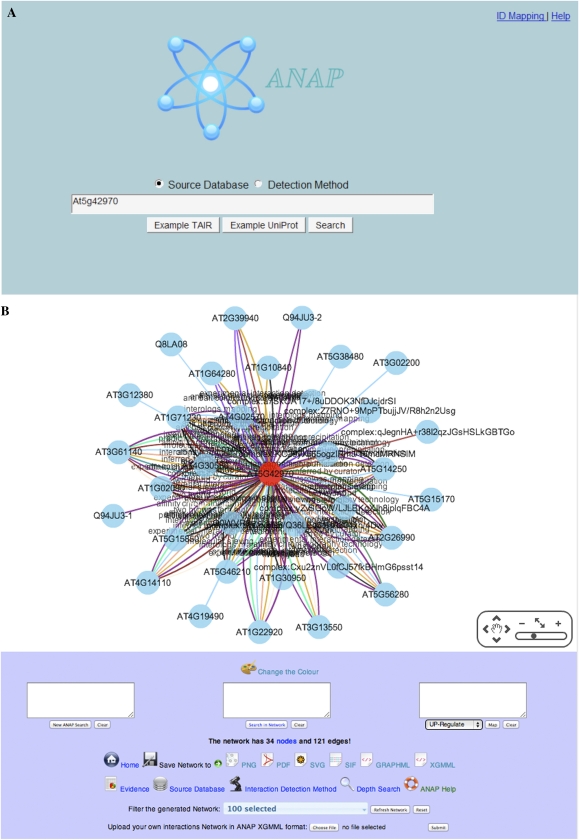
ANAP has an intuitive graphical user interface that allows the user to easily construct molecular networks using single or multiple starting proteins as inputs; the results are displayed showing the proteins that interact with the initial query protein(s). Figure 1A shows the ANAP tool interface, which includes ID Mapping and a Help link. The user enters the Arabidopsis TAIR AGI code(s) or the protein UniProt ID(s) into the central search box, with the option of selecting two types of node relationship: “Source Database” and “Interaction Detection Method.” The selection of node relationship does not affect the overall network that is generated but rather the presentation of the links between the nodes; Source Database lists the database information used to generate the links, while Interaction Detection Method presents the experimental technique that has been used to generate the relationship.
Figure 1.
(Legend appears on following page.)
Figure 1B shows the whole framework of the ANAP output, which includes several useful functions to enable the user to easily extract extensive information from the resultant network. This framework includes the network map in the center of the main panel, a “Change the Color” button underneath, network information about numbers of nodes and interactions, and a panel for searching and mapping data onto the network. There is a panel for saving the resultant network, another panel for useful information, which includes links to the supporting evidence for the interactions, and a Simple Interaction Format (SIF; Cytoscape format) file containing the Source Database and Interaction Detection Method. This panel also contains a “Depth Search” button, which supports the indirect interaction search option. Moreover, there are another two panels at the bottom of the framework, one is “network filtering,” which is useful for simplifying the output of a complex network and allows users to toggle between different databases and different interaction detection methods to generate networks; the other is “upload network,” which is useful for reanalyzing the generated network and making the input nodes remain in their original positions.
Single Protein Searches
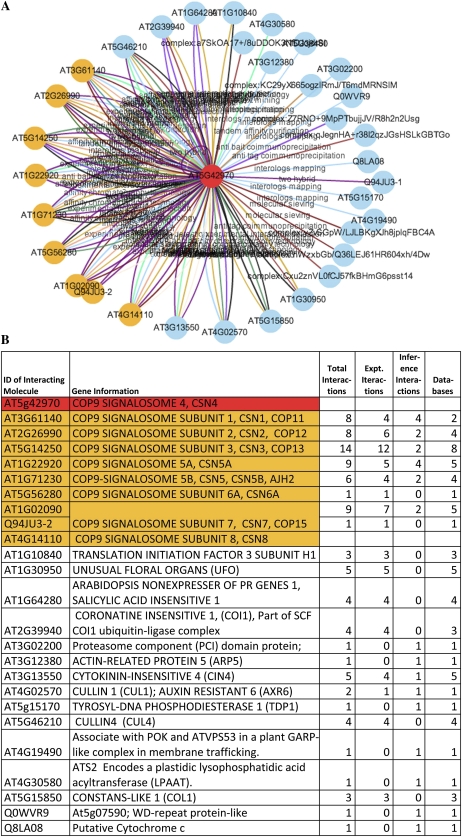
The locus AT5G42970 (which encodes subunit 4 of the COP9 signalosome [CSN] complex) was used as an example for analysis using the ANAP tool. Figure 2 shows the resulting network of 34 nodes and 130 edges, based on the direct protein interactions generated after selecting the option of Interaction Detection Method. The query protein AT5G42970 is marked in red in the center of the figure, and each associated protein is linked by a uniquely colored line, based on the interaction detection method and the rendering rules from the complete list of all interaction detection methods (Supplemental Data Set S1).
Figure 2.
Network generated using the COP9 signalosome protein (AT5G42970). A, Network map based on direct protein interactions and the node relationship of the Interaction Detection Method. The query protein is marked in red, and each interaction detection method is indicated by a unique linking colored line. B, Table generated from the “evidence” from the COP9 interaction network. The table shows the numbers of interactions detected by ANAP, which of these are from experimental data and from inference-based approaches, and the total number of databases supporting the interactions. Members of the COP9 signalosome are shown in orange in both A and B.
The CSN is a highly conserved protein complex that is associated with the ubiqutin-proteolytic breakdown pathway. In eukaryotes, it is formed of nine subunits, of which subunit 4 (AT5G42970) is one member (Schwechheimer and Isono, 2010). Searching ANAP with AT5G42970 identified 34 nodes; the gene identities and functions of these are shown in Figure 2B. The nine components of the CSN (COP9 subunit 4 and eight others) were all identified and are highlighted in orange (Fig. 2). To construct the network, ANAP has utilized data from multiple sources, comprising both predicted interactions and experimental evidence; the numbers of each for these proteins are shown in Figure 2B. This wide range of data provides valuable support for any interactions; for example, a large number of interactions were seen between COP9 subunit 4 and the other well-established components of the CSN. The other proteins that have been identified in the network range from those with established roles in ubiquitination pathways (Schwechheimer and Isono, 2010) to other developmental processes that are regulated by ubiquitin proteolysis. It is also very easy to go directly from the identified proteins to PubMed sources to aid in characterizing the network and further interrogate the validity of the predicted parts of the networks. The multiple sources of data accessed by ANAP offer excellent opportunities to confirm known networks but also to extend these further to identify novel targets. The range and depth of the data utilized for network generation, therefore, provide a valuable mechanism to assess the validity of such predictions prior to follow-up experimental analysis.
Table II shows an example of five evidence records generated when searching using AT5G42970 based on direct protein interactions. In addition, Supplemental Data Set S2 lists the complete relevant evidence records for the AT5G42970 protein network. The user can dynamically interact with the network by using the mouse-over function on the nodes; this shows the protein’s UniProt ID, TAIR AGI code, and a short description of the predicted protein function. Additionally, there are links to the relevant locus details, which are visible when the node is double clicked. Moreover, each node in the network has a direct link to the AtGenExpress Visualization Tool, the Arabidopsis 1,001 Genomes GBrowse, the Protein Knowledgebase, the Kyoto Encyclopedia of Genes and Genomes, and the Ensembl Genome Browser. A similar feature is also seen with the edges in the network, which highlight the interaction method when the mouse hovers over each edge. ANAP provides the opportunity for the user to select a node(s) of interest in the resultant network and to use this to construct a new network and extract the evidence data. Furthermore, users can also search for specific protein(s) in the resultant network; such proteins are marked in blue when in the resultant network and marked in fuchsia when it is the same as the query protein(s). Using AT5G42970, a network was constructed based on the same configuration as the network in Figure 2 by selecting the option of Source Database (Supplemental Fig. S1). The edges of each source database are indicated by a unique color based on the rendering rules for the complete list of all source databases (Supplemental Data Set S1).
Table II. Five evidence records generated when searching ANAP using protein AT5G42970 based on direct protein interactions.
Ath, Arabidopsis.
| Name Molecule A | Name Molecule B | Interaction Detection Method | Species Molecule A | Species Molecule B | PubMed Identifier | Source Database |
| AT1G02090 | AT5G42970 | Two hybrid | Ath | Ath | 12615944 | IntAct |
| AT1G10840 | AT5G42970 | Affinity chromatography technology | Ath | Ath | 15548739 | BioGRID |
| AT1G22920 | AT5G42970 | Predictive text mining | Ath | Ath | 10521526 | STRING |
| AT1G30950 | AT5G42970 | Anti-tag coimmunoprecipitation | Ath | Ath | 12724534 | APID |
| AT4G19490 | AT5G42970 | Interolog mapping | Ath | Ath | 18508856 | InteroPORC |
There is also an added feature that allows users to easily identify the indirect interactions of the original protein using the “Depth Search” button. Supplemental Figure S2 shows the network constructed based on the indirect protein interaction data generated for AT5G42970. This approach is useful for recognizing new potential interactions in the network (Jansen et al., 2003) to assign putative functions to less well-characterized proteins and to provide more comprehensive understanding of the query protein at the system level with the help of each cluster in the constructed network.
Multiple Protein Searches
Currently, more and more researchers are employing transcriptomic, next-generation sequencing and many high-throughput technologies in the fields of molecular, cell, and developmental biology to decipher novel biological phenomena. By using bioinformatics-based approaches, lists of key genes can be further classified to confirm candidates by biological experimentation. However, for such gene selection and functional analysis to be effective, particularly at a protein level, these data sets require supplementation and detailed analysis. Therefore, it is critical to produce protein interaction networks using multiple proteins as a way of visualizing and analyzing all the interactions simultaneously to aid in functional analysis. ANAP supports such multiple protein searches and protein interaction network construction, so that users can submit targets as TAIR AGI code, UniProt ID, or a combination of these identifiers into the ANAP tool. Such networks, therefore, provide valuable information establishing links between proteins, which are likely to represent functional and regulatory conservation.
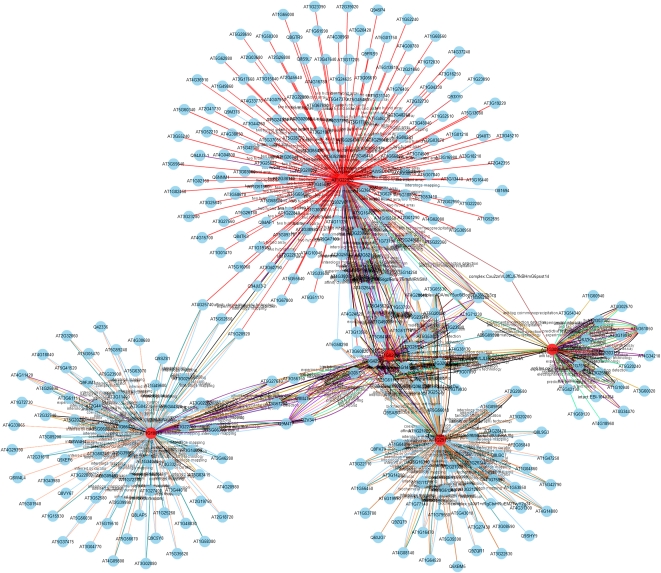
Figure 3 shows the network generated by searching using five proteins (AT1G02090, AT1G10840, AT1G22920, AT1G29150, and AT1G30950) from the AT5G42970 (COP9 signalosome complex) ANAP interaction network. This was constructed based on direct protein interactions, with the option of selecting based on Interaction Detection Method. Each of the query proteins is marked as a red node, and each interaction detection method is allocated a unique color. Several clusters from each query protein can be easily recognized within the network graph (Fig. 3).
Figure 3.
Network result generated by using a multiple protein search (five proteins searched: AT1G02090, AT1G10840, AT1G22920, AT1G29150, and AT1G30950) based on direct protein interactions and the node relationship Interaction Detection Method. Each query protein is marked in red, and each interaction detection method is indicated by a uniquely colored line. Several node clusters can be identified, with each query protein evident in the network graph. [See online article for color version of this figure.]
DISCUSSION
The Current Challenges of Integrating Protein Interaction Networks
Protein interaction networks can give a system-level view that is vital for the detailed analysis of complex biological systems (Jansen et al., 2003). However, providing mechanisms to integrate protein interaction data that have been generated from various sources poses significant challenges. For instance, two proteins may only interact during a certain developmental stage and/or in a specific tissue; however, most of the currently available protein interaction data do not provide temporal or spatial specificity. Furthermore, these data sets have frequently been generated in ectopic expression systems and thus may not represent the genuine interactions occurring in vivo. These limitations reduce the accuracy of the established networks, although such problems can be lessened by the successful integration of the increasing amounts of protein interaction data that have been generated by different approaches. The importance of data integration is now being fully appreciated, and there is a general emphasis toward the development of standards for large data sets with defined specific formats, which include PSI-MI (Kaiser, 2002) for protein interactions and BioPAX (Demir et al., 2010) and SBML (Hucka et al., 2003) for pathway standards. Several other approaches utilize controlled vocabularies with a defined glossary of terms for types of interactions (Côté et al., 2006) and the use of a specific protein identifiers, which are constant in all the available protein interaction databases to facilitate easier integration. There is also a need for these same standards to be established in published scientific journals to further enhance the effectiveness of text mining to supplement the ANAP integrated protein interaction data set.
Interaction with Other Resources
Defining protein function is an essential requirement for effective, functional network characterization. Moreover, recent studies have shown that protein interaction networks are able to give a good prediction of protein function (Jansen et al., 2003; Sharan et al., 2007). Therefore, bridging target genes from transcriptomic data, or next-generation sequencing data, with the help of Gene Ontology term enrichment to the protein interaction network can provide added substance for network characterization (Maere et al., 2005).
Currently, ANAP provides the function of mapping up- and down-regulated transcriptomic data, next-generation sequencing data, and other biology-based results onto the generated network. For transcriptomic mapping, the nodes are colored in red or green, which represent the up- or down-regulated genes in the network (Supplemental Fig. S3). The node can also be highlighted in blue if customized gene list data (any interesting data that users want to overlay onto the ANAP network) are mapped onto the network nodes. This makes the ANAP tool very flexible for the user to identify specific proteins and transcriptomic regulatory relationships within the network. The node is colored in fuchsia or turquoise if the mapped customized data also exist in the up- or down-regulated transcriptomics data. Moreover, ANAP provides another seven colors (olive, orange, purple, yellow, maroon, navy, and teal) in the mapping function for users to integrate data such as different subcellular localizations or other biology-based data, rather than only using this to indicate differing expression levels. Another strength of the ANAP tool is the ability for the user to be able to import the resultant networks into Cytoscape and other software for subsequent additional analysis (Shannon et al., 2003; Kohl et al., 2011; Smoot et al., 2011). The user can import the SIF, GRAPHML, or XGMML file generated by ANAP into Cytoscape. The Cytoscape mapping functions can then be used to integrate different resources and plugins for analyzing existing networks, inferring new networks, functional enrichment of networks, etc. This tool also supports import into other network analysis tools, such as Network Workbench (GRAPHML; NWB, 2006).
CONCLUSION
In this paper, the Web-based ANAP tool has been designed and implemented for Arabidopsis protein interaction network analysis. ANAP currently integrates approximately 201,699 unique protein interaction pairs into a tool that has a well-designed, simple-to-use, intuitive interface for biologists that can be exported to Cytoscape. Thus, it can be widely used for Arabidopsis protein interaction network construction and analysis. This is particularly valuable where large numbers of genes of interest have been selected from microarray and next-generation sequencing experiments and where only limited information is known. Case studies using single protein searches and multiple protein searches from the COP9 signalosome complex (Figs. 2 and 3; Supplemental Figs. S1 and S2) and the cytokinin regulatory pathway (ANAP user guide; Supplemental Fig. S3) have demonstrated the consistently good performance of ANAP for Arabidopsis protein interaction network analysis. The current ANAP framework provides a novel, intuitive, and easy-to-interpret tool that will greatly aid biologists in understanding plant developmental networks, which will allow them to decipher their specific biological network interactions far more quickly than by using biological techniques alone. Furthermore, ANAP has been designed to easily add features for extending functionality as the tool develops. Future work is planned to extend this tool to integrate the protein interaction data with metabolic pathway data, gene coexpression data, and other types of interactions to decipher biological problems more effectively.
MATERIALS AND METHODS
Data Sets
Arabidopsis (Arabidopsis thaliana) protein interaction data sets integrated into ANAP include APID (8,014 pairs; Prieto and De Las Rivas, 2006), BIND (1,545 pairs; Bader et al., 2003), BioGRID (5,862 pairs; Stark et al., 2006, 2011), ChEMBL (54 pairs; Overington, 2009), DIP (403 pairs; Xenarios et al., 2000, 2001, 2002), IntAct (16,286 pairs; Aranda et al., 2010), InteroPORC (14,722 pairs; Michaut et al., 2008), iRefIndex (18,362 pairs; Razick et al., 2008), MINT (499 pairs; Ceol et al., 2010), MolCon (116 pairs; Aranda et al., 2010), and STRING (21,5358 pairs; Jensen et al., 2009; Szklarczyk et al., 2011). All these data were collected from the PSICQUIC Registry of the European Bioinformatics Institute (Aranda et al., 2010), which is an accurate and frequently used resource for many projects, such as Bio::Homology::InterologWalk, Cytoscape, EnVision 2, IMEx Consortium, IntAct, Reactome, Taverna, etc. (http://code.google.com/p/psicquic/wiki/WhoUsesPsicquic).
In a recent ANAP update, we also integrated 5,664 confirmed binary interactions between 2,661 proteins from the Arabidopsis Interactome Mapping Consortium (2011), which is a recently published high-throughput Arabidopsis yeast two-hybrid data set.
Access Availability
ANAP is implemented in HTML, Shell, AWK, PHP, and JavaScript with the support of the Cytoscape Web, which allows the developer to embed dynamic networks into HTML (Lopes et al., 2010). The tool is open access for any use and available at http://gmdd.shgmo.org/Computational-Biology/ANAP. The top right corner of the index page includes a Help link, which is very useful to new users. The Help page contains a “Video Tutorial,” “Frequently Asked Questions,” and a “User Guide.” If users have questions regarding using ANAP or some problems in understanding the terms or concepts, please refer to the Help page.
Generally, ANAP is updated with new interaction data every 3 months; however, we have developed a semiautomatic formatting and updating program for ANAP. This has been rigorously tested with random access checks and manual checks to ensure stable and accurate integration of new data. In addition, we have established a log analysis tool to analyze access to ANAP.
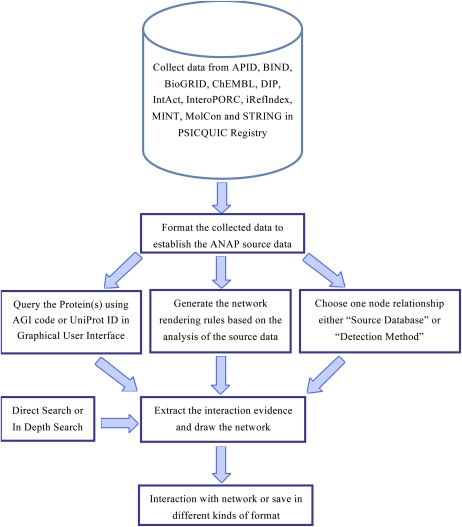
Flow Chart of the ANAP Tool
The main modules in ANAP connect together data collection, data integration, and network viewing. The architecture of the ANAP pipeline is shown in the flow chart in Figure 4. We first searched the Arabidopsis protein interaction data based on the mnemonic (ARATH), taxon identifier (3702), scientific name (Arabidopsis thaliana), common name (Mouse-ear cress) and other names [Arabidopsis thaliana (L.) Heynh., Arabidopsis thaliana (thale cress), Arabidopsis thaliana, thale cress, and thale-cress] used in current protein interaction databases. The collected protein interaction data were then formatted to establish the ANAP database source (Supplemental Data Set S3); the graphical user interface was designed to support querying the protein(s) using a TAIR AGI code or UniProt ID. At the same time, the network rendering rules (Supplemental Data Set S1), based on the statistical analysis of the source data, were generated. The option is provided to select Source Database and Interaction Detection Method for the user to choose the desired node relationship. ANAP then produces the resultant network and extracts the interaction evidence. In addition, ANAP generates query keywords that extract the connecting proteins for in-depth searching. Finally, users can interact with the network and save it in different formats, including network maps or as network data.
Figure 4.
Flow chart of the ANAP tool. The main modules in ANAP include data collection, data integration, and network visualization. Details of each module are described in the text. [See online article for color version of this figure.]
Protein Interaction Data Format
A protein interaction data format was designed to convert and then integrate the 11 Arabidopsis data sets. Initially, there were numerous issues associated with independently integrating the protein interaction data from the 11 different databases. Different programs were written to collect and format each database; however, these posed problems for subsequent automated continuous updates. In addition, each database had a different data access method, which meant that integrating all the Arabidopsis data was difficult. Furthermore, each database contained very different formats for the interaction evidence. We found an excellent recent resource named PSICQUIC (Aranda et al., 2010), which had integrated raw data from about 22 protein interaction databases. However, after searching and checking extensive random data from each of the 11 Arabidopsis available databases, we found that they did not format well (Supplemental Data Set S3), which made it unsuitable for protein interaction network-based analysis. Taking Interaction Detection Method, for example, the raw PSICQUIC data have 149 unique methods while the formatted ANAP has 89 unique methods, and among these 149 unique methods, many, such as “two hybrid,” “2 hybrid,” and “two-hybrid-test” have the same ontology code, “MI:0018.”
Furthermore, the raw data collected from PSICQUIC have 17 fields (Aranda et al., 2010), but most of them are blank, including “Links Molecule A,” “Links Molecule B,” “Alt. Identifiers Molecule A,” “Alt. Identifiers Molecule B,” “Interaction Type,” ” Interaction AC,” and “Confidence Value.”
Based on the protein interaction network analysis and the current proteomics standard PSI-MI (Kaiser, 2002), the following seven fields were created: “Name Molecule A,” “Name Molecule B,” “Interaction Detection Method,” “PubMed Identifier,” “Species Molecule A,” “Species Molecule B,” and “Source Database” (Table II).
Name Molecule A and Name Molecule B represent the TAIR AGI code, protein complex name, or small molecule name. Interaction Detection Method represents the methods used to support the protein interactions, such as yeast two-hybrid, coimmunoprecipitation, or electrophoretic mobility shift assays, etc. If the interaction has been published, then the PubMed Identifier will also be provided. Species Molecule A and Species Molecule B describe the species name of two molecules, since some Arabidopsis proteins may interact with proteins from other species. Source Database describes which database contains the interaction data. Since all integrated protein interaction data, including TAIR, UniProt, GeneID, RefSeq, and Gene Symbol, etc., use different protein names, it was necessary to convert all these IDs to the accepted standard TAIR AGI code for the network-based analysis. Standard mapping conversions to AGI can be carried out using a variety of tools, including DAVID (Dennis et al., 2003; Huang et al., 2009), UniProt ID Mapping (Jain et al., 2009), and bioDBnet (Mudunuri et al., 2009), but there are issues. When using the UniProt ID Mapping tool, it autofilters the repetitive IDs and ignores the IDs that cannot be converted to TAIR IDs, which is not suitable to format a large amount of data. Also, DAVID is not well supported for the plant community, whereas bioDBnet offers the best method to covert IDs to the TAIR Arabidopsis AGI code. However, the conversion data were not always current with bioDBnet. Based on these findings, a combined approach was employed that first used bioDBnet and subsequently converted the unmapped IDs using data downloaded from the newest different identifier annotations from UniProt and the National Center for Biotechnology Information.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ANAP network result for searching using protein AT5G42970 based on direct protein interactions and the node relationship for the Source Database.
Supplemental Figure S2. Network result of searching protein AT5G42970 based on the indirect protein interactions and the node relationship of the Interaction Detection Method.
Supplemental Figure S3. ANAP network mapping of the Cytokinin microarray data set.
Supplemental Data Set S1. Color legend showing the interaction detection methods and the database source for the ANAP data and networks.
Supplemental Data Set S2. Evidence list from the network result generated by searching using protein AT5G42970, based on direct protein interactions and the node relationship of the Interaction Detection Method.
Supplemental Data Set S3. Summary of the PSICQUIC data format, including the summary of PSICQUIC raw data and a summary of the formatted ANAP data, Interaction Detection Method, Ontology Code, Species Molecule A and B, and taxonomic ID used in the formatted ANAP data.
Supplementary Material
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. (2010) The IntAct molecular interaction database in 2010. Nucleic Acids Res 38: D525–D531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res 31: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Jensen LJ, von Mering C, Ramani AK, Lee I, Marcotte EM. (2004) Protein interaction networks from yeast to human. Curr Opin Struct Biol 14: 292–299 [DOI] [PubMed] [Google Scholar]
- Brandão MM, Dantas LL, Silva-Filho MC. (2009) AtPIN: Arabidopsis thaliana Protein Interaction Network. BMC Bioinformatics 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. (2010) MINT, the Molecular Interaction Database: 2009 update. Nucleic Acids Res 38: D532–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté RG, Jones P, Apweiler R, Hermjakob H. (2006) The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li P, Li G, Xu F, Zhao C, Li Y, Yang Z, Wang G, Yu Q, Li Y, et al. (2008) AtPID: Arabidopsis thaliana Protein Interactome Database—an integrative platform for plant systems biology. Nucleic Acids Res 36: D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, et al. (2010) The BioPAX community standard for pathway data sharing. Nat Biotechnol 28: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. (April 3, 2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: http://dx.doi.org/10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. (2007) A predicted interactome for Arabidopsis. Plant Physiol 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. (2003) A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science 302: 449–453 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. (2009) STRING 8: a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. (2002) Proteomics: public-private group maps out initiatives. Science 296: 827. [DOI] [PubMed] [Google Scholar]
- Kohl M, Wiese S, Warscheid B. (2011) Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 696: 291–303 [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. (2010) Cytoscape Web: an interactive Web-based network browser. Bioinformatics 26: 2347–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Michaut M, Kerrien S, Montecchi-Palazzi L, Chauvat F, Cassier-Chauvat C, Aude JC, Legrain P, Hermjakob H. (2008) InteroPORC: automated inference of highly conserved protein interaction networks. Bioinformatics 24: 1625–1631 [DOI] [PubMed] [Google Scholar]
- Mudunuri U, Che A, Yi M, Stephens RM. (2009) bioDBnet: the biological database network. Bioinformatics 25: 555–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NWB Team (2006) Network Workbench Tool. Indiana University, Northeastern University, and University of Michigan. http://nwb.slis.indiana.edu [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington J. (2009) ChEMBL: an interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J Comput Aided Mol Des 23: 195–198 [DOI] [PubMed] [Google Scholar]
- Prieto C, De Las Rivas J. (2006) APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res 34: W298–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. (2008) iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Isono E. (2010) The COP9 signalosome and its role in plant development. Eur J Cell Biol 89: 157–162 [DOI] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. (2000) A network of protein-protein interactions in yeast. Nat Biotechnol 18: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Ulitsky I, Shamir R. (2007) Network-based prediction of protein function. Mol Syst Biol 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. (2011) The BioGRID Interaction Database: 2011 update. Nucleic Acids Res 39: D698–D704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Xenarios I, Fernandez E, Salwinski L, Duan XJ, Thompson MJ, Marcotte EM, Eisenberg D. (2001) DIP: the Database of Interacting Proteins. 2001 update. Nucleic Acids Res 29: 239–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. (2000) DIP: the database of interacting proteins. Nucleic Acids Res 28: 289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwínski L, Duan XJ, Higney P, Kim SM, Eisenberg D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30: 303–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.