Abstract
Chloroplast 93-kD heat shock protein (Hsp93/ClpC), an Hsp100 family member, is suggested to have various functions in chloroplasts, including serving as the regulatory chaperone for the ClpP protease in the stroma and acting as a motor component of the protein translocon at the envelope. Indeed, although Hsp93 is a soluble stromal protein, a portion of it is associated with the inner envelope membrane. The mechanism and functional significance of this Hsp93 membrane association have not been determined. Here, we mapped the region important for Hsp93 membrane association by creating various deletion constructs and found that only the construct with the amino-terminal domain deleted, Hsp93-ΔN, had reduced membrane association. When transformed into Arabidopsis (Arabidopsis thaliana), most atHsp93V-ΔN proteins did not associate with membranes and atHsp93V-ΔΝN failed to complement the pale-green and protein import-defective phenotypes of an hsp93V knockout mutant. The residual atHsp93V-ΔN at the membranes had further reduced association with the central protein translocon component Tic110. However, the degradation of chloroplast glutamine synthetase, a potential substrate for the ClpP protease, was not affected in the hsp93V mutant or in the atHSP93V-ΔN transgenic plants. Hsp93-ΔN also had the same ATPase activity as that of full-length Hsp93. These data suggest that the association of Hsp93 with the inner envelope membrane through its amino-terminal domain is important for the functions of Hsp93 in vivo.
Chloroplasts are structurally complex organelles that perform diverse functions (Leister, 2003; Block et al., 2007). They are composed of three membranes, the outer and inner envelope membranes and the thylakoid membranes, and these membranes enclose three aqueous compartments, the intermembrane space, stroma, and thylakoid lumen. Although chloroplasts have their own genome, most chloroplast proteins are encoded by the nuclear genome and are translated in the cytosol as a precursor protein with an N-terminal extension called the transit peptide. Transit peptides direct the import of proteins into chloroplasts through the translocon complex located in the chloroplast envelope. Translocon components of the outer membrane are called Toc (for translocon at the outer envelope membrane of chloroplasts) proteins and those in the inner envelope membrane are called Tic (for translocon at the inner envelope membrane of chloroplasts) proteins. Three Toc components, Toc159, Toc34, and Toc75, form the Toc core complex. Toc159 and Toc34 function as receptors that recognize the precursors targeting to chloroplasts. Toc75 forms a protein-conducting channel across the outer membrane. Three proteins, Tic20, Tic21, and Tic110, have been suggested to function as the channel for precursor translocation across the inner membrane. Tic110 has also been shown to function as the stromal-side receptor for transit peptides and as a scaffold for assembling other stromal translocon components. Tic40 is a cochaperone that coordinates the actions of Tic110 and Hsp93 during protein translocation across the inner membrane (for review, see Jarvis, 2008; Kessler and Schnell, 2009; Kovács-Bogdán et al., 2010; Li and Chiu, 2010).
In chloroplasts, both stromal 93-kD heat shock protein (Hsp93/ClpC) and 70-kD heat shock protein (Hsp70) have been shown to be important for protein import, and both have been proposed to function as the motors that drive protein translocation across the envelope into the stroma (Akita et al., 1997; Nielsen et al., 1997; Constan et al., 2004; Kovacheva et al., 2005, 2007; Shi and Theg, 2010; Su and Li, 2010). However, evidence directly demonstrating the motor function for either chaperone is still lacking. Hsp93 belongs to the Hsp100 subfamily of AAA+ proteins (for ATPase associated with various cellular activities). It has an N-terminal domain (N domain) and two ATPase domains (D1 and D2 domains), which are separated by a spacer (Schirmer et al., 1996). Most Hsp93 proteins are found in the stroma. However, a significant portion of Hsp93 molecules are associated with the inner envelope membrane and can be coisolated with other Tic components (Moore and Keegstra, 1993; Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998; Su and Li, 2010). In Arabidopsis (Arabidopsis thaliana), Hsp93 is encoded by two genes, atHSP93III (At3g48870; ClpC2) and atHSP93V (At5g50920; ClpC1), and atHsp93V is the major functional form. Knockout mutants of hsp93III are indistinguishable from the wild type, while hsp93V mutants are pale green and defective in protein import into chloroplasts (Constan et al., 2004; Kovacheva et al., 2005). Double knockout of both genes causes lethality, indicating that Hsp93 is essential (Kovacheva et al., 2007). In addition, Hsp93 has been proposed to act as a regulatory subunit of the ClpP protease in chloroplasts (Shanklin et al., 1995; Desimone et al., 1997; Sokolenko et al., 1998; Halperin et al., 2001). Interestingly, although Hsp93 was not detected in the ClpP core complex isolated from chloroplasts, the land plant chloroplast ClpP complex contains two additional peripheral subunits, ClpT1 and ClpT2 (previously named ClpS1 and ClpS2), that have high sequence similarity to the N-terminal portion of Hsp93 (Peltier et al., 2004). Hsp93 may also be involved in the biogenesis of photosystems (Sjögren et al., 2004), regulating chlorophyll b biosynthesis (Nakagawara et al., 2007), iron homeostasis (Wu et al., 2010), and antagonizing the function of the VAR2 protease (Park and Rodermel, 2004). It cannot, therefore, be excluded that the pale-green and protein import-defective phenotypes of hsp93V mutants are secondary effects due to the reduction of these other functions. One major distinction between these functions is that a function in protein import requires Hsp93 to be localized in the inner envelope membrane with other translocon components, while the other proposed functions are localized inside the stroma.
We are interested in the molecular function of Hsp93 during protein import into chloroplasts. Here, we investigated how Hsp93 tethers to the inner envelope membrane and whether this membrane association is important for its in vivo functions. Our results show that the N-domain-deleted mutant of pea (Pisum sativum) Hsp93 (Hsp93-ΔN) had greatly reduced membrane association. Transgenic plants expressing atHsp93V-ΔN failed to complement the pale-green and protein import-defective phenotypes of an hsp93V mutant but did not affect the degradation of glutamine synthetase (GS2), a proposed substrate for the ClpP protease. Immunoprecipitation by anti-Tic110 antibodies showed that N-domain deletion of atHsp93V further reduced the interaction between atTic110 and atHsp93V-ΔN in the inner envelope membrane. These data indicate that the N domain is important for membrane association and essential for the in vivo functions of Hsp93.
RESULTS
N-Domain Deletion Reduces the Hsp93 Membrane Association
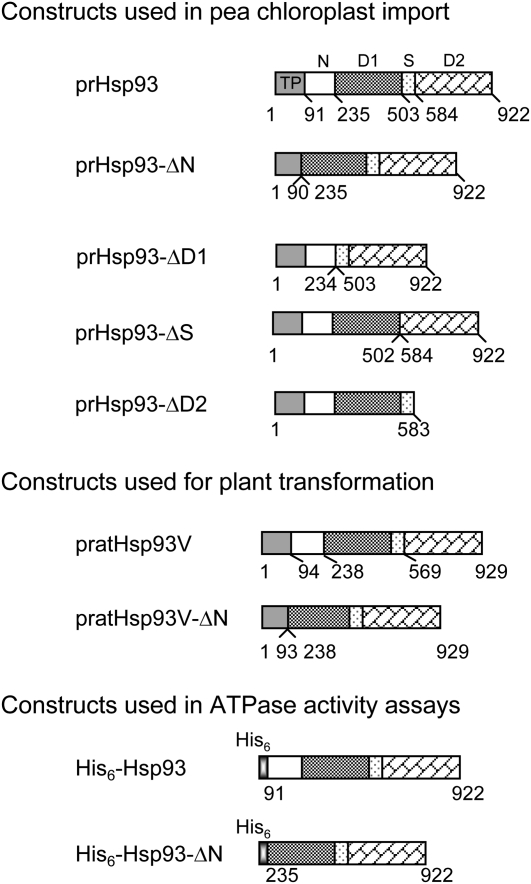
To investigate whether the membrane association of Hsp93 is really important for its function, we first tried to identify the region needed for its membrane association. We made plasmid constructs encoding the pea Hsp93 precursor protein (prHsp93) with the N domain, the D1 domain, the spacer region (referred to as the S region herein), or the D2 domain deleted (designated as prHsp93-ΔN, prHsp93-ΔD1, prHsp93-ΔS, and prHsp93-ΔD2, respectively; Fig. 1). [35S]Met-labeled precursors were synthesized by in vitro transcription and translation and incubated with isolated pea chloroplasts under import conditions. After import, a lower Mr mature protein was produced for all constructs and the mature protein was thermolysin resistant (Fig. 2A, lane 4), indicating that these proteins were located within chloroplasts. These data indicate that these truncated forms of prHsp93 were successfully imported into chloroplasts.
Figure 1.
Schematic representation of constructs used in this study. Numbers below each construct indicate the amino acid residue numbers, with the first amino acid of the precursor protein as 1. Proteins from Arabidopsis are specified with “at.” All others are from pea. D1 and D2, The ATPase I and ATPase II domains, respectively; N, the N domain; pr, precursor form; S, the spacer region between the D1 and D2 domains; TP, transit peptide.
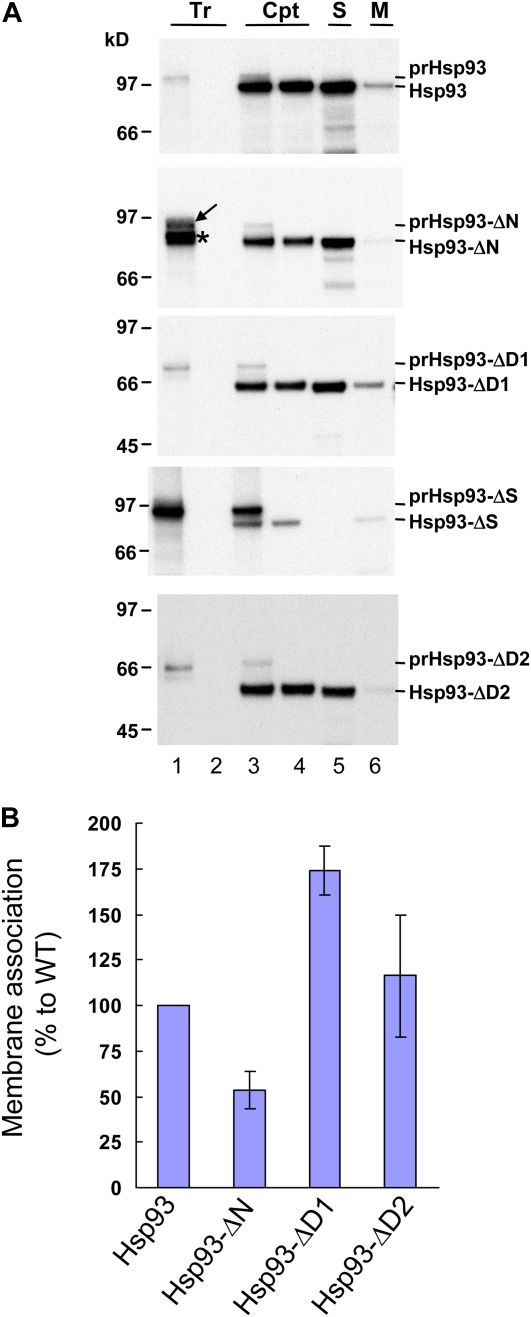
Figure 2.
The Hsp93 N domain is important for Hsp93 membrane association. A, In vitro-translated [35S]Met-prHsp93 and various deletion mutants (Tr; lane 1 of all panels) were treated with thermolysin directly (lane 2) or incubated with isolated pea chloroplasts (Cpt) under import conditions for 25 min. After import, a small portion of the chloroplasts were centrifuged through a 40% Percoll cushion to reisolate intact chloroplasts (lane 3). The rest of the reactions were pelleted and resuspended, and the chloroplasts were further digested with thermolysin. After digestion, intact chloroplasts were reisolated through a 40% Percoll cushion (lane 4). Some chloroplasts were further lysed hypotonically and separated into soluble (S; lane 5) and membrane (M; lane 6) fractions by centrifugation. The proteins in the soluble fraction were precipitated by TCA and dissolved with the protein extraction buffer. The membrane fractions were resuspended with the same volume of protein extraction buffer. Protein concentrations were then determined for all samples. Ten micrograms of proteins was loaded in all the chloroplast and soluble fraction lanes (lanes 3–5). For each membrane fraction, the same volume as its corresponding soluble fraction was loaded, so each pair of membrane and soluble fractions shown were derived from the same amount of chloroplasts. Samples were analyzed by SDS-PAGE and fluorography. All Tr lanes contain 20% of the in vitro-translated precursors used for the chloroplast (Cpt) lanes. B, The amount of imported mature proteins in the soluble and membrane fractions (lanes 5 and 6 of A) was quantified. The percentage in the membrane for each construct was calculated and then normalized to the wild type (WT). Data shown are means ± sd (n = 3). Data for prHsp93-ΔS were not included because no signal was obtained in the soluble fraction. [See online article for color version of this figure.]
From the in vitro translation system, prHsp93-ΔN was synthesized as three major protein products (Fig. 2A, an 87-kD protein marked by an arrow and an 83-kD doublet of proteins marked by an asterisk). To verify which of the translation products was the true precursor form, we performed chloroplast-binding experiments under low-temperature conditions to test which protein could bind to chloroplasts. The binding of other precursors to chloroplasts was also tested as controls (Supplemental Fig. S1). The 87-kD protein was the only protein that could bind to chloroplasts in the translation products of prHsp93-ΔN (Supplemental Fig. S1, lane 4). The 83-kD doublet of proteins most likely resulted from internal initiations during in vitro translation; thus, their transit peptides were truncated and they failed to bind to chloroplasts.
To investigate whether the deletion of individual domains in Hsp93 affected the localization of Hsp93 within chloroplasts, after the import reactions, chloroplasts were lysed hypotonically and separated into soluble and membrane fractions by centrifugation. As shown in Figure 2, only deletion of the N domain reduced the membrane association to about half of that of wild-type Hsp93. All the other deletion mutants have either similar or higher membrane association compared with the wild type. For prHsp93-ΔS, all of the imported mature Hsp93-ΔS was localized in the membrane fraction, and we could not detect signals significantly above background in the soluble fraction. Thus, the quantification data of imported Hsp93-ΔS were not included in Figure 2B. These data suggest that the N domain of Hsp93 is important for its membrane association. The S region between the two ATPase domains may be important for Hsp93 stromal localization, or deletion of this region may cause aggregation of prHsp93-ΔS after removal of the transit peptide; thus, all imported mature Hsp93-ΔS was detected in the membrane fraction.
atHsp93V-ΔN Cannot Complement the hsp93V Mutant
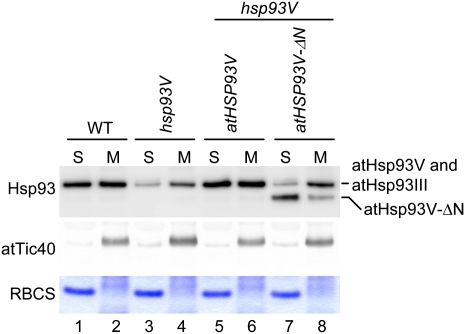
To investigate whether the membrane association is important for Hsp93 function in vivo, we transformed atHsp93V or an atHsp93V N-domain deletion mutant (atHsp93V-ΔN) into an Arabidopsis hsp93V knockout mutant (the Wassilewskija allele from the University of Wisconsin, as reported by Constan et al. [2004]). The cDNAs encoding the precursor forms of atHsp93V and atHsp93V-ΔN (Fig. 1) were cloned into the binary vector pCHF1 under the control of the cauliflower mosaic virus 35S promoter and transformed into the hsp93V mutant. Several independent transformants were obtained, and plants homozygous for a single insertion of the transgene were selected. The genotypes of these transgenic plants were confirmed by genomic PCR (Supplemental Fig. S2). No significant difference was observed among different lines of each transgene. Two representative lines for each transgene were selected for further characterization. As shown in Figure 3A, the atHSP93V transgene restored the atHsp93 protein level in the hsp93V mutant to the wild-type level (Fig. 3A, lanes 3 and 4). The level of Tic40 was analyzed as a loading control, since it has been shown that the protein levels of major Toc/Tic components are not affected in the hsp93V mutant (Constan et al., 2004; Kovacheva et al., 2007). The atHSP93V-ΔN transgene was also successfully expressed, and the atHsp93 protein level in the atHSP93V-ΔN transgenic plants, when adding together the amounts of atHsp93V-ΔN and atHsp93III, was comparable to the total amount of atHsp93 in the wild type (Fig. 3A, lanes 5 and 6; for quantification, see Supplemental Fig. S3).
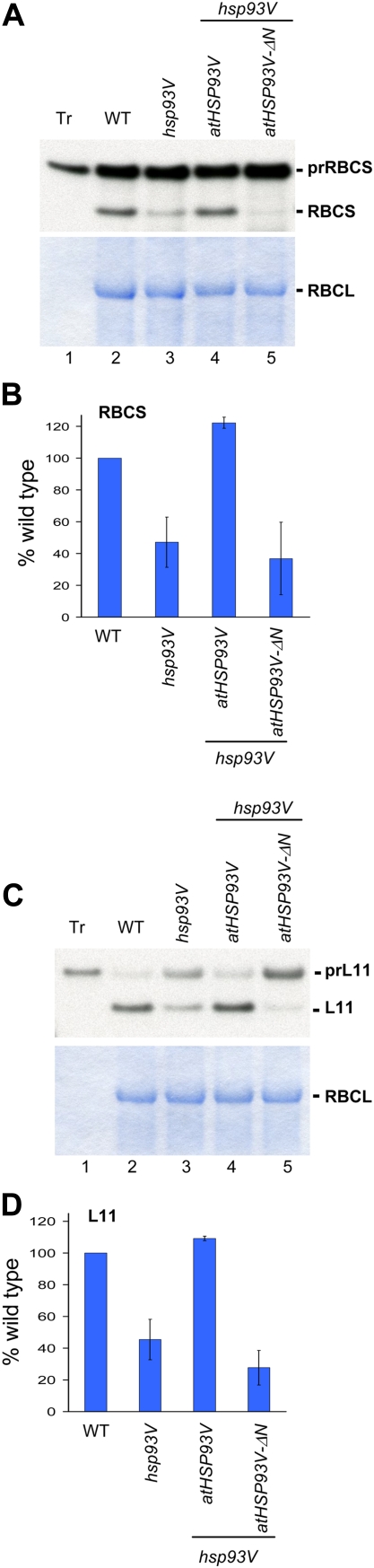
Figure 3.
atHsp93V-ΔN cannot complement the visible phenotypes of an hsp93V mutant. A, Total proteins were extracted from 14-d-old seedlings of the wild type (WT), hsp93V, and two independent lines of hsp93V mutant transformed with atHSP93V or atHSP93V-ΔN grown on MS plates. Thirty micrograms of proteins from each plant was analyzed by immunoblotting using antibodies against Hsp93 and atTic40. The amount of atTic40 was analyzed as a loading control. B and C, Visible phenotypes of plants grown on an MS plate under a 16-h photoperiod for 14 d (B) or grown on soil under a 12-h photoperiod for 26 d (C). D, Chlorophyll contents of seedlings grown on MS plates under a 16-h photoperiod for 15 d. Data shown are means ± sd (n = 3).
All the atHSP93V transgenic plants appeared indistinguishable from the wild type under various growth conditions (Fig. 3, B and C), indicating that the atHSP93V transgene fully complemented the pale-green and smaller-size phenotypes of the hsp93V mutant. It also fully restored the hsp93V mutant chlorophylls to the wild-type level (Fig. 3D). In comparison, all the atHSP93V-ΔN transgenic plants remained the same as the hsp93V mutant (Fig. 3, B and C). Chlorophyll quantification also showed that atHsp93V-ΔN could not rescue the chlorophyll-deficient phenotype of hsp93V (Fig. 3D).
We next assayed the effect of atHsp93V and atHsp93V-ΔN in rescuing the import defects of the hsp93V mutant. As shown in Figure 4, the import of prRBCS and prL11 into chloroplasts isolated from the hsp93V mutant was approximately 50% of that of wild-type chloroplasts. In the atHSP93V transgenic plants, the import defect was fully complemented (Fig. 4, A and C, lane 4). In contrast, the atHSP93V-ΔN transgenic plants still had the same deficiency as the hsp93V mutant (Fig. 4, A and C, lane 5).
Figure 4.
atHsp93V-ΔN cannot rescue the protein-import defect of an hsp93V mutant. In vitro-translated [35S]Met-prRBCS (A and B) or [35S]Met-prL11 (C and D) were imported into chloroplasts isolated from 14-d-old plate-grown plants of the indicated genotypes for 5 min. Line 2 of the atHSP93V transformants and line 11 of the atHSP93V-ΔN transformants shown in Figure 3 were used. Samples were analyzed by SDS-PAGE, stained with Coomassie blue, and dried for fluorography. Twenty micrograms of proteins was loaded in all lanes of the import samples. Tr, In vitro-translated precursors before import. In B and D, the amount of imported mature proteins was quantified and the amount imported into the wild type (WT) was set as 100%. Data shown are means ± sd of at least three independent experiments. [See online article for color version of this figure.]
Mutations in atHsp93V Do Not Affect the Degradation of a Potential ClpP Substrate
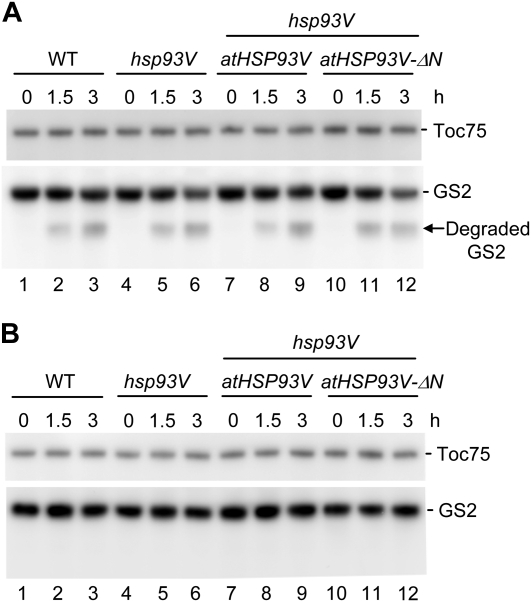
Because Hsp93 is also proposed to function as a regulatory subunit for the ClpP protease complex in the stroma (Halperin and Adam, 1996; Desimone et al., 1997; Sokolenko et al., 1998; Halperin et al., 2001), an in organello proteolytic assay (Sjögren et al., 2006) was conducted to investigate the degradation of the chloroplast GS2. GS2 has been shown to be a potential substrate for ClpP, because its amount increased in a mutant that only has 10% of the wild-type level of intact ClpP core complex (Stanne et al., 2009). An antibody against GS2 is also commercially available. For the assay, an equal number of chloroplasts isolated from wild-type, hsp93V mutant, atHSP93V transgenic, and atHSP93V-ΔN transgenic plants were incubated in the presence of ATP at 22°C under constant light as described (Sjögren et al., 2006). The amount of Toc75 was used as a control, because Toc75 is localized in the outer envelope membrane of chloroplasts and should not be a substrate for the stromal ClpP protease. As shown in Figure 5A, a progressive decrease of GS2 proteins was observed in chloroplasts during the 3-h incubation in the presence of ATP and light. A lower Mr fragment appeared after 1.5 h and increased in amount after 3 h (Fig. 5A, arrow). This fragment was about 31 kD and was the same size as the degraded GS2 reported previously (Palatnik et al., 1999). This fragment was not observed when chloroplasts were incubated under the same conditions but without ATP and light (Fig. 5B), further supporting that it is a likely GS2 degradation product by ClpP. Nonetheless, the degradation pattern and rate of GS2 were similar in the wild type and the hsp93V mutant, and the atHSP93V and atHSP93V-ΔN transgenic plants showed no significant change from the wild type either (Fig. 5A). These results suggest that knocking out the major functional form of Hsp93 in Arabidopsis, atHsp93V, does not have a significant effect on the ClpP protease activity and, further, that the addition of atHsp93V or atHSP93V-ΔN into the mutant also has no additional effect.
Figure 5.
atHsp93V-ΔN does not affect the degradation of chloroplast GS2. Chloroplasts isolated from 27-d-old plate-grown plants of the indicated genotypes were incubated in the presence of ATP and an ATP regeneration system under constant light (A) or in the absence of ATP and the ATP regeneration system in the dark (B). Line 2 of the atHSP93V transformants and line 11 of the atHSP93V-ΔN transformants shown in Figure 3 were used. At 0, 1.5, and 3 h, an equal volume of chloroplast suspensions was taken and analyzed by immunoblotting using antibodies against Toc75 and chloroplast GS2. A total of 1.5 × 106 chloroplasts were loaded in each lane. The amount of Toc75 was analyzed as a loading control. The arrow indicates the degraded GS2 fragment. WT, Wild type.
The Hsp93 N-Terminal Deletion Mutant Has Reduced Membrane and Tic110 Association in Vivo
To investigate whether the atHsp93V-ΔN protein in the transgenic plants also had similar reductions in membrane association as the in vitro imported Hsp93-ΔN, chloroplasts isolated from wild-type, hsp93V, and atHSP93V transgenic and atHSP93V-ΔN transgenic plants were lysed and separated into soluble and membrane fractions. As shown in Figure 6, the distribution of atHsp93 proteins in the atHSP93V transgenic plants was similar to that in the wild type (Fig. 6, lanes 1, 2, 5, and 6). In comparison, the majority of atHsp93V-ΔN was found in the soluble fraction in the atHSP93V-ΔN transgenic plant chloroplasts (Fig. 6, lanes 7 and 8; the membrane association of atHsp93V-ΔN was 55.5% ± 1.3% of atHsp93 in the wild type).
Figure 6.
atHsp93V-ΔN has reduced membrane association in vivo. Chloroplasts isolated from 14-d-old plate-grown plants of the indicated genotypes were lysed hypotonically and separated into soluble (S) and membrane (M) fractions by centrifugation. The proteins in the soluble fraction were precipitated by TCA and dissolved with the protein extraction buffer. The membrane fractions were resuspended with the same volume of protein extraction buffer. Protein concentrations were then determined for all samples. Eight micrograms of proteins was loaded in all the soluble-fraction lanes. For each membrane fraction, the same volume as its corresponding soluble fraction was loaded so that each pair of membrane and soluble fractions shown were derived from the same amount of chloroplasts. Samples were analyzed by SDS-PAGE. The top half of the gel was analyzed by immunoblotting with antibodies against Hsp93 and atTic40. The bottom half of the gel was stained with Coomassie blue to reveal the location of RBCS. WT, Wild type. [See online article for color version of this figure.]
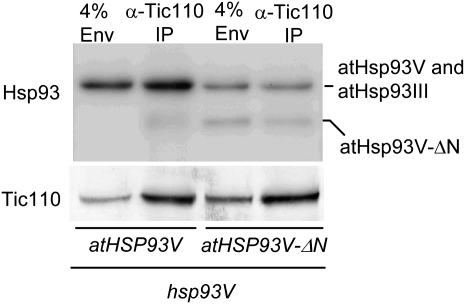
In the atHSP93V-ΔN transgenic plant chloroplasts, there was still a small residual amount of atHsp93V-ΔN associated with the membranes. To investigate the reason why this small amount of membrane-associated atHsp93V-ΔN did not slightly rescue the import defect of the hsp93V mutant, we analyzed how much of the membrane-associated atHsp93V-ΔN was associated with the translocon by performing immunoprecipitation with the anti-Tic110 antibody. Chloroplasts were isolated from the atHSP93V and atHSP93V-ΔN transgenic plants, cross-linked with 1 mm dithiobis-9-succinimidylpropionate (DSP), and lysed. An envelope-enriched membrane fraction (Su and Li, 2010) was collected, solubilized, and immunoprecipitated with an anti-Tic110 antibody (Tu et al., 2004). In the atHSP93V transgenic plant chloroplasts, about 4.9% ± 0.2% of the envelope-associated-atHsp93 was immunoprecipitated by the anti-Tic110 antibody (Fig. 7). In comparison, in the atHSP93V-ΔN transgenic plant chloroplasts, of the envelope-associated atHsp93V-ΔN, only about 2.0% ± 1.0% of atHsp93V-ΔN was immunoprecipitated in the same conditions. These data indicate that the N-domain deletion reduces Hsp93 membrane association, and of those Hsp93-ΔN molecules that could still tether to the membrane, their association with Tic110 was further decreased to about half. Thus, in the atHSP93V-ΔN transgenic plants, Tic110 only has about 25% of the wild-type level of atHsp93V associated with it.
Figure 7.
atHsp93V-ΔN has reduced association with Tic110. Chloroplasts isolated from 14-d-old plate-grown plants of the indicated genotypes were cross-linked with DSP. Eight hundred microliters of chloroplasts, in the concentration of 1 mg chlorophyll mL−1, was used for the cross-linking. Chloroplasts were lysed, and the enriched envelope membrane fraction was collected and solubilized in 800 μL of immunoprecipitation buffer containing 1% n-decyl-β-d-maltopyranoside. The clarified supernatant of solubilized membranes was immunoprecipitated with the anti-Tic110 antibody. The immunoprecipitates (IP) were analyzed by SDS-PAGE followed by immunoblotting with antibodies as indicated on the left. The loading of the immunoprecipitate lanes was adjusted to make the amount of Tic110 precipitated in the two transgenic lines equal (equivalent to 207 and 120 μL of solubilized membranes from the atHSP93V and atHSP93V-ΔN transgenic lines, respectively, on the gel shown). 4% Env, Four percent of the solubilized membranes used in the corresponding immunoprecipitate lane. The atHSP93V-ΔN transgenic line is pale green and contains a higher number of chloroplasts per milligram of chlorophyll and thus a higher concentration of Tic110 in the solubilized membranes.
N-Domain Deletion Does Not Affect the ATPase Activity of Hsp93
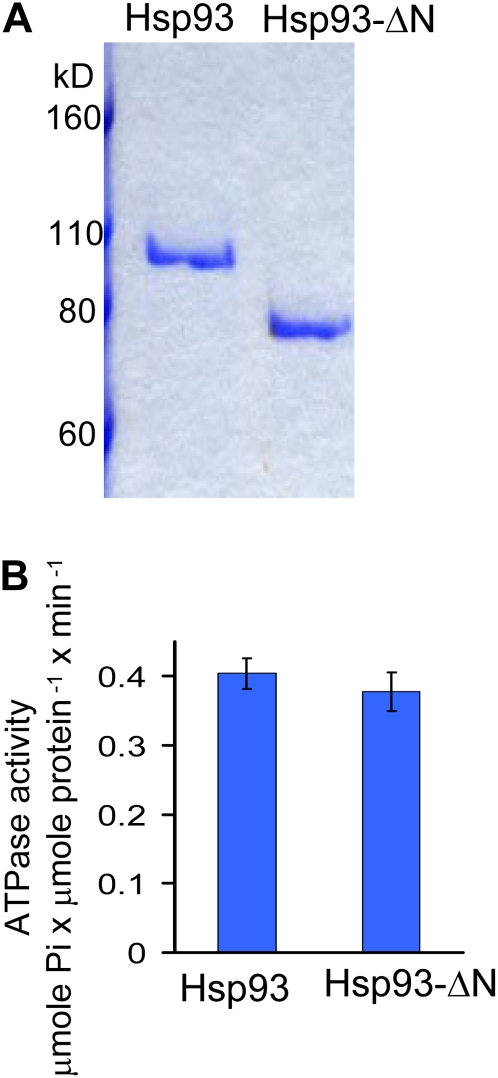
We next investigated whether the N-domain deletion also had an effect on the ATPase activity of Hsp93. We expressed both Hsp93 (Chou et al., 2006) and the deletion mutant Hsp93-ΔN (Fig. 1) as His6-tagged recombinant proteins in Escherichia coli and purified them (Fig. 8A). As shown in Figure 8B, deletion of the Hsp93 N domain did not affect the ATPase activity of Hsp93.
Figure 8.
N-domain deletion does not affect the ATPase activity of Hsp93. A, His6-Hsp93 and His6-Hsp93-ΔN were expressed and purified from the soluble fraction of E. coli and analyzed by SDS-PAGE and Coomassie blue staining. B, His6-Hsp93 and His6-Hsp93-ΔN have similar ATPase activities. Data shown are means ± sd of two independent experiments. [See online article for color version of this figure.]
DISCUSSION
To function as a motor for chloroplast protein import, Hsp93 needs to be localized at the inner envelope membrane with other translocon components. However, it was not clear whether the membrane association of Hsp93 is really required for its function, since the majority of Hsp93 is in the stroma. Here, we first identified that the N domain of Hsp93 is important for its membrane association. We then showed that the N-domain deletion mutant, Hsp93-ΔN, has reduced membrane association in vivo but has normal ATPase activity and accumulation level in the stroma. We also determined that this mutant could not complement the growth and protein-import defects of an hsp93V mutant. Our results indicate that the membrane association of Hsp93 is indeed important for its functions in vivo.
Although Hsp93 indeed possesses the conserved ClpP-binding tripeptide, an I/L-G-F motif with an upstream K/R residue (Kim et al., 2001; Weibezahn et al., 2004), this motif is located in the D2 domain of Hsp93 and therefore is retained in the atHsp93V-ΔN protein. Thus, the failure of the stromal atHsp93V-ΔN proteins to complement the hsp93V mutant phenotypes is less likely due to a loss of potential interaction between Hsp93 and ClpP. Furthermore, the degradation of a ClpP protease substrate, GS2, was not affected in the atHSP93V-ΔN transgenic plants. However, our data indicate that even the hsp93V mutant, which shows protein-import defects, degrades GS2 normally. The mutant also shows normal degradation of the mistargeted 33-kD protein of the oxygen-evolving complex OEC33, another potential substrate of the ClpP protease (Sjögren et al., 2004). Therefore, functional evidence for Hsp93 involvement in ClpP protease activity in higher plant chloroplasts is not yet available. The in vivo functional relationship between the ClpP complex and Hsp93 remains to be investigated.
The mammalian p97 (yeast Cdc48) is also a member of the AAA+ protein family, and like Hsp93, it consists of two ATPase domains preceded by an additional N domain. Although p97/Cdc48 is an abundant cytosolic protein, a significant portion of it is associated with the endoplasmic reticulum (ER) membrane (Ye et al., 2003). p97/Cdc48 is the motor component for the retrotranslocation of misfolded proteins from the ER to the cytosol for degradation (Vembar and Brodsky, 2008). p97 is recruited to the ER membrane complex for retrotranslocation mostly through the interaction of the p97 N domain with the cytosolic domain of the ER membrane protein VIMP, and this interaction is critical for the function of p97 in retrotranslocation (Ye et al., 2003, 2004). Our data suggest a similar scenario for Tic110 and Hsp93: the N domain of Hsp93 mediates most of the interaction between Hsp93 and Tic110 and is critical for the function of Hsp93 in protein import into chloroplasts. Other domains of Hsp93 may also interact with other proteins in the inner membrane, since the membrane association of Hsp93-ΔN is not totally abolished. For example, the two ATPase domains may have some interactions with Tic40, since it has been shown that part of Tic40 can stimulate ATP hydrolysis by Hsp93 (Chou et al., 2006).
Deletion of the Hsp93 N domain reduced the membrane association of Hsp93-ΔN to half of that of Hsp93. For the residual Hsp93-ΔN proteins associated with membranes, their association with Tic110 was further reduced to half that of wild-type Hsp93. Therefore, in the atHSP93V-ΔN transgenic plants, the amount of atHsp93V-ΔN associated with Tic110 was about 25% of the atHsp93V associated with Tic110 in the atHSP93V transgenic plants. Although atHsp93V is the major functional form and the hsp93V mutant shows defects in growth and chloroplast protein import, it has been shown that when atHsp93III is overexpressed, it can fully complement the hsp93V mutant phenotypes. Furthermore, the hsp93III/hsp93V double knockout is lethal and an hsp93V knockout/hsp93III knockdown mutant shows a more severe chlorotic phenotype than the hsp93V single knockout mutant (Kovacheva et al., 2007). These data indicate that atHsp93V and atHsp93III are redundant in function and that a sufficient level of Hsp93, higher than the level provided by endogenous atHsp93III, is required for normal plant growth. The atHSP93V-ΔN transgenic plants has restored the level of Hsp93 in the stroma but only provided 25% of Hsp93 to Tic110. We thus suggest that insufficient association with Tic110 is most likely the major reason for the failure of atHSP93V-ΔN to complement the hsp93V mutant. It is also possible that a shuttling between an inner membrane-associated state and a soluble stroma-localized state and a balanced and sufficient amount in each state may be important for the function of Hsp93 during protein import. A 75% reduction in its ability to associate with Tic110 will severely hamper this shuttle cycle and render Hsp93-ΔN nonfunctional.
However, it cannot be excluded that the Hsp93 N domain has other functions at the inner membrane in addition to its association with Tic110. It also cannot be excluded that Hsp93 has other unknown interacting partners in the stroma and that deletion of the Hsp93 N domain has reduced interactions with these partners. However, a function in protein import at the inner membrane is most likely upstream of other functions in the stroma, and a defect in protein import should be one of the most pleiotropic defects in chloroplast biogenesis. Thus, currently available data support that a defect in chloroplast protein import is most likely the major cause for the hsp93V mutant phenotypes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
For growing pea seedlings (Pisum sativum ‘Green Arrow’; De Bruyn Seed), the imbibed seeds were grown on vermiculite for 7 to 10 d under a 12-h photoperiod at 20°C with a light intensity of approximately 150 μmol m−2 s−1. Wild-type and mutant Arabidopsis (Arabidopsis thaliana) plants used in this study were the Wassilewskija ecotype. Sterilized Arabidopsis seeds were plated on 0.3% Gelrite-solidified Murashige and Skoog (MS) medium containing Gamborg’s B5 vitamin and 2% Suc. After a 2- to 3-d cold stratification, seeds were grown in growth chambers under a 16-h photoperiod with a light intensity of approximately 70 μmol m−2 s−1 at 22°C. Soil-grown Arabidopsis plants were grown under a 12- or 16-h photoperiod with a light intensity of approximately 100 μmol m−2 s−1.
Translation of Precursor Proteins
The cDNA encoding pea prHsp93 (accession no. L09547) was cloned into a plasmid under the control of the SP6 promoter. The plasmid was then used as a template to generate all the prHsp93 deletion mutants, except prHsp93-ΔD2, by inverse PCR using the primers listed in Supplemental Table S1. The prHsp93-ΔD2 construct was generated by mutating residue 584 to a stop codon (for primers, see Supplemental Table S1) using the QuickChange II site-directed mutagenesis kit (Agilent Technologies). All constructs were confirmed by sequencing. [35S]Met-labeled precursor proteins were in vitro transcribed/translated in the TNT wheat germ lysate system (Promega) using the SP6 or T3 RNA polymerase.
Protein Import and Postimport Analyses
Chloroplast isolation from pea and Arabidopsis was conducted as described (Perry et al., 1991), except that the grinding buffer for Arabidopsis was modified to 50 mm HEPES-KOH (pH 8.0), 330 mm sorbitol, 2 mm EDTA, and 0.5% bovine serum albumin, and a domestic blender was used instead of the Polytron to homogenize the Arabidopsis leaf tissue. Isolated chloroplasts were adjusted to 1 mg chlorophyll mL−1 in import buffer (330 mm sorbitol and 50 mm HEPES-KOH, pH 8.0) for import assays. 35S-labeled precursor proteins were incubated with isolated pea or Arabidopsis chloroplasts in the presence of 3 mm ATP in import buffer at room temperature for 5 or 25 min as indicated. After import, intact chloroplasts were reisolated through a 40% Percoll cushion at 4°C, washed once with import buffer, and dissolved in protein extraction buffer containing 300 mm Tris-HCl, pH 8.5, 1 mm EDTA, 8% SDS (w/v), and 1 mm phenylmethylsulfonyl fluoride. Thermolysin digestion of precursor proteins and chloroplasts after import was performed as described (Perry et al., 1991). To separate chloroplasts into soluble and membrane fractions, chloroplasts after import were lysed with a hypotonic buffer (25 mm HEPES-KOH, pH 7.5, and 5 mm MgCl2) at a chloroplast concentration of 0.25 mg chlorophyll mL−1, freeze thawed once, and then separated into soluble and membrane fractions by ultracentrifugation at 100,000g for 45 min at 4°C. Soluble fractions were precipitated by 10% TCA (w/v), washed with 100% ice-cold acetone, and dissolved in the protein extraction buffer. Membrane fractions were washed with the hypotonic buffer once, collected by another ultracentrifugation at 100,000g for 15 min at 4°C, and the pellets were resuspended in the protein extraction buffer. Protein concentrations of the dissolved samples were measured with the bicinchoninic acid kit (Pierce). Samples were analyzed by SDS-PAGE. Quantification of gel bands was performed using the Fuji FLA5000 phosphorimager (Fujifilm).
Plant Transformation
The cDNA fragment encoding atHSP93V (At5g50920) was amplified from Arabidopsis leaf total first-strand cDNA by reverse transcription-PCR with the primer pair atHsp93V/F7-PstI and atHsp93V/R7-KpnI (Supplemental Table S1). The PCR product was cloned into the pGEM-T vector (Promega) and named pGEM-T-atHsp93V. The plasmid was used as a template to generate the mutant construct of atHsp93V without the N domain, named pGEM-T-atHsp93V-ΔN, by inverse PCR with the primer pair atHsp93V-delN-F and atHsp93V-delN-R (Supplemental Table S1). The coding regions of atHsp93V and atHsp3V-ΔN were further amplified by PCR using the primer pair atHsp93V/F8-KpnI and atHsp93V/R8-PstI (Supplemental Table S1), digested, and cloned into the KpnI/PstI sites of the binary vector pCHF1 (Hajdukiewicz et al., 1994). The resulting plasmids were named pCHF1-atHsp93V and pCHF1-atHsp93V-ΔN, respectively, and transformed into Agrobacterium tumefaciens GV3101. The hsp93V mutant plants were transformed by the floral spray method (Chung et al., 2000). Transgenic plants harboring the introduced atHSP93V and atHSP93V-ΔN transgenes were screened on MS medium containing 100 μg mL−1 G418. The genotypes of the transgenic plants were confirmed by genomic PCR. Primers used for the atHSP93V endogenous wild-type copy and the transgenes were atHsp93V-F8-KpnI and atClpC-V-R1, and primers used for the T-DNA insertion of the hsp93V mutant were JL202 and atHsp93V-R (Supplemental Table S1).
Protein Extraction and Immunoblotting
Total proteins from Arabidopsis leaves were extracted by the protein extraction buffer (300 mm Tris-HCl, pH 8.5, 1 mm EDTA, 8% SDS [w/v], and 1 mm phenylmethylsulfonyl fluoride). Chloroplasts isolated from Arabidopsis were lysed with the hypotonic buffer at a chloroplast concentration of 0.25 mg chlorophyll mL−1, freeze thawed once, and separated into soluble and membrane fractions as described above. Samples were analyzed by SDS-PAGE and immunoblotting. The purified pea His6-Hsp93-ΔN proteins prepared for the ATPase assays (see below) were used to raise anti-Hsp93 antibodies in rats and used for all immunoblot analyses of Hsp93. For immunoblot analyses of Tic110 and Tic40, the rat anti-atTic110 antibodies (Su and Li, 2010) and the mouse anti-atTic40 antibodies (Chou et al., 2006) were used. Immunoblotting was performed by electroblotting the proteins onto Immobilon-P membranes (Millipore). Immunoblots of Hsp93 in Figures 6 and 7 were hybridized with horseradish peroxidase-conjugated secondary antibodies, detected by the Immobilon Western Chemiluminescent HRP system (Millipore), and visualized and quantified with the UVP BioSpectrum 600 Image system (Ultra Violet Products). All the Tic110 and Tic40 immunoblots and the Hsp93 immunoblot in Figure 3A were visualized using alkaline phosphatase-conjugated secondary antibodies and the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate colorimetric system. The primary antibodies were used at a 1:2,000 dilution. The secondary antibodies used for the Chemiluminescent HRP system were used at a 1:50,000 dilution. For the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate detection system, the secondary antibodies were used at a 1:1,000 dilution.
Chlorophyll Quantification
Fifteen-day-old wild-type, hsp93V mutant, atHSP93V, and atHSP93V-ΔN transgenic seedlings grown on MS medium were weighed and ground with 80% acetone. Ground tissue was spun at 3,000g for 5 min to remove debris. The amount of chlorophyll in the supernatant was determined as described (Lichtenthaler, 1987).
In Organello Proteolytic Assay
Intact chloroplasts were isolated from 27-d-old plate-grown plants as described above, resuspended with grinding buffer to a chloroplast concentration of 1 mg chlorophyll mL−1, and further counted with a phase-contrast microscope (Olympus BH2) using a hemocytometer. An equal number of chloroplasts were retrieved, pelleted by centrifugation at 1,020g for 5 min, and resuspended to 1.5 × 106 chloroplasts μL−1 in a resuspension buffer (330 mm sorbitol, 5 mm MgCl2, 10 mm NaHCO3, and 20 mm HEPES-NaOH, pH 8) containing 5 mm Mg-ATP, 2.5 mm phosphocreatine, and 0.5 mg mL−1 creatine phosphokinase as an ATP regeneration system (modified from Sjögren et al. [2006]). Chloroplasts were incubated under a light intensity of approximately 60 μmol m−2 s−1 at 22°C. For control assays, an equal number of chloroplasts were resuspended in resuspension buffer without ATP and the ATP regeneration system and incubated at 22°C in the dark. An equal amount of chloroplasts were taken at 0, 1.5, and 3 h and analyzed by SDS-PAGE and immunoblotting using antibodies against Toc75 (Tranel et al., 1995) and GS2 (Agrisera; AS08 296). The primary antibodies were used at 1:4,000 and 1:5,000 dilutions for Toc75 and GS2, respectively. The secondary antibodies for the Chemiluminescent HRP system were used at a 1:50,000 dilution.
Immunoprecipitation by the Anti-Tic110 Antibody
The immunoprecipitation assays were conducted as described (Su and Li, 2010). In brief, chloroplasts isolated from Arabidopsis were cross-linked with 1 mm DSP at 4°C for 15 min in the dark, the cross-linking was terminated by adding Gly to a final concentration of 50 mm, and the chloroplasts were incubated at 4°C for another 15 min to quench the DSP. To prepare a membrane fraction enriched with envelope membranes, chloroplasts after cross-linking were washed once with import buffer, resuspended in import buffer, lysed by two freeze-thaw cycles, and centrifuged at 3,000g for 3 min to remove most of the thylakoid membranes. The supernatant was again centrifuged at 100,000g for 30 min at 4°C. The pellet was solubilized in immunoprecipitation buffer (50 mm HEPES-KOH, pH 8.0, 150 mm NaCl, 4 mm MgCl2, 1% n-decyl-β-d-maltopyranoside [w/v], and 10% glycerol) at 4°C for 30 min and clarified by centrifugation at 100,000g for 15 min. Rabbit antibodies against atTic110 (Tu et al., 2004) were added to the clarified supernatant, incubated for 4 h at 4°C, and precipitated using protein A-agarose (Pierce). The precipitates were washed with the immunoprecipitation buffer three times, eluted by SDS sample buffer, and analyzed by SDS-PAGE and immunoblotting.
Hsp93 Protein Expression, Purification, and ATPase Assays
pET28a-His6-Hsp93 (Chou et al., 2006) was used as a template to amplify the fragment corresponding to Hsp93 without the N domain by PCR using the primer pair mHsp93-delN-F-NdeI and mHsp93-delN-R-NotI (Supplemental Table S1). The PCR product was digested, cloned into pET28a (Invitrogen), and named pET28a-His6-Hsp93-ΔN. Plasmids were transformed into Escherichia coli strain BL21 (DE3), and protein induction was conducted with 1 mm isopropyl-1-thio-β-d-galactopyranoside at 37°C for 3 h. His6-Hsp93 and His6-Hsp93-ΔN were purified by TALON resins (Clontech Laboratories) with 50 mm imidazole, dialyzed with 50 mm Tris, pH 8, and concentrated by Amicon Ultra-15 (Millipore). ATPase activity assays and quantification were performed as described (Chou et al., 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers L09547 (pea Hsp93) and At5g50920 (atHsp93V).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Association of prHsp93 and the deletion mutants with chloroplasts under binding conditions.
Supplemental Figure S2. Genotyping of atHSP93V and atHSP93V-ΔN transgenic plants.
Supplemental Figure S3. Wild-type and atHSP93V-ΔN transgenic plants contain an equal amount of atHsp93 proteins.
Supplemental Table S1. Sequences of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth Keegstra for the anti-Toc75 antibody and the hsp93V mutants. We also thank Harry Wilson and AndreAna Pena for English editing.
References
- Akita M, Nielsen E, Keegstra K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MA, Douce R, Joyard J, Rolland N. (2007) Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth Res 92: 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Chu CC, Chen LJ, Akita M, Li HM. (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J Cell Biol 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MH, Chen MK, Pan SM. (2000) Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res 9: 471–476 [DOI] [PubMed] [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K. (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimone M, Weiss-Wichert C, Wagner E, Altenfeld U, Johanningmeier U. (1997) Immunochemical studies on the Clp-protease in chloroplasts: evidence for the formation of a ClpC/P complex. Bot Acta 110: 234–239 [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z. (1996) Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol 30: 925–933 [DOI] [PubMed] [Google Scholar]
- Halperin T, Ostersetzer O, Adam Z. (2001) ATP-dependent association between subunits of Clp protease in pea chloroplasts. Planta 213: 614–619 [DOI] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell D. (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8: 230–233 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P. (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kovács-Bogdán E, Soll J, Bölter B. (2010) Protein import into chloroplasts: the Tic complex and its regulation. Biochim Biophys Acta 1803: 740–747 [DOI] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Li HM, Chiu CC. (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Moore T, Keegstra K. (1993) Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol Biol 21: 525–537 [DOI] [PubMed] [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A. (2007) Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J 49: 800–809 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Carrillo N, Valle EM. (1999) The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol 121: 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rodermel SR. (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc Natl Acad Sci USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy J, van Wijk KJ. (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Perry SE, Li HM, Keegstra K. (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. (1996) HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci 21: 289–296 [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM. (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK. (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Stanne TM, Zheng B, Sutinen S, Clarke AK. (2006) Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko A, Lerbs-Mache S, Altschmied L, Herrmann RG. (1998) Clp protease complexes and their diversity in chloroplasts. Planta 207: 286–295 [DOI] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LL, Koussevitzky S, Clarke AK. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Chen LJ, Smith MD, Su YS, Schnell DJ, Li HM. (2004) Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Wu H, Ji Y, Du J, Kong D, Liang H, Ling HQ. (2010) ClpC1, an ATP-dependent Clp protease in plastids, is involved in iron homeostasis in Arabidopsis leaves. Ann Bot (Lond) 105: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429: 841–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.