Abstract
Amino acids serve as transport forms for organic nitrogen in the plant, and multiple transport steps are involved in cellular import and export. While the nature of the export mechanism is unknown, overexpression of GLUTAMINE DUMPER1 (GDU1) in Arabidopsis (Arabidopsis thaliana) led to increased amino acid export. To gain insight into GDU1’s role, we searched for ethyl-methanesulfonate suppressor mutants and performed yeast-two-hybrid screens. Both methods uncovered the same gene, LOSS OF GDU2 (LOG2), which encodes a RING-type E3 ubiquitin ligase. The interaction between LOG2 and GDU1 was confirmed by glutathione S-transferase pull-down, in vitro ubiquitination, and in planta coimmunoprecipitation experiments. Confocal microscopy and subcellular fractionation indicated that LOG2 and GDU1 both localized to membranes and were enriched at the plasma membrane. LOG2 expression overlapped with GDU1 in the xylem and phloem tissues of Arabidopsis. The GDU1 protein encoded by the previously characterized intragenic suppressor mutant log1-1, with an arginine in place of a conserved glycine, failed to interact in the multiple assays, suggesting that the Gdu1D phenotype requires the interaction of GDU1 with LOG2. This hypothesis was supported by suppression of the Gdu1D phenotype after reduction of LOG2 expression using either artificial microRNAs or a LOG2 T-DNA insertion. Altogether, in accordance with the emerging bulk of data showing membrane protein regulation via ubiquitination, these data suggest that the interaction of GDU1 and the ubiquitin ligase LOG2 plays a significant role in the regulation of amino acid export from plant cells.
Amino acids are the main form of organic nitrogen transported in the xylem and the phloem in plants (Peoples and Gifford, 1990). Besides a role in nitrogen transfer between plant organs, amino acids are important to coordinate shoot and root metabolism in response to environmental conditions. Cycling of amino acids between shoots and roots, successively transported by the phloem and the xylem, has been proposed to carry information about the nitrogen status of roots or shoots to the other organ system. In particular, amino acids have been shown to inhibit root nitrate uptake, adjusting inorganic nitrogen uptake to shoot demand (Miller et al., 2007).
Amino acid cycling involves successive transfers across the plasma membrane from symplasm (phloem and cytosol) to apoplasm (xylem and cell wall) and vice versa. Because membranes are fairly impermeable to these solutes, both import and export are catalyzed by integral membrane proteins. Identified in early physiological studies (Li and Bush, 1990, 1992), amino acid importers are proton gradient-dependent transporters with broad amino acid specificities (Tegeder and Rentsch, 2010). In stark contrast to importers, molecular mechanisms of plasma membrane amino acid export remain largely unknown. While export across the plasma membrane has been measured at the physiological level (Secor and Schrader, 1984; De Jong et al., 1997; Lesuffleur and Cliquet, 2010), no transporter has yet been identified that mediates this process (for review, see Okumoto and Pilot, 2011). The first cloned plant amino acid exporter, GAMMA AMINO BUTYRIC ACID PERMEASE (GABP), mediates γ-aminobutyrate transport from the cytosol to the mitochondrion (Dündar and Bush, 2009; Michaeli et al., 2011). Because of its localization at the mitochondrial membrane, GABP is not expected to mediate amino acid export at the plasma membrane.
The first gene suggested to be involved in plasma membrane amino acid export is GLUTAMINE DUMPER1 (GDU1). The gdu1-1D mutant, which overexpresses GDU1, was isolated in an activation-tag screen for plants with altered hydathode function. gdu1-1D exhibited high free amino acid content in the phloem, xylem, and guttation stream, leading to Gln crystallization at the hydathodes (Pilot et al., 2004). The Gdu1D phenotype also entailed plant size reduction, constitutive necrotic lesions, and resistance to toxic concentrations of amino acids (Pilot et al., 2004; Pratelli and Pilot, 2007; Liu et al., 2010). gdu1-1D was later found to constitutively export amino acids from plant cells (Pratelli et al., 2010). GDU1 encodes a small protein with a single transmembrane domain. Paralogs are found in Arabidopsis (Arabidopsis thaliana; named GDU2–GDU7), and homologs are present in higher and lower plant genomes (Pratelli and Pilot, 2006). The physiological function of the GDU family is unknown, but overexpression of any GDU gene causes phenotypes reminiscent of Gdu1D (Pratelli et al., 2010). Apart from the membrane domain, GDU proteins also share a VIMAG domain (representing the amino acids Val-Ile-Met-Ala-Gly). The log1-1 allele of GDU1 (GDU1G100R, in which the VIMAG Gly is mutated to Arg) was shown to suppress the Gdu1D phenotype (Pratelli and Pilot, 2006), suggesting that the VIMAG domain is essential for GDU1 function. The molecular basis for this suppression was not determined.
In spite of its effect on amino acid export, the predicted structure of GDU1 makes it unlikely to be a transporter (Pilot et al., 2004), and it had been suggested that GDU1 could be a transporter subunit (Pratelli et al., 2010) by analogy to mammalian heteromeric amino acid transporters (Palacín et al., 2005). Small, single transmembrane domain proteins have also been shown to be involved in the organization of large membrane protein complexes (for review, see Zickermann et al., 2010). Finding interacting partners of GDU1, therefore, is a necessary step to gain insight about its role. To this end, we performed yeast two-hybrid and ethyl-methanesulfonate (EMS) suppression screens. We report here that both of these approaches uncovered the same RING-type ubiquitin E3 ligase, subsequently named LOSS OF GDU2 (LOG2). In a survey of Arabidopsis RING finger-containing E3s, LOG2 was shown to exhibit in vitro E3 activity, but the biological function of LOG2 was not investigated in that study (Stone et al., 2005).
E3 ligases facilitate the covalent attachment of the small protein ubiquitin to other proteins (ubiquitination). Ubiquitination is an efficient and highly specific mechanism by which intracellular protein activity, localization, and/or stability are governed in eukaryotes. Following ATP-dependent activation by ubiquitin-activating enzyme (UBA or E1) and transthioesterification to an ubiquitin-conjugating enzyme (UBC or E2), substrate ubiquitination is catalyzed by E3 ubiquitin ligases (E3s; Deshaies and Joazeiro, 2009). E3s bind protein substrates and E2 ubiquitin thioesters in a conformation that facilitates ubiquitin transfer to substrates. In the case of RING-type E3s, the RING domain enables interaction with E2s (Joazeiro et al., 1999). In addition to promoting nuclear and cytosolic protein degradation via the 26S proteasome, ubiquitination by E3s can also regulate plasma membrane protein abundance via lysosomal/vacuolar proteolysis (Komander, 2009; Léon and Haguenauer-Tsapis, 2009).
Here, we report that the ubiquitin ligase LOG2 interacts with GDU1 in vitro and in planta and that reduction of LOG2 expression suppresses the Gdu1D phenotype. In addition, the previously described log1-1 mutation abolishes GDU1 interaction with LOG2. Altogether, these data support a model whereby LOG2 and its interaction with GDU1 are required for the increased amino acid export observed upon GDU1 overexpression.
RESULTS
GDU1 Interacts with LOG2, a RING Domain-Containing Protein
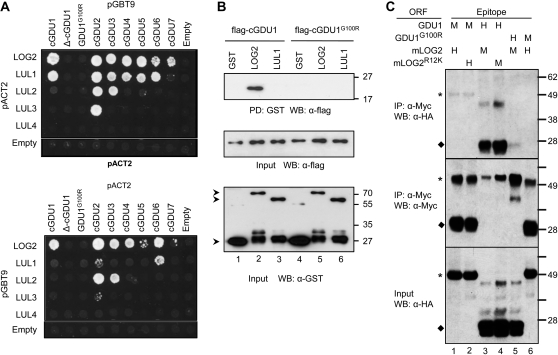
The physiological consequences of GDU1 overexpression have been studied extensively, but the mechanism by which GDU1 activates amino acid efflux remains unclear. GDU1 is unlikely to be a transporter; thus, it must interact with other proteins to activate efflux. A screen for interacting proteins was performed using a yeast two-hybrid strategy, in which the region C terminal to the putative transmembrane domain of GDU1 (cGDU1; amino acids 61–158) was used as bait against an Arabidopsis cDNA library. Three clones, whose inserts contained partial open reading frames of genes At3g09770 and At5g03200 (Supplemental Fig. S1, A and C), were shown to restore yeast prototrophy when coexpressed with cGDU1 in the yeast two-hybrid screen. At3g09770 and At5g03200 encode members of the same subfamily of RING finger ubiquitin E3 ligases (Kraft et al., 2005; Stone et al., 2005). At3g09770 and At5g03200 were named LOG2 (see below) and LOG2-LIKE UBIQUITIN LIGASE1 (LUL1), respectively. The three other paralogs were designated LUL2 (At3g53410), LUL3 (At5g19080), and LUL4 (At3g06140). Full-length LOG2 and LUL1 also interacted with cGDU1 in the yeast two-hybrid assay (Fig. 1A). Yeast coexpressing LUL1 and cGDU1 grew slower than yeast coexpressing LOG2 and cGDU1, while the proteins were expressed at similar levels (data not shown). This observation was consistent with the activity of the lacZ reporter gene. β-Galactosidase activity of yeast cells was 0.14 ± 0.13 nmol o-nitro phenol (ONP) h−1 0.10−7 cells when cGDU1 was coexpressed with LUL1 and 3.02 ± 0.18 nmol ONP h−1 0.10−7 cells when coexpressed with LOG2, suggesting that the interaction between cGDU1 with LUL1 was weaker compared with that of LOG2.
Figure 1.
Interaction assays between GDU family members and the E3 ubiquitin ligases of the LOG2 family. A, yeast two-hybrid interaction of the cytosolic domain of the GDU proteins (cGDU) with LOG2 and LULs. The panels show swapping of inserts between the bait (pGBT9) and prey (pACT2) plasmids. Yeast coexpressing the protein pairs were grown for 4 d on medium selecting for protein interaction, lacking Leu, Trp, adenine, and His. All yeast grew on a medium lacking Leu and Trp, selecting for the plasmids only (data not shown). B, GST pull-down (PD) assay of flag-cGDU1 or flag-cGDU1G100R with GST-LOG2, GST-LUL1, or GST alone. Top, pulled-down samples; middle, cGDU input; bottom, GST protein input. Arrowheads indicate GST (approximately 27 kD) or full-length GST-tagged LOG2 and LUL1. C, Coimmunoprecipitation (IP) assay after the expression of GDU1 or GDU1G100R and mLOG2 or mLOG2R12K in transiently infiltrated N. benthamiana leaves. Top, Myc coimmunoprecipitation samples probed with α-HA (H); middle, Myc coimmunoprecipitation samples probed with α-Myc (M); bottom, HA-protein input. Asterisks and diamonds indicate LOG2 and GDU1 proteins, respectively. Numbers on the right in B and C indicate molecular mass in kD. ORF, Open reading frame; WB, western blot.
In addition to the presence of a C-terminal C3HC4 RING finger domain and a predicted N-terminal myristoylation site (see below), the five LOG2 subfamily members contain a functionally uncharacterized region N terminal to the RING finger that was previously designated Domain Associated with RING2 (DAR2; Supplemental Fig. S1C; Stone et al., 2005). LOG2-like genes are present in the genomes of diverse taxa, including monocots, lower plants, and mammals, but surprisingly not in fungi. While LOG2 function has not been studied in plants, the mammalian homolog MAHOGUNIN RING FINGER1 (MGRN1) plays an unclear role in the down-regulation of melanocortin signaling and the maturation of endosomes (He et al., 2003; Kim et al., 2007; Cooray et al., 2011). MGRN1 contains a large C-terminal domain not found in plant LOG2/LUL proteins (Supplemental Fig. S1C) that seems to play an important role in endosomal trafficking (Kim et al., 2007), suggesting divergence in function from plant LOG2/LULs.
The interaction of cGDU1 with LOG2 and LUL1 was tested by glutathione S-transferase (GST) in vitro pull-down assays. The interaction of GST-LOG2 with cGDU1 was the only one to be detected (Fig. 1B, top panel, lane 2), indicating that cGDU1 binds directly to LOG2 and more strongly to LOG2 than to LUL1 in this assay. The interaction between GDU1 and LOG2 was then tested in planta using proteins transiently expressed in Nicotiana benthamiana leaves. Wild-type LOG2 was expressed at levels too low to be suitable for coimmunoprecipitation experiments (data not shown). Hypothesizing that autoubiquitination may contribute to protein instability, we generated a LOG2 mutant with a catalytically inactive RING domain (LOG2CC354+357AA, called mLOG2; see below and Fig. 2C), which could successfully be expressed at high levels. Full-length GDU1 and mLOG2 with C-terminal Myc or hemagglutinin (HA) tags were then coexpressed in N. benthamiana leaves, and the Myc-fused protein was immunoprecipitated. Independent of which protein contained the Myc epitope tag, GDU1 and mLOG2 coimmunoprecipitated (Fig. 1C, lanes 1 and 3), showing that the interactions detected by yeast two-hybrid and in vitro assays are observed in planta as well.
Figure 2.
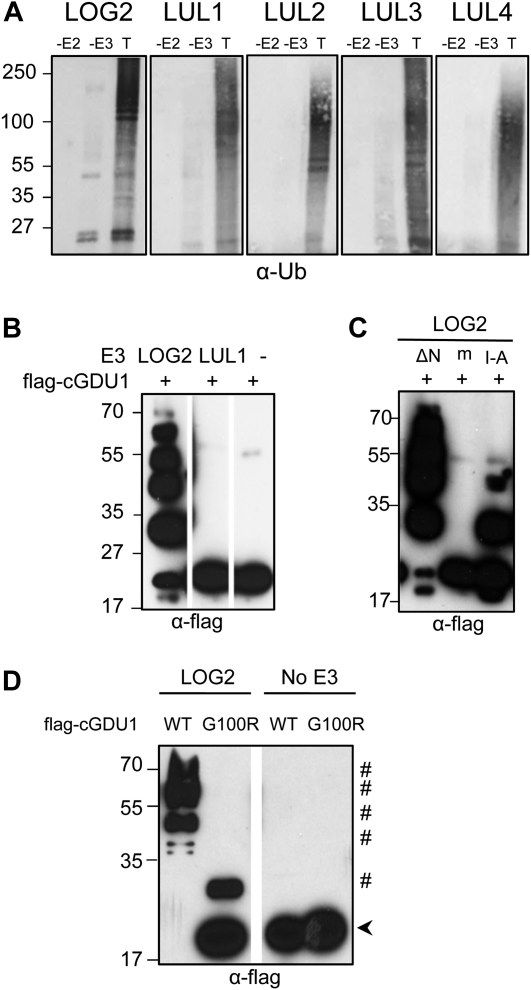
Ubiquitin ligase activity tests of LOG2 and LULs. A, In vitro ubiquitination assay of GST-LOG2 and GST-LUL1 to -4 visualized with an anti-ubiquitin antibody. −E2 and −E3, Omission of ubiquitin-conjugating enzyme (E2) and the indicated ubiquitin ligase (E3) from the assay, respectively; T, all ubiquitin pathway components were present in the assay. B, In vitro ubiquitination assay of flag-cGDU1 by GST-LOG2, GST-LUL1, or no added E3, visualized with anti-flag antibody. C, In vitro ubiquitination assay of flag-cGU1 by truncated and mutant forms of GST-LOG2. ΔN, LOG2 truncation lacking amino acids N terminal to DAR2; M, LOG2CC354/347AA (RING-dead); I-A, LOG2I321A (RING-E2 interaction impaired). D, In vitro ubiquitination assay of either flag-cGDU1 or flag-cGDU1G100R by LOG2-V5. The arrowhead indicates nonubiquitinated flag-cGDU1; the pound symbol indicates ubiquitinated flag-cGDU1. Numbers on the left indicate molecular mass in kD. WT, Wild type.
The GDU1-LOG2 Interaction Is Affected by a Suppressor Mutation in GDU1
It had previously been reported that the EMS-generated log1-1 mutant, corresponding to a G100R substitution in the VIMAG motif of GDU1, suppresses multiple features of the Gdu1D phenotype (Pratelli and Pilot, 2006, 2007). Although the C-terminal domains of the seven Arabidopsis GDUs show limited sequence conservation outside the VIMAG domain, all cGDUs could interact with LOG2 in yeast, and most could interact with LUL1 (Fig. 1A). This suggested that the interaction with LOG2 is mediated through the VIMAG domain. To test the hypothesis that the G100R mutation alters the interaction with LOG2, the interactions of cGDU1G100R (log1-1) and Δ-cGDU1 (in which the whole VIMAG domain has been deleted) with LOG2 and LULs were tested using the yeast two-hybrid system. While these mutations did not alter protein abundance (data not shown), neither log1-1 nor Δ-cGDU1 could interact with LOG2 or LUL1 (Fig. 1A). Consistent with these results, flag-tagged cGDU1G100R failed to interact with GST-LOG2 in a GST pull-down assay (Fig. 1B, lane 5), and coimmunoprecipitation of GDU1G100R and mLOG2 expressed in N. benthamiana was greatly reduced (Fig. 1C, lanes 5 and 6). These data indicate that the GDU1-LOG2 interaction requires the conserved VIMAG domain of GDU1 and that the suppression of the Gdu1D phenotype observed in log1-1 plants (Pratelli and Pilot, 2006) may result from the loss of interaction with LOG2.
cGDUs Interact with LOG2 and LUL Proteins
To systematically characterize interactions between the LOG2/LUL and GDU families, yeast two-hybrid assays were conducted between the C-terminal domains of all Arabidopsis GDUs and LOG2/LUL proteins. In contrast to LOG2, cGDU-LUL interactions were typically specific for a pair of bait-prey combinations (Fig. 1A). LUL2 and LUL3 interactions were observed only for cGDU2 or cGDU3, and LUL4 did not interact with any cGDU. As expected, deletion of the VIMAG motif from cGDU1 abolished all LUL interactions. The interaction between GDU and LOG2-like proteins appears to be facilitated by the VIMAG motif of the GDUs and a conserved domain among LOG2 homologs (most likely DAR2; see below).
LOG2/LULs Exhibit E3 Ligase Activity, but cGDU1 Is Ubiquitinated Exclusively by LOG2 in Vitro
LOG2 had previously been shown to polymerize ubiquitin in vitro (Stone et al., 2005). To examine the E3 activities of the LOG2 paralogs, GST fusions of LOG2 and LUL1 to -4 were expressed in Escherichia coli, purified, and assayed with ubiquitin pathway proteins (E2, E1, and ubiquitin). Mirroring previously published results with LOG2 (Kraft et al., 2005; Stone et al., 2005), LUL1 to -4 were active, forming high-Mr ubiquitinated proteins in the presence of all ubiquitin pathway components, while omission of the E2 prevented their production (Fig. 2A).
Only the proteins demonstrated to interact with GDU1 (i.e. LOG2 and LUL1 [Fig. 1]) were tested for ubiquitination of cGDU1 using in vitro assays. Only LOG2 could ubiquitinate cGDU1 above the background of the E3 reaction (Fig. 2B). Substitution to Ala of two zinc-coordinating Cys residues (mLOG2) or an Ile residue in the RING domain (LOG2I321A) was previously described for other RING E3s to abolish E3 activity or hinder association with E2s, respectively (Lorick et al., 1999; Brzovic et al., 2003). As predicted, these modifications of LOG2 led to abolished or compromised cGDU1 ubiquitination, respectively (Fig. 2C). Interestingly, truncation of the nonconserved 125-amino acid region N terminal to DAR2 did not impede cGDU1 ubiquitination by LOG2 (Fig. 2C). cGDU1G100R (log1-1) was then assayed with or without LOG2-V5-His6 in the presence of ubiquitin pathway components. While cGDU1 formed ladders in the presence of LOG2, only weak monoubiquitination of cGDU1G100R was observed (Fig. 2D). These data show that the decreased interaction of cGDU1G100R with LOG2 (Fig. 1B) results in decreased in vitro ubiquitination of cGDU1G100R and suggest that the specificity determinants of the GDU1-LOG2 interaction lie in the DAR2 of LOG2.
Similar to GDU1, LOG2 Is Expressed in the Vasculature
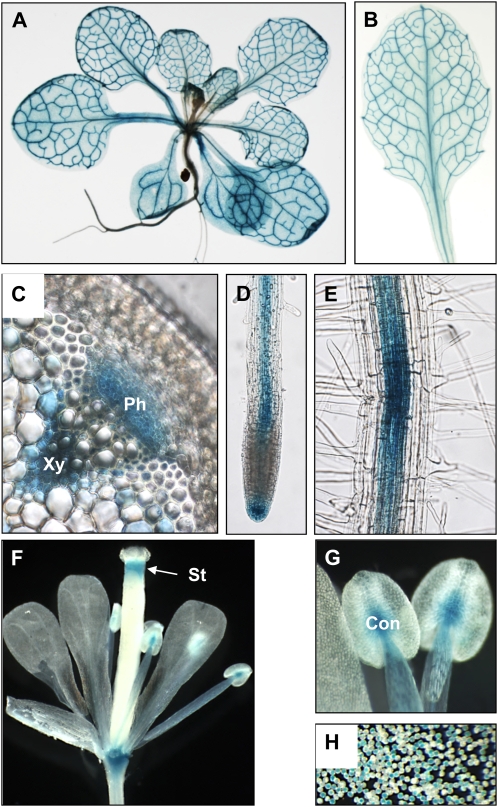
The interaction between GDU1 and LOG2, confirmed so far in vitro and after coexpression in planta, is physiologically relevant only if the two proteins are expressed in the same cell types. To investigate LOG2 expression pattern, the LOG2 promoter region fused to the coding sequence of the uidA gene from E. coli, encoding GUS, was introduced into the Arabidopsis genome. GUS activity was detected in vascular tissues of roots, leaves, and stems (Fig. 3, A–D), and more precisely in both phloem and xylem parenchyma cells, as revealed by stem cross-sections (Fig. 3C). The LOG2 promoter was active in all shoot cells: a light background staining appeared in leaves and in nonvascular parenchyma cells of the stems (Fig. 3, A and C). Examination of lightly stained plants showed that this background staining did not result from diffusion of the product of the histochemical reaction out of the vascular tissues (data not shown). The presence of staining in roots up to the division zone indicated that the LOG2 promoter is active in the root phloem (Fig. 3D). Interestingly, the LOG2 promoter was only active in cells from the root stele and root tip; even with longer reaction time, no staining could be observed in the cortex and epidermis (Fig. 3, D and E). In reproductive organs, strong GUS activity was detected in the style, connective tissue, and the base of the flower (Fig. 3, F and G), which persisted along the development of the silique (data not shown). Finally, the LOG2 promoter was active in pollen grains (Fig. 3H). The observed expression pattern is in good agreement with quantitative reverse transcription (RT)-PCR results (Supplemental Fig. S2), the AtGenExpress data set (Schmid et al., 2005), and the Arabidopsis Gene Expression Database (Cartwright et al., 2009), where stronger expression was detected in vasculature-rich tissues (stems), pollen, columella, and phloem and xylem cells (data not shown). The expression pattern of LOG2 largely overlaps with that of GDU1, shown to be expressed in the vasculature of roots, stems, and petioles (Pilot et al., 2004).
Figure 3.
LOG2 promoter-GUS analysis. A, Rosette. B, Leaf. C, Stem cross-section. D and E, Root. F, Flower. G, Stamen. H, Pollen grain. Con, Connective; Ph, phloem; St, style; Xy, xylem.
GDU1 and LOG2 Localize to Microsomes and Are Enriched in the Plasma Membrane
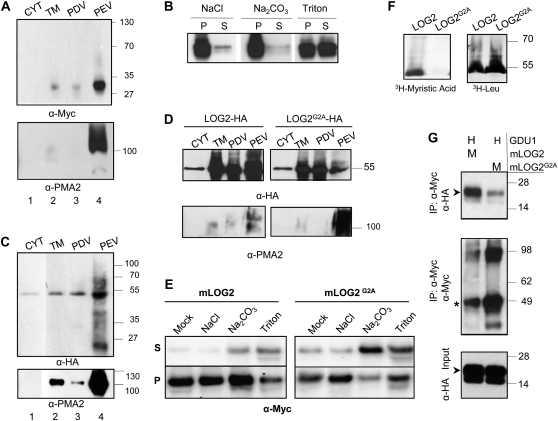
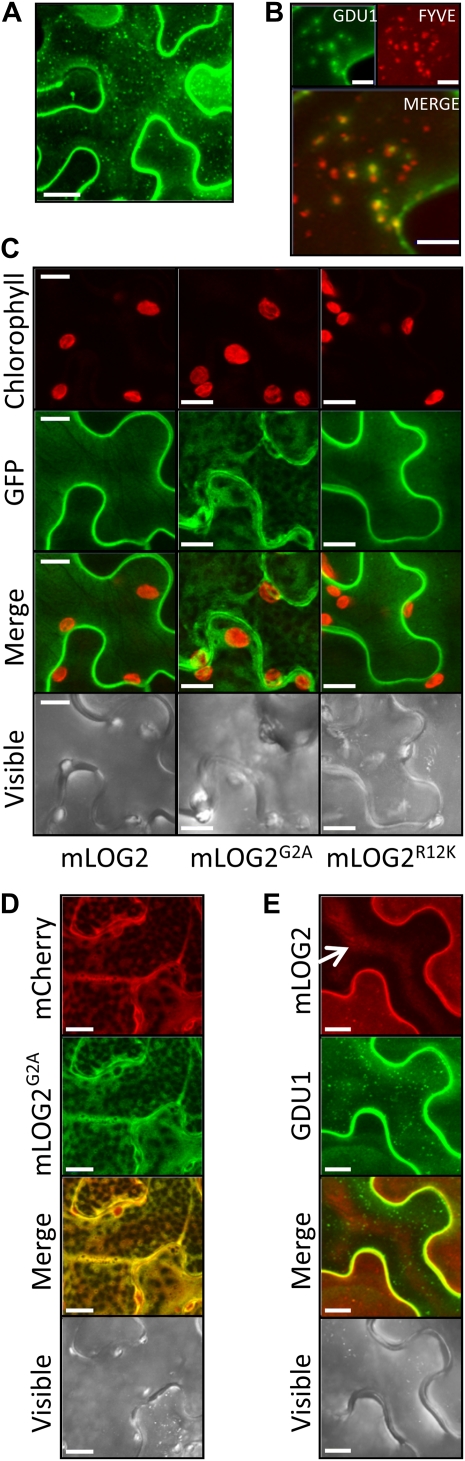
GDU1 is predicted to be a single-pass transmembrane domain protein (Pilot et al., 2004). In order to interact with GDU1, LOG2 needs to be recruited to the same membrane(s). Confocal imaging of Arabidopsis protoplasts transiently expressing GDU1-GFP had suggested that GDU1 was targeted to the plasma membrane (Pilot et al., 2004). We created transgenic Arabidopsis that overexpressed Myc-tagged GDU1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. One line, designated 35S-GDU1-Myc, showed stable expression of GDU1-Myc over several generations and recapitulated the smaller leaf size and overaccumulation of amino acids seen in the activation-tagged gdu1-1D line (Supplemental Fig. S3; Supplemental Table S1). To examine the subcellular localization of GDU1, rosette leaf proteins were fractionated into cytosolic and total microsome fractions, after which total microsomes were further processed into plasma membrane-depleted vesicles (PDVs) and plasma membrane-enriched vesicles (PEVs). GDU1-Myc was highly enriched in the microsomal fraction compared with the soluble fraction, confirming membrane localization (Fig. 4A, lanes 1 and 2). Similar to the plasma membrane-localized PMA2 used as a control (Dambly and Boutry, 2001; Elmore and Coaker, 2011), GDU1 was enriched in PEVs compared with PDVs (Fig. 4A, lanes 3 and 4), indicating enrichment at the plasma membrane. Consistent with these results, GDU1-GFP transiently expressed in N. benthamiana epidermal cells localized at the plasma membrane and in small dots (Fig. 5A), which could be labeled with the endosome marker FYVE-GFP (Voigt et al., 2005; Fig. 5B). Limited overlap was found with fluorescent markers specific to the Golgi apparatus, and no localization could be detected in mitochondria, chloroplasts, the endoplasmic reticulum, lysosomes, or the cytosol (data not shown).
Figure 4.
Analysis of membrane localization and the association of GDU1 and LOG2. A and C, Subcellular fractionation of transgenic Arabidopsis expressing GDU1-Myc (A) or LOG2-HA (C). The fractionation of plasma membrane-localized PMA2 in respective preparations is shown at the bottom. CYT, Cytosolic fraction; TM, total microsomes. B, Sensitivity of microsomal GDU1-Myc expressed in Arabidopsis to 1 m NaCl, 0.1 m Na2CO3, pH 11.5, or 1% (v/v) Triton X-100. P, Pellet; S, supernatant. D, Subcellular fractionation of LOG2-HA and LOG2G2A-HA after transient expression in N. benthamiana. E, Sensitivity of microsomal LOG2 and LOG2G2A to 1 m NaCl, 0.1 m Na2CO3, pH 11.5, or 1% (v/v) Triton X-100 after transient expression in N. benthamiana. F, In vitro transcription/translation assay reactions incubated with either [3H]myristic acid (left) or [3H]Leu (right) using plasmids expressing either wild-type LOG2 or LOGG2A. G, Coimmunoprecipitation (IP) after transient expression of GDU1-HA (H) and mLOG2-Myc or mLOG2G2A-Myc (M) in N. benthamiana leaves using an anti-Myc antibody. Top, Myc coimmunoprecipitation samples probed with α-HA; middle, Myc coimmunoprecipitation samples probed with α-Myc; bottom, HA-protein input. Asterisks and arrowheads indicate LOG2 and GDU1 proteins, respectively. Numbers on the right indicate molecular mass in kD.
Figure 5.
Subcellular localization of GDU1, LOG2, and LOG2 variants. All proteins were transiently expressed in N. benthamiana epidermis cells. Images are maximum projections of optical sections of the abaxial side of cells, obtained by confocal microscopy. A, Localization of GDU1-GFP protein. B, Colocalization of GDU1-GFP and FYVE-RFP, an endosome marker. C, Localization of mLOG2-GFP, mLOG2G2A-GFP, and mLOG2R12K-GFP. D, Colocalization of mLOGG2A with cytosolic mCherry. E, Change of localization of GDU1-GFP when coexpressed with mLOG2-mCherry. The middle cell (arrow) expressed mLOG2-mCherry at a low level, while the two lateral cells expressed mLOG2-mCherry at higher levels. All three cells expressed GDU1-GFP. The central fluorescence of the middle cell is potentially due to internal reflection. Bars = 20 μm (A), 5 μm (B) or 10 μm (C to E).
To explore the membrane association of GDU1, microsomes from 35S-GDU1-Myc leaves were isolated and treated with NaCl, alkaline sodium carbonate, or Triton X-100 detergent (i.e. reagents that extract peripheral, lumenal, or integral membrane proteins, respectively; Santoni et al., 1999; Rolland et al., 2006). GDU1 was retained in the microsomal pellet after incubation with salt or base, whereas it was solubilized by the detergent (Fig. 4B). In accordance with earlier characterizations of protein-membrane interactions (Santoni et al., 1999; Rolland et al., 2006), this result indicates that GDU1 is an integral membrane protein.
LOG2 was found to interact directly with cGDU1 in vitro (Fig. 1B) and to coimmunoprecipitate with GDU1 in planta (Fig. 1C), but unlike GDU1, LOG2 lacks a predicted transmembrane domain. To determine whether LOG2 is membrane associated, microsomes, PDVs, and PEVs were prepared from Arabidopsis stably expressing LOG2-HA under the control of the CaMV 35S promoter (Fig. 4C) or after transient expression in N. benthamiana leaves (Fig. 4D, left). Mirroring the localization profile of GDU1, LOG2 was found primarily in the total microsomal fraction and enriched in PEVs. In accordance with these data, expression in N. benthamiana epidermal cells of LOG2-GFP led to a continuous fluorescence at the cell periphery, suggesting plasma membrane localization (Fig. 5C, left). This pattern was identical to the pattern obtained with BRI1, a known plasma membrane protein (Friedrichsen et al., 2000; Supplemental Fig. S4). Microsomal LOG2-Myc expressed in N. benthamiana was, like GDU1, resistant to NaCl, but some protein was released with alkaline sodium carbonate and Triton X-100 detergent, indicating a weaker association with the membrane than GDU1 (Fig. 4E, left).
Myristoylation of LOG2 Is Important for Its Membrane Localization and the GDU1-LOG2 Interaction
We reasoned that the membrane association of LOG2 could result from covalent lipid modifications such as myristoylation, prenylation, or palmitoylation. Myristoylator (Bologna et al., 2004) and The MYR Predictor (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm) predicted N-myristoylation sites for LOG2 and LUL1-4. Moreover, in vitro myristoylation of LUL1 was recently demonstrated (Yamauchi et al., 2010). To experimentally verify that LOG2 could be myristoylated, LOG2 and the corresponding G2A mutant (bearing a substitution known to inhibit N-terminal myristoylation; Gordon et al., 1991) were expressed in rabbit reticulocyte lysate-coupled transcription-translation systems in the presence of [3H]myristic acid or [3H]Leu. These lysates contain the enzymes necessary for myristoylation (Heuckeroth et al., 1988). Both proteins were expressed at similar levels, and LOG2 incorporated [3H]myristic acid. As expected, LOG2G2A was not myristoylated (Fig. 4F). Similar to LUL1 and LOG2, LUL3 was also myristoylated in a Gly-2-dependent manner (Supplemental Fig. S5), suggesting that all LOG2-like proteins are myristoylated in planta.
To determine whether myristoylation affects LOG2 localization in planta, plasma membrane vesicles were prepared from N. benthamiana transiently expressing mLOG2-HA or mLOG2G2A-HA. While both proteins were found primarily in the microsomal fraction, mLOG2G2A was depleted from PEVs compared with wild-type mLOG2 (Fig. 4D), suggesting that LOG2 myristoylation is important for localization to the plasma membrane. Extraction of mLOG2G2A with sodium carbonate and Triton X-100 was markedly enhanced compared with mLOG2 (Fig. 4E), showing that the G2A mutation reduced the strength of binding to the membrane. While wild-type LOG2-GFP located exclusively at the plasma membrane in N. benthamiana, LOG2G2A-GFP also localized in the cytoplasm (Fig. 5C, middle). In accordance with biochemical data (Fig. 4D), the fluorescence pattern of LOG2G2A-GFP overlapped extensively with cytosolic mCherry (Fig. 5D), suggesting that suppression of myristoylation prevented a fraction of LOG2 from being anchored to the membrane.
To assess the effect of the G2A mutation on the GDU1-LOG2 interaction in planta, GDU1-HA was coexpressed with mLOG2-Myc or mLOG2G2A-Myc in N. benthamiana, and Myc-tagged proteins were immunoprecipitated as in Figure 1C. GDU1 coimmunoprecipitated with less efficiency with mLOG2G2A than mLOG2 (Fig. 4G), indicating that myristoylation enhances the LOG2 interaction with GDU1 in planta. The localization of the LOG2-GDU1 interaction was further studied by expressing in N. benthamiana mLOG2 and GDU1 fused to mCherry and GFP, respectively. LOG2-expressing Agrobacterium strains were infiltrated at low density, leading to heterogeneous expression in the epidermis, while GDU1-expressing Agrobacterium was infiltrated at a density enabling expression in all cells. GDU1 localized at the plasma membrane and in endosomes in cells expressing mLOG2 at low levels (Fig. 5E, middle cell) but mainly at the plasma membrane in cells expressing mLOG2 at higher levels (Fig. 5E, lateral cells). This change in localization pattern suggests that the interaction of LOG2 and GDU1 stabilized GDU1 localization at the plasma membrane.
T-DNA Disruption of LOG2 Suppresses the Gdu1D Phenotype Conferred by GDU1 Overexpression
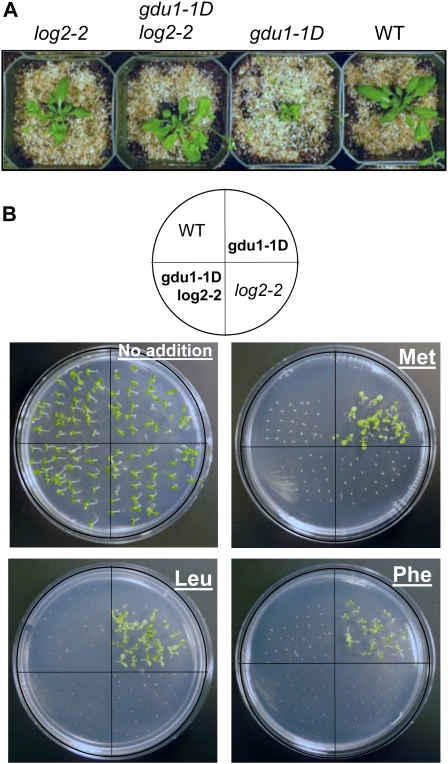
The observation that disruption of the LOG2-GDU1 interaction by the log1-1 mutation suppresses the Gdu1D phenotype (Pratelli and Pilot, 2006) suggested that LOG2 is involved in the pathway altered by GDU1 overexpression. To test this hypothesis, gdu1-1D was crossed with a plant carrying a T-DNA insertion (log2-2) in the first intron of LOG2 (Supplemental Fig. S1B). Reduction of LOG2 wild-type mRNA in the homozygous log2-2 mutant was confirmed by RT-PCR (Supplemental Fig. S6). In log2-2 gdu1-1D double homozygous plants, GDU1 mRNA levels remained unaffected by the log2-2 mutation (Supplemental Figs. S7, A and B, and S8C). Five-week-old log2-2 and wild-type plants had similar rosette sizes, while as previously observed, gdu1-1D individuals exhibited characteristically smaller rosettes (Fig. 6A). In contrast to gdu1-1D segregants, gdu1-1D log2-2 double mutant plants developed rosettes similar in size to wild type (Fig. 6A) and grew to normal height (data not shown), indicating suppression of the Gdu1D growth phenotype in the log2-2 background.
Figure 6.
Suppression of the Gdu1D phenotype by T-DNA insertion in the LOG2 gene. A, gdu1-1D was crossed to plants harboring a T-DNA in the first intron of LOG2 (log2-2). Five-week old F3 plants were recovered from F2 parents with the indicated genotypes. B, gdu1-1D, log2-2, and log2-2 gdu1-1D seeds were sown on medium containing 10 mm of the indicated amino acid or no added amino acid (top left plate). Each plate is oriented with quadrants as shown in the model above. Clockwise from top left: the wild type (WT), gdu1-1D, log2-2, and log2-2 gdu1-1D double homozygote. Experiments were repeated three or more times with 25 seeds from each line. Representative images are shown.
gdu1-1D plants have been shown to be more tolerant to toxic concentrations of amino acids compared with the wild type (Pratelli and Pilot, 2007). To determine whether this other aspect of the Gdu1D phenotype is suppressed in the gdu-1D log2-2 double mutant, the susceptibilities of wild type, log2-2, gdu1-1D, and gdu1-1D log2-2 seedlings to 10 mm Phe, Met, and Leu were compared. Wild-type, log2-2 and gdu1-1D log2-2 plants did not grow in the presence of these amino acids, while most gdu1-1D plants developed green cotyledons (Fig. 6B). The phenotype of the soil-grown plants and amino acid tolerance assays indicate that the loss of LOG2 expression suppresses all characterized aspects of the Gdu1D phenotype.
Expression of Artificial MicroRNA Targeting LOG2 Suppressed the Gdu1D Phenotype
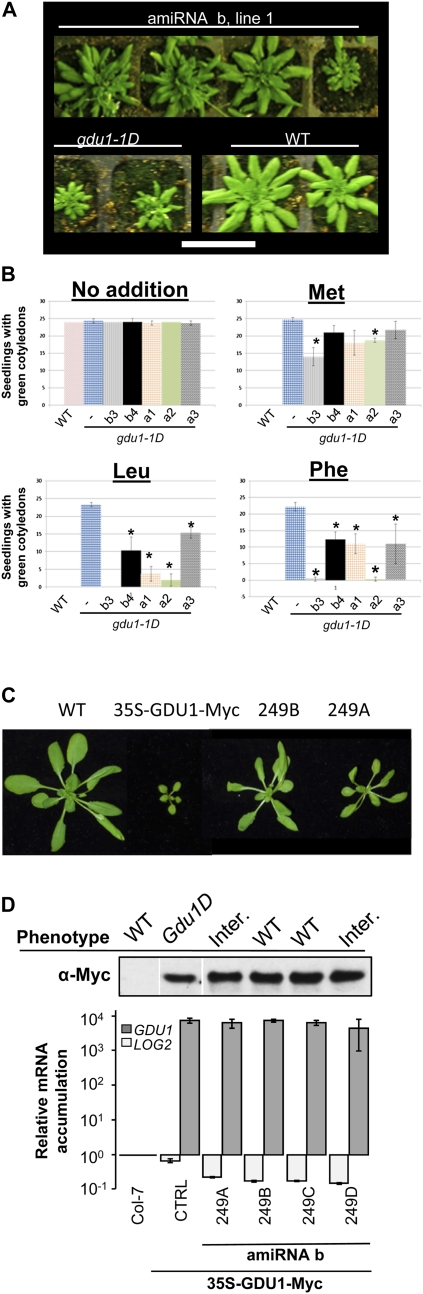
Of the available T-DNA insertion lines, only the log2-2 T-DNA was found to repress LOG2 transcript accumulation. To confirm the results obtained with the log2-2 line, four artificial microRNAs (amiRNAa, -b, -c, and -d) directed against the LOG2 transcript were created (Supplemental Fig. S1B). The amiRNAs were expressed in the gdu1-1D line under the control of the cassava vein mosaic virus (CsVMV) promoter (Verdaguer et al., 1998), a viral promoter of activity comparable to that of the CaMV 35S. The CsVMV promoter was used to avoid silencing of GDU1, driven in gdu1-1D by the 35S enhancer. Similar results were obtained with amiRNA expression as seen with the log2-2 mutation in the gdu1-1D background. Transformants created using LOG2-amiRNAa and -b had a phenotype similar to wild-type plants in the T2 generation at the expected 75% ratio (Fig. 7A). This phenotypic change occurred while GDU1 transcripts remained at levels similar to the progenitor line (Supplemental Fig. S8A). Amino acid tolerance to Leu, Phe, and Met was tested for five lines (three from LOG2-amiRNAa and two from LOG2-amiRNAb). The tolerance of the transformants was intermediate between gdu1-1D and the wild type, with lines a2 and b3 showing almost wild-type susceptibility on Leu and Phe (Fig. 7B; Supplemental Fig. S8B).
Figure 7.
Suppression of the Gdu1D phenotype by LOG2-directed amiRNAs in two different Gdu1D overexpression backgrounds. A, Phenotypic segregation on soil of T2 plants expressing LOG2-amiRNA in the gdu1-1D background. Bar = 5 cm. B, Growth of seedlings expressing one of two LOG2-amiRNAs in the gdu1-1D background was assessed for the presence of green, expanded cotyledons after 14 d of growth on 10 mm Met, Leu, and Phe. Error bars represent sd. Asterisks indicate significant differences from gdu1-1D alone (−) on the same medium (t test with Bonferroni correction for multiple comparisons; P < 0.01). Experiments were repeated three or more times with 25 seeds from each line. C, Phenotype of plants overexpressing LOG2-miRNAb in the 35S-GDU1-Myc background. D, For each line overexpressing LOG2-miRNAb in the 35S-GDU1-Myc background, the phenotype of about 10 progeny was recorded (WT, wild type; Inter., intermediate between Gdu1D and the wild type), GDU1 protein levels were estimated by western blotting using an anti-Myc antibody (top), and GDU1 and LOG2 mRNA contents were measured by quantitative RT-PCR. Accumulation is expressed relative to the levels in the wild type (Col-7), and error bars represent the results of two independent experiments. CTRL, Control (35S-GDU1-Myc background only, no amiRNA present). [See online article for color version of this figure.]
While the expression of LOG2-directed amiRNAs did suppress the Gdu1D phenotype, it was not resolved if the suppression affected GDU1 protein content. The 35S-GDU1-Myc line (see above) was used to probe GDU1 protein accumulation in a parallel experiment. amiRNAb was expressed in the 35S-GDU1-Myc line. Similar to the above results, expression of LOG2-amiRNAb in 35S-GDU1-Myc suppressed the Gdu1D phenotype in most of the transformants. Four independent transformation lines (lines 249A to -D) were chosen and studied for GDU1-Myc protein accumulation, LOG2 and GDU1 mRNA contents, and amino acid levels. Lines 249A and -D exhibited a mild Gdu1D phenotype, while lines 249B and -C showed wild-type morphology (Fig. 7C). Expression of LOG2-amiRNAb led to an 80% decrease in LOG2 mRNA in the four lines (Fig. 7D). GDU1 mRNA content was not significantly changed compared with the untransformed control, with about 5,000-fold overaccumulation compared with the wild type (Fig. 7D). Amino acid contents were reduced in the suppressed lines but were not equivalent to those in the wild type (Supplemental Table S1). Significantly, GDU1-Myc protein content was only slightly increased by the expression of LOG2-amiRNAb (Fig. 7D).
An R12K Substitution in LOG2 Suppresses the Gdu1D Phenotype but Not the GDU1-LOG2 Interaction
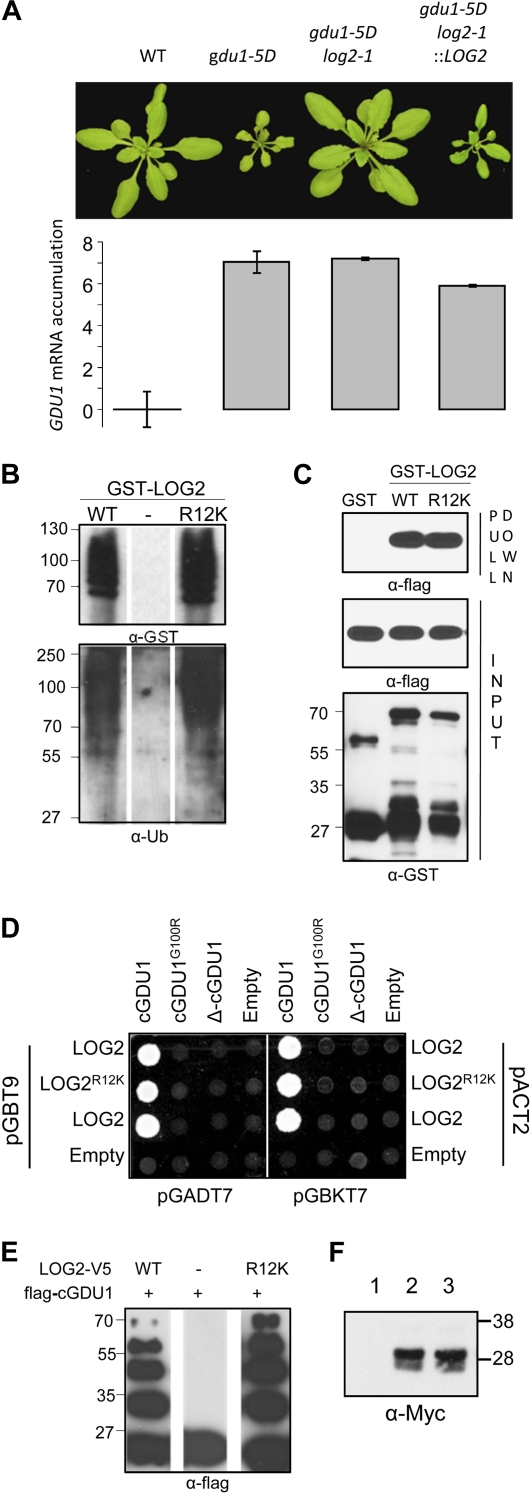
A suppressor mutation, log2-1, was isolated in the same screening that led to the isolation of the log1-1 mutation (Pratelli and Pilot, 2006; Fig. 8A; Supplemental Text S1). Analyses of phenotypic segregation after crosses of log2-1 with the wild type and the gdu1-5D parental line validated the hypothesis that log2-1 is a single recessive mutation, independent of the log1-1 mutation (Supplemental Text S1). Positional cloning of log2-1 showed that the mutation was very close to the LOG2 gene (Supplemental Fig. S9). Sequencing of the LOG2 gene in log2-1 revealed the presence of a G-to-A mutation 35 bp after the ATG, leading to an Arg-to-Lys mutation (R12K; Supplemental Fig. S1B). Transformation of the log2-1 gdu1-5D double mutant by a genomic fragment containing the wild-type LOG2 gene complemented the log2-1 mutation. This complementation proved that suppression of the Gdu1D phenotype in the log2-1 gdu1-5D double mutant resulted from the R12K mutation in LOG2 (Fig. 8A).
Figure 8.
Analysis of the effect of the log2-1 mutation on LOG2 protein properties. A, Complementation of the log2-1 mutation. A wild-type (WT) genomic fragment (8,511-bp XhoI-PstI from bacterial artificial chromosome F11F8) was cloned into a hygromycin resistance-conferring binary vector and inserted into the genome of the log2-1 gdu1-5D double mutant. GDU1 mRNA content was estimated by quantitative RT-PCR and is given as the double difference between the qPCR cycle threshold (Ct) of GDU1 and Actin2 mRNAs obtained in the wild type and the mutants (ΔΔCt). Errors bars are from two biological replicates. B, GST-LOG2 and GST-LOG2R12K ubiquitination assays without substrate. C, GST pull-down assay using flag-cGDU1 and GST-LOG2 or GST-LOG2R12K. D, Yeast two-hybrid interaction assay of LOG2R12K with cGDU1, cGDU1G100R, or ΔcGDU1. E, In vitro ubiquitination assay with LOG2-V5 or LOG2R12K-V5 and flag-cGDU1. F, GDU1-Myc accumulation in 35S-GDU1-Myc and the 35S-GDU1-Myc log2-1 double mutant. Lane 1, Wild type; lane 2, 35S-GDU1-Myc; lane 3, 35S-GDU1-Myc log2-1. Numbers on the side of western blots indicate molecular mass in kD. [See online article for color version of this figure.]
The fact that log2-1 is recessive suggested a loss-of-function mutation, but, despite extensive trials, no difference in functional properties has been found to date between LOG2R12K and LOG2: the R12K mutation did not impair the ubiquitin ligase activity of the protein (Fig. 8B) or the ability to ubiquitinate cGDU1 in vitro (Fig. 8E). In addition, yeast two-hybrid, GST pull-down, and in planta coimmunoprecipitation assays showed that LOG2R12K interacted with GDU1 very similarly to LOG2 (Figs. 1C and 8, C and D). LOG2R12K-GFP also localized to the plasma membrane like wild-type LOG2-GFP in transiently transformed N. benthamiana leaves (Fig. 5C, right). The R12K mutation had no effect on GDU1-Myc accumulation when log2-1 was introduced into the 35S-GDU1-Myc line (Fig. 8F). Leading to significant suppression of the Gdu1D phenotype with no detectable effect on the LOG2-GDU1 interaction, the nature of the defect in LOG2R12K is currently unknown.
DISCUSSION
LOG2 Is Necessary for the Gdu1D Phenotype
The GDU1 protein has been suggested to be involved directly or indirectly in the control of amino acid export, potentially interacting with an amino acid exporter (Pilot et al., 2004; Pratelli et al., 2010). Parallel attempts at characterizing GDU1 function using EMS suppressor and yeast two-hybrid screens led to the isolation of LOG2, an E3 ubiquitin ligase. Subsequent biochemical analyses showed that LOG2 and GDU1 interacted in in vitro pull-down and in planta coimmunoprecipitation assays (Figs. 1, B and C, and 2B). We also establish here the functional significance of this interaction in multiple assays. Previous characterization of the log1-1 suppressor allele (GDU1G100R) validated the relevance of the conserved GDU VIMAG motif (Pratelli and Pilot, 2006), but the molecular mechanism of the suppressor effect was not explained. When tested by yeast two-hybrid, GST pull-down, in planta coimmunoprecipitation, and in vitro ubiquitination assays, the log1-1 mutation diminished the interaction between LOG2 and GDU1 (Figs. 1, A–C, and 2D, respectively). This indicated that the phenotypic suppression conferred by the log1-1 allele could result from impaired interaction with LOG2. Genetic interaction assays with plants affected in LOG2 expression further support this conclusion. Reduction of the Gdu1D phenotype was observed in three independent genetic contexts: in gdu1-1D plants homozygous for the log2-2 T-DNA (Fig. 6), upon expression of LOG2-directed amiRNAs in either gdu1-1D or 35S-GDU1-Myc backgrounds (Fig. 7), and in gdu1-5D plants homozygous for the log2-1 loss-of-function allele (LOG2R12K; Fig. 8). The nature of phenotypic suppression was similar in all three cases: plant size increased (Figs. 6A, 7, A and C, and 8A), amino acid sensitivity decreased (Figs. 6B and 7B; Supplemental Fig. S8B), and amino acid accumulation decreased (Supplemental Table S1). Notably, the suppression effects observed in multiple LOG2 mutation/GDU1 overexpressor combinations reinforce the hypothesis that LOG2 is necessary for the development of the Gdu1D phenotype upon GDU1 overexpression. These findings presage a role for LOG2 in amino acid homeostasis.
Localization of LOG2 and GDU1 at the Plasma Membrane
LOG2 and GDU1 were found almost exclusively in the microsomal membrane fraction and were particularly enriched at the plasma membrane. GDU1 was previously postulated to be an integral membrane protein (Pratelli and Pilot, 2006), and the sensitivity of GDU1 to a high-ionic-strength buffer, high pH, and detergent, typical of integral membrane proteins, confirmed this hypothesis (Fig. 5B). Although LOG2 is not predicted to contain a membrane-spanning domain, LOG2 could be myristoylated in vitro (Fig. 4F), a lipid modification that can anchor otherwise cytosolic proteins to membranes (Resh, 1999). Inhibition of LOG2 myristoylation did not completely abolish membrane association per se, as most LOG2G2A still partitioned into microsomes (Fig. 4, D and E). However, LOG2G2A was significantly depleted from plasma membrane-enriched vesicles (Fig. 4D), its association with microsomes was markedly more sensitive to extraction (Fig. 4E), and the fluorescence signal of C-terminally GFP-tagged LOG2 was no longer confined to the plasma membrane (Fig. 5C, left and center panels). Possible reasons for the partial membrane retention of LOG2G2A include its ability to bind to integral membrane proteins and/or the presence of an N-terminal poly-Arg tract (Supplemental Fig. S1C). Polybasic regions can facilitate the binding of soluble proteins to the negatively charged intracellular leaflets of lipid bilayers via electrostatic interactions (Murray et al., 1998). However, other lipid modifications or undetected reentrant loops could account for persistent membrane anchoring. The observed weakened in planta interaction between GDU1 and LOG2G2A (Fig. 4G) hints that myristoylation may potentiate the interaction of LOG2 with GDU1, possibly by shifting LOG2 into the same membrane microdomains as GDU1.
Role of LOG2 with Respect to GDU1
Multiple tagged forms of LOG2 ubiquitinated cGDU1 (Fig. 2, B–D), an observation consistent with the possibility that GDU1 is a substrate of LOG2 in vivo. Polyubiquitination will often destine a substrate protein for degradation, either via the 26S proteasome or, in the case of many mammalian and yeast plasma membrane proteins, the vacuole or lysosome upon endocytosis (Jehn et al., 2002; Korolchuk et al., 2010). Such ubiquitin ligases are thus negative regulators of the activity of the substrate protein. Our data do not support the hypothesis that LOG2 negatively regulates GDU1. Mutations that inhibit GDU1 (log1-1) association with LOG2, or that eliminate LOG2 (log2-2), would be expected to enhance the Gdu1D phenotype if LOG2 were a negative regulator of GDU1. However, the opposite was observed: in a GDU1 overexpression background, log1-1 and log2-1 homozygotes are phenotypically wild type (Pratelli and Pilot, 2006; Fig. 8A). GDU1 protein overaccumulation does not cause the Gdu1D phenotype in the absence of LOG2 or when GDU1 is unable to interact with LOG2. In conclusion, LOG2 appears to be a required component of the pathway affected by GDU1 overexpression.
Our data are consistent with a model in which either LOG2 activates GDU1 via ubiquitination or GDU1 acts as an adaptor protein for LOG2 to recognize an unidentified substrate (for review, see Woelk et al., 2007; Léon and Haguenauer-Tsapis, 2009). Examples for both hypotheses in other systems have been reported. In mammals, activation of a kinase in the inflammatory response requires its monoubiquitination (Hinz et al., 2010). Plasma membrane-associated E3 ligase adaptors have also been described. For instance, the adaptor Grb2 binds to the RING E3 c-Cbl and facilitates its association with activated plasma membrane epidermal growth factor receptor in mammals, resulting in ubiquitination of the latter (Huang and Sorkin, 2005). Similar E3 adaptors have also been described in yeast (Léon and Haguenauer-Tsapis, 2009).
LOG2-Associated Proteins Probably Contribute to Amino Acid Export
By analogy to mammalian heteromeric amino acid transporters (Palacín et al., 2005), GDU1 has been hypothesized to be a subunit of an amino acid exporter (Pilot et al., 2004; Pratelli et al., 2010). One viable hypothesis is that GDU1 overexpression alters the localization of an unknown amino exporter through a LOG2-dependent pathway, either by promoting the degradation of a yet-to-be-identified negative regulator of the exporter or by directly influencing the movement of the transporter to the cell surface. The log2-1 mutation suppresses the Gdu1D phenotype without discernibly affecting its subcellular localization, ubiquitin ligase activity, or interaction with GDU1 (Figs. 5C, right panel, and 8, B–D). It is possible that this mutation alters the recognition/interaction with as-yet-undiscovered LOG2 substrate(s). While an Arg-to-Lys mutation is often regarded as conservative, Lys residues are common sites of modification such as acetylation, methylation, and ubiquitination. Modification of Lys-12 of LOG2R12K could disrupt interactions between LOG2R12K and its substrate(s) or other interacting proteins, leading to suppression of the Gdu1D phenotype. This hypothesis suggests that all the proteins involved in the GDU1 pathway are not discovered. The identification of these proteins and their role in membrane protein trafficking or amino acid export will be the subject of further investigations.
MATERIALS AND METHODS
Plant Lines, Transformations, and Crosses
Transgenic Arabidopsis (Arabidopsis thaliana; ecotypes Columbia-0 [Col-0] and Col-7) plants were procured via the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101 (pMP90). Seed harboring the log2-2 T-DNA (Syngenta Arabidopsis Insertion Library accession 729_A08; Sessions et al., 2002) was obtained from the Arabidopsis Biological Resource Center. gdu1-1D log2-2 double homozygote plants were identified in the F2 generation of a cross between gdu1-1D pollen and log2-2 ovules via PCR of genomic DNA (for primer sequences, see Supplemental Table S2). Phenotypic analyses of gdu1-1D log2-2 and gdu1-1D amiRNA-LOG2 double mutant plants were performed on homozygous F3 and T4 individuals, respectively (except for segregation of the GDU1 overexpression phenotype of gdu1-1D amiRNA-LOG2 double mutant plants on soil, which was carried out on T2 individuals). 35S-GDU1-Myc log2-1 double homozygotes were identified in the F2 generation of a cross between the pollen of the log2-1 single mutant, obtained from a cross of the log2-1 gdu1-5D double mutant with Col-7, and 35S-GDU1-Myc ovules via analysis of antibiotic resistance and phenotype. For transient expression, leaves of 5-week-old Nicotiana benthamiana plants were infiltrated with a suspension of A. tumefaciens carrying constructs of interest according to Batoko et al. (2000). To assess amino acid sensitivity, surface-sterilized seeds were sown on agar plates comprising 1× Murashige and Skoog salts (Murashige and Skoog, 1962), 1% (w/v) Suc, 2.5 mm MES, 0.5 μg mL−1 pyroxidine-Cl, 0.1 μg mL−1 myoinositol, 0.8% (w/v) BactoAgar (BD Biosciences), pH 5.7, and 10 mm amino acids. Plants were grown for 14 d in an incubator (24 h of light, 50 μmol m−2 s−1, 22°C).
Cloning and Constructs
Primer sequences used for cloning and site-directed mutagenesis are listed in Supplemental Table S2. For the yeast two-hybrid screen, the region encoding the C-terminal domain of GDU1 (residues 61–158) was cloned into pGBKT7 or pGBT9 (Clontech) using specific primers and the EcoRI and PstI sites. For the yeast two-hybrid interaction matrix, cDNAs encoding the C-terminal soluble part of the GDUs and the full sequences of LOG2 and LULs were amplified by PCR with primers containing the attB Gateway sequences, cloned into pDONRZeo (Invitrogen), sequenced, and transferred into pACT2 and pGBT9 vectors, which were previously made Gateway compatible by insertion of the Gateway cassette (R. Pratelli and G. Pilot, unpublished data). To obtain GST- and V5-tagged LOG2 and LUL1, the coding sequences were amplified by PCR, cloned into pDONR201 (Invitrogen) using the Gateway technology, and recombined into pDEST15 and pET-DEST42 (Invitrogen), respectively. flag-cGDU1 was produced similarly with the destination vector pEAK2 (Kraft, 2007). Site-directed mutagenesis was performed with the QuikChange kit (Stratagene) to create RING-dead LOG2 (LOG2C354/C557AA; mLOG2) RING-weak LOG2 (LOG2I321A), and myristoylation-inhibited LOG2 (LOG2G2A). For the in vitro myristoylation assay, LOG2 was recombined into pEXP2 and pET-DEST42 (Invitrogen). GFP and mCherry fusion constructs were created by Gateway cloning into pPWGTkan and pPWMTkan, derivatives of pJHA212K (Yoo et al., 2005), where the Gateway cassette was inserted between the CaMV 35S promoter and the eGFP or mCherry coding sequence (R. Pratelli and G. Pilot, unpublished data). The HA fusion construct used for Arabidopsis transformation was obtained by Gateway cloning into pGWB14 (Nakagawa et al., 2007). HA and Myc fusions used for transient assays in N. benthamiana were obtained using vectors similar to pPWGTkan, where a double HA tag or a triple Myc tag replaced the eGFP. The GDU1 VIMAG domain was deleted by cloning next to each other in pBluescript two PCR fragments corresponding to the regions upstream and downstream of the domain and sharing an EcoRI site at the exact place of the VIMAG domain. The resulting construct was used as a template for PCR toward Gateway cloning into pDONR221 (Invitrogen). amiRNAs were designed following the guidelines found in WMD3 (http://wmd3.weigelworld.org/; Schwab et al., 2010). The primers corresponding to pRS300 (Schwab et al., 2010) used for amplification of the miRNAs contained the Gateway attB sites. The final PCR fragment was cloned into pDONRZeo (Invitrogen), sequenced, and transferred into the pSWsNkan binary vector, another derivative of pJHA212K (Yoo et al., 2005, R. Pratelli and G. Pilot, unpublished data), between the CsVMV promoter (Verdaguer et al., 1998) and the terminator of the small subunit of the Rubisco from pea (Pisum sativum; accession no. X00806). For log2-1 complementation, a XhoI-PstI 8.5-kb genomic DNA fragment from bacterial artificial chromosome F11F8 containing the wild-type LOG2 gene was cloned into the pTkan binary vector, a derivative of pJHA212K.
Yeast Two-Hybrid Screening and Interaction Assays
For screening, the bait plasmids were cointroduced along with an Arabidopsis cell cDNA library (Németh et al., 1998) into yeast strain AH109 (Clontech) using the TRAFO protocol (Gietz and Schiestl, 2007). About 2 million transformants were selected on SC medium lacking Leu, Trp, His, and adenine (2% Glc, 6.7 g L−1 yeast nitrogen base without amino acid [BD Biosciences], pH 6.3, and dropout amino acid mix). Plasmids were extracted from the colonies displaying auxotrophy for the four amino acids and introduced back into yeast together with the cGDU1 construct. The inserts of the eight plasmids restoring yeast growth on selective medium were then sequenced. For interaction matrices, yeast strains AH109 (MATa) and Y187 (MATα) were transformed as described above with the prey and bait vectors, respectively, and selected on medium lacking Trp or Leu. Several colonies were scrapped and resuspended together in 250 μL of water. Prey-bait pairs were mixed together (10 μL of each suspension and 50 μL of water), and 5 μL was spotted on YPDA (1% [w/v] yeast extract, 2% [w/v] bacto peptone, 2% [w/v] Glc, 80 mg L−1 adenine, and 1.5% agar) for mating. After overnight growth at 30°C, the cell spots were scrapped and resuspended in 100 μL of water, and 5 μL was spotted on SC medium lacking Trp and Leu. After growth at 30°C for 2 d, the spots were scrapped and resuspended in 100 μL of water, and 5 μL was spotted on SC medium lacking Trp, Leu, adenine, and His. Growth was assessed after 2, 3, and 6 d to identify positive interactions. β-Galactosidase activity of yeast was measured using a protocol obtained from Clontech (manual no. PT3024-1). Briefly, cells grown to an optical density at 600 nm of 0.5 were harvested by centrifugation, resuspended in 100 mm Na2HPO4, pH 7, 10 mm KCl, 1 mm MgSO4, and 5 mm β-mercaptoethanol (Z buffer), and subjected to three freeze-thaw cycles in liquid N2 and at 37°C. The broken cell suspension (100 μL) was added to 900 μL of 800 μg mL−1 ortho-nitrophenyl-β-galactoside in Z buffer and incubated for several hours at 37°C. The reaction was stopped by the addition of 400 μL of 1 m Na2CO3, and optical density at 420 nm was measured.
Recombinant Protein Expression and Purification
GST-, V5-His6-, and His6-flag fusion proteins were expressed in BL21-pLysS Escherichia coli essentially as described by Kraft et al. (2005) with a few modifications. The lysis buffer comprised 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 1 mm dithiothreitol (DTT), 0.1% (v/v) Nonidet P-40, and 0.5× Complete Protease Inhibitors (Roche Diagnostics) for GST-tagged proteins and was supplemented with 20 mm imidazole for His-tagged proteins. Cells were lysed by sonication. For GST proteins, glutathione bead slurries were brought to 20% (v/v) glycerol after the final wash, flash frozen in liquid nitrogen, and stored at −80°C. His-tagged proteins were eluted from nickel Sepharose beads, brought to 20% glycerol, and flash frozen. His6-flag-cGDU1 was purified likewise, except that buffers contained 50 mm K2HPO4-KH2PO4, pH 5.75, in place of Tris-HCl. To further purify cGDU1, eluted protein was centrifuged through a 30-kD NMWL Amicon concentrator (Millipore), and the eluate was concentrated and buffer exchanged in a 5-kD NMWL Amicon concentrator with 50 mm K2HPO4-KH2PO4, pH 5.75, 150 mm KCl, 1 mm DTT, 0.1% Nonidet P-40, and 0.5× Complete Protease Inhibitors. The final retentate was brought to 20% glycerol and flash frozen.
GST Pull-Down Assays
In vitro pull-downs were performed as described by Gilkerson et al. (2009) with the following differences: GST proteins on beads were washed once in 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1 mm DTT, 1% (v/v) Nonidet P-40, and 0.5× Complete Protease Inhibitors (wash buffer) prior to being mixed with a soluble prey protein in 400 μL of wash buffer and incubated at 4°C for 2 h. Proteins were eluted by suspending beads in 60 μL of elution buffer (wash buffer + 50 mm glutathione) and shaking at 4°C for 15 min.
Ubiquitination Assays
In vitro ubiquitination assays were conducted essentially according to Hsia and Callis (2010) with slight modifications: 4 μg of bovine ubiquitin (Sigma-Aldrich), approximately 2 μg of GST- or V5-His6-LOG2, and approximately 1.5 μg of His6-flag-cGDU1 were used. Reactions were quenched with 10 μL of 5× Laemmli sample buffer (200 mm Tris, pH 6.8, 32% [v/v] glycerol, 6.4% [w/v] SDS, 0.32% [w/v] bromophenol blue, and 200 mm DTT), boiled for 5 min, and separated via SDS-PAGE. Proteins were visualized by western blot using anti-flag-linked horseradish peroxidase (Sigma-Aldrich) and anti-GST (Santa Cruz Biotechnology) according to the manufacturers’ recommendations or anti-ubiquitin antibodies. Anti-ubiquitin antibodies were raised against bovine full-length ubiquitin prepared according to Haas and Bright (1985) by Aves Labs, affinity purified, and used at 1:5,000 dilution.
In Planta Coimmunoprecipitation Assay
Proteins were transiently expressed in N. benthamiana leaves. Three days after infiltration, 500 mg of leaves was ground with 1.5 mL of extraction buffer on ice (50 mm Tris-HCl, pH 7.3, 150 mm NaCl, 10 mm MgCl2, 0.5% Nonidet P-40, 10 mm DTT, and 1× Complete Protease Inhibitors). Homogenate was centrifuged at 10,000g and 4°C for 15 min. The supernatant was filtered through several layers of Miracloth (EMD Biochemicals) and quantitated using the Bradford assay (Fermentas). Proteins were coimmunoprecipitated use the ProFound c-Myc Tag IP/Co-IP Kit (Pierce): 3 mg of proteins was placed on a rotary wheel overnight at 4°C in a coimmunoprecipitation spin column with cMyc-agarose; after washing, the proteins were eluted three times with 10 μL of the supplied elution buffer and neutralized by 1.5 μL of 1 m Tris, pH 9.5. One microliter of coimmunoprecipitation eluate and 10 μg of total proteins were analyzed by SDS-PAGE (4%–12% polyacrylamide MES gel; Invitrogen) and western blotting. Proteins were transferred on a nitrocellulose membrane (GE Healthcare) and detected using anti-cMyc (clone A-14; Santa Cruz; 1:10,000) or anti-HA (clone 3F10; Roche Diagnostics; 1:5,000) primary antibodies, anti-rabbit or anti-rat (Thermo Scientific) secondary antibodies, and the ECL-Plus western-blotting detection system (GE Healthcare).
Subcellular Fractionation
Endomembranes were prepared from Arabidopsis and N. benthamiana according to Liu et al. (2009) with the following modifications: 5 to 30 g of 8-d-old Arabidopsis seedlings, 5-week-old Arabidopsis rosette leaves, or N. benthamiana leaves 3 d after infiltration were homogenized with a Waring blender in 60 mL of homogenization buffer (50 mm MOPS-KOH, pH 6.8, 5 mm EDTA, 0.33 m Suc, 2 mm ascorbic acid, 1.5 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, and 0.2% [w/v] polyvinylpolypyrrolidone). The sensitivity of GDU1 or LOG2 membrane association to detergent, salt, and pH was examined as described by Phan et al. (2008) with a 1-h incubation on ice. PEVs and PDVs were purified from total microsomes according to Larsson et al. (1994): upper phases 4 and 5 were combined to afford PEVs, while lower phase 1 was extracted five times with fresh upper phase to afford PDVs. Plasma membrane enrichment and depletion were qualitatively assessed with the α-PMA2 antibody (M. Boutry, Université Catholique de Louvain). Protein concentration was assessed by the Bradford or bicinchoninic acid protein assays.
Localization of Expression and Imaging
About 3 kb of the region upstream from LOG2 ATG was amplified by PCR and cloned using BamHI and PstI in pUTkan (Pratelli et al., 2010). GUS histochemical staining was performed as described (Pratelli et al., 2010). N. benthamiana epidermis cells were visualized with the Zeiss LSM510 META confocal system on an Axio Observer.Z1 microscope using a C-Apochromat 40× water-immersion, numerical aperture 1.2 objective (Carl Zeiss), a 488-nm argon multiline gas laser, and a 543-nm helium-neon gas laser, with band-pass 505 to 550 and long-pass 560 emission filters, respectively. Serial images were captured and processed by the Zen 2009 software (Carl Zeiss) using maximal projection.
Nucleic Acid Isolation and PCR
Genomic DNA was extracted from Arabidopsis plants using the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). Total RNA was extracted either with the RNeasy kit (Qiagen) or by 1 mL of TRI Reagent (Sigma-Aldrich). cDNAs were synthesized using the SuperScript III system (Invitrogen) according to the manufacturer’s instructions. For quantitative PCR, the efficiency-calibrated method was implemented (Pfaffl, 2001). Five microliters of primer mix (1 μm each) and 5 μL of the RT product (made from 2 μg of total RNA) diluted 50 times were mixed with 10 μL of 2× SYBR Green PCR Master Mix (Applied Biosystems) and subjected to the following cycles: 50°C for 2 min, 95°C for 10 min, followed by 40 times at 95°C for 15 s, 55°C for 15 s, and 72°C for 1 min (in a 7300 Real Time PCR System; Applied Biosystems).
Amino Acid Quantitation
Tissues were frozen in liquid nitrogen, freeze dried, weighed, and ground with three 3-mm glass beads in an Ultramat Amalgamator (SDI, Inc.). Amino acids were extracted from the dry powder by 200 μL of 10 mm HCl and 200 μL of chloroform. After vortexing for 2 min, the solution was centrifuged for 5 min at 16,000g, and 150 μL of the supernatant was dried under vacuum. The dried extract was solubilized in 500 μL of 50% acetonitrile in water and 0.05% heptafluorobutyric acid, and the metabolites were separated by ion-pairing liquid chromatography and analyzed by mass spectrometry (Supplemental Text S2).
The locus numbers of the genes studied in this article are as follows: GDU1, At4g31730;GDU2, At4g25760; GDU3, At5g57685; GDU4, At2g24762; GDU5, At5g24920; GDU6, At3g30725; GDU7, At5g38770; LOG2, At3g09770; LUL1, At5g03200; LUL2, At3g53410; LUL3, At5g19080; LUL4, At3g06140.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure and alignment of LOG2 and LUL1 proteins.
Supplemental Figure S2. Accumulation of LOG2 mRNA in the organs of the plant.
Supplemental Figure S3. Phenotype of the 35S-GDU1-Myc line.
Supplemental Figure S4. Co-localization of mLOG2 and BRI1 in N. benthamiana epidermis cells.
Supplemental Figure S5. LUL3 can be myristoylated in vitro.
Supplemental Figure S6. Suppression of wild type LOG2 transcript accumulation in log2-2.
Supplemental Figure S7. GDU1 transcript accumulation in a gdu1-1D log2-2 double mutant.
Supplemental Figure S8. GDU1 transcript accumulation and amino acid sensitivity phenotypes of gdu1-1D plants overexpressing the LOG2-amiRNA.
Supplemental Figure S9. Positional cloning of log2-1.
Supplemental Table S1. Free amino acid content of plants overexpressing LOG2-amiRNA.
Supplemental Table S2. Sequence of the oligonucleotides used for this study.
Supplemental Text S1. EMS mutagenesis and positional cloning.
Supplemental Text S2. LC-MS analysis details.
Supplementary Material
Acknowledgments
We thank members, past and present, of the Callis laboratory for helpful discussion and V. Kam for technical assistance. We thank the University of California-Davis Controlled Environment Facility for assistance with the propagation of transgenic plants at the University of California-Davis. We thank K. Harich for running the liquid chromatography-tandem mass spectrometry for amino acid analyses, K. DeCourcy for help with confocal microscopy, and the College of Agriculture and Life Sciences for providing facilities for growing plants at Virginia Tech. We thank J.M. Elmore and M. Boutry for anti-PMA antibodies and J.M. Elmore for assistance with the plasma membrane enrichment procedure.
References
- Batoko H, Zheng HQ, Hawes C, Moore I. (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna G, Yvon C, Duvaud S, Veuthey AL. (2004) N-terminal myristoylation predictions by ensembles of neural networks. Proteomics 4: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, III, Fukuda M, Ohta T, Klevit R. (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright DA, Brady SM, Orlando DA, Sturmfels B, Benfey PN. (2009) Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25: 2581–2587 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooray SN, Guasti L, Clark AJL. (2011) The E3 ubiquitin ligase Mahogunin ubiquitinates the melanocortin 2 receptor. Endocrinology 152: 4224–4231 [DOI] [PubMed] [Google Scholar]
- Dambly S, Boutry M. (2001) The two major plant plasma membrane H+-ATPases display different regulatory properties. J Biol Chem 276: 7017–7022 [DOI] [PubMed] [Google Scholar]
- De Jong A, Koerselman-Kooij JW, Schuurmans J, Borstlap AC. (1997) The mechanism of amino acid efflux from seed coats of developing pea seeds as revealed by uptake experiments. Plant Physiol 114: 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dündar E, Bush DR. (2009) BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta 229: 1047–1056 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Coaker G. (2011) The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol Plant 4: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Gilkerson J, Hu J, Brown J, Jones A, Sun TP, Callis J. (2009) Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics 181: 945–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JI, Duronio RJ, Rudnick DA, Adams SP, Gokel GW. (1991) Protein N-myristoylation. J Biol Chem 266: 8647–8650 [PubMed] [Google Scholar]
- Haas AL, Bright PM. (1985) The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem 260: 12464–12473 [PubMed] [Google Scholar]
- He L, Lu XY, Jolly AF, Eldridge AG, Watson SJ, Jackson PK, Barsh GS, Gunn TM. (2003) Spongiform degeneration in mahoganoid mutant mice. Science 299: 710–712 [DOI] [PubMed] [Google Scholar]
- Heuckeroth RO, Towler DA, Adams SP, Glaser L, Gordon JI. (1988) 11-(Ethylthio)undecanoic acid: a myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J Biol Chem 263: 2127–2133 [PubMed] [Google Scholar]
- Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C. (2010) A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol Cell 40: 63–74 [DOI] [PubMed] [Google Scholar]
- Hsia MM, Callis J. (2010) BRIZ1 and BRIZ2 proteins form a heteromeric E3 ligase complex required for seed germination and post-germination growth in Arabidopsis thaliana. J Biol Chem 285: 37070–37081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Sorkin A. (2005) Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol Biol Cell 16: 1268–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. (2002) c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem 277: 8033–8040 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. (2007) Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell 18: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584: 1393–1398 [DOI] [PubMed] [Google Scholar]
- Kraft E. (2007) An investigation of the ubiquitin conjugating enzymes and RING E3 ligases in Arabidopsis thaliana. PhD thesis. University of California, Davis [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S. (1994) Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol 228: 451–469 [Google Scholar]
- Léon S, Haguenauer-Tsapis R. (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp Cell Res 315: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Lesuffleur F, Cliquet J-B. (2010) Characterisation of root amino acid exudation in white clover (Trifolium repens L.). Plant Soil 333: 191–201 [Google Scholar]
- Li ZC, Bush DR. (1990) Δ-pH dependent amino acid transport into plasma membrane vesicles isolated from sugar beet leaves. I. Evidence for carrier-mediated, electrogenic flux through multiple transport systems. Plant Physiol 94: 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZC, Bush DR. (1992) Δ-pH dependent amino acid transport into plasma membrane vesicles isolated from sugar beet (Beta vulgaris L.) leaves. II. Evidence for multiple aliphatic, neutral amino acid symports. Plant Physiol 96: 1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Ji Y, Bhuiyan NH, Pilot G, Selvaraj G, Zou J, Wei Y. (2010) Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22: 3845–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Fait A, Lagor K, Nunes-Nesi A, Grillich N, Yellin A, Bar D, Khan M, Fernie AR, Turano FJ, et al. (2011) A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J 67: 485–498 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Shen Q, Smith SJ. (2007) Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J Exp Bot 59: 111–119 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio-assays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Murray D, Hermida-Matsumoto L, Buser CA, Tsang J, Sigal CT, Ben-Tal N, Honig B, Resh MD, McLaughlin S. (1998) Electrostatics and the membrane association of Src: theory and experiment. Biochemistry 37: 2145–2159 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Okrész L, Stabel S, et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Pilot G. (2011) Amino acid export in plants: a missing link in nitrogen cycling. Mol Plant 4: 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacín M, Nunes V, Font-Llitjós M, Jiménez-Vidal M, Fort J, Gasol E, Pineda M, Feliubadaló L, Chillarón J, Zorzano A. (2005) The genetics of heteromeric amino acid transporters. Physiology (Bethesda) 20: 112–124 [DOI] [PubMed] [Google Scholar]
- Peoples MB, Gifford RM. (1990) Long-distance transport of carbon and nitrogen. In DT Dennis, DH Turpin, eds, Plant Physiology, Biochemistry and Molecular Biology. Longman Scientific and Technical, Harlow, UK, pp 434–447 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan NQ, Kim SJ, Bassham DC. (2008) Overexpression of Arabidopsis sorting nexin AtSNX2b inhibits endocytic trafficking to the vacuole. Mol Plant 1: 961–976 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB. (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G. (2006) The plant-specific VIMAG domain of Glutamine Dumper1 is necessary for the function of the protein in Arabidopsis. FEBS Lett 580: 6961–6966 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Pilot G. (2007) Altered amino acid metabolism in Glutamine Dumper1 plants. Plant Signal Behav 2: 182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G. (2010) Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol 152: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Rolland N, Ferro M, Ephritikhine G, Marmagne A, Ramus C, Brugière S, Salvi D, Seigneurin-Berny D, Bourguignon J, Barbier-Brygoo H, et al. (2006) A versatile method for deciphering plant membrane proteomes. J Exp Bot 57: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Santoni V, Doumas P, Rouquié D, Mansion M, Rabilloud T, Rossignol M. (1999) Large scale characterization of plant plasma membrane proteins. Biochimie 81: 655–661 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Warthmann N, Weigel D. (2010) Directed gene silencing with artificial microRNAs. In BC Meyers, PJ Green, eds, Plant MicroRNAs, Vol 592. Humana Press, New York, pp 71–88 [Google Scholar]
- Secor J, Schrader LE. (1984) Characterization of amino acid efflux from isolated soybean cells. Plant Physiol 74: 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D. (2010) Uptake and partitioning of amino acids and peptides. Mol Plant 3: 997–1011 [DOI] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Fux CI, Beachy RN, Fauquet C. (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Samaj J, Hlavacka A, Ueda T, Preuss M, Nielsen E, Mathur J, Emans N, Stenmark H, et al. (2005) Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur J Cell Biol 84: 609–621 [DOI] [PubMed] [Google Scholar]
- Woelk T, Sigismund S, Penengo L, Polo S. (March 13, 2007) The ubiquitination code: a signalling problem. Cell Div 2: http://dx.doi.org/10.1186/1747-1028-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S, Fusada N, Hayashi H, Utsumi T, Uozumi N, Endo Y, Tozawa Y. (2010) The consensus motif for N-myristoylation of plant proteins in a wheat germ cell-free translation system. FEBS J 277: 3596–3607 [DOI] [PubMed] [Google Scholar]
- Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, Lee JS, Weigel D, Ahn JH. (2005) The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221: 523–530 [DOI] [PubMed] [Google Scholar]
- Zickermann V, Angerer H, Ding MG, Nübel E, Brandt U. (2010) Small single transmembrane domain (STMD) proteins organize the hydrophobic subunits of large membrane protein complexes. FEBS Lett 584: 2516–2525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.