Abstract
Papain-like cysteine proteases (PLCPs) are a large class of proteolytic enzymes associated with development, immunity, and senescence. Although many properties have been described for individual proteases, the distribution of these characteristics has not been studied collectively. Here, we analyzed 723 plant PLCPs and classify them into nine subfamilies that are present throughout the plant kingdom. Analysis of these subfamilies revealed previously unreported distinct subfamily-specific functional and structural characteristics. For example, the NPIR and KDEL localization signals are distinctive for subfamilies, and the carboxyl-terminal granulin domain occurs in two PLCP subfamilies, in which some individual members probably evolved by deletion of the granulin domains. We also discovered a conserved double cysteine in the catalytic site of SAG12-like proteases and two subfamily-specific disulfides in RD19A-like proteases. Protease activity profiling of representatives of the PLCP subfamilies using novel fluorescent probes revealed striking polymorphic labeling profiles and remarkably distinct pH dependency. Competition assays with peptide-epoxide scanning libraries revealed common and unique inhibitory fingerprints. Finally, we expand the detection of PLCPs by identifying common and organ-specific protease activities and identify previously undetected proteases upon labeling with cell-penetrating probes in vivo. This study provides the plant protease research community with tools for further functional annotation of plant PLCPs.
Proteases determine the fate of all proteins. Proteolysis is not only needed to cycle proteins back into amino acids but also to activate and inactivate proteins by processing. By doing so, proteases are involved in almost any biological phenomenon, ranging from the cell cycle to programmed cell death to immunity (van der Hoorn, 2008). Plant genomes encode for about 500 to 800 proteases (García-Lorenzo et al., 2006; van der Hoorn, 2008). The vast majority of plant proteases have unknown functions. An important limitation in the annotation of plant protease functions is the fact that many protease families are rather large and presumed to contain proteases that act redundantly, making it difficult to study protease functions using single gene knockout. To annotate functions to proteases of large families, we need to subclassify these families into smaller units that can be studied further (e.g. using reverse genetics).
Here, we focus on a functional subclassification of plant papain-like cysteine proteases (PLCPs). PLCPs belong to clan CA, family C1A in the Merops protease database (Rawlings et al., 2010). The global protein structure of PLCPs is a papain-like fold of two domains: an α-helix and a β-sheet domain (Turk et al., 2001). The two domains (lobes) are linked to each other in such a way that a deep cleft is formed that acts as the substrate-binding groove and contains the catalytic triad Cys-His-Asn. Substrate specificity is accomplished by the substrate-binding pockets along the substrate-binding groove that bind amino acid side chains at positions 2 and 3 before the cleavage site (P3 and P2 positions; Turk et al., 2001).
PLCPs are very stable enzymes and often are found in proteolytically harsh environments such as the apoplast, the vacuole, and lysosomes. To target these locations, PLCPs are encoded as preproproteins and carry various targeting signals. The signal peptide ensures that the proprotease enters the endomembrane system, whereas the autoinhibitory prodomain prevents premature activation of the protease. The prodomain is proteolytically removed in cis (intramolecular) or in trans (intermolecular) at the destination. Some PLCPs carry a signal for retention in the endoplasmic reticulum (ER) at the C terminus (KDEL; Than et al., 2004), whereas other PLCPs carry a signal at the N terminus of the proprotease for vacuolar targeting (NPIR; Ahmed et al., 2000). Some PLCPs also carry a C-terminal granulin-like domain, which shares homology to granulins in animals, which are growth hormones released upon wounding (Bateman and Bennett, 2009). Although these motifs and features have been noted for individual proteases, their distribution within the PLCP superfamily has not yet been studied.
A first classification of PLCPs was made by Beers and colleagues (2004). Using an assembly of 138 plant PLCPs (including 30 Arabidopsis [Arabidopsis thaliana] PLCPs), the proteases were phylogenetically classified into eight subfamilies. This subclassification was supported by conserved positions of introns in the corresponding genes. A similar subfamily structure was found during a subclassification of poplar (Populus spp.) and Arabidopsis PLCPs (García-Lorenzo et al., 2006) and during comparative genomic analysis of PLCPs in different taxons of the plant kingdom (Martinez and Diaz, 2008). The latter study named plant PLCPs according to their closest animal counterparts: cathepsin B, H, F, and L.
Here, we extend the phylogenetic analysis of plant PLCPs by taking advantage of the extensive number of publicly available sequences. The broad phylogenetic analysis of an increased number of PLCPs allows us to detect conserved features within each subfamily that can be used as classifiers for new plant PLCPs. Furthermore, we study selected representatives of different subfamilies encoded by Arabidopsis to characterize the proteases biochemically using transient expression and protease activity profiling (van der Hoorn et al., 2004). Finally, we extend the detection of PLCP activities in Arabidopsis using protease activity profiling with new probes on extracts of various organs and in vivo labeling. These tools and observations provide an important framework for the functional classification of plant PLCPs.
RESULTS
Plant PLCPs Are Phylogenetically Divided into Nine Subfamilies
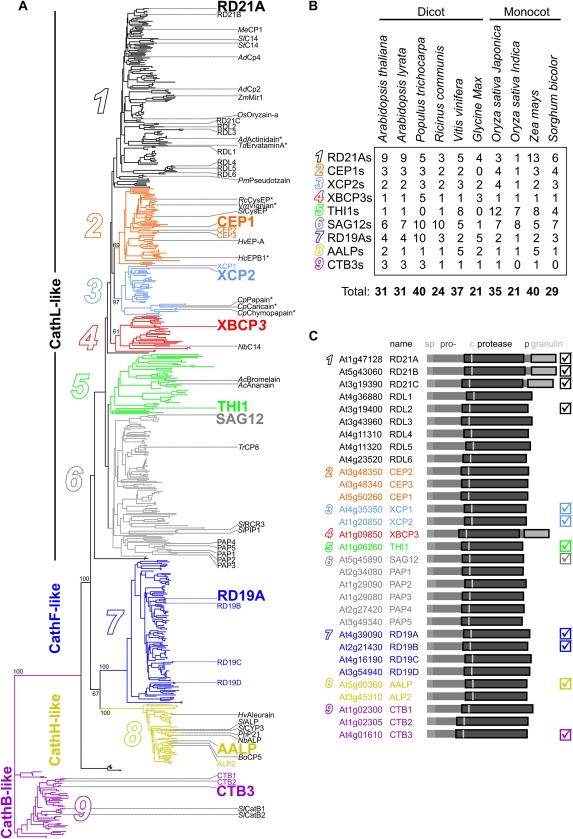
PLCP sequences were retrieved from the National Center for Biotechnology Information (NCBI) and The Institute for Genomic Research (TIGR) by BLAST searches using each of the 138 PLCPs reported by Beers et al. (2004). Redundant sequences, truncated sequences, and sequences not having the catalytic motif [SCG][WC][AST][FV] were removed, resulting in a PLCP database of 723 unique PLCP sequences. Phylogenetic analysis of the proprotease domain shows that these PLCPs fall into nine subfamilies, numbered 1 to 9 (Fig. 1A; Supplemental Fig. S1). This classification is similar to the previous classification (Beers et al., 2004), except that the C1A-1 superfamily with the granulin-containing proteases splits into subfamilies 1 and 4, which is supported by further bioinformatic analysis (see below).
Figure 1.
Phylogenetic subclassification of plant PLCPs. A, The unrooted phylogenetic tree of 723 plant PLCPs is subdivided into nine PLCP subfamilies (1–9). Arabidopsis PLCPs are indicated in the first column in color, and other studied PLCPs are indicated in the second column in black. Asterisks indicate that a crystal structure is available. Type members for each subfamily are shown in larger font. Key bootstrap values are indicated. The annotated phylogenetic tree with readable entries is available as Supplemental Figure S1. B, Distribution of PLCPs over subfamilies for plant species with more than 20 sequenced PLCPs. C, Nomenclature and subclassification of Arabidopsis PLCPs. The ATG accession codes of genes encoding putative PLCPs are followed by given names. The domain structure consists of a signal peptide (sp), prodomain (pro), protease domain with catalytic Cys (c), and in some cases a Pro-rich domain (p) and a granulin domain. The PLCPs studied biochemically in this work are marked in the right column. [See online article for color version of this figure.]
Subfamily 9 contains cathepsin B-like PLCPs, which are clearly separated from the other PLCPs. Subfamilies 7 (RD19A-like proteases; also called cathepsin F-like proteases) and 8 (aleurain-like proteases; also called cathepsin H-like proteases) are also relatively distinct from the remaining PLCPs. The remaining subfamilies, 1 to 6, are more related to each other and have also been classified as cathepsin L-like proteases (Martinez and Diaz, 2008).
Previously studied plant PLCPs can be found in different subfamilies (Fig. 1A, names on the right). These studies include several Arabidopsis PLCPs: RD21A (Yamada et al., 2001); XCP1 and XCP2 (Avci et al., 2008); XBCP3 (Funk et al., 2002); SAG12 (Noh and Amasino, 1999; Otegui et al., 2005); RD19A (Bernoux et al., 2008); AALP (Ahmed et al., 2000; Watanabe et al., 2004); and cathepsin B-like proteases (CTBs; McLellan et al., 2009). Other studied PLCPs include papain (Konno et al., 2004), maize (Zea mays) Mir1 (Pechan et al., 2000), tomato (Solanum lycopersicum) RCR3, PIP1, and C14 (Krüger et al., 2002; Rooney et al., 2005; Shabab et al., 2008; van Esse et al., 2008; Kaschani et al., 2010), and bean (Phaseolus vulgaris) CysEP (Greenwood et al., 2005).
Representatives of subfamilies 1, 2, and 3 have been crystallized (Fig. 1A, marked with asterisks). These studies concern papain, chymopapain, and caricain from papaya (Carica papaya; Drenth et al., 1968; Katerelos et al., 1996; Maes et al., 1996), CysEP of castor bean (Ricinus communis; Than et al., 2004), actinidin of kiwi (Actinidia deliciosa; Varughese et al., 1992), ervatamin-A of crape jasmine (Ervatamia coronaria; Ghosh et al., 2008), and EP-B2 of barley (Hordeum vulgare; Bethune et al., 2006).
Six subfamilies contain 32 to 67 PLCPs, whereas subfamilies 1, 6, and 7 are twice as large, having 157, 105, and 125 PLCPs, respectively. The relatively large size of these three subfamilies is corroborated by the larger number of PLCPs encoded by sequenced genomes (Fig. 1B). Each plant species seems to have multiple copies for members of subfamilies 1, 6, and 7. The total number of PLCPs per plant genome is between 20 and 40 genes, and nearly each subfamily is represented in sequenced plant species (Fig. 1B). This indicates that plants carry a PLCP repertoire that is conserved throughout angiosperm evolution.
Importantly, also the Arabidopsis genome encodes PLCPs of each subfamily (Fig. 1C). Studies on Arabidopsis PLCPs, therefore, may provide information that is representative for the different subfamilies. We consequently refer to each of the nine subfamilies with a type member from Arabidopsis: subfamily 1 (contains RD21A-like proteases); subfamily 2 (CEP1-like); subfamily 3 (XCP2-like); subfamily 4 (XBCP3-like); subfamily 5 (THI1-like); subfamily 6 (SAG12-like); subfamily 7 (RD19A-like); subfamily 8 (AALP-like); and subfamily 9 (CTB3-like; Fig. 1A, larger names).
PLCP Sequences Carry Subfamily-Specific Functional Motifs
We next investigated the frequency of functional motifs in each of the nine PLCP subfamilies. Prediction of the signal peptide using SignalP (Emanuelsson et al., 2007) shows that 70% to 100% of the members of each subfamily carry a predicted signal peptide (Fig. 2A, first column). The protein sequences for the other PLCPs may not have been complete, may carry a signal peptide that is not recognized by SignalP, or do not have a signal peptide. Nevertheless, this finding indicates that the vast majority of the members of each PLCP subfamily enter the endomembrane system. This observation is in agreement with a recent study on the frequent occurrence of signal peptides of proteases of Arabidopsis and rice (Oryza sativa; Goulet et al., 2012).
Figure 2.
Conserved functional motifs in PLCP subfamilies. A, Positions and frequencies of functional motifs in each PLCP subfamily. SP, N-terminal signal peptide, predicted by SignalP; NPIR, vacuolar targeting signal at the N terminus of the prodomain; ERFNIN, structural motif in the prodomain (ExxxRxxxFxxNxxx{I/V}xxxN; allows one mismatch); Triad, the catalytic triad: Cys-His-Asn; KDEL, C-terminal retrieval signal for localization to the ER ({K/H}DEL); Granulin, C-terminal granulin domain containing the Cys pattern Cx5Cx5CCCx7Cx4CCx6CCx5CCx6Cx6C. B, Presence of the granulin domain in PLCP subfamilies 1 and 4. Granulin-containing proteases are indicated with black or red lines, and proteases lacking a granulin domain are indicated in gray or pink. Shown are only the trees of subfamilies 1 and 4 from Figure 1A. C, The phylogenetic tree of the granulin domain branches into the same subfamilies as the proprotease tree. Note that the subfamily 4 PLCPs (red; XBCP3-like) are grouped together. D, Conserved structural features of the granulin domain. Consensus sequences of the granulin domain of subfamilies 1 and 4 are aligned with those of RD21A and hGrnA. Disulfide bridging has been determined for hGrnA and is shown at the top. E, Illustration of the structure of the granulin domain of RD21A, modeled on hGrnA (2jye). The numbering of the disulfide bridge residues corresponds to that in D.
Subfamilies 1 to 6 and 8 carry the ERFNIN motif (ExxxRxxxFxxNxxx{I/V}xxxN; one mismatch allowed) in the prodomain (Fig. 2A; Supplemental Fig. S2). This motif provides the core structure of the autoinhibitory prodomain and was originally identified as a plant-specific feature (Karrer et al., 1993). The ERFNIN motif is absent from CTBs (subfamily 9), whereas RD19A-like proteases (subfamily 7, cathepsin F-like) carry a conserved ERFNAQ motif instead of the ERFNIN motif (Supplemental Fig. S2).
Nearly all PLCPs possess the catalytic triad (Cys-His-Asn; indicating that all these proteins are functional Cys proteases; Fig. 2A; Supplemental Fig. S2). Exceptions are three highly homologous soybean (Glycine max) proteins in subfamily 4 (XBCP3-like) that carry Gly-His-Asn (GI:1199563, GI:129353, and GI:3097321). These proteins are the P34 proteins of soybean that cause allergy and have been targeted for silencing, but their endogenous function is so far unknown (Herman et al., 2003). Furthermore, Ipomoea nil (ipnitc1650) and japonica rice (GI:13365804) sequences from subfamily 5 (THI1-like) carry a Ser-His-Asn motif. Their different phylogenetic origins indicate that these replacements occurred independently. Since Ser can act as a nucleophile, it might be that these two proteins still have proteolytic activity.
Subcellular targeting of the subfamilies is predicted by the presence of two different motifs. First, the vacuolar targeting sequence NPIR at the N terminus of the prodomain is reported for some PLCPs, including AALP, which served as a model protein for vacuolar targeting (Holwerda et al., 1992; Ahmed et al., 2000). Interestingly, NPIR is found in over 70% of AALP-like proteases (subfamily 8) but not in any of the other PLCP subfamilies (Fig. 2A; Supplemental Fig. S2). A second targeting signal is the C-terminal KDEL or HDEL motif, which mediates the retrieval of soluble proteins to the ER. This feature has been described (e.g. for bean CysEP; Schmid et al., 1999). Importantly, the C-terminal KDEL or HDEL occurs in 70% of CEP1-like proteases (subfamily 2) and in none of the other subfamilies (Fig. 2A). The majority of the other 30% of the CEP1-like proteases carry a C-terminal sequence that is similar: KDEM, TDEL, SDEL, or KETQ (Supplemental Fig. S2). These subfamily-specific distributions of targeting signals have not been reported before and indicate that AALP-like and CEP1-like proteases function in the ER and vacuole, respectively.
The Granulin-Containing PLCPs
Some of the PLCPs carry a C-terminal extension, consisting of a Pro-rich domain followed by a granulin-like domain. Granulins were originally described in the animal kingdom and are growth hormones that are released upon wounding (Bateman and Bennett, 2009). Granulins are usually encoded as an array of granulin domain (progranulin) that can be processed. The fusion of the granulin domain with a PLCP, however, only occurs in the plant kingdom, and granulin domains not fused to PLCPs have not been detected in plants.
The plant granulin-containing PLCPs are specific to RD21A-like proteases (subfamily 1) and XBCP3-like proteases (subfamily 4; Fig. 2A). However, not all PLCPs of these two subfamilies carry a granulin domain. There are two possible scenarios for the polymorphic nature of the granulin domain in these subfamilies. In the loss-of-domain scenario, the granulin-lacking PLCPs evolved from PLCP-granulin fusions by domain deletion. In the gain-of-domain scenario, the PLCP-granulin fusions evolved by domain fusion. Interestingly, when plotted on the phylogenetic trees of the subfamilies, the granulin-lacking proteases are scattered throughout both subfamilies (Fig. 2). This indicates that the granulin polymorphism evolved by the loss-of-domain scenario.
The phylogenetic tree based on the prodomain and protease domains also indicates that the proprotease domains of RD21A-like and XBCP3-like proteases are distinct from each other (Figs. 1A and 2B). This subdivision is different from the preceding analysis made by Beers et al. (2004), who placed both subfamilies in superfamily C1A-1. To investigate if our subdivision is correct, we performed phylogenetic analysis of the granulin domains only. This analysis shows that the granulin domains fall into two distinct groups: one group contains all granulin domains from RD21A-like proteases (subfamily 1) and the second group contains all granulin domains from XBCP3-like proteases (subfamily 4; Fig. 2, C and D). Thus, the separation of granulin-containing PLCPs into the two subfamilies based on the pro and protease domains is supported by phylogenetic separation based on the granulin domain. Furthermore, these data support the loss-of-domain scenario, as they indicate that the proprotease domains have coevolved with the granulin domains in two distinct subfamilies.
The granulin domain is approximately 60 amino acids long and contains 14 Cys residues in a remarkably conserved pattern (Cx5Cx5CCCx7Cx4CCx6CCx5CCx6Cx6C; Fig. 2D). Alignment with human granulin A (hGrnA) reveals the same pattern of Cys residues, but the plant granulins have one additional pair of Cys residues that reside in a region that is absent in hGrnA (Fig. 2D). The crystal structure of hGrnA has been resolved by NMR (2jye; Tolkatchev et al., 2008). We modeled the granulin domain of RD21A using 2jye as a template to predict the disulfide bridging in plant granulins. hGrnA folds as series of three pairwise antiparallel β-sheets that are connected by six disulfide bridges (Tolkatchev et al., 2008). The two extra Cys residues in RD21A are in close proximity, suggesting that they are making a disulfide bond that stabilizes a plant-specific loop that is absent in hGrnA (Fig. 2E; Tolkatchev et al., 2001). This analysis indicates that the disulfide bridging in the plant granulin domains is as follows: 1-3, 2-7, 4-9, 5-6, 8-11, 10-13, and 12-14 (Fig. 2D). This intense disulfide bridging probably serves to provide stability to the granulin domain in proteolytic environments.
Structural Features Are Subfamily Specific
We further investigated class-specific structural features in the PLCP subfamilies. These structural features are illustrated with a crystal structure of papain (1ppp; Kim et al., 1992), which belongs to XCP2-like PLCPs (subfamily 3) and represents subfamilies 1 to 8. CTB3-like proteases (subfamily 9) are illustrated with a crystal structure of human cathepsin B (4csb; Turk et al., 1995; Fig. 3A). PLCPs fold in two domains: an α-helix domain (left) and a β-sheet domain (right), forming the substrate-binding cleft and the catalytic triad in between (Fig. 3A). PLCPs of all subfamilies carry two pairs of highly conserved Cys residues that make disulfide bridges 1 and 2 in the α-helix domain (Fig. 3). Cys residues for disulfide bridge 3 are present in all subfamilies, except for CTB3-like proteases (subfamily 9), which lack the first Cys residue, leaving one conserved unpaired Cys residue in the C terminus (Fig. 3). Interestingly, RD19A-like proteases (subfamily 7) carry two distinct extra pairs of highly conserved Cys residues. Modeling of RD19A using papain as a template indicates that these four Cys residues make Cys bridges 4 and 5, which stabilize the α-helix and β-sheet domains, respectively (Fig. 3). Four additional disulfide bridges (Cys bridges 6–9) stabilize the α-helix domain and are highly conserved and unique to CTB3-like proteases (subfamily 9; Fig. 3). Notably, we identified a double Cys in the catalytic site of SAG12-like proteases (subfamily 6; Fig. 3B). Furthermore, AALP-like proteases (subfamily 8) carry a conserved Cys in the C terminus of both the prodomain and the protease domain.
Figure 3.
Conserved structural features in PLCP subfamilies. A, Positions of disulfide bridges and PGSs mapped onto the crystal structure of papain (1ppp) and cathepsin B (CathB; 3k9m). Cartoon models show the enzymes from the side with the α-helix domain (left) and β-sheet domain (right) and the catalytic triad (dotted spheres) with the catalytic Cys (cyan sticks) on top. The color code is as follows: α-helix (cyan); β-sheet (purple); loop (pink); extendable loop (blue); disulfide bridge (red); PGS (green). Numbers 1 to 9 correspond to conserved putative disulfide brides, summarized in B and C. B, Summary of the positions of putative disulfide bridges and PGSs in the mature protease domain. Positions are indicated for catalytic residues C, H, and N (black dashed lines), PGSs (NxS/T; green lines), and putative disulfide bridges (red lines). C, Frequency of conserved putative disulfide bridges, a double catalytic Cys (CCW), and PGS (NxS/T) in the different subfamilies. [See online article for color version of this figure.]
We next investigated the location of putative N-glycosylation sites (PGSs; with sequence NxS/T). Many PLCPs have one or two PGSs at nonconserved positions (data not shown). Three PGSs, however, are conserved at subfamily-specific positions. PGS1 and PGS3 are common for XCP2-like proteases (subfamilies 3) and CTB3-like proteases (subfamily 9), whereas PGS2 occurs in RD19A- and AALP-like proteases (subfamilies 7 and 8, respectively; Fig. 3, A and B). All PGSs are in the α-helix domain, but at different positions. The positions of PGS1 and PGS3 are different when plotted onto the crystal structures of papain and cathepsin B, respectively (Fig. 3A).
Polymorphic Labeling Profiles for Arabidopsis PLCPs
To characterize the PLCPs biochemically, we selected 10 Arabidopsis enzymes as representatives of different subfamilies and cloned them into binary vectors for Agrobacterium tumefaciens-mediated transient overexpression in Nicotiana benthamiana (agroinfiltration; Van der Hoorn et al., 2000; Voinnet et al., 2003). Epitope tags were not included in the constructs, since they are frequently degraded in the proteolytic environment created when proteases are overexpressed (data not shown).
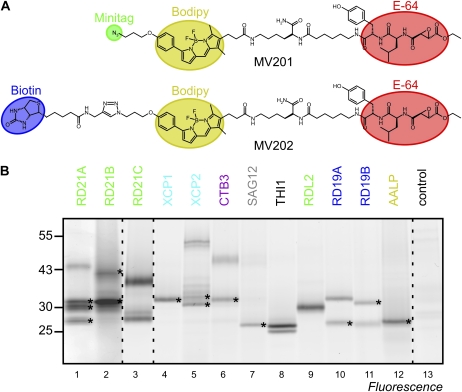
To quantitatively display the activities of different PLCP isoforms, we designed and synthesized MV201, an activity-based probe that labels the active site Cys residue of PLCPs. MV201 is a fluorescent derivative of PLCP inhibitor E-64 and is similar to the biotinylated activity-based probe DCG-04 (Greenbaum et al., 2000), which has been frequently used to determine PLCP activities in plants (van der Hoorn et al., 2004; Rooney et al., 2005; Gilroy et al., 2007; Martínez et al., 2007; Tian et al., 2007; Shabab et al., 2008; van Esse et al., 2008; Song et al., 2009; Kaschani et al., 2010; Lampl et al., 2010). Labeling with the fluorescent MV201 is detected and quantified from protein gels by fluorescent scanning.
Labeling of extracts of agroinfiltrated leaves revealed that all 10 tested PLCPs react with MV201 (Fig. 4B). Interestingly, the fluorescent signals are highly polymorphic between the different PLCPs (Fig. 4B). Single signals were detected only for XCP1 and RDL2 at 30 kD and for SAG12 and AALP at 25 kD (Fig. 4B, lanes 4, 9, 7, and 12, respectively). Two signals were detected for THI1 at 15 kD, two for CTB3 at 30 and 40 kD, and two for both RD19A and RD19B at 25 and 30 kD (Fig. 4B, lanes 6, 8, 10, and 11, respectively). A single 30-kD signal appeared for XCP2 (Supplemental Fig. S3), but a ladder of signals at 30 to 40 kD and 50-kD signals appeared when frozen leaf extracts were used (Fig. 4B, lane 5). Labeling of the other PLCPs did not display mobility shifts between fresh extracts or frozen extracts. Three or more signals appeared for RD21A, RD21B, and RD21C (Fig. 4B, lanes 1–3). Signals in the 40-kD regions correspond to the granulin-containing proteases, and the 25- to 30-kD signals are caused by different isoforms of the mature protease domains (Yamada et al., 2001).
Figure 4.
Protease activity profiling of representative PLCPs. A, Structures of MV201 and MV202. The E-64-based inhibitor group (red) contains an epoxide reactive group and a dipeptide carrying Leu (P2) and Tyr (P3) and is linked to the BODIPY fluorescent group (yellow) and either an azide minitag (green) or biotin (blue). B, PLCPs react with MV201. PLCPs were overexpressed in N. benthamiana by agroinfiltration in the presence of p19 silencing inhibitors. Extracts of agroinfiltrated leaves were labeled with 2 μm MV201 at pH 6 for 1 h, and labeled protein was detected from protein gels using fluorescence scanning. Asterisks indicate that, for identification purposes, proteins were labeled with MV202 and identified by in-gel digestion with trypsin and MS. This gel is a representative of at least three independent labeling experiments. [See online article for color version of this figure.]
PLCP Labeling Occurs at Distinct pH Ranges
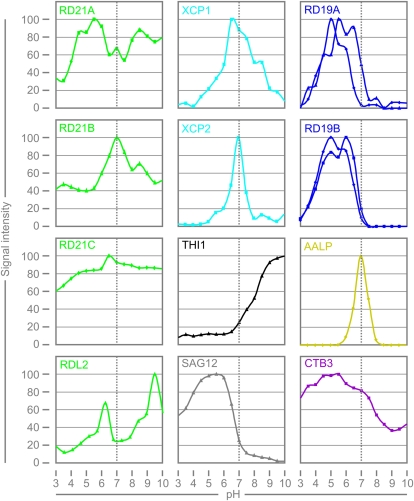
Since PLCPs act at different locations inside and outside the cell, they are exposed to different microenvironments that differ (e.g. in pH). To test if PLCP labeling depends on pH, we performed protease activity profiling at different pH levels and quantified the labeled signals. This analysis revealed distinct differences in the pH dependencies of PLCP labeling (Fig. 5). RD21A, RD21B, RD21C, RDL2, and CTB3 can be labeled at any pH to different degrees, but labeling of the other PLCPs is pH sensitive. Labeling of AALP occurs at a narrow, neutral pH range. Labeling of XCP1 and XCP2 also occurs at neutral pH, but the range of labeling is wider. In contrast, SAG12, RD19A, and RD19B can only be labeled at acidic pH (pH 4–6.5), whereas THI1 labeling increases significantly at basic pH (pH 7–10). These distinct pH sensitivities are remarkably different and may reflect that these proteases act at various locations in the cell where their activities are specified by controlled pH.
Figure 5.
pH-dependent labeling of PLCPs. N. benthamiana leaves overexpressing different PLCPs were labeled with 2 μm MV201 for 1 h at different pH levels. Fluorescent signals were quantified from protein gels by fluorescence scanning and plotted against pH. Each pH series was repeated at least once with similar outcome. [See online article for color version of this figure.]
Inhibitory Fingerprinting Displays Common and Unique Inhibitor Sensitivities
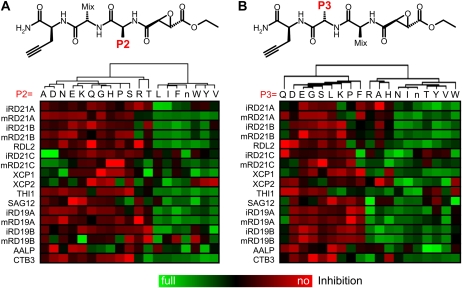
To determine if the PLCPs have distinct sensitivities for inhibitors, we screened a peptide-epoxide library carrying different amino acids at the P2 and P3 positions for their ability to prevent labeling with MV201. Two libraries were synthesized and tested. The P2 library contains fixed amino acids at the P2 position (Fig. 6A) and a mixture of amino acids at the P3 position, whereas the P3 library contains fixed amino acids at the P3 position and a mixture of amino acids at the P2 position (Fig. 6B). All natural amino acids except Cys and Met were included at both the fixed and the mixed positions, and the nonnatural nor-Ile (n) was included to replace Met. To determine inhibitor specificity, protease-containing extracts were preincubated with the compound mixtures and then labeled with MV201. Fluorescent signals were quantified from protein gels, and a heat map of the inhibition was calculated for the most abundant protease-derived signals (Fig. 6).
Figure 6.
PLCPs have distinct inhibitor sensitivities. Inhibitory fingerprinting is shown using P2 (A) and P3 (B) scanning epoxide libraries. These libraries contain a fixed amino acid at the P2 or P3 position, respectively, and an isokinetic mixture of 19 amino acids at the P3 or P2 position, respectively (top). Extracts containing different PLCPs were preincubated with a 10 μm epoxide library for 30 min and then labeled with 2 μm MV201 for 2 h. Labeled proteins were quantified from fluorescent gels and normalized relative to the median signal. The data were quantified and clustered based on the similarity of inhibition profiles. Similar inhibition data were obtained with repetition experiments.
The amino acid at the P2 position has a strong effect on the inhibitory ability of the peptide epoxides. Generally, peptide epoxides carrying hydrophobic amino acids (L, I, F, n, W, Y, V) at the P2 position are good inhibitors for most PLCPs, whereas peptide epoxides carrying small or hydrophilic amino acids (A, D, E, K, Q, G, H, P, S, R, T) at the P2 position are poor inhibitors (Fig. 6A). A few PLCPs (RD21C, XCP1, XCP2, and AALP) make some exceptions to these general rules.
The amino acid at the P3 position also has a strong effect on the inhibitor ability of peptide epoxides, but here the general rules are different. Peptide epoxides carrying various amino acids (N, I, n, T, Y, V, W) are generally good inhibitors, whereas those carrying others (D, E, G, S, L, K, P, F) are generally poor inhibitors (Fig. 6B). Peptide epoxides carrying three of the amino acids (R, A, H) are good inhibitors of RD19As and poor inhibitors of RD21As.
If a protease has two different isoforms, then the inhibitory profile is generally similar between the two isoforms. For example, intermediate and mature isoforms of RD21A behave similarly for P2 and P3 scanning libraries (Fig. 6), indicating that these isoforms cannot be discriminated using inhibitors. The isoforms are also indistinguishable for RD21B, RD21C, RD19A, and RD19B. However, some exceptions are noted for RD19B (P2 library) and RD21C (P3 library).
When inhibitory fingerprints of different proteases within one family are compared, there are both similarities and clear differences. RD21A and RD21B, for example, have similar inhibitory fingerprints, and also RD19A and RD19B behave similar to each other (Fig. 6). The similarity of inhibitory sensitivities of these protease pairs indicates that also the substrate specificity may be similar. Differences between proteases in the same subfamily are also observed. RD21C, for example, is different from RD21A/B, and XCP1 is different from XCP2 (Fig. 6). The differences in inhibitor sensitivity indicate that these proteases may have different substrate specificities.
Detection of Common and Organ-Specific PLCP Activities
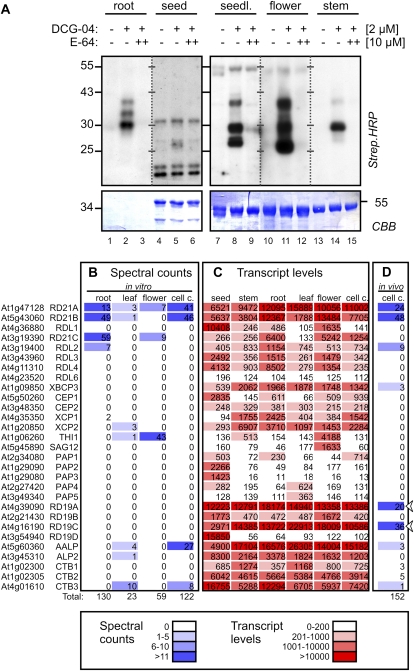
Protease activity profiling on extracts of various Arabidopsis organs with DCG-04 revealed that PLCP activities can be detected in all organs to different degrees (Fig. 7A). Specific signals are absent in the no-probe control and upon pretreatment with the PLCP inhibitor E-64. Interestingly, activity profiles seem similar but differ in details between the different organs (Fig. 7A). Root extracts generated signals at 30, 35, and 40 kD, whereas seed extracts only generated a 25-kD signal (Fig. 7A, lanes 2 and 5, respectively). Seedling extracts generated doublet signals at 25 and 30 kD and a single signal at 40 kD (Fig. 7A, lane 8). Flower extracts generated doublets at 20 and 30 kD and one signal at 40 kD (Fig. 7A, lane 11). Extracts from stem generated a strong 30-kD signal and weaker signals at 25 and 40 kD (Fig. 7A, lane 14).
Figure 7.
PLCP activity and expression in different organs and in vivo. A, Activity profiles of PLCPs of different organs. Crude extracts of various organs were labeled with 2 μm DCG-04, either with or without a 30-min pretreatment with E-64 (10 μm), for 2.5 h at pH 6. Biotinylated proteins were detected on protein blots using streptavidin-horseradish peroxidase. CBB, Coomassie Brilliant Blue. B, Spectral counts of labeled PLCPs detected in different organs. Extracts from various organs were labeled with MV202 at pH 6, and labeled proteins were purified on avidin beads, separated on protein gels, excised, digested with trypsin, and analyzed by LC-MS/MS. The spectrum of peptides with greater than 95% confidence was counted for each PLCP; these are summarized in Supplemental Table S1. C, Transcript levels of all PLCPs in different organs. These data were extracted from Genevestigator (Hruz et al., 2008). D, In vivo labeling of PLCPs reveals activities of RD19A and RD19B. Arabidopsis cell cultures were labeled with 5 μm MV201. Proteins were extracted and coupled to Bio≡ using click chemistry. Biotinylated proteins were purified on avidin beads, separated on denaturing acrylamide gels, excised, digested with trypsin, and analyzed by LC-MS/MS. The spectrum of peptides with greater than 95% confidence was counted for each PLCP. [See online article for color version of this figure.]
We determined the identities of the labeled proteins from root, leaf, and flower by enriching the DCG-04-labeled proteins on streptavidin beads. Biotinylated proteins were separated on protein gels, stained, excised, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Spectra with high-quality scores for being peptides from PLCPs were counted and are summarized in Figure 7B and Supplemental Table S1. We could identify RD21A in root, leaf, and flower (Fig. 7B), consistent with the 40-kD signal seen in the activity profiles (Fig. 7A). In contrast, AALP was detected in leaves but not in roots and flowers (Fig. 7B), consistent with the absence of a 25-kD signal from activity profiles from these organs (Fig. 7A). Interestingly, the majority of the spectral counts from flowers are from THI1 (Fig. 7B). The abundance of this protease may explain the strong 20-kD signal in the flower activity profile (Fig. 7A, lane 11), which is consistent with the size obtained by labeling transiently expressed THI1 (Fig. 4B). RD21B and RD21C were detected in roots (Fig. 7B) and may explain the 30- and 35-kD signals in the root activity profiles (Fig. 7A).
The identified PLCPs from different organs are consistent with the mRNA levels of PLCPs in different organs (Hruz et al., 2008). For instance, RD21A and AALP are expressed in root, leaf, stem, and flower but not in seed (Fig. 7C). This is consistent with the MS data and the presence of 40- and 25-kD signals in the activity profiles (Fig 7, A and B). THI1 is highly expressed in flowers (Fig. 7C), hence its identification and the flower-specific 20-kD signal (Fig. 7, A and B). RD21B and RD21C are highly expressed in roots (Fig. 7C), where they were identified by MS (Fig. 7B). Thus, the PLCPs that were identified by MS are also highly expressed in the different organs. However, not all highly expressed proteases were also identified by MS. The expression of RD19A and RD19C, for example, is notably high in all plant tissues, yet the corresponding proteases have not been identified in any of the MS experiments, despite the fact that RD19A can react with DCG-04 when overexpressed by agroinfiltration (Fig. 4B).
In Vivo Labeling Expands PLCP Detection
The MS experiments described above were performed on extracts at chosen labeling conditions (e.g. pH 6). Since the compartmentalization is lost in leaf extracts, it is possible that PLCPs are artificially activated and inactivated. To investigate which PLCPs are active in living tissue, we labeled cell cultures in vivo using MV201, which carries an azide minitag (Fig. 4A). After labeling, azide-labeled proteins were biotinylated with alkyne-biotin using “click chemistry” (Kaschani et al., 2009b), and the resulting biotinylated proteases were purified and analyzed by MS. Interestingly, besides RD21A, RD21B, and AALP, which were also detected after labeling cell culture extracts, we also detected RDL2, RD19A, and RD19B (Fig. 7D; Supplemental Table S1). Thus, RD19A and RD19B activities can be detected in living tissues but not in tissue extracts, unless they are overexpressed. These data indicate that in vivo labeling greatly expands the number of PLCPs that can be detected.
DISCUSSION
Despite the importance of protein degradation, plant PLCPs have not been extensively characterized before. Here, we exploited the large number of publicly available sequences to subclassify plant PLCPs into nine subfamiles, revealing subfamily-specific features. Some features, like NPIR, KDEL, and ERFNIN, were noted earlier, but their distribution in the PLCP superfamily has not been analyzed before. Other features, like the double Cys in the catalytic site of SAG12-like proteases and the additional disulfides in RD19A-like proteases, have not been noted before and might be important for the function of these subfamilies. Biochemical analysis of representatives from the different subfamilies shows that these proteases exist in different active isoforms, have a distinct pH requirement, and show different sensitivities toward inhibitors. Finally, activity profiling on various Arabidopsis organ extracts reveals different PLCP activities, and even more PLCPs were detected when profiling was performed in vivo. These observations and functional tools are instrumental for further research of the PLCP superfamily.
Classifiers of PLCP Subfamilies
We divided the plant PLCP superfamily into nine subfamilies, based on phylogenetic analysis and conserved functional and structural features. The facts that these subfamilies appear in previous studies (Beers et al., 2004), are conserved between poplar and Arabidopsis (García-Lorenzo et al., 2006), and also exist in moss and green algae (Martinez and Diaz, 2008) indicate that this subclassification is valid and has biological relevance. A number of obvious subfamily-specific sequence features can be used as classifiers for newly identified PLCPs (Table I). If the PLCP carries a KDEL or HDEL ER-retrieval signal or the NPIR vacuolar targeting signal, then it probably belongs to CEP1-like (subfamily 2) or AALP-like (subfamily 8) proteases, respectively. The presence of the double Cys in the catalytic site (CCW) is indicative of SAG12-like proteases (subfamily 6). The presence of the ERFNAQ motif in the prodomain and the presence of two additional disulfide bridges are classifiers for RD19A-like proteases (subfamily 7). The absence of ERFNIN/ERFNAQ motifs in the prodomain and the presence of a distinct set of six disulfide bridges are indicative for CTB3-like proteases (subfamily 9). Also, the presence of conserved putative N-glycosylation sites (PGS1–PGS3) could aid in quick subclassification.
Table I.
Sequence-based classifiers of plant PLCP subfamilies
| Subfamily | Classifiers |
| RD21A-like | C-terminal granulin domain |
| CEP1-like | {K/H}DEL ER-retrieval signal at the C terminus |
| XCP2-like | Carries a conserved PGS in the TGNL{S/T} motif in the protease domain |
| XBCP3-like | C-terminal granulin domain |
| THI1-like | No distinct features noted |
| SAG12-like | Carries an extra Cys before the catalytic Cys (CGCCWAFS motif) |
| RD19A-like | ERFNAQ instead of ERFNIN in the prodomain; two extra disulfide bridges in the protease domain (4 and 5); conserved PGS in the VxNF{S/T} motif in the middle of the protease domain |
| AALP-like | NPIR vacuolar targeting signal at the N terminus of the prodomain; extra Cys in the ATC motif in the C terminus of the protease domain; extra Cys in the CSAT motif at the C terminus of the prodomain; conserved PGS in the VNIT motif in the middle of the protease domain |
| CTB3-like | No ERFNIN/ERFNAQ motif in the prodomain; no disulfide bridge 3: unpaired Cys in the ECGIE motif in the C terminus; four extra disulfide bridges (6–9); PGS located in the nonconserved loop between disulfide bridges 6 and 2 |
However, not all PLCPs can be classified using these features. Even within each subfamily, there are some exceptions for these classifiers. The presence of the granulin domain, for example, would classify the PLCP to RD21A-like (subfamily 1) or XBCP3-like (subfamily 4) proteases, but these families also contain many PLCPs that do not carry a granulin domain. In case none of the classifiers are present in the sequence, a BLAST search to identify the closest PLCP subfamily is needed to classify the PLCP.
Similarities and Differences of PLCPs and Animal Cathepsins
Some of the features described above are absent in the type members of the C1A family, the human cathepsins. Human cathepsin F (Q9UBX1) is closest to RD19A-like proteases (subfamily 7) and shares a signal peptide, ERFNAQ motif, and catalytic residues but lacks the extra disulfide bridges 4 and 5 and conserved PGS2. Human cathepsin H (P09668) is most closely related to AALP-like proteases (subfamily 8) and shares a signal peptide, ERFNIN motif, two unpaired Cys residues, and PGS2 but lacks the NPIR vacuolar targeting signal. Furthermore, human cathepsin B (P07858) is related to CTB3-like proteases (subfamily 9) and shares the signal peptide, a prodomain lacking an ERFNIN motif, catalytic residues, and all Cys residues but lacks conserved PGS3. Finally, human cathepsin L (P07711) is closest to subfamilies 1 to 6 and contains a signal peptide, ERFNIN motif, catalytic residues, and conserved Cys residues but lacks a granulin domain, ER-retrieval motif, or conserved PGS1. In conclusion, although the similarities with human cathepsins are high, plant PLCPs have distinct targeting signals and conserved glycosylation sites that are absent in human cathepsins.
Putative Biological Roles of Subfamily-Specific Features
The subfamily-specific features are of biological relevance. The importance of NPIR and KDEL/HDEL localization signals is evident, since they target the proteases to specific cellular locations. We speculate that the presence of the additional Cys residue in the catalytic site of SAG12-like proteases may also have functional relevance. The extra Cys may act as an alternative catalytic residue or redox sensor (Weerapana et al., 2010). It is interesting that also the defense-related protease RCR3 and PIP1 of tomato belong to the SAG12 subfamily and carry a CCW motif. The additional disulfide bridges in RD19A-like protease and CTB3-like proteases may increase their stability in a proteolytic environment. This would indicate that the microenvironment of these proteases might be different from that of the other proteases.
PLCPs Have Distinct Biochemical Properties
Our biochemical studies reveal that PLCPs exist in multiple active isoforms. The molecular basis and the biological relevance of these isoforms are not clear, but we anticipate that these isoforms also exist under native conditions in Arabidopsis. Different active RD21A isoforms, for example, have been detected in Arabidopsis previously (Yamada et al., 2001).
Labeling of the different PLCPs depends strongly on pH. pH-dependent labeling reflects a property of the protease and not of the probe. Some caution is needed while interpreting these results, since the absence of labeling could be caused by protease degradation, even though these proteases are thought to sustain proteolytic environments. The distinct pH dependency is remarkable and probably reflects the fact that these proteases act in different microenvironments inside the cell. Subfamily-specific biochemical properties are evident from the facts that PLCPs within the family behave similarly (e.g. XCP1/XCP2 and RD19A/RD19B) and PLCPs from different subfamilies have distinct pH-sensitive labeling profiles.
Inhibitory fingerprinting revealed that most PLCPs have common sensitivity for inhibitors carrying certain amino acids at the P2 and P3 positions. There are, however, also some differences in inhibitor sensitivity between PLCPs for these inhibitors. Some of these differences are common to different members of the subfamily, while others may distinguish different members of the same subfamily. Similar observations were made by inhibitory fingerprinting of human cathepsins (Greenbaum et al., 2002). The distinct inhibitor fingerprints give an impression of the differences of the substrate-binding sites of the proteases and therefore are of functional relevance. However, this study should be followed up by screening a next-generation inhibitor library having fixed P2 and P3 positions to confirm the general observations made here. The specificity of peptide epoxides could significantly increase when both P2 and P3 positions are fixed. These studies may also result in the development of activity-based probes that are more specific for one particular PLCP subfamily.
Detecting More PLCP Activities by Protease Activity Profiling
Protease activity profiling only displays a fraction of the 30 PLCPs that are encoded by the Arabidopsis genome. Obviously, many PLCPs are not detected because the corresponding genes are not expressed in the tissues used in these studies. However, some PLCPs are highly expressed in leaf tissue but were not detected by protease activity profiling. RD19A, for example, is highly expressed in many tissues, yet it was never identified when labeling tissue extracts (van der Hoorn et al., 2004; van Esse et al., 2008; this paper). Detecting RD19A activity is of particular interest, since this protease interacts with the bacterial type III effector PopP2 and relocalizes to the nucleus (Bernoux et al., 2008). The detection of RD19A by protease activity profiling on living cells is remarkable and is consistent with previous findings. RD19A was also detected by in vivo labeling with vinyl-sulfone probes (Kaschani et al., 2009b). These vinyl-sulfone probes cause relatively strong labeling of PLCPs in vivo but not in vitro (Gu et al., 2010), consistent with the labeling of RD19A in living cells. These data indicate that RD19A is active in living tissues but its activity is suppressed in extracts. Alternatively, the probe may concentrate in RD19A-containing vesicles, causing enhanced RD19A labeling in vivo. Besides RD19A, several other PLCP activities were detected upon in vivo labeling.
In conclusion, we have subdivided the plant PLCP superfamily into nine functional subfamilies. In silico analysis revealed several subfamily-specific features that can be used as classifiers. Biochemical characterization of representatives of the subfamilies indicates pH-dependent activities and inhibitor sensitivities that are subfamily specific. Several new PLCP activities were detected in extracts of various Arabidopsis organs and upon in vivo labeling. These discoveries will greatly facilitate the further functional characterization of this important protease family in plants.
MATERIALS AND METHODS
Construction of the Plant PLCP Database
Plant PLCP sequences were retrieved from the TIGR database (Childs et al., 2007) using local BLAST searches with the 31 Arabidopsis (Arabidopsis thaliana) PLCP protein sequences (Altschul et al., 1990). Nucleotide sequences were retrieved and translated in all six open reading frames. Open reading frames smaller than 100 amino acids and sequences not containing the characteristic CWAF-like sequence ([S|C|G][W|C][A|S|T][F|V]) were removed. Identical sequences were kept if they were from different species. This resulted in a TIGR PLCP database with 317 PLCP sequences.
An NCBI PLCP database was generated by BLASTing the protein sequences from Beers et al. (2004) against the NCBI protein database, restricting the search to plant species. All the obtained GI numbers were pooled, duplicate entries were removed, and sequences were retrieved from the NCBI batch download site. Arabidopsis sequences, all sequences from x-ray projects, sequences that were too short, and sequences not containing the CWAF sequence ([S|C|G][W|C][A|S|T][F|V]) were removed, resulting in an NCBI PLCP database containing 794 plant PLCP sequences.
The TIGR PLCP database and the NCBI PLCP database were combined and supplemented with the 31 Arabidopsis PLCP sequences. We also added and replaced several sequences that were of particular interest to our research but were not present in the screened databases (e.g. LeCatB1, LeCatB2, LeALP, NbC14; Shabab et al., 2008; Kaschani et al., 2010). Next, we removed duplicate entries and submitted the resulting 1,106 sequences to a PFAM search (Finn et al., 2010). All sequences not containing the Peptidase_C1 domain were removed from the database. This step was included to ensure that only PLCPs would enter the subsequent phylogenetic analysis. The remaining database contained 1,034 entries. Next, we counted the distance from the CWAF sequence to the C and N termini. Database entries with an N-terminal distance of less than 50 and a C-terminal distance of less than 150 were removed. These sequences were considered to be fragments and not useful for the phylogenetic analysis (Baldauf, 2003). Also, we noticed that several sequences were exceptionally long. These sequences were also removed or trimmed.
The remaining database with 789 unique plant PLCP entries was then used to generate a first alignment with ClustalW2 (Larkin et al., 2007; settings were as follows: pairwise alignment, gap opening 35, gap extension 0.75; multiple alignment, gap opening 15, gap extension 0.3, delay divergence 25%). Sixty-six sequences did not align well and had to be removed (723 sequences remained). A second alignment with ClustalW2 resulted in a better alignment. In order to remove the phylogenetically uninformative gaps in the alignment (Talavera and Castresana, 2007), the alignment was submitted to the trimAl program (Capella-Gutiérrez et al., 2009) with the setting –gt 0.8. This setting removed gap columns if they were found in more than 20% of the sequences. The obtained alignment was then refined once more by resubmitting to ClustalW2. A bootstrap tree was generated from this alignment using ClustalW2 (standard settings, correct for multiple substitutions, bootstrap 100). The tree was annotated and edited in TreeDyn version 198.3 (Chevenet et al., 2006).
Chemicals and Antibiotics
All chemicals were supplied by Sigma, Roth, Merck, and Promega. DCG-04 was synthesized as described previously (Greenbaum et al., 2000). E-64d was purchased from Sigma.
Synthesis of Azido-BODIPY-DCG04 (MV201)
To a solution of Azido-BODIPY-OSu (Verdoes et al., 2008; 59 mg, 0.105 mmol) and N,N-diisopropylethylamine (20 μL, 0.115 mmol, 1.1 equivalents) in dimethylformamide (DMF; 1 mL), a solution of DCG-04 amine (Greenbaum et al., 2002; 78 mg, 0.115 mmol, 1.1 equivalents) in DMF (1 mL) was added, and the reaction mixture was stirred for 16 h before being concentrated in vacuo. Purification by flash column chromatography (5% methanol in dichloromethane [DCM] → 10% methanol in DCM) afforded the title compound (110 mg, 98 μmol, 93%) as a deep-red solid. 1H NMR (400 MHz, CDCl3/MeOD [for deuterated methanol]): δ ppm 7.87 (d, J = 8.9 Hz, 2H), 7.63 (s, 1H), 7.25 to 7.20 (m, 1H), 7.17 to 7.10 (m, 1H), 7.05 to 6.95 (m, 5H), 6.73 (d, J = 8.5 Hz, 2H), 6.57 (d, J = 4.1 Hz, 1H), 4.47 (t, J = 7.5 Hz, 1H), 4.41 (t, J = 7.3 Hz, 1H), 4.33 to 4.23 (m, 3H), 4.13 (t, J = 6.0 Hz, 2H), 3.67 (d, J = 1.8 Hz, 1H), 3.57 (d, J = 1.8 Hz, 1H), 3.55 (t, J = 6.6 Hz, 2H), 3.21 to 3.11 (m, 3H), 3.04 (dd, J1 = 13.5, J2 = 6.8 Hz, 1H), 2.97 (dd, J1 = 13.8, J2 = 7.6 Hz, 1H), 2.86 (dd, J1 = 13.7, J2 = 7.5 Hz, 1H), 2.75 (t, J = 7.5 Hz, 2H), 2.53 (s, 3H), 2.31 (t, J = 7.5 Hz, 2H), 2.25 (s, 3H), 2.19 (t, J = 7.5 Hz, 2H), 2.08 (p, J = 6.3 Hz, 2H), 1.82 to 1.71 (m, 1H), 1.66 to 1.50 (m, 6H), 1.49 to 1.34 (m, 6H), 1.33 (t, J = 7.1 Hz, 3H), 1.22 to 1.11 (m, 2H), 0.91 (dd, J1 = 13.7, J2 = 6.0 Hz, 6H).
Synthesis of Azido-BODIPY-DCG04 (MV202; MVB093)
To a solution of MV201 (15 mg, 13.3 μmol) in tert-butanol (0.5 mL) was added propargylamine-biotine (Verdoes et al., 2008; 7.5 mg, 26.6 μmol, 2 equivalents), CuSO4 (250 μL of 5.32 mm in water, 1.33 μmol, 10 mol %), and sodium ascorbate (250 μL of 10.64 mm in water, 2.66 μmol, 15 mol %). After the addition of toluene (0.5 mL), the solution was stirred vigorously at 80°C for 6 h. Excess toluene was added, and the mixture was concentrated in vacuo before purification by flash column chromatography (acetone → 5% water in acetone) to afford the title compound (17.4 mg, 12.4 μmol, 93%). 1H NMR (400 MHz, MeOD) δ = 7.91 to 7.81 (m, 3H), 7.42 (s, 1H), 7.25 to 7.08 (m, 5H), 7.07 (d, J = 4.1, 1H), 7.01 (d, J = 8.4, 2H), 6.94 (d, J = 8.8, 2H), 6.69 (d, J = 8.4, 2H), 6.60 (d, J = 4.1, 1H), 4.61 (t, J = 6.6, 2H), 4.50 to 4.36 (m, 5H), 4.31 to 4.17 (m, 4H), 4.03 (t, J = 5.8, 2H), 3.66 (d, J = 1.7, 1H), 3.57 (d, J = 1.7, 1H), 2.99 (m, 9H), 2.75 (t, J = 7.3, 2H), 2.50 (s, 3H), 2.38 (m, 2H), 2.25 (s, 3H), 2.19 (m, 4H), 1.81 to 1.11 (m, 38H), 0.91 (m, 6H). 13C NMR (101 MHz, MeOD) δ 177.14, 176.10, 175.95, 174.77, 173.68, 173.16, 168.73, 168.39, 160.78, 157.29, 141.84, 136.60, 131.89, 131.37, 129.92, 129.36, 129.21, 128.92, 127.20, 126.30, 124.73, 119.14, 116.25, 115.24, 65.72, 63.29, 63.23, 61.60, 56.98, 56.51, 54.40, 54.22, 53.42, 53.20, 49.64, 49.43, 49.21, 49.00, 48.79, 48.57, 48.45, 48.36, 41.65, 41.05, 40.34, 40.18, 38.22, 36.92, 36.65, 36.55, 35.62, 32.72, 30.91, 29.95, 29.85, 29.68, 29.41, 27.41, 26.68, 26.48, 25.89, 24.26, 23.31, 22.04, 21.34, 14.37, 9.62.
Synthesis of the Peptide Epoxide Library
Rink resin (0.63 g, 0.8 mmol g−1, 0.5 mmol) was treated with 20% piperidine in DMF for 20 min. After washing the resin with DMF (three times) and DCM (three times), it was reacted with Fmoc-propargylglycine (0.78 g, 1.25 mmol, 2.5 equivalents), O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluorophosphate (0.47 g, 1.25 mmol, 2.5 equivalents), and N,N-diisopropylethylamine (0.26 mL, 1.5 mmol, 3 equivalents) for 16 h. The P2 and P3 positional scanning libraries were synthesized following literature procedures (Ostresh et al., 1994; Nazif and Bogyo, 2001). In short, the P2 and P3 libraries were made up of 19 sublibraries containing each of the natural amino acids (without Cys and Met, plus nor-Leu) at the designated constant position. At the variable position, an isokinetic mixture of those same Fmoc-protected amino acids was coupled (Ostresh et al., 1994) in a 10-fold excess over the resin load with diisopropylcarbodimide and hydroxybenzotriazole as the condensating agents in DMF. The resin-bound, Fmoc-protected peptide libraries were deprotected with 20% piperidine in DMF for 20 min. The resin was washed with DMF (three times) and DCM (three times) before being capped with the warhead (2S,3S)-3-(ethoxycarbonyl)oxirane-2-carboxylic acid (Willems et al., 2010). The libraries were cleavage from the resin by treatment with TFA:TIS (97.5:2.5, v/v) for 2 h. The peptides were precipitated in ice-cold ether and lyophilized to dryness. The crude peptide libraries were dissolved in dimethyl sulfoxide (10 mm stock) based on average weights for each mixture.
Plant Material
Arabidopsis plants (ecotype Columbia) were grown under a 12-h light regime in growth cabinets at day/night temperatures of 24°C/20°C, respectively. Four- to five-week old plants were used for protein extraction for further experiments. The Arabidopsis cell culture (ecotype Landsberg erecta) was obtained from the Sainsbury Laboratory (John Innes Center) and maintained according to the method described by Kaffarnik et al. (2009). Nicotiana benthamiana was grown in a climate chamber under a 14-h light regime at 18°C (night) and 22°C (day). Four- to six-week-old plants were used for infiltration experiments.
Molecular Cloning
A summary of the cloning is provided in Supplemental Tables S2 and S3. The following plasmids have been described previously: pRH80 (Van der Hoorn et al., 2000); pRH385 (van der Hoorn et al., 2005); pTP5 and pFK26 (Shabab et al., 2008); and pMOG800 (Honée et al., 1998). The construction of the RD21A-expressing binary plasmid (pRH628) has been described elsewhere (Wang et al., 2008). The open reading frames encoding full-length PLCPs were amplified from cDNA clones or cDNA isolated from various Arabidopsis organs using primers summarized in Supplemental Tables S2 and S3. PCR fragments were subcloned into pRH80 or pFK26 (Supplemental Table S2). The cloned PCR fragments were verified by sequencing. Correct expression cassettes were shuttled into binary vectors as summarized in Supplemental Table S2, resulting in a collection of binary plasmids that all carry a T-DNA with a 35S-driven PLCP open reading frame, followed by a terminator of the potato PI-II gene.
Agroinfiltration
Transient overexpression of PLCPs by agroinfiltration was achieved as described previously (Shabab et al., 2008). Overnight-grown Agrobacterium tumefaciens cultures (strain GV3101) carrying binary plasmids were centrifuged, and bacteria were resuspended in infiltration buffer (10 mm MES, pH 5, 10 mm MgCl2, and 1 mm acetosyringone). Cultures were incubated for 2 to 4 h at room temperature. The optical density at 600 nm was adjusted to 2, and cultures carrying PLCP-expressing vectors were mixed with cultures carrying p19-expressing vectors. These cultures were infiltrated into fully expanded leaves of 4- to 6-week-old N. benthamiana plants. Infiltrated leaves were harvested after 3 to 5 d post infiltration.
Protease Activity Profiling of Extracts
Protein of agroinfiltrated leaves was extracted by grinding three fully expanded leaves in 5 mL of water in a mortar. Extracts of Arabidopsis organs were made by grinding tissues in 1 mL of water. The extracts were cleared by centrifugation (5 min at 13,000g), and extracts were divided into aliquots and stored at −80°C. Protein concentrations were determined by using the Reducing Agent Compatible/Detergent Compatible protein assay (Bio-Rad) following the manufacturer’s instructions.
In vitro labeling was carried out by incubating 100 μg of protein in a 100-μL final volume containing 25 mm sodium acetate buffers (pH < 7) or 50 mm Tris buffers (pH > 7) in the presence of 2 mm dithiothreitol. DCG-04, MV201, or MV202 was added at a 2 μm final concentration, and the sample was incubated for 2.5 h in the dark under gentle agitation. Equal volumes of dimethyl sulfoxide were added to the no-probe control. A pretreatment with 10 μm E-64 or epoxide peptides for 30 min before adding the probes was done during inhibition assays. Incubation was stopped by adding 20 μL of 4× SDS-PAGE loading buffer containing β-mercaptoethanol, and the samples were heated for 10 min at 90°C. The samples were shortly centrifuged (1 min at 13,000g) and separated by 12% SDS-PAGE (10 μg of protein per lane). The gels were washed with water, and fluorescently labeled proteins were detected using a Typhoon 8600 scanner (Molecular Dynamics) with a TAMRA filter and excitation and emission of 532 and 580 nm, respectively. Fluorescence signals were quantified using ImageQuant (Molecular Dynamics). To detect biotinylated proteins, the proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore). Membranes were incubated with streptavidin-horseradish peroxidase (Ultrasensitive; Sigma; dilution of 1:3,000), and signals were detected using enhanced chemiluminescence (Super Signal Femto/Pico substrate; Thermo Fisher Scientific) and x-ray films (Kodak).
Affinity Purification and Identification of Labeled Proteins
Purification was usually done by incubating approximately 5 mg mL−1 protein. Proteins of 4- to 5-week-old plants were extracted by grinding rosette leaves, roots, and flowers in an ice-cold mortar with 3 mL of water and subsequent centrifugation for 10 min at 20,000g. The supernatant was labeled with 10 μm MV202 in labeling buffer (50 mm NaOAc, pH 6) and 1 mm dithiothreitol for 2 h at room temperature under gentle agitation. The labeled plant extracts were applied to a phosphate-buffered saline (PBS)-equilibrated Econo-Pac 10DG-size exclusion column (Bio-Rad). Desalted samples were incubated with 100 μL of avidin beads (Sigma) for 1 h at room temperature under gentle agitation. The beads were collected by centrifugation (10 min at 2,000 rpm) and were washed twice with 1 mL of 0.1% SDS, twice with 1 mL of 6 m urea, once with 1 mL of 50 mm Tris, pH 8, containing 0.1% Tween 20, once with 1 mL of 0.1% Tween 20, and once with 1 mL of water. After washing with water three times, the beads were boiled in 50 μL of 1× SDS-PAGE loading buffer containing β-mercaptoethanol and separated on a 12% one-dimensional SDS-PAGE gel. Labeled proteins were visualized by in-gel detection of fluorescence on a Typhoon 8600 fluorescence scanner (GE Healthcare). Fluorescent bands were excised with a razor blade and placed into a 1.5-mL vial. The slices were washed twice with 500 μL of 100 mm ammonium bicarbonate solution (Sigma) for 15 min. Proteins were reduced for 30 min at 62°C with 10 mm Tris(2-carboxyethyl)-phosphine (Sigma) and alkylated with 55 mm iodoacetamide in the dark for 30 min at room temperature. Gel fragments were washed three times for 15 min with 500 μL of a 50:50 mixture of acetonitrile with 100 mm ammonium bicarbonate and two times with 50 μL of 100% acetonitrile to dehydrate the gel slices. The gel fragments were dried using an Eppendorf SpeedVac for 5 min. Dry gel slices were incubated for 10 min with 20 μL of 25 mm ammonium bicarbonate containing 10 ng μL−1 trypsin (Promega) at room temperature. The quelled gel pieces were covered with 25 mm ammonium bicarbonate, vortexed briefly, and incubated overnight (12–16 h) at 37°C under vigorous shaking. The supernatant was transferred to a new vial, and the gel slices were covered with 5% formic acid (Agros Organics) and incubated for 15 min at room temperature to inactivate the trypsin. Gel slices were washed three times (100, 75, and 50 μL) for 5 min with 100% acetonitrile. All supernatants were combined and concentrated in an Eppendorf SpeedVac to a final volume of approximately 10 μL. Tryptic peptides were analyzed using a Thermo Scientific LTQ XL mass spectrometer and annotated as described previously (Kaschani et al., 2009a).
Protease Activity Profiling in Vivo
Twenty milliliters of a 7-d-old Arabidopsis cell culture (ecotype Landsberg erecta; Kaffarnik et al., 2009) was allowed to settle down. The medium was replaced by an equal amount of fresh medium. One milliliter of the cell suspension was then transferred to a multiwell plate and incubated for 2 h in the presence of 5 μm MV201. The cells were washed three times with cell culture medium. The cell culture was transferred to a fresh Eppendorf tube and ground in water (final volume of approximately 250–300 μL). The protein concentration was determined, and approximately 1 mg of protein precipitated with acetone and dissolved in 1 mL (1× PBS, pH 7.4) supplemented with 1% SDS. The biotin-alkyne affinity tag was then chemically attached to the azide group of MV201-labeled proteins by click chemistry (Kaschani et al., 2009b). The click reaction mix was passed over a 10DG desalting column (Bio-Rad). The eluted proteome was diluted with 1× PBS to a final volume of 8.5 mL and supplemented with avidin agarose beads (Sigma). Affinity purification and MS were done as described above.
The following Arabidopsis proteins from the TAIR10 database have been studied experimentally in this paper: RD21A (At1g47128), RD21B (At5g43060), RD21C (At3g19390), RDL2 (At3g19400), XBCP3 (At1g09850), XCP1 (At4g35350), XCP1 (At1g20850), THI1 (At1g06260), SAG12 (At5g45890), RD19A (At4g39090), RD19B, (At2g21430), RD19C (At4g16190), AALP (At5g60360), ALP2 (At3g45310), and CTB3 (At4g1610).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of 723 plant PLCPs.
Supplemental Figure S2. Alignments of protein sequences of each subfamily.
Supplemental Figure S3. Labeled XCP2 is 30 kD in fresh extracts.
Supplemental Table S1. Spectral count of identified peptides.
Supplemental Table S2. Binary plasmids.
Supplemental Table S3. Primers.
Supplementary Material
Acknowledgments
We thank Chintha Raju for technical assistance.
References
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV. (2000) The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 149: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. (2008) Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J 56: 303–315 [DOI] [PubMed] [Google Scholar]
- Baldauf SL. (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19: 345–351 [DOI] [PubMed] [Google Scholar]
- Bateman A, Bennett HPJ. (2009) The granulin gene family: from cancer to dementia. Bioessays 31: 1245–1254 [DOI] [PubMed] [Google Scholar]
- Beers EP, Jones AM, Dickerman AW. (2004) The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65: 43–58 [DOI] [PubMed] [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, Brière C, de Wit PJGM, Marco Y, Deslandes L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20: 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. (2006) Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem Biol 13: 637–647 [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP. (2007) The TIGR Plant Transcript Assemblies database. Nucleic Acids Res 35: D846–D851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth J, Jansonius JN, Koekoek R, Swen HM, Wolthers BG. (1968) Structure of papain. Nature 218: 929–932 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V, Kositsup B, Zhao C, Beers EP. (2002) The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol 128: 84–94 [PMC free article] [PubMed] [Google Scholar]
- García-Lorenzo M, Sjödin A, Jansson S, Funk C. (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Chakraborty S, Chakrabarti C, Dattagupta JK, Biswas S. (2008) Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria. FEBS J 275: 421–434 [DOI] [PubMed] [Google Scholar]
- Gilroy EM, Hein I, van der Hoorn R, Boevink PC, Venter E, McLellan H, Kaffarnik F, Hrubikova K, Shaw J, Holeva M, et al. (2007) Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J 52: 1–13 [DOI] [PubMed] [Google Scholar]
- Goulet C, Khalf M, Sainsbury F, D’Aouost MA, Michaud D. (2012) A protease activity-depleted environment for heterologous proteins migrating towards the leaf cell apoplast. Plant Biotechnol J 10: 83–94 [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. (2000) Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol 7: 569–581 [DOI] [PubMed] [Google Scholar]
- Greenbaum DC, Arnold WD, Lu F, Hayrapetian L, Baruch A, Krumrine J, Toba S, Chehade K, Brömme D, Kuntz ID, et al. (2002) Small molecule affinity fingerprinting: a tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol 9: 1085–1094 [DOI] [PubMed] [Google Scholar]
- Greenwood JS, Helm M, Gietl C. (2005) Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc Natl Acad Sci USA 102: 2238–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Kolodziejek I, Misas-Villamil JC, Shindo T, Colby T, Verdoes M, Richau KH, Schmidt J, Overkleeft HS, van der Hoorn RAL. (2010) Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J 62: 160–170 [DOI] [PubMed] [Google Scholar]
- Herman EM, Helm RM, Jung R, Kinney AJ. (2003) Genetic modification removes an immunodominant allergen from soybean. Plant Physiol 132: 36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda BC, Padgett HS, Rogers JC. (1992) Proaleurain vacuolar targeting is mediated by short contiguous peptide interactions. Plant Cell 4: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honée G, Buitink J, Jabs T, Sijbolts F, Apotheker M, Weide R, Sijen T, Stuiver M, De Wit PJGM. (1998) Induction of defense-related responses in Cf9 tomato cells by the AVR9 elicitor peptide of Cladosporium fulvum is developmentally regulated. Plant Physiol 117: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v4: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffarnik FA, Jones AM, Rathjen JP, Peck SC. (2009) Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics 8: 145–156 [DOI] [PubMed] [Google Scholar]
- Karrer KM, Peiffer SL, DiTomas ME. (1993) Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci USA 90: 3063–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Gu C, Niessen S, Hoover H, Cravatt BF, van der Hoorn RAL. (2009a) Diversity of serine hydrolase activities of unchallenged and Botrytis-infected Arabidopsis thaliana. Mol Cell Proteomics 8: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, Ilyas M, Win J, Kamoun S, van der Hoorn RAL. (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol 154: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Verhelst SH, van Swieten PF, Verdoes M, Wong CS, Wang Z, Kaiser M, Overkleeft HS, Bogyo M, van der Hoorn RAL. (2009b) Minitags for small molecules: detecting targets of reactive small molecules in living plant tissues using ‘click chemistry.’ Plant J 57: 373–385 [DOI] [PubMed] [Google Scholar]
- Katerelos NA, Taylor MAJ, Scott M, Goodenough PW, Pickersgill RW. (1996) Crystal structure of a caricain D158E mutant in complex with E-64. FEBS Lett 392: 35–39 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Yamamoto D, Matsumoto K, Inoue M, Ishida T, Mizuno H, Sumiya S, Kitamura K. (1992) Crystal structure of papain-E64-c complex: binding diversity of E64-c to papain S2 and S3 subsites. Biochem J 287: 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno K, Harayama C, Nakamura M, Tateishi K, Tamura Y, Hattori M, Kohno K. (2004) Papain protects papaya trees from herbivorous insects: role of cysteine protease in latex. Plant J 37: 370–378 [DOI] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JDG. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Lampl N, Budai-Hadrian O, Davydov O, Joss TV, Harrop SJ, Curmi PMG, Roberts TH, Fluhr R. (2010) Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21). J Biol Chem 285: 13550–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Maes D, Bouckaert J, Poortmans F, Wyns L, Looze Y. (1996) Structure of chymopapain at 1.7 A resolution. Biochemistry 35: 16292–16298 [DOI] [PubMed] [Google Scholar]
- Martínez DE, Bartoli CG, Grbic V, Guiamet JJ. (2007) Vacuolar cysteine proteases of wheat (Triticum aestivum L.) are common to leaf senescence induced by different factors. J Exp Bot 58: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Martinez M, Diaz I. (2008) The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol Biol 8: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H, Gilroy EM, Yun BW, Birch PR, Loake GJ. (2009) Functional redundancy in the Arabidopsis cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol 183: 408–418 [DOI] [PubMed] [Google Scholar]
- Nazif T, Bogyo M. (2001) Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc Natl Acad Sci USA 98: 2967–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Ostresh JM, Winkle JH, Hamashin VT, Houghten RA. (1994) Peptide libraries: determination of relative reaction rates of protected amino acids in competitive couplings. Biopolymers 34: 1681–1689 [DOI] [PubMed] [Google Scholar]
- Otegui MS, Noh YS, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ. (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41: 831–844 [DOI] [PubMed] [Google Scholar]
- Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. (2000) A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12: 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. (2010) MEROPS: the peptidase database. Nucleic Acids Res 38: D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HC, Van’t Klooster JW, van der Hoorn RAL, Joosten MHAJ, Jones JDG, de Wit PJGM. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Gietl C. (1999) Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc Natl Acad Sci USA 96: 14159–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabab M, Shindo T, Gu C, Kaschani F, Pansuriya T, Chintha R, Harzen A, Colby T, Kamoun S, van der Hoorn RAL. (2008) Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 20: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RAL, Kamoun S. (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci USA 106: 1654–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577 [DOI] [PubMed] [Google Scholar]
- Than ME, Helm M, Simpson DJ, Lottspeich F, Huber R, Gietl C. (2004) The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. J Mol Biol 336: 1103–1116 [DOI] [PubMed] [Google Scholar]
- Tian M, Win J, Song J, van der Hoorn RAL, van der Knaap E, Kamoun S. (2007) A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, Bennett HPJ, Bateman A, Ni F. (2008) Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci 17: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkatchev D, Xu P, Ni F. (2001) A peptide derived from the C-terminal part of a plant cysteine protease folds into a stack of two β-hairpins, a scaffold present in the emerging family of granulin-like growth factors. J Pept Res 57: 227–233 [DOI] [PubMed] [Google Scholar]
- Turk D, Podobnik M, Popovic T, Katunuma N, Bode W, Huber R, Turk V. (1995) Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: a basis for the design of specific epoxysuccinyl inhibitors. Biochemistry 34: 4791–4797 [DOI] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J 20: 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Laurent F, Roth R, De Wit PJGM. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol Plant Microbe Interact 13: 439–446 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Leeuwenburgh MA, Bogyo M, Joosten MHAJ, Peck SC. (2004) Activity profiling of papain-like cysteine proteases in plants. Plant Physiol 135: 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL, Wulff BB, Rivas S, Durrant MC, van der Ploeg A, de Wit PJGM, Jones JDG. (2005) Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17: 1000–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse HP, Van’t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJGM, Thomma BPHJ. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varughese KI, Su Y, Cromwell D, Hasnain S, Xuong NH. (1992) Crystal structure of an actinidin-E-64 complex. Biochemistry 31: 5172–5176 [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Hillaert U, Willems LI, van der Linden WA, Sae-Heng M, Filippov DV, Kisselev AF, van der Marel GA, Overkleeft HS. (2008) Azido-BODIPY acid reveals quantitative Staudinger-Bertozzi ligation in two-step activity-based proteasome profiling. ChemBioChem 9: 1735–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gu C, Colby T, Shindo T, Balamurugan R, Waldmann H, Kaiser M, van der Hoorn RAL. (2008) Beta-lactone probes identify a papain-like peptide ligase in Arabidopsis thaliana. Nat Chem Biol 4: 557–563 [DOI] [PubMed] [Google Scholar]
- Watanabe E, Shimada T, Tamura K, Matsushima R, Koumoto Y, Nishimura M, Hara-Nishimura I. (2004) An ER-localized form of PV72, a seed-specific vacuolar sorting receptor, interferes the transport of an NPIR-containing proteinase in Arabidopsis leaves. Plant Cell Physiol 45: 9–17 [DOI] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468: 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems LI, Verdoes M, Florea BI, van der Marel GA, Overkleeft HS. (2010) Two-step labeling of endogenous enzymatic activities by Diels-Alder ligation. ChemBioChem 11: 1769–1781 [DOI] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. (2001) A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol 127: 1626–1634 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.