Abstract
As our understanding of the dynamics of lipid droplets (LDs) in animal, plant, and fungal cells is rapidly evolving, still little is known about the formation and turnover of these organelles in microalgae. Yet with the growing importance of algal feedstock for the production of biofuels and high-value lipids, there is a need to understand the mechanisms of LD dynamics in microalgae. Thus, we investigated the proteins associated with LDs of the emerging heterokont model alga Nannochloropsis sp. and discovered an abundant hydrophobic lipid droplet surface protein (LDSP) with unique primary sequence but structural similarities to other LD proteins. LDSP abundance in Nannochloropsis cells closely tracked the amount of triacylglycerols during conditions of oil accumulation and degradation. Functional characterization of LDSP in an Arabidopsis (Arabidopsis thaliana) OLEOSIN1-deficient mutant allowed a separation of its physical and structural properties in its interaction with LDs from its physiological or biochemical activities. Although LDSP presence in Arabidopsis predictably affected LD size, it could not reverse the physiological impact of OLEOSIN deficiency on triacylglycerol hydrolysis during germination.
Nannochloropsis sp. is a marine microalga rich in triacylglycerols (TAGs). It belongs to the class of Eustigmatophytes, which includes freshwater and marine unicellular coccoid photosynthetic organisms. Eustigmatophytes are grouped with economically and ecologically important classes like oomycetes, diatoms, and brown algae into the heterokont division (van den Hoek et al., 1995). Species of the genus Nannochloropsis have come into focus as a potential feedstock for biofuels (Hu et al., 2008; Rodolfi et al., 2009), particularly in view of their potential amenability to molecular engineering (Kilian et al., 2011). Nannochloropsis relies on TAGs as its main energy storage compound that accumulates in periods of illumination and is rapidly turned over in the dark (Sukenik and Carmeli, 1990). This naturally high TAG content is further increased during stress conditions, such as high light or salinity, but especially nitrogen (N) deprivation increases the lipid content to 50% of the dry weight of the cells with high productivity (Rodolfi et al., 2009; Pal et al., 2011). This TAG is stored in cytosolic lipid droplets (LDs), which are always present in the cells but vary in size and number (Sheehan et al., 1998).
Research efforts over the last couple of decades have changed the perception of LDs from static, energy-dense particles to dynamic organelles found across kingdoms (Beller et al., 2010a; Fujimoto and Parton, 2011; Murphy, 2001). But independent of the respective phylogeny, all LDs share common features: a hydrophobic core typically comprised of TAGs or sterols surrounded by a monolayer of polar glycerolipids into which LD-associated proteins are embedded. Despite these similarities, LDs are versatile organelles that vary in size, shape, and function depending on the cell type.
Typically, in cells of animals and fungi, LDs are coated by proteins of the perilipin (Plin) family (Kimmel et al., 2010). Presently, five different types of Plins have been described (Brasaemle, 2007). Their abundance is tissue specific and affects the abundance of TAGs (Brasaemle, 2007; Beller et al., 2010b). Plin proteins play crucial roles in regulating TAG hydrolysis by protein-protein interactions (Subramanian et al., 2004; Miyoshi et al., 2006; Wang et al., 2009), suggesting physiological and regulatory functions beyond mere structural roles.
Plin-like proteins are not found in plants. Instead, the most abundant LD proteins belong to the oleosin and caleosin families. Oleosins likely occur in all seed plants, but in desiccation-tolerant oil seeds, they are particularly abundant and can represent up to 10% of the cellular protein content (Murphy, 2001). They play a critical role in determining the size and stability of LDs in Arabidopsis (Arabidopsis thaliana) seed (Siloto et al., 2006; Shimada et al., 2008). All oleosins share a characteristic domain of 70 to 80 hydrophobic and nonpolar amino acids immersed into the hydrophobic TAG core (Li et al., 2002; Abell et al., 1997, 2002, 2004; Pons et al., 2009). In the center of this hydrophobic stretch lies a conserved motif containing three Pro residues that are crucial for the localization of oleosins to LDs (Abell et al., 1997). This motif is also found in caleosins, which in contrast to oleosins contain a short hydrophobic stretch of about 30 amino acids. Whereas oleosins are restricted to seed and root cells (Naested et al., 2000), caleosins occur in almost all plant tissues and orthologs are present in algae and fungi.
Oleosins are not present in green algae, but a major LD protein was identified applying proteomics in Chlamydomonas and shown to modulate LD size (Moellering and Benning, 2010). Orthologs of the major LD protein are present in other green algae (Peled et al., 2011). A wide variety of parameters affects the abundance of LDs in Chlamydomonas (Kropat et al., 2011), and there is ample evidence that turnover of LDs plays crucial roles in cellular lipid or carbon homeostasis.
Here, we present the identification and functional analysis of a heterokont LD protein from Nannochloropsis showing partial restoration of the wild-type phenotype when present in the Arabidopsis OLEOSIN1-deficient mutant oleo1.
RESULTS
LDSP Is the Predominant LD-Associated Protein in N-Deprived Cells
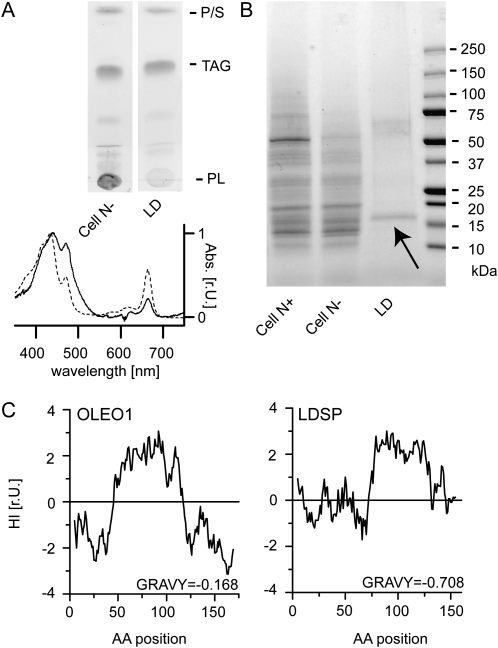
The purified LD fractions from N-deprived Nannochloropsis cells were highly enriched in TAG and showed an accumulation of carotenoids and depletion of chlorophyll as is typical for LDs of other algae (Moellering and Benning, 2010; Peled et al., 2011; Fig. 1A). The protein fraction of LDs showed a characteristic pattern when analyzed by SDS-PAGE (Fig. 1B), with one predominant protein band at approximately 17 kD indicated with an arrow in Figure 1B. Mass spectrometry was employed, and the data were interpreted by comparing spectral data to a Nannochloropsis protein database translated from open reading frames obtained by genomic and RNA sequencing. A 16.8-kD protein was identified, supported by six unique peptides that cover 44% of the predicted amino acid sequence, which was the most abundant protein in the sample based on spectral counts (Supplemental Tables S1 and S2). This protein was tentatively designated as a lipid droplet surface protein (LDSP). Sequence analysis of LDSP using BLASTP (Altschul et al., 1997) revealed no known orthologs. However, the strong enrichment of LDSP in the LD-associated protein fraction, its size, and its hydrophobicity resemble properties of the plant seed LD proteins of the oleosin family (Fig. 1C). Similar to oleosins, LDSP of Nannochloropsis possesses an uninterrupted sequence of 62 amino acids with nonpolar side chains. Even though this is shorter than the hydrophobic region of oleosins, it theoretically allows the formation of a hairpin or similar structure sufficiently long to reach beyond the phospholipid monolayer surrounding the TAGs. A Pro knot motif as found in oleosins was not present in LDSP. However, its relative Pro content (8.9%) is higher than that of OLEO1 (2.9%), with five Pro residues found in the hydrophobic region alone. Another difference between the two proteins is the overall hydrophobicity, expressed as the grand average hydrophobicity (Kyte and Doolittle, 1982), which is considerably higher in LDSP (0.708) compared to the overall less hydrophobic OLEO1 (−0.168).
Figure 1.
LDSP is the predominant protein in LD Fractions. A, Thin layer chromatogram of nonpolar lipids showing TAG enrichment in N-deprived cells (cell N-) and LD fractions (top); 10 μg fatty acid was loaded per lane. P, Pigments; S, sterols; PL, polar lipids. Absorption spectrum of cell N- (dashed line) and LD fractions (solid line) showing enrichment of carotenoids by increased absorption in the blue light and depletion of chlorophyll. B, Coomassie blue-stained SDS-PAGE gel of Nannochloropsis total protein in the N-replete state (Cell N+), after 40 h of N deprivation (Cell N-), and the LD fraction (LD). Arrow indicates 17-kD protein submitted to mass spectrometry. C, Kyte-Doolittle hydropathy index (HI) with window size of 9; r.U., relative units.
LDSP Abundance Parallels TAG Content
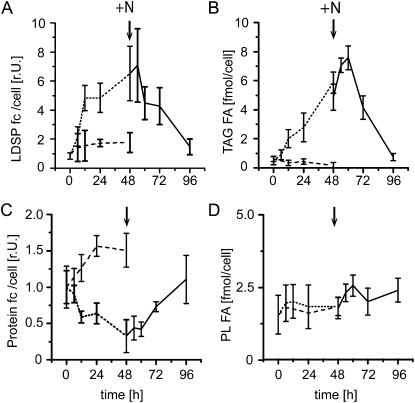
Polyclonal antibodies were raised against LDSP and used to determine its abundance in Nannochloropsis cells (Supplemental Fig. S1B). N deprivation was used to induce TAG accumulation, which resulted in cessation of growth (Fig. 2; Supplemental Fig. S1). Upon resupply of N, TAG was degraded and cells resumed dividing. Even though the total protein content per cell declined to half of its initial value during N deprivation, the content of LDSP increased approximately 6-fold on a per cell basis. Following resupplying of N, LDSP content specifically declined as did the TAG content. However, while the TAG content per cell continued to increase for another 12 h following N resupply, LDSP content began to decrease at an earlier time point. These observations suggested that LDSP plays an important role in the formation and stabilization of LDs in Nannochloropsis in ways comparable to that of plant oleosins.
Figure 2.
LDSP content correlates with TAG content and increases during N starvation even though total protein content per cell is decreasing. A, LDSP content per cell following N deprivation and recovery as determined by immunodetection. fc, Fold change. B, TAG content per cell as determined by thin-layer chromatography and gas chromatography-flame ionization detection. C, Total protein content per cell. D, Polar lipid content per cell. Dotted line, N deprivation; solid line, recovery after resupply of N; dashed line, control. Arrows indicate time of N resupply. Three independent repeats were averaged with standard deviations indicated. r.U., Relative units.
LDSP Localizes to LDs in Arabidopsis and Reduces LD Size in oleo1
To investigate the function of LDSP, we expressed the respective cDNA under the control of the Arabidopsis OLEO1 promoter producing LDSP (6xHis C-terminally tagged) and LDSP fused at the C terminus with enhanced GFP (eGFP) in the T-DNA insertion oleo1 mutant of Arabidopsis. Seeds (T1) of the transgenic lines were screened for high eGFP fluorescence in case of the fusion protein or for the presence of LDSP in seed (T2) protein extracts using immunoblotting. Because OLEO1 is a structural, highly abundant protein in mature embryos (Siloto et al., 2006), we expected that high levels of LDSP would be required for successful complementation. Further analysis focused on three lines that met this criterion: the two LDSP-His lines R5-5 and R3-4 and the LDSP-eGFP line Y13 (Supplemental Fig. S2).
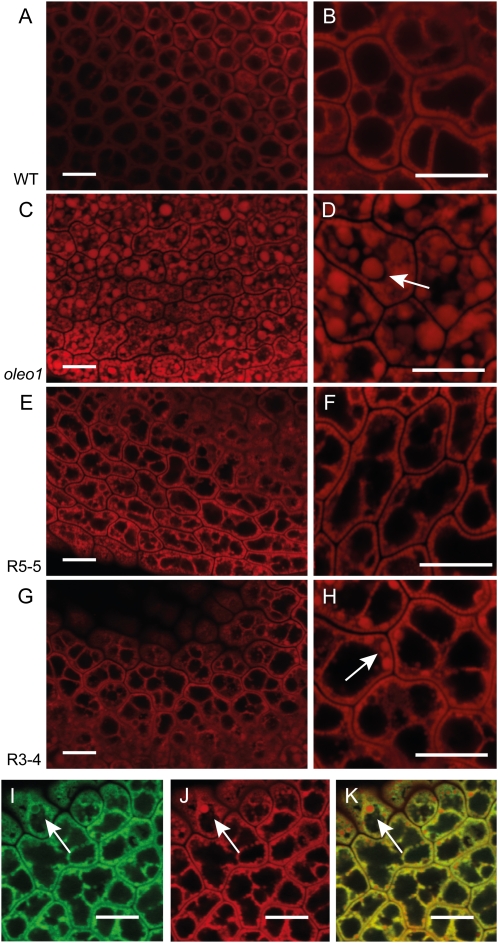
Confocal microscopy of mature embryos of line Y13 showed a strong eGFP fluorescence signal visible in the periphery of the Nile Red-stained LDs (Fig. 3, I–K). Fluorescence associated with eGFP emanating from other cell organelles was not detected above background levels. Nile Red fluorescence was recorded in the red narrow channel at shorter wavelength (575–625 nm) to minimize the fluorescence signal from membrane-associated dye (Greenspan et al., 1985). Embryos of the oleo1 mutant are characterized by an increase in LD size compared to wild-type plants (Siloto et al., 2006; Fig. 3, A–D). This phenotype was mostly reversed in the presence of LDSP-His, as shown in Figure 3, E to H, even though the two lines did not show LD size reduction to the same extent. Whereas line R5-5 produced embryos with no oversized LDs, in line R3-4, smaller LDs were found interspersed with larger LDs. This result can be explained by lower expression levels of the LDSP-His encoding construct in R3-4 compared to R5-5 as determined by immunoblotting (Supplemental Fig. S2). A similar dose effect was observed in the T2 generation of line Y13, for which low- and high-fluorescent seeds were independently examined (Supplemental Fig. S3).
Figure 3.
LDSP is associated with LDs in Arabidopsis seed and reduces LD size when introduced into the oleo1 mutant. Confocal microscopy of mature wild-type embryos (A and B), oleo1 embryos (C and D), and two independent LDSP-His lines, R5-5 (E and F) and R3-4 (G and H), showing Nile Red fluorescence in the red channel. GFP fluorescence (I), Nile Red fluorescence (J), and merged channels (K) of LDSP-eGFP line Y13 showed the localization of LDSP at the surface of LDs. Bars = 10 μm. Arrows point out bigger LDs with clearly visible ring-shaped GFP fluorescence signal.
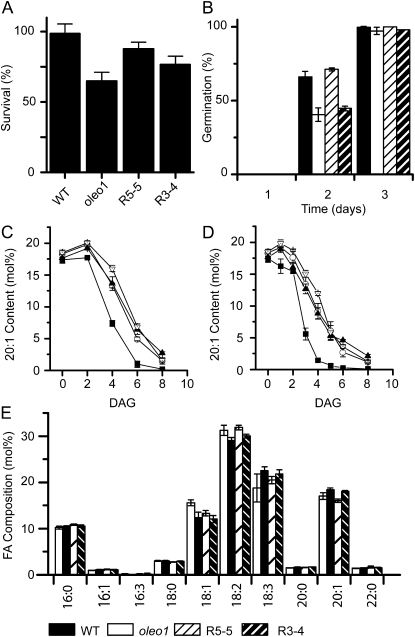
The lack of OLEO1 causes not only larger but also less stable LDs in mature embryos (Shimada et al., 2008). LDs in the oleo1 mutant fuse upon exposure to physical stress, such as freezing, leading to embryo death during vernalization. As a result, reduced germination rates were observed for oleo1 after freezing treatment (Shimada et al., 2008). The described reduction of LD size in the presence of LDSP led to the hypothesis that smaller LDs will be less prone to coalescence. It was therefore tested whether LDSP is stabilizing and preventing fusion of the LDs during harsh conditions in the absence of native OLEO1 by monitoring the germination rates after storage of the seed at −80°C for 5 d (Fig. 4A). In the LDSP-producing lines R5-5 and R3-4, the impact of the freezing treatment led to 88% and 77% survival, respectively, compared to 99% in the wild type and 65% in oleo1, consistent with a stabilization of plant LDs by LDSP. A gradual rescue of the oleo1 phenotype was also observed in the fatty acid profile, where the line R5-5 resembled the wild type, whereas the fatty acid profile of R3-4 was not different from oleo1. These differences in the fatty acid profile were minor but consistent with previous reports (Shimada et al., 2008).
Figure 4.
LDSP expression rescues the germination defect after freezing and the fatty acid composition of the ole1 mutant but not the delay in degradation of seed TAG in ole1 seed. A, Survival of wild-type, ole1, R5-5, and R3-4 plants after freezing treatment (8 d at −80°C) followed by 3 d at 7°C in the dark (vernalization), measured as percentage of germinated seedlings 8 d after transfer to light relative to control seed (stored at room temperature for 8 d). B, Germination at room temperature without prior vernalization performed as in A. Decrease of 20:1 fatty acid content during germination without (C) and with (D) prior vernalization (DAG, days after germination). Black squares, the wild type; white circles, oleo1; black triangles, R5-5; white triangles, R3-4. E, Fatty acid composition of mature seed in the wild type, oleo1, R5-5, and R3-4. Three replicates were averaged and sd determined.
LDSP Does Not Rescue the Delay of TAG Degradation in oleo1 Seedlings
Following germination, TAG hydrolysis is initiated early during the seedling establishment phase to provide energy (Kelly et al., 2011). Oversized LDs in the oleo1 seedlings delay TAG hydrolysis during seedling establishment, which may be due to their reduced surface to volume ratio and thereby decreased access of lipases to TAGs sequestered in the LDs. We tested the degradation of TAG during germination by monitoring the presence of cis-11-eicosenoic acid (20:1), a fatty acid that is predominantly esterified to TAG and not to membrane lipids (Lemieux et al., 1990). Indeed, in oleo1 seedlings 20:1 was detected later following germination than in wild-type plants (Fig. 4C). By fitting a sigmoidal function to the curve, a comparison of the power p (as a measure of the slope of the decline) and the infliction point x(0) was possible and revealed a statistically significant shift in both parameters between the wild-type and the oleo1 seedlings (two-sample t test, P < 0.05; Supplemental Table S4). Surprisingly, both LDSP expression lines showed no change in 20:1 content during germination compared to oleo1. To validate this result, the TAG content was confirmed by thin-layer chromatography and gas chromatography-flame ionization detection at selected times (Supplemental Fig. S4).
As part of the oleo1 phenotype, germination (defined as radicle emergence) was shown to be delayed for dormant seeds (Siloto et al., 2006). To exclude that the delay in TAG degradation was due to a delay in germination, we repeated the experiment with vernalized seeds to break dormancy. We were not able to detect a delay in germination after a vernalization period in oleo1 or oleo1 producing LDSP (Supplemental Table S3). The examination of TAG contents revealed earlier degradation in oleo1 seedlings and LDSP expression lines R3-4 and R5-5 (Fig. 4D) compared to dormant seed as defined by the infliction point of the curve (Supplemental Table S4). Nevertheless, TAG degradation of wild-type seeds was still occurring faster with complete hydrolysis about 2 d earlier than of oleo1 mutant seed and LDSP producing lines, and no differences were observed between the degradation curves for R5-5, R3-4, and oleo1.
DISCUSSION
LDSP of Nannochloropsis
The high abundance of LDSP in the LD fractions corresponding to high cellular TAG content, the long hydrophobic stretch of amino acids in LDSP, and its high grand average hydrophobicity score are consistent with LDSP functioning as a major structural component of LDs in Nannochloropsis. This hypothesis is strongly supported by the results from ectopic LDSP production in Arabidopsis embryos, showing that it not only localizes to LDs but also reduces LD size and prevents LD coalescence in the absence of OLEO1, as shown by increased tolerance to freezing. This effect of LDSP on plant LDs is likely attributable to its hydrophobic properties that, like OLEO1, allow it to insert into the surface of LDs. Plant oleosins also have been ectopically produced in yeast cells and localized to LDs (Beaudoin et al., 2000). This study also provided evidence for the involvement of components of the secretory pathway in oleosin targeting from the endoplasmic reticulum to the LD in vivo. Whether this is the case for LDSP as well, or if it localizes to the LD merely due to its high hydrophobicity, remains to be determined. It should be pointed out that oleosins have the ability to stabilize artificial LDs (Peng et al., 2004), suggesting that they are capable of spontaneous insertion into a lipid matrix.
Besides their basic similarities, the LDs of microalgae and plants have to function in different physiological contexts. In oil seeds, TAG is accumulating during embryogenesis. The mature embryo then undergoes desiccation and stays dormant until germination is initiated. By contrast, microalgal cells are exposed to rapid changes in their environment, such as temperature, light, and nutrient availability, that promote the synthesis or hydrolysis of TAGs. In Nannochloropsis, LDSP was always detectable, and the TAG content was rarely observed to be below 15% of the total fatty acid content (Fig. 2). However, as the stress becomes severe, for example, during prolonged N deprivation, the cell cycle of microalgae is arrested and the cells assume a quasi-dormant state similar to that of embryo cells of dormant plant seeds. Whether LDs formed in actively dividing cells of Nannochloropsis differ from those formed following N deprivation-induced cell arrest and whether LDSP plays a regulatory role in TAG accumulation and hydrolysis remain to be investigated.
Evolution of LD Proteins
Interestingly, orthologs of LDSP were not detected in any of the heterokont genomes available at this time; however, immunodetection of a protein of the same size using the LDSP-specific antibodies described here was successful in all Nannochloropsis species tested (N. oceanica, N. gaditana, N. granulata, and N. salina; Supplemental Fig. S5). Eustigmatophytes stand out from other heterokonts by a variety of ultrastructural and biochemical features and therefore have been proposed to have diverged early from other heterokont classes (van den Hoek et al., 1995). Distinct primary sequences but partially overlapping functions imply that LD proteins evolved independently multiple times (convergent evolution) in plants and unikonts (animals, fungi, and amoebozoa) and, given the unique primary sequence of LDSP, also among heterokonts. Common structural features beyond the primary sequence must provide a common denominator for the presence of different LD-associated proteins in different organismal groups, while at the same time permitting the recruitment of proteins through protein-protein interactions to the LDs in a species-specific manner. Thus, while effects on the structure of LDs is likely similar, the more intricate functions of Plin-like proteins, such as the recruitment of TAG-degrading enzymes to LDs, seem to require a species-specific protein as the lack of biochemical complementation through production of Nannochloropsis LDSP in the Arabidopsis oleo1 mutant suggests.
LDSP Structure and Function
Even though at present true orthologs of LDSP cannot be identified based on primary sequence similarity, partial functional convergence between LDSP and plant oleosins suggests a possible similarity of the higher order structures of both proteins. LDSP and plant oleosins are fairly unique with their extended, central sequence of hydrophobic residues, potentially sufficiently long to form a secondary structure submerged into the TAG core of LDs as already predicted for oleosins (Abell et al., 1997, 2002, 2004; Li et al., 2002; Kim and Huang, 2003). The Pro knot motif found in oleosins is not apparent in LDSP, but five Pro residues are present in the hydrophobic region of LDSP. Whether they play a role in forming a central structure immersed into the TAG core of the LD remains to be seen. The secondary structure prediction of the hydrophobic region of OLEO1 is still under discussion. Even though bioinformatics tools predict an α-helical structure, circular dichroism and Fourier transform infrared spectroscopy suggested a β-sheet conformation (Li et al., 2002). Including LDSP in future structure function analyses may provide the necessary insights to address this still unanswered question.
A Role for Oleosin in TAG Hydrolysis during Seed Germination
The heterologous expression of an LDSP-encoding cDNA in the Arabidopsis oleo1 mutant did not only provide insights into the function of LDSP, but shed some light on the physiological roles of oleosin as well. By replacing OLEO1 with a foreign protein unrelated at the primary sequence level, but comparable hydropathy profile and possible related structural features, hints were obtained pointing toward possible oleosin-specific binding partners or regulatory events that beg to be explored in future. If the volume to surface ratio of the LDs would be the only critical parameter determining the rate of TAG hydrolysis, one would expect a gradual enhancement in TAG turnover as LD size decreases. However, we did not observe measurable changes in TAG degradation during seedling establishment in the transgenic lines, even though they showed decreased size of their LDs compared to oleo1. This may indicate an additional role of OLEO1 that cannot be fulfilled by LDSP or the presence of a regulatory factor that is inhibited by LDSP.
Biotechnological Applications for LDSP
LDSP might be used in biotechnological applications similar to those conceived for oleosins (Capuano et al., 2007). Oleosins are employed in molecular farming, allowing the targeting of proteins or peptides to LDs or the endoplasmic reticulum (Pons et al., 2009), and have been proposed to stabilize emulsions and artificial LDs. The localization of LDSP to LDs in plants suggests that it might localize to any LD, delivering a given protein to a LD due to its inherent hydrophobic characteristics. Thus, LDSP might provide an additional reagent with diverging properties in the tool box of bioengineers.
MATERIALS AND METHODS
Strains and Culture Conditions
Nannochloropsis oceanica strain W2J3B was provided by Aurora Algae, and strains CCMP1779, 531, 369, 529, and 1775 were obtained from the Provasoli Guillard National Center for Marine Algae and Microbiota (www.bigelow.org). The strains were grown in batch culture in brackish artificial seawater (17 g/L Tropic Marin salt; www.tropic-marin.com) enriched with f/2 nutrients, trace elements, and vitamins (Andersen et al., 2005), 40 mm bicarbonate, and 15 mm Tris buffer (pH 7.6) to prevent carbon limitation. Cultures were kept in Erlenmeyer or Fernbach Flasks filled to 20% and 35% of their maximum volume, respectively, in continuous light at a photon flux density of 85 μmol m−2 s−1 with shaking at 105 rpm at 22°C. Cell density was determined using a Z1 Coulter Counter (Beckman-Coulter) with a 50-μm aperture according to the manufacturer’s instructions. For N deprivation experiments, cells were harvested at a density of 10 to 20 million cells/mL by centrifugation (10 min at 6,000g) and washed with N-free medium before resuspension in N-free medium.
Arabidopsis (Arabidopsis thaliana ecotype Columbia-2) and the T-DNA insertion line SM_3_29875 (oleo1) were grown in 16/8-h light/dark cycles at 22°C and 18°C, respectively, on agar-solidified medium or on soil as previously described (Xu et al., 2005).
Isolation of LDs and Analysis of LD-Associated Proteins
LDs were purified from Nannochloropsis cells deprived of N for 36 to 40 h. Two liters of cell suspension with an OD540 of 0.3 to 0.5 equivalent to 20 to 40 × 106 cells/mL were concentrated by centrifugation, resuspended in 20 mL ice-cold breaking buffer (Jolivet et al., 2004) containing Complete Protease Inhibitor tablets (Roche Diagnostics), and homogenized with a French pressure cell at 20,000 p.s.i. before subjecting the cell homogenate to Suc density centrifugations (Jolivet et al., 2004). The LD-rich floating phase was recovered, resuspended in 15 mm phosphate buffer (pH7.6) containing 0.2 m Suc and 0.1% Tween 20, and incubated for 15 min at room temperature with occasional mixing. Lipids and proteins were extracted after two additional washing steps without detergent.
Extraction, gel electrophoresis, and mass spectrometry analysis of LD proteins were performed as previously described (Moellering and Benning, 2010). The transcriptome of N. oceanica CCMP1779 was sequenced using Illumina (55-bp directional single-end reads) from RNA samples of N-replete (log-phase) and N-deprived (38 h) cells. Transcripts were assembled de novo and after mapping to a draft genome. A data set of 12,012 annotated protein models used for the identification of the peptides will be available upon request.
Sequence Analysis of LDSP
Calculation of hydropathy by Kyte and Doolittle (1982) and structural predictions were made using the online tools provided by the Pasteur Institute, France (mobyle.pasteur.fr).
Lipid Analysis
Lipids were extracted from lyophilized material according to the method by Folch et al. (1957) and analyzed by gas-liquid chromatography as previously described (Castruita et al., 2011).
Immunodetection of LDSP
The peptide antibody against LDSP was produced by Covance Nonclinical Services using the C-terminal peptide [H]-CVPALDKVLANKKVAKFLLK-[NH2]. For immunoblot analysis, cell pellets were extracted with sample buffer to yield 50,000 cells/μL. Extracts were diluted 10-, 20-, and 50-fold, and 20 μL was loaded onto a 15% SDS-PAGE gel. Gel electrophoresis and electroblotting was performed with a Bio-Rad Mini Protean system according to the manufacturer’s instructions (Bio-Rad Life Sciences). Anti-LDSP antibody was used in a 1:10,000 dilution. Quantification was performed densitometrically using ImageJ software (National Institutes of Health) on immunoblots with immunoreactive proteins visualized using alkaline phosphatase (Blake et al., 1984).
Production of LDSP in Arabidopsis
The coding sequence of LDSP (GenBank accession no. JQ268559) was amplified from cDNA and fused with 1.5 kb of the promoter sequence of OLEO1 from wild-type Arabidopsis by PCR and assembled with or without eGFP coding sequence in the p35S-OCS-BM plasmid prior to transfer into the binary vector pLH9000 using the SfiI restriction enzyme (both plasmids were obtained from the DNA Cloning Service; www.dna-cloning.de). Plants were transformed by the floral dip method (Clough and Bent, 1998) and selected on kanamycin. PCR primers used are listed in Supplemental Table S5.
Confocal Microscopy
Arabidopsis embryos were prepared from freshly harvested seeds according to Perry and Wang (2003) and fixed in 2% paraformaldehyde overnight before microscopy analysis. Nile Red was used to stain LDs (Greenspan et al., 1985) before mounting as described by Johnson and Nogueira Araujo (1981). Preparations were examined with an Olympus Fluoview FV10i confocal microscope using the supplied image acquisition software. Images were generally recorded with the ×60 objective lens (unless stated differently) with or without additional ×8 digital zoom at a 1024 × 1024-pixel resolution. The preset filter options were used to record the eGFP signal with 473-nm excitation and 510-nm emission wavelength and Nile Red in the red-narrow channel with 559-nm excitation and 600-nm emission wavelength. Laser intensities were kept below 10%.
Germination Assays
The germination assays were performed as previously described (Siloto et al., 2006). Four batches of 100 seeds each were analyzed per line from a mixed batch of five to nine individual plants per line grown side-by-side. For freezing treatments, seed batches were stored in polypropylene tubes at −80°C for 5 d, followed by 3 d vernalization at 7°C in the dark.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JQ268559 and JQ301469.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Chlorophyll content and immunodetection of LDSP.
Supplemental Figure S2. Detailed characterization of Arabidopsis transgenic lines.
Supplemental Figure S3. Dose effect of LDSP on LD size in transgenic Arabidopsis lines.
Supplemental Figure S4. TAG content of Arabidopsis transgenic lines during germination.
Supplemental Figure S5. Presence of LDSP like proteins in different Nannochloropsis sp.
Supplemental Table S1. Proteins identified by mass spectrometry.
Supplemental Table S2. List of LDSP peptides.
Supplemental Table S3. Germination of Arabidopsis transgenic lines after 3 d of vernalization.
Supplemental Table S4. Statistical analysis of 20:1 fatty acid content of Arabidopsis transgenic lines.
Supplemental Table S5. Oligonucleotide sequences.
Supplementary Material
Acknowledgments
We thank Dough Whitten at the Michigan State University Proteomics Facility for performing electrospray ionization-liquid chromatography-tandem mass spectrometry analysis, Hideki Takahashi at Michigan State University for access to the confocal microscope, Gregg Howe at Michigan State University for access to the fluorescence dissecting microscope, Michigan State University-Research Technology Support Facility for access to the Beckman Coulter Z1 particle counter, Blair Bullard at Michigan State University for technical assistance, and Maurice M. Moloney and Rodrigo Siloto at the University of Calgary for the homozygous oleo1 line.
References
- Abell BM, Hahn M, Holbrook LA, Moloney MM. (2004) Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J 37: 461–470 [DOI] [PubMed] [Google Scholar]
- Abell BM, High S, Moloney MM. (2002) Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J Biol Chem 277: 8602–8610 [DOI] [PubMed] [Google Scholar]
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. (1997) Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9: 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RA, Berges JA, Harrison PJ, Watanabe MM. (2005) Appendix A. In RA Andersen, ed, Algal Culturing Techniques, Ed 1. Elsevier Academic Press, San Diego, pp 429–538 [Google Scholar]
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA. (2000) In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J 23: 159–170 [DOI] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jäckle H, Kühnlein RP. (2010b) PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab 12: 521–532 [DOI] [PubMed] [Google Scholar]
- Beller M, Thiel K, Thul PJ, Jäckle H. (2010a) Lipid droplets: a dynamic organelle moves into focus. FEBS Lett 584: 2176–2182 [DOI] [PubMed] [Google Scholar]
- Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. (1984) A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem 136: 175–179 [DOI] [PubMed] [Google Scholar]
- Brasaemle DL. (2007) Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 48: 2547–2559 [DOI] [PubMed] [Google Scholar]
- Capuano F, Beaudoin F, Napier JA, Shewry PR. (2007) Properties and exploitation of oleosins. Biotechnol Adv 25: 203–206 [DOI] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan WH, Cokus S, Loo JA, Benning C, et al. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509 [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. (2011) Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol 3: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Johnson GD, Nogueira Araujo GM. (1981) A simple method of reducing the fading of immunofluorescence during microscopy. J Immunol Methods 43: 349–350 [DOI] [PubMed] [Google Scholar]
- Jolivet P, Roux E, D’Andrea S, Davanture M, Negroni L, Zivy M, Chardot T. (2004) Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 42: 501–509 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ. (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian O, Benemann CS, Niyogi KK, Vick B. (2011) High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci USA 108: 21265–21269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Huang AHC. (2003) Oleosins and plastid-lipid-associated proteins in Arabidopsis. In N Murata, M Yamada, I Nishida, H Okuyama, J Sekija, W Hajime, eds, Advanced Research on Plant Lipids: Proceedings of the 15th International Symposia on Plant Lipids. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 147–150 [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. (2010) Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res 51: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J 66: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J. (1990) Mutants of Arabidopsis with alterations in seed lipid fatty-acid composition. Theor Appl Genet 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Li M, Murphy DJ, Lee KH, Wilson R, Smith LJ, Clark DC, Sung JY. (2002) Purification and structural characterization of the central hydrophobic domain of oleosin. J Biol Chem 277: 37888–37895 [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Souza SC, Zhang HH, Strissel KJ, Christoffolete MA, Kovsan J, Rudich A, Kraemer FB, Bianco AC, Obin MS, et al. (2006) Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem 281: 15837–15844 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40: 325–438 [DOI] [PubMed] [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Pal D, Khozin-Goldberg I, Cohen Z, Boussiba S. (2011) The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl Microbiol Biotechnol 90: 1429–1441 [DOI] [PubMed] [Google Scholar]
- Peled E, Leu S, Zarka A, Weiss M, Pick U, Khozin-Goldberg I, Boussiba S. (2011) Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae). Lipids 46: 851–861 [DOI] [PubMed] [Google Scholar]
- Peng CC, Chen JCF, Shyu DJH, Chen MJ, Tzen JTC. (2004) A system for purification of recombinant proteins in Escherichia coli via artificial oil bodies constituted with their oleosin-fused polypeptides. J Biotechnol 111: 51–57 [DOI] [PubMed] [Google Scholar]
- Perry SE, Wang H. (2003) Rapid isolation of Arabidopsis thaliana developing embryos. Biotechniques 35: 278–280, 282 [DOI] [PubMed] [Google Scholar]
- Pons L, Battaglia-Hsu SF, Orozco-Barrios CE, Ortiou S, Chery C, Alberto JM, Arango-Rodriguez ML, Dumas D, Martinez-Fong D, Freund JN, et al. (2009) Anchoring secreted proteins in endoplasmic reticulum by plant oleosin: the example of vitamin B12 cellular sequestration by transcobalamin. PLoS ONE 4: e6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. (2009) Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng 102: 100–112 [DOI] [PubMed] [Google Scholar]
- Sheehan J, Dunahay T, Benemann J, Roessler P. (1998). A look back at the U.S. Department of Energyaposs Aquatic Species Program: biodiesel from algae. In Closeout Report NREL/TP-580-24190, U.S. Department of Energyaposs Office of Fuels Development, National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, et al. (2004) Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem 279: 42062–42071 [DOI] [PubMed] [Google Scholar]
- Sukenik A, Carmeli Y. (1990) Lipid synthesis and fatty acid composition in Nannochloropsis sp. (Eustigmatophyceae) grown in a light-dark cycle. J Phycol 26: 463–469 [Google Scholar]
- van den Hoek C, Mann D, Jahns H. (1995) Heterokontophyta: class Eustigmatophyta. In C van den Hoek, D Mann, H Jahns, eds, Algae: An Introduction to Phycology. University Press, Cambridge, UK, pp 130–133 [Google Scholar]
- Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. (2009) Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell 8: 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich JE, Awai K, Benning C. (2005) Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.