Abstract
Cyanobacteria are oxygenic photosynthetic prokaryotes that are the progenitors of the chloroplasts of algae and plants. These organisms harvest light using large membrane-extrinsic phycobilisome antenna in addition to membrane-bound chlorophyll-containing proteins. Similar to eukaryotic photosynthetic organisms, cyanobacteria possess thylakoid membranes that house photosystem (PS) I and PSII, which drive the oxidation of water and the reduction of NADP+, respectively. While thylakoid morphology has been studied in some strains of cyanobacteria, the global distribution of PSI and PSII within the thylakoid membrane and the corresponding location of the light-harvesting phycobilisomes are not known in detail, and such information is required to understand the functioning of cyanobacterial photosynthesis on a larger scale. Here, we have addressed this question using a combination of electron microscopy and hyperspectral confocal fluorescence microscopy in wild-type Synechocystis species PCC 6803 and a series of mutants in which phycobilisomes are progressively truncated. We show that as the phycobilisome antenna is diminished, large-scale changes in thylakoid morphology are observed, accompanied by increased physical segregation of the two photosystems. Finally, we quantified the emission intensities originating from the two photosystems in vivo on a per cell basis to show that the PSI:PSII ratio is progressively decreased in the mutants. This results from both an increase in the amount of photosystem II and a decrease in the photosystem I concentration. We propose that these changes are an adaptive strategy that allows cells to balance the light absorption capabilities of photosystems I and II under light-limiting conditions.
Photosynthetic organisms use antenna systems to capture light energy and transfer the energy to reaction centers where photochemistry occurs. Antenna complexes are structurally diverse and highly specialized among the various classes of photosynthetic organisms and are optimized to allow the maximal absorption of light energy available in their environmental niche (Blankenship, 2002). Cyanobacteria are photosynthetic prokaryotes that use a combination of membrane-intrinsic and -extrinsic antennas to harvest sunlight over a wide spectral range. Green-orange (550–650 nm) light excitation is principally absorbed by open-chain tetrapyrroles termed phycobilins that are covalently bound to a protein scaffold to form phycobilisomes. In Synechocystis species PCC 6803 (hereafter, Synechocystis 6803), phycobilisomes are hemidiscoidal structures composed of rods of the pigment phycocyanin (PC) and an allophycocyanin (APC) core (MacColl, 1998). Phycobilisomes are functionally flexible and can associate with both photosystem (PS) I and PSII. They can serve to increase the absorption cross-section of the approximately 40 chlorophylls of PSII (Tang and Diner, 1994) or transfer excitation to PSI directly (Mullineaux, 1992).
Synechocystis 6803 are spherical cells of approximately 1.5 to 2 μm in diameter that have been extensively studied by electron microscopy, so their intracellular organization is well characterized (Liberton et al., 2006; van de Meene et al., 2006). These cells have thylakoid membranes organized primarily as three or more concentric layers around the cell periphery, with a central cytoplasmic region largely devoid of thylakoid membranes. Thylakoid membranes are pairs of membrane bilayers that enclose a lumenal space that allows for chemiosmotic energy coupling of the photosynthetic and respiratory electron transfer chains. Phycobilisomes are found associated with the cytoplasmic side of the thylakoid membranes, so that the thylakoids are separated by a space sufficient for two rows of phycobilisomes offset from each other (Anderson and Toole, 1998). The amenability of Synechocystis 6803 to genetic manipulation has resulted in the generation of a number of strains in which phycobilisomes are modified to varying degrees and has allowed for the functioning of phycobilisomes to be probed at the molecular level (Sidler, 2004).
While thylakoid membrane arrangement has been studied in some strains of cyanobacteria (for review, see Liberton and Pakrasi, 2008; Nevo et al., 2009) and crystal structures have shown, with exquisite detail, photosynthetic complexes at high resolution (El-Mohsnawy et al., 2010; Umena et al., 2011), it is not sufficiently understood how these complexes are organized in the membrane. In Synechocystis 6803, freeze-fracture analysis has shown that particles likely corresponding to PSII complexes could be either in parallel rows or randomly arranged according to the state 1-state 2 transition (Olive et al., 1997). Single-particle analysis showed rows of PSII in membrane patches isolated from Synechocystis 6803, leading to a model of how the photosynthetic and other membrane protein complexes are arranged in the thylakoid membrane (Folea et al., 2008). However, it is unclear how the organization of these membrane regions can be extrapolated to the whole cell level and, furthermore, how uniform such an arrangement is among different strains of cyanobacteria.
Recently, hyperspectral confocal fluorescence microscopy (HCFM) and multivariate analysis were used to investigate living Synechocystis 6803 wild-type cells, a PSI-deletion mutant, and a mutant deficient in a component of the light-independent NADPH/protochlorophyllide oxidoreductase that could not synthesize chlorophyll in darkness (Vermaas et al., 2008). This combination of mutants allowed for the separation and location of spectral components attributed to carotenoid, PSII, PSI, PC, APC, and APC-B, the terminal emitter of energy from the phycobilisome (Glazer, 1984). In wild-type cells, heterogeneity was found between the thylakoid layers, with phycobilisomes and PSII most prevalent along the outer thylakoids while PSI was enriched along the inner thylakoids. This study demonstrated a new technique to assess pigment distribution in vivo and circumvented the necessity to purify membrane components for analysis.
Here, we exploit the combination of HCFM and electron microscopy to correlate the effect of phycobilisome antenna size on thylakoid organization and pigment localization in the model cyanobacterium Synechocystis 6803. We examined the wild-type strain of Synechocystis 6803 and a series of mutants in which phycobilisomes are partially truncated or absent. With the high spatial resolution of electron microscopy, we show that the thylakoid membrane organization is severely altered when phycobilisomes are truncated, in that the membranes form layers with little long-range curvature. Using high-content HCFM and multivariate curve resolution (MCR) analysis, we resolve the locations and relative concentrations of phycobilisomes, PSII, and PSI in diffraction-limited optical sections of living cells. We performed three-dimensional (3D) volume reconstruction of individual cells to highlight the global arrangement of these complexes in the thylakoid membrane. We show that there are distinct regions of PSII (and phycobilisome) enrichment over PSI and that, as the phycobilisome antenna is truncated, the PSII-to-PSI segregation becomes exacerbated. Importantly, our multivariate image analysis provides relative quantification of the fluorescence intensities for PSII and PSI on a per cell basis. These studies extend over a large population of cells for the wild type and the mutants and result in a statistically significant decrease in the PSI:PSII ratio as the phycobilisomes are increasingly truncated. Together, these data suggest a strategy in which a lack of light-harvesting capability by phycobilisomes is offset by a rearrangement of photosystems within the thylakoid membrane.
RESULTS
Transmission Electron Microscopy Characterization of Mutant Strains
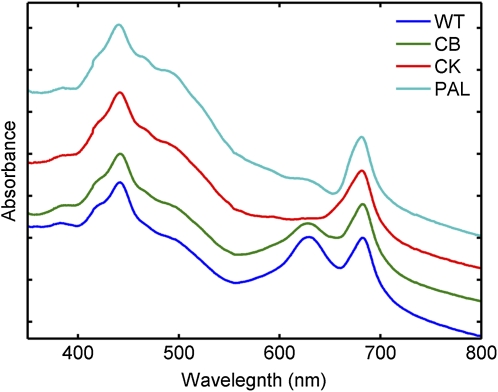
We examined the wild type and CB, CK, and PAL mutant strains of Synechocystis 6803 that have previously been described (Ajlani and Vernotte, 1998; Arteni et al., 2009). In the wild-type strain, phycobilisomes contain a core composed of trimeric APC discs stacked into three cylinders. Radiating from the core are PC-containing rods of stacked hexameric discs. These components are connected by linker polypeptides that function in connecting and stabilizing the structure and also in energy transfer (MacColl, 1998). The CB strain lacks the rod-linker polypeptide and therefore contains only one PC hexamer per rod, the CK strain contains only the APC core, lacking assembled rods, and the PAL strain cannot assemble any functional phycobilisomes. Absorption spectra from these strains show the expected decrease in A625 corresponding to a decrease in PC in the mutants (Fig. 1).
Figure 1.
Whole cell absorption spectra of the wild type (WT), CB, CK, and PAL. Note that in the mutants, the absorbance in the 625-nm region is progressively decreased, indicating a decrease in PC. Each spectrum was normalized to the same value at an optical density at 750 nm and then offset by 0.2 absorbance units for clarity.
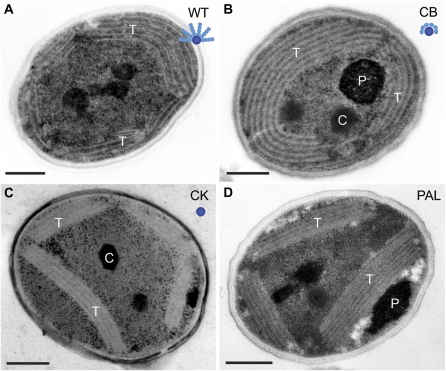
Analysis of transmission electron microscopy (TEM) thin sections revealed that the intracellular organization of the thylakoid membranes was altered as a result of phycobilisome modification, with the more severe phycobilisome mutation corresponding with more significant alteration to the membranes (Fig. 2). In wild-type Synechocystis 6803 cells, thylakoid membranes are generally arranged as concentric layers that tend to follow the curvature of the plasma membrane, so that the central cytoplasmic region of the cell is largely devoid of thylakoids (Fig. 2A; Liberton et al., 2006; van de Meene et al., 2006). Furthermore, when thylakoids are present in the cell interior, they often form arching loops of membrane layers (Mullineaux, 1999; Liberton et al., 2006). When compared with wild-type cells, it was apparent that the thylakoid membranes in the phycobilisome mutants did not display the same degree of long-range membrane curvature, with decreased thylakoid curvature observed in the CK and PAL cell lines, where phycobilisome disruption was greatest (Fig. 2, C and D). In these mutants, the thylakoids form multilayered bands of membranes that do not necessarily follow the plasma membrane but span the cell interior as straight, dense membrane layers. In contrast, wild-type and CB cells had thylakoid membrane layers that appeared more flexible, and typically followed the shape of the cell envelope (Fig. 2, A and B). Genetic mutation attenuating the overall size of phycobilisomes has resulted in thylakoid layers that are more closely spaced compared with the wild-type strain (Westerman et al., 1994; Olive et al., 1997). We measured the cytoplasmic distance between thylakoids in these mutants and found that it was decreased in the mutants consistent with the progression of phycobilisome attenuation. The interthylakoid distance was approximately 40 nm in wild-type cells, approximately 32 nm in both CB and CK cells, and approximately 20 nm in PAL cells.
Figure 2.
Thin-section electron micrographs of Synechocystis 6803 wild type and phycobilisome antenna mutants. A, Wild-type (WT) strain with intact phycobilisomes. B, CB mutant with one PC hexamer per rod. C, CK mutant that contains only the APC core. D, PAL mutant that lacks the assembled phycobilisomes. Labeled are thylakoid membranes (T), polyphosphate bodies (P), and carboxysomes (C). A cartoon model of the phycobilisome structure in each strain is shown as an inset. The APC core is represented by the central blue circle, and the PC discs are shown as teal ellipsoids. Bars = 250 nm. [See online article for color version of this figure.]
Assignment of Spectral Signatures
Since thin-section TEM images revealed altered thylakoid morphology in the antenna mutants, we sought to investigate the global changes in pigmentation with HCFM in intact, unfixed cells. HCFM records an entire emission spectrum (500–800 nm) at every spatial voxel in the image, yielding a single spectral image containing four dimensions of data: three spatial dimensions and one spectral dimension. While simple band integration approaches, such as a conventional confocal microscopy with a filter cube, can be used to reach some qualitative conclusions about the images, mathematically isolating the pure spectral components responsible for the fluorescence data can provide quantitative relationships between component concentration and spatial location within the cell. To this end, we employed MCR algorithms (Bro and De Jong, 1997; Van Benthem et al., 2002; Haaland et al., 2003; Keenan and Kotula, 2004) to analyze the spectral images (see “Materials and Methods”).
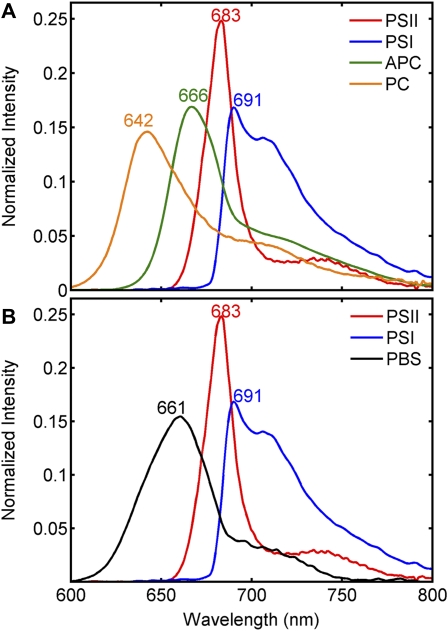
With 488-nm excitation, carotenoids and, to a lesser extent, phycobilisomes are preferentially excited over chlorophyll. However, bulk fluorescence measurements show that emission was observed from phycobilisomes and chlorophyll due to energy transfer (Supplemental Fig. S1). Two separate combinations of the spectral images were developed to delineate fluorescence from all Synechocystis 6803 cell lines investigated. In the first set, CB, CK, and PAL were combined into a composite data set, and MCR was used to develop a model with spectral components ascribed to PC, APC, PSII, and PSI (Fig. 3A) as well as a carotenoid component (Supplemental Fig. S2). Carotenoids have extremely short fluorescence lifetimes and therefore low-fluorescence quantum yields. Despite this, a weak-fluorescence component convolved with resonance-enhanced Raman bands characteristic of carotenoid was observed in the 500- to 600-nm detection range. This component is similar to previous observations of carotenoid signatures with HCFM (Vermaas et al., 2008). The identity of the PC and APC components was corroborated by their presence or absence in the antenna mutants (see below). Note that in Figure 3A, the component assigned to APC has a subtle shoulder on the long-wavelength side of the emission peak. A second derivative plot of this band revealed a minimum at 680 nm (data not shown) and likely reflects emission from APC-B (also referred to as APC680). The fluorescence band shape and emission maxima of the PC and APC fluorescence components are similar to the bulk room-temperature fluorescence from phycobilisome or phycobilin complexes purified from the CB and CK mutants (Supplemental Fig. S3). The fluorescence component ascribed to PSII was based upon previous results (Vermaas et al., 2008) as well as the fluorescence of purified PSII from this same strain (Gounaris et al., 1989). The PSI component was assigned based on its comparison with the fluorescence of either purified PSI from Synechococcus PCC 7942 (Andrizhiyevskaya et al., 2002) or purified thylakoids devoid of phycobilisomes in a ΔPSII mutant from Synechocystis 6803 (Wittmershaus et al., 1998). The MCR spectral components described above were used to model the spectral variance of this data set essentially to the noise floor of the microscope (Jones et al., 2008) and is evident by the structureless character of the unmodeled residuals (data not shown).
Figure 3.
MCR models for the wild type and phycobilisome mutants. A, MCR-derived pure spectral components for CB, CK, and PAL. B, MCR model for the wild type and PAL, where PSII and PSI have been constrained and are spectrally identical to their counterparts in A. The data have been normalized to unit length. For details on how the models were determined, see “Materials and Methods.”
The MCR model developed for CB, CK, and PAL could not fit the spectral data for wild-type cells without a significant increase in the spectral residuals in the 630- to 670-nm range, showing that a linear combination of the PC and APC components identified in the previous model could not adequately describe the fluorescence emission from phycobilisomes in wild-type cells. Therefore, a second MCR model was developed using a composite data set from images of the wild type and the phycobilisome-deficient mutant, PAL. To generate this model, the shapes of the carotenoid, PSII, and PSI components were held constant (equality constrained), and a new component was allowed to model the remaining spectral variance. By constraining carotenoid, PSII, and PSI to be spectrally identical in both models, the subsequent concentration maps generated from the two models can be directly compared. This approach yielded a single, broad component with an emission maximum at 661 nm situated between the PC and APC bands (Fig. 3B). Again, based upon its emission maximum position and shape of the fluorescence spectra from isolated phycobilisomes from wild-type cells (Supplemental Fig. S3), we assigned this component to intact phycobilisomes (PBS). The likely reason that the emission of PBS in this model did not separate into its PC and APC substituents is because the emission of these two components covaried in all voxels of this image data set and therefore could not be separated as unique fluorophores using factor analysis-based methods.
Pigment Localization
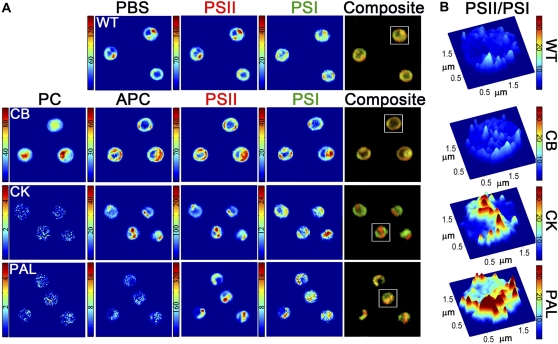
MCR-generated concentration maps for pigments from representative two-dimensional (2D) image sections from the wild type, CB, CK, and PAL are shown in Figure 4A. In wild-type cells, PBS, PSII, and PSI are generally colocalized. The morphology of these spectral components has similarity to the TEM image shown in Figure 2A and elsewhere (Liberton et al., 2006; van de Meene et al., 2006), where the thylakoid membranes follow the cell periphery or can span the central cytoplasm. There are regions of PSII and PBS enrichment compared with PSI that are highlighted in the red-green composite images (Fig. 4A, right panel). We speculate that these regions originate from the domain structures of PSII adapted to state 1, where PBS excitation is preferentially transferred to PSII (Vernotte et al., 1990; Olive et al., 1997; Folea et al., 2008). A volume-rendered movie of the spatial location of PSII and PSI within wild-type cells is shown in Supplemental Movie S1.
Figure 4.
A, MCR-derived concentration maps for PC, APC, PBS, PSII, and PSI components within each cell type. Each image box is 10 × 10 μm. Note that the color scale was constructed from 0 to maximum fluorescence intensity and is unique for each image. A red-green composite image was constructed to highlight the spatial segregation between PSII (red) and PSI (green) and is displayed in the right-most panel of A. The fluorescence intensities presented here have been calculated at the 99.5% confidence interval (see “Materials and Methods”). B, Surface plot of the per-pixel PSII-PSI ratio for the representative cells boxed in the composite images in A. Note that the color scale in B is uniform for each cell type. WT, Wild type.
In the CB mutant, where phycobilisomes form with only one PC hexamer per APC disc, the PC component was found not only in the thylakoid associated with APC but also in the cell cytoplasm (Fig. 4A). Phycobilisomes attach to the cytoplasmic side of thylakoid membranes (Mullineaux, 2008), and since this mutant lacks the rod linker polypeptides necessary to assemble stacks of hexameric PC discs, it is likely that nonassembled PC accumulates in the cytoplasm. A volume-rendered movie highlighting the spatial location of the PC component compared with PSII is shown in Supplemental Movie S2. The phycobilisome core composed of APC is colocalized in the same voxels as PSII and PSI. This is similar to the localization of PBS, PSII, and PSI in wild-type cells. Furthermore, the morphology of these components follows the thylakoid membrane pattern observed in TEM micrographs, with the thylakoid membranes following the curvature of the cell, underlying the plasma membrane (Fig. 2B; Supplemental Movie S3).
In the CK mutant, where the phycobilisomes have been truncated to only possess the APC core, the PC component is completely absent from these cells and is indistinguishable from the baseline noise (Fig. 4A). This observation reinforces the assignment of the PC spectral component. In the CK mutant, APC and PSII appear in bright patches while PSI is more evenly distributed within the cell. These bright patches are also clearly visible in 3D reconstructions of the cell volume (Supplemental Movie S4). Interestingly, many of the 2D images and 3D reconstructions revealed that most CK cells exhibit a single PSII- and APC-enriched region. These regions were at least several pixels in diameter (several hundred nanometers), suggesting that there are large areas within the thylakoids where PSII and PBS are significantly more concentrated compared with PSI. The contour of the thylakoids from the TEM image of CK showed marked distinction from wild-type or CB cells, with the thylakoids forming thick bands, and, as mentioned above, the cytoplasmic spacing between thylakoid layers was reduced. These bands of thylakoids generally follow the cell periphery but can also extend through the cytoplasm (Fig. 2C). A similar thylakoid organization has been observed by TEM in a different PC-less mutant of Synechocystis PCC 6803 (Olive et al., 1997).
The PAL mutant, which fails to assemble phycobilisomes, completely lacks the PC and APC components (Fig. 4A). TEM images revealed that this mutant forms dense bands of thylakoids similar to the CK antenna mutant; however, the spacing between thylakoid layers was further decreased. As in the CK mutant, PSII fluorescence was observed in distinct foci, but in PAL cells, the signal from PSII showed the greatest fluorescence intensity among all strains examined. PSI was found colocalized in these same regions but also in regions separate from the bright PSII emission (Fig. 4A).
Due to the noted heterogeneity of the PSII and PSI distribution between cell types, the per-pixel ratio of PSII to PSI was calculated and a representative cell for each cell type is shown in Figure 4B. The ratio of the fluorescence intensity for the PSII and PSI components should serve as a proxy to the relative quantity of these complexes in the thylakoid membranes within each spatial voxel. For example, the CK cell shown in Figure 4B has a central cell region where the PSII:PSI fluorescence ratio is very large. Given the intensity values for PSII and PSI from this same cell (Fig. 4A), we can conclude that this central region is significantly enriched in PSII but diminished in PSI compared with other parts of the cell. Contrasting this observation to the wild-type cell (Fig. 4B), where the PSII:PSI fluorescence ratio is markedly smaller and more uniform throughout the image, indicates far less variance in the PSII:PSI distribution in the wild-type cell.
We noted that as the antenna was sequentially truncated, the ratio of PSI to PSII fluorescence was generally decreased. Therefore, we sought to obtain statistical information on a per-cell basis. For this approach, 2D optical slices of individual cells were segmented by applying a mask to exclude pixels outside of the cell area, and each cell was indexed. The mean cell intensity for each spectral component was then determined by summing the fluorescence intensity for every pixel within the mask and then dividing by the number of pixels that composed the mask. Because the cells are likely randomly oriented on the microscope slide, a 2D image through the center of a cell should be sufficient to quantify the average fluorescence intensities, if a large enough population of cells is analyzed. In total, this was done for 332 cells across the four different strains.
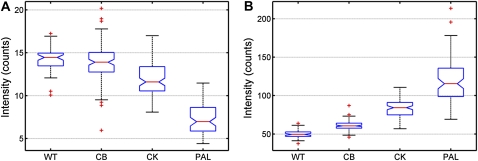
To determine the significance of the mean fluorescence intensities for PSI and PSII across the wild-type and mutant strains, a nested ANOVA (see “Materials and Methods”) was conducted. This nested analysis compared the variance across the strains (wild type, CB, CK, and PAL) with variances associated with the biological replicates within each strain and the variances between individual cells within each biological replicate. The variance between the strains was greater than the variances associated with the biological replicates and the cell-to-cell variance for both the PSI and PSII mean fluorescence intensities. These results are statistically significant at the greater than 99% level. A graphic representation of the mean fluorescence intensities is given in Figure 5 by using a statistical box plot. Since the notches in each box plot do not overlap with the notches of the other box plots, it can be concluded with greater than 95% confidence that the PSI and PSII intensities between the strain types are significantly different. Interestingly, not only is the fluorescence intensity for PSII progressively increased in the antenna mutants, but the PSI emission is constituently decreased. Although the PSI fluorescence decrease was subtle between the different strains, it was significant based upon the statistical analysis described above.
Figure 5.
Statistical box plots of the mean intensities for PSI (A) and PSII (B) from all cells for each cell type. The red center line represents the median of these values, and the 95% confidence band is denoted by notches. The bottom and top of the box represent the 25th and 75th percentiles, respectively. The dotted extended lines indicate ±2.7σ (99.3% data coverage), and the red crosses represent statistical outliers. n = 332 cells. WT, Wild type. [See online article for color version of this figure.]
DISCUSSION
High-resolution structural data have shed light on the structure-function relationships of the individual complexes associated with oxygenic photosynthesis (i.e. PSII, PSI, light-harvesting antenna, and components of the electron transport chain; Blankenship, 2002; Nelson and Yocum, 2006; Fromme and Grotjohann, 2011). Despite these finding, how these individual complexes come together and are arranged in the membrane is not well understood, but such knowledge is necessary for a deep understanding of photosynthesis on the macro scale. Progress has been made in understanding the supramolecular organization of these complexes in the thylakoid membrane of plants (for review, see Dekker and Boekema, 2005), where PSI and PSII are organized into so-called supercomplexes and are spatially separated in the stroma lamellae and grana stacks, respectively. However, this is not the case for cyanobacteria, where stroma lamellae and grana stacks are absent (Liberton et al., 2006; van de Meene et al., 2006).
In this study, we have used TEM and HCFM to investigate changes in thylakoid morphology as well as the spatial location of PSI, PSII, and phycobilisomes in the Synechocystis 6803 wild type and a series of antenna mutants with increasingly truncated phycobilisomes. TEM images of thin sections of the wild type, CB, CK, and PAL revealed that long-range thylakoid membrane curvature was substantially decreased in the CK and PAL mutants and that the thylakoid membranes did not necessarily follow the cell envelope, as is generally the case in wild-type cells. As expected, the interthylakoid spacing was decreased in the mutants and was coordinated with phycobilisome size, but the cytoplasmic distance between thylakoid layers was larger than those reported for similar mutants (Olive et al., 1997).
With a smaller phycobilisome antenna, photon flux delivered to PSII is decreased compared with wild-type cells. Given the large changes in thylakoid morphology in these mutants as noted above, we used HCFM and MCR to spectrally resolve, spatially locate, and quantify emission from PSI, PSII, and phycobilisomes in these same cell types. Wild-type PBS could not be described by a combination of the PC and APC components assigned from the CB and CK mutants. There are likely two contributing factors to this observation. First, phycobilisome emission from the CB and CK mutants is fundamentally unique compared with the wild type. This is expected considering the differences in the fluorescence band shapes of the phycobilisome complexes from the strains (Supplemental Fig. S3). Second, in the wild-type cells studied here, emission from the phycobilisomes was constant or covaried in all voxels; thus, MCR analysis could not resolve such an emission feature as multiple fluorophores. The MCR model developed for this work is proposed to represent emission from the photosynthetic complexes PSII, PSI, and PBS in wild-type cells, while the CB and CK cells possess APC and PC components due to their unique phycobilisome composition. In our previous work (Vermaas et al., 2008), chromophores within wild-type phycobilisomes (APC, APC-B, and PC) could be spectrally isolated, because the strains and growth conditions used in that study were altered in such a manner to allow the underlying components to vary independently. This allowed the MCR analysis to resolve individual components for PC, APC, and APC-B.
In wild-type cells, we observed regions of the cell where PSII and phycobilisomes were enriched over PSI (Fig. 4A; Supplemental Movie S1). TEM images of cyanobacterial thylakoids have suggested that PSII can be enriched in domains (Mörschel and Schatz, 1987; Vernotte et al., 1990; Olive et al., 1997) or even crystalline arrays of multiple rows of PSII double dimers (Folea et al., 2008) under state 1 conditions, where the phycobilisomes are primarily associated with PSII (Mullineaux, 2008). The function of these domains or arrays is unknown but they have been proposed to act in preventing phycobilisome excitation from transferring to PSI by spatially separating the two complexes. In this study, we cultured all cells under low-intensity fluorescent light, and cells remained under dim room lighting before and during imaging. Under such conditions, phycobilisomes are speculated to be in state 1. Indeed, for both the wild type and the mutants studied here, the phycobilisome component was always colocalized with PSII, with the obvious exception of PAL and the PC component in the CB cell type (see below). The dynamics of thylakoid reorganization under state 1 and state 2 will be the subject of future experimentation.
The CB mutant was an unusual case in that substantial amounts of PC were found in the cytoplasm (Supplemental Movie S2). Comparing the ratio of A625 to A680 (phycobilins to chlorophyll) in whole cells (Fig. 1) suggests that the CB mutant possesses fewer phycobilins compared with the wild type. Despite this, there is greater emission in the phycobilisome spectral region, suggesting that many of these pigments are uncoupled from energy acceptors (Supplemental Fig. S1). The spatial location of PSI and PSII within the cell was similar to that in wild-type cells (Fig. 4).
Analysis of the CK cells revealed regions where PSII and APC emission was very intense and largely devoid of PSI. These regions were several hundred nanometers in diameter and suggest a remarkably different organization of complexes within the thylakoid membrane compared with wild-type cells (Fig. 4). Although physical separation of PSII and PSI seems unusual for a cyanobacterial cell, it is not unprecedented. In the chlorophyll d-containing cyanobacterium Acaryochloris marina, phycobilisomes are replaced by phycobiliproteins that are relatively small and are composed of individual rods of four hexameric units of PC or PC and APC (Marquardt et al., 1997, 2000). In an ultrastructure study of this same organism, Chen et al. (2009) noted thylakoid regions largely devoid of the phycobiliproteins and, because of their close association with PSII, suggested that there was physical separation of PSI and PSII in the membrane. Whether there is any analogy to the proposal of Chen et al. (2009) and the CK mutant presented here remains to be determined.
Interestingly, when phycobilisomes are completely absent in the PAL mutant, there are regions of greatest PSII emission intensity, with PSI also found in these same general areas (Fig. 4A). This result contrasts with that of the CK mutant and is more similar to the localization of PSII and PSI observed in the wild type and CB. As noted earlier, the interthylakoid distance is coordinated with the size of the phycobilisomes between the membranes (Olive et al., 1997). PAL, having no phycobilisomes, has the most densely packed thylakoid membranes (Fig. 2D). The increase in PSII intensity likely originates from a greater number of complexes fluorescing within the diffraction-limited confocal volume (250 nm × 250 nm × 600 nm). In other words, there are a greater number of PSII complexes per unit of membrane area. Olive et al. (1997) used freeze-fracture studies of Synechocystis 6803 thylakoids to reveal particles exposed on the external-fractured face that are ascribed to PSII dimers. Compared with the wild type, a PC-less mutant had 32% more PSII particles and a mutant that completely lacked phycobilisomes possessed 109% more PSII particles on a per area basis. In our study here, mean fluorescence intensities from similar mutant strains followed this trend nearly identically (Fig. 5).
Finally, we sought to quantify the amount of PSII and PSI on a per cell basis for each cell type based on 2D images. A dramatic increase in the mean per-cell PSII fluorescence was observed as the antenna was progressively truncated. Moreover, a subtle yet statistically significant decrease in PSI emission was noted in these same cells (Fig. 5). A similar trend has been observed with bulk 77K fluorescence measurements of Synechocystis 6803 mutants with truncated phycobilisome antennas (Ajlani et al., 1995). In that same study, the functional chlorophyll antenna size of PSII was found to be identical between the wild type and the antenna mutants, suggesting that the number of PSII centers must be increased in the mutants. Furthermore, the PSI:PSII ratio in cyanobacteria is sensitive to the light intensity and quality that the organism is cultured in. Typical PSI:PSII ratios for cyanobacterial cells grown under moderate to low fluorescent light range from 2 to 5 (Fujita et al., 1994). The PSI:PSII ratio in the PAL mutant has been reported to be 0.7 (Stadnichuk et al., 2009), and the PSII concentration was reported to increase 5-fold over wild-type cells when cells were grown at 40 μmol photons m−2 s−1. Lastly, as demonstrated by Fujita and Murakami (1987), the PSI:PSII ratio becomes greater when electron carriers between the two photosystems are reduced. In other words, when PSII is more sensitized, the plastoquinone pool and the cytochrome b6/f complex are to a greater extent reduced. This results in an increase in the amount of PSI to ensure linear electron transport between the two photosystems and to minimize the possibility of photodamage (Andersson and Barber, 2004). Taken together, these observations strongly suggest that as the phycobilisome antenna is progressively truncated, PSII is less sensitized. To compensate for the excitation loss to PSII, its concentration is increased. Our results show that not only does the PSII concentration increase, but the amount of PSI is constituently decreased.
In conclusion, long-range thylakoid membrane curvature and cytoplasmic spacing are significantly decreased in phycobilisome mutants of Synechocystis 6803, as revealed by TEM. We have spectrally resolved, spatially localized, and quantified emission profiles of PSII, PSI, and the light-harvesting antenna in individual, living Synechocystis 6803 cells. The organization of these complexes is altered in response to diminished phycobilisomes, with PSII segregated into large patches separate from PSI. The PSI:PSII ratio is decreased in the mutants, suggesting an adaptive strategy to balance light excitation between the two photosystems. These results demonstrate the flexibility of Synechocystis 6803 cells to modulate their PSII:PSI ratios in response to light sensitization.
MATERIALS AND METHODS
Growth of Cyanobacterial Cultures
Wild-type and mutant strains of Synechocystis species PCC 6803 were cultured photoautotrophically on BG-11 medium (Allen, 1968) at 30°C under 25 μmol photons m−2 s−1. Antenna mutants were maintained with added antibiotics as described previously (Ajlani and Vernotte, 1998; Ughy and Ajlani, 2004). For all experiments, antibiotics were omitted and samples were grown for 3 to 5 d.
Purification of Phycobilisomes
Phycobilisomes were purified similar to the procedure described by Ajlani et al. (1995). Cells from 1 L of culture were washed twice with 0.8 m potassium phosphate buffer, pH 7.0, and broken by vortexing with glass beads. Triton X-100 was added to a final concentration of 2%, followed by incubation for 30 min. After pelleting debris and unbroken cells, the supernatant was loaded onto a Suc gradient prepared with 3 mL of 1.0 m, 2.5 mL of 0.75 m, 2.5 mL of 0.5 m, and 2 mL of 0.25 m Suc in 0.8 m potassium phosphate buffer. After centrifugation for 18 h in a Beckman SW41 rotor at 35,000 rpm, the phycobilin-containing bands were isolated from the gradient.
Electron Microscopy
Cells were prepared for electron microscopy essentially as described (Liberton et al., 2011). Culture aliquots (approximately 15 mL) were centrifuged, and the pellet was resuspended in a small volume, transferred to planchettes with 100- to 200-μm-deep wells, and frozen in a Baltec High Pressure Freezer (Bal-Tec). Samples were freeze substituted in 2% osmium/acetone (3 d at –80°C, 15 h at –60°C, slow thaw to room temperature) and embedded in Spurr’s resin. Thin sections (approximately 80 nm) were stained with uranyl acetate and lead citrate. Digital images were viewed and collected using a LEO 912 transmission electron microscope operating at 120 kV and a ProScan digital camera. Intracellular distances were measured on magnification-calibrated images using the Soft Imaging Systems iTEM software integrated into the LEO 912 system. Distances reported are averages of 20 to 25 measurements per cell type.
Steady-State Spectroscopy
Absorbance measurements were made on a Beckman Coulter DU800 spectrophotometer. Fluorescence measurements were performed on a Photon Technology International fluorometer. Excitation was at 488 nm, and emission was collected from 500 to 850 nm with a sampling interval of 1 nm and a bandwidth of 4 nm.
HCFM
Cells grown to the midexponential phase, as described above, were harvested by gentle centrifugation, applied to agar-coated slides, and sealed with a no. 1.5 coverslip. At least 24 cells were imaged for each two to three biological replicates for each cell type. The imaging approach employed in this work involved a hyperspectral confocal fluorescence microscope described previously (Sinclair et al., 2006). The general imaging methodology has been described previously (Vermaas et al., 2008). Briefly, 488-nm laser light (6 μW cm−2) was focused through a 60× oil-immersion objective (Nikon Plan Apochromat; numerical aperture 1.4) to a diffraction-limited spot. Fluorescence was collected through the same objective, dispersed by a custom-made prism spectrometer, and detected by an electron-multiplied CCD array (Andor Technologies). Each single voxel fluorescence spectrum was integrated for 240 μs, and the image was sampled at one-half the diffraction-limited lateral resolution (250 nm). 3D reconstruction of cell volumes was achieved by collecting 13 successive 2D images at an axial spacing of 0.4 μm and volume rendering using VOXX2 (http://www.indiana.edu/∼voxx/index.html).
Data Processing
Data processing and image analysis were performed essentially as described (Vermaas et al., 2008). Spectral image data were combined into a composite image and analyzed with in-house-written MCR algorithms (Bro and De Jong, 1997; Van Benthem et al., 2002; Haaland et al., 2003; Keenan and Kotula, 2004; Baker, 2008) and software (Jones et al., 2008; Haaland et al., 2009). MCR is an iterative numerical analysis method implemented using constrained alternating least squares (Van Benthem and Keenan, 2004) and generates spectral components that combine in a linear additive manner to recreate noise-free spectroscopic data. The MCR model is applied to the spectral images on a per pixel basis to generate concentration maps that give the relative concentration and location of spectral components within the image. Error associated with the inherent read and Poisson noise of the microscope system was estimated as described previously (Vermaas et al., 2008) and used to generate spectral data at the 99.5% confidence interval. Five spectral components were used to describe the spectral variance associated from 2D images of CB, CK, and PAL and were assigned to carotenoid, PC, APC, PSII, and PSI. When wild-type cells were included in the analysis, a poor fit resulted in the region of the phycobilisomes (630–670 nm), suggesting that the PC and APC components cannot adequately describe wild-type phycobilisomes. Therefore, separate analyses utilizing wild-type and PAL cell images were combined into a composite data set and a second MCR model was developed. The carotenoid, PSII, and PSI components from the first analysis was spectrally constrained to allow for direct comparison of relative concentrations between the two models, and a fourth spectral component was allowed to model the remaining variance. This resulted in a spectral component that was ascribed to intact phycobilisomes from wild-type cells. PSII:PSI ratio images were calculated by taking the ratio of the MCR-generated concentration maps of these components on a per pixel basis. In the PSII:PSI ratio image data shown in Figure 4B, there were about three to four pixels per cell, particularly around the cell edge, where the intensity of PSI was only slightly above the noise. The small PSI intensity value resulted in a PSII:PSI ratio value that was artificially inflated. These three to four pixels were replaced by the average intensity of the neighboring pixels before taking the ratio of PSII to PSI.
To obtain statistics on a per-cell basis, individual cells were masked from 2D images containing several cells using a custom-written Matlab graphical user interface for cell identification and segmentation. Briefly, cells were identified using a depth-first search/Moore neighborhood algorithm (implemented using the bwconncomp function in Matlab) that assigns pixels to the same cell based on a user-defined integrated spectral threshold. A minimal pixel (area) threshold was also set to eliminate indexing portions of cells or other small, noncellular artifacts and was applied at the same value to all spectral images. Appressed cells were manually segmented within the software by the user. The quality of the segmentation was visually compared against the integrated intensity image. After the cells had been indexed, each pixel in the cell was mapped to the spectral domain, and the mean per-cell component intensity was calculated by summing the component intensity across all pixels in the cell and then dividing by the total number of pixels that constitute the cell. In total, 60 spectral images (332 cells) were analyzed. Nested ANOVA statistical analysis of the mean fluorescence intensities collected from the 332 cells in the spectral images was performed using the N-way ANOVA function (nanova) found in Matlab (Mathworks; version 7.10.0.499, R2010a). A more complete description of nested ANOVA can be found elsewhere (Snedecor and Cochran, 1967). The box plots shown in Figure 5 were generated in Matlab using the box-plot function with the notch option turned on to show the 95% confidence levels.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Steady-state fluorescence from whole cells of Synechocystis 6803 wild type and mutants excited with 488-nm light.
Supplemental Figure S2. MCR-identified spectral component corresponding to carotenoid pigments.
Supplemental Figure S3. Isolation of phycobilisomes from Synechocystis 6803 wild type and phycobilisome mutants.
Supplemental Movie S1. Volume-rendered animation of a wild-type cell.
Supplemental Movie S2. Volume-rendered animation of a CB cell.
Supplemental Movie S3. Volume-rendered animation of a CB cell.
Supplemental Movie S4. Volume-rendered animation of a CK cell.
Supplemental Movie S5. Volume-rendered animation of a PAL cell.
Supplementary Material
Acknowledgments
We thank Dr. Ghada Ajlani for her gift of the mutant strains. We thank all members of the Pakrasi and Timlin laboratories for insightful discussions of the project. We thank Howard Berg for TEM assistance, Dr. Edward Thomas for critical insight into statistical analysis of the image data, Dr. Michael Sinclair for developing and maintaining the hyperspectral microscope, Drs. David Melgaard, Michael R. Keenan, and Mark Van Benthem and Mr. Greg Poulter for MCR algorithm and software development, and Michelle Raymer for assistance with making movies.
References
- Ajlani G, Vernotte C. (1998) Construction and characterization of a phycobiliprotein-less mutant of Synechocystis sp. PCC 6803. Plant Mol Biol 37: 577–580 [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C, DiMagno L, Haselkorn R. (1995) Phycobilisome core mutants of Synechocystis PCC 6803. Biochim Biophys Acta 1231: 189–196 [Google Scholar]
- Allen MM. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Anderson LK, Toole CM. (1998) A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol Microbiol 30: 467–474 [DOI] [PubMed] [Google Scholar]
- Andersson B, Barber J. (2004) Mechanisms of photodamage and protein degradation during photoinhibition of photosystem II. In NR Baker, ed, Photosynthesis and the Environment, Vol 5. Springer, Dordrecht, The Netherlands, pp 101–121 [Google Scholar]
- Andrizhiyevskaya EG, Schwabe TME, Germano M, D’Haene S, Kruip J, van Grondelle R, Dekker JP. (2002) Spectroscopic properties of PSI–IsiA supercomplexes from the cyanobacterium Synechococcus PCC 7942. Biochim Biophys Acta 1556: 265–272 [DOI] [PubMed] [Google Scholar]
- Arteni AA, Ajlani G, Boekema EJ. (2009) Structural organisation of phycobilisomes from Synechocystis sp. strain PCC6803 and their interaction with the membrane. Biochim Biophys Acta 1787: 272–279 [DOI] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Blankenship RE. (2002) Molecular Mechanisms of Photosynthesis. Blackwell Science, Oxford [Google Scholar]
- Bro R, De Jong S. (1997) A fast non-negativity-constrained least squares algorithm. J Chemometr 11: 393–401 [Google Scholar]
- Chen M, Floetenmeyer M, Bibby TS. (2009) Supramolecular organization of phycobiliproteins in the chlorophyll d-containing cyanobacterium Acaryochloris marina. FEBS Lett 583: 2535–2539 [DOI] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ. (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- El-Mohsnawy E, Kopczak MJ, Schlodder E, Nowaczyk M, Meyer HE, Warscheid B, Karapetyan NV, Rögner M. (2010) Structure and function of intact photosystem 1 monomers from the cyanobacterium Thermosynechococcus elongatus. Biochemistry 49: 4740–4751 [DOI] [PubMed] [Google Scholar]
- Folea IM, Zhang P, Aro E-M, Boekema EJ. (2008) Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett 582: 1749–1754 [DOI] [PubMed] [Google Scholar]
- Fromme P, Grotjohann I. (2011) Structure of Cyanobacterial Photosystems I and II In GA Peschek, C Obinger, G Renger, eds, Bioenergetic Processes of Cyanobacteria. Springer, Dordrecht, The Netherlands, pp 285–335 [Google Scholar]
- Fujita Y, Murakami A. (1987) Regulation of electron transport composition in cyanobacterial photosynthetic system: stoichiometry among photosystem I and II complexes and their light-harvesting antennae and cytochrome b6/f complex. Plant Cell Physiol 28: 1547–1553 [Google Scholar]
- Fujita Y, Murakami A, Aizawa K, Ohki K. (1994) Short-term and long-term adaptation of the photosynthetic apparatus: homeostatic properties of thylakoids. In DA Bryant, ed, The Molecular Biology of Cyanobacteria, Vol 1. Springer, Dordrecht, The Netherlands, pp 677–692 [Google Scholar]
- Glazer AN. (1984) Phycobilisome a macromolecular complex optimized for light energy transfer. Biochim Biophys Acta 768: 29–51 [Google Scholar]
- Gounaris K, Chapman DJ, Barber J. (1989) Isolation and characterisation of a Dl/D2/cytochrome b-559 complex from Synechocystis 6803. Biochim Biophys Acta 973: 296–301 [Google Scholar]
- Haaland DM, Jones HDT, Van Benthem MH, Sinclair MB, Melgaard DK, Stork CL, Pedroso MC, Liu P, Brasier AR, Andrews NL, et al. (2009) Hyperspectral confocal fluorescence imaging: exploring alternative multivariate curve resolution approaches. Appl Spectrosc 63: 271–279 [DOI] [PubMed] [Google Scholar]
- Haaland DM, Timlin JA, Sinclair MB, Van Benthem MH, Martinez MJ, Aragon AD, Werner-Washburne M. (2003) Multivariate curve resolution for hyperspectral image analysis: applications to microarray technology. In RM Levenson, GH Bearman, A Mahadevan-Jansen, eds, Spectral Imaging: Instrumentation, Applications, and Analysis. SPIE, San Jose, CA, p 55–66 [Google Scholar]
- Jones HDT, Haaland DM, Sinclair MB, Melgaard DK, Van Benthem MH, Pedroso MC. (2008) Weighting hyperspectral image data for improved multivariate curve resolution results. J Chemometr 22: 482–490 [Google Scholar]
- Keenan MR, Kotula PG. (2004) Accounting for Poisson noise in the multivariate analysis of ToF-SIMS spectrum images. Surf Interface Anal 36: 203–212 [Google Scholar]
- Liberton M, Austin JR, II, Berg RH, Pakrasi HB. (2011) Unique thylakoid membrane architecture of a unicellular N2-fixing cyanobacterium revealed by electron tomography. Plant Physiol 155: 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberton M, Berg RH, Heuser J, Roth R, Pakrasi HB. (2006) Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227: 129–138 [DOI] [PubMed] [Google Scholar]
- Liberton M, Pakrasi HB. (2008) Membrane systems in cyanobacteria. In A Herrero, E Flores, eds, The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Horizon Scientific Press, Norwich, UK, pp 217–287 [Google Scholar]
- MacColl R. (1998) Cyanobacterial phycobilisomes. J Struct Biol 124: 311–334 [DOI] [PubMed] [Google Scholar]
- Marquardt J, Mörschel E, Rhiel E, Westermann M. (2000) Ultrastructure of Acaryochloris marina, an oxyphotobacterium containing mainly chlorophyll d. Arch Microbiol 174: 181–188 [DOI] [PubMed] [Google Scholar]
- Marquardt J, Senger H, Miyashita H, Miyachi S, Mörschel E. (1997) Isolation and characterization of biliprotein aggregates from Acaryochloris marina, a prochloron-like prokaryote containing mainly chlorophyll d. FEBS Lett 410: 428–432 [DOI] [PubMed] [Google Scholar]
- Mörschel E, Schatz GH. (1987) Correlation of photosystem-II complexes with exoplasmatic freeze-fracture particles of thylakoids of the cyanobacterium Synechococcus. Planta 172: 145–154 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (1992) Excitation energy transfer from phycobilisomes to photosystem I in a cyanobacterium. Biochim Biophys Acta 1100: 285–292 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (1999) The thylakoid membranes of cyanobacteria: structure, dynamics and function. Aust J Plant Physiol 26: 671–677 [Google Scholar]
- Mullineaux CW. (2008) Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res 95: 175–182 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF. (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nevo R, Chuartzman SG, Tsabari O, Reich Z, Charuvi D, Shimoni E. (2009) Architecture of Thylakoid Membrane Networks. In H Wada, N Murata, eds, Lipids in Photosynthesis: Essential and Regulatory Functions. Springer Science and Business Media, Dordrecht, The Netherlands, pp 295–328 [Google Scholar]
- Olive J, Ajlani G, Astier C, Recouvreur M, Vernotte C. (1997) Ultrastructure and light adaptation of phycobilisome mutants of Synechocystis PCC 6803. Biochim Biophys Acta 1319: 275–282 [Google Scholar]
- Sidler WA. (2004) Phycobilisome and phycobiliprotein structures. In DA Bryant, ed, The Molecular Biology of Cyanobacteria, Vol 1. Springer, Dordrecht, The Netherlands, pp 139–216 [Google Scholar]
- Sinclair MB, Haaland DM, Timlin JA, Jones HDT. (2006) Hyperspectral confocal microscope. Appl Opt 45: 6283–6291 [DOI] [PubMed] [Google Scholar]
- Snedecor GW, Cochran WG. (1967) Statistical Methods. Iowa State University Press, Ames, IA [Google Scholar]
- Stadnichuk I, Lukashev E, Elanskaya I, Boichenko V, Bukhov N. (2009) Increase in the rate of photosynthetic linear electron transport in cyanobacteruim Synechocystis sp. PCC 6803 lacking phycobilisomes. Russ J Plant Physiol 56: 439–444 [Google Scholar]
- Tang X-S, Diner BA. (1994) Biochemical and spectroscopic characterization of a new oxygen-evolving photosystem II core complex from the cyanobacterium Synechocystis PCC 6803. Biochemistry 33: 4594–4603 [DOI] [PubMed] [Google Scholar]
- Ughy B, Ajlani G. (2004) Phycobilisome rod mutants in Synechocystis sp. strain PCC6803. Microbiology 150: 4147–4156 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Van Benthem MH, Keenan MR. (2004) Fast algorithm for the solution of large-scale non-negativity-constrained least squares problems. J Chemometr 18: 441–450 [Google Scholar]
- Van Benthem MH, Keenan MR, Haaland DM. (2002) Application of equality constraints on variables during alternating least squares procedures. J Chemometr 16: 613–622 [Google Scholar]
- van de Meene AM, Hohmann-Marriott MF, Vermaas WF, Roberson RW. (2006) The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol 184: 259–270 [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ, Timlin JA, Jones HDT, Sinclair MB, Nieman LT, Hamad SW, Melgaard DK, Haaland DM. (2008) In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc Natl Acad Sci USA 105: 4050–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernotte C, Astier C, Olive J. (1990) State 1-state 2 adaptation in the cyanobacteria Synechocystis PCC 6714 wild type and Synechocystis PCC 6803 wild type and phycocyanin-less mutant. Photosynth Res 26: 203–212 [DOI] [PubMed] [Google Scholar]
- Westerman M, Ernst A, Brass S, Böger P, Wehrmeyer W. (1994) Ultrastructure of cell wall and photosynthetic apparatus of the phycobilisome-less Synechocystis sp. strain BO 8402 and phycobilisome-containing derivative strain BO 9201. Arch Microbiol 162: 222–232 [Google Scholar]
- Wittmershaus BP, Tran TD, Panaia B. (1998) Fluorescence from low-energy chlorophylls in photosystem I of Synechocystis sp. PCC 6803 at physiological temperatures. Photosynth Res 57: 29–39 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.