Abstract
Two-component signaling elements play important roles in plants, including a central role in cytokinin signaling. We characterized two-component elements from the monocot rice (Oryza sativa) using several complementary approaches. Phylogenetic analysis reveals relatively simple orthologous relationships among the histidine kinases in rice and Arabidopsis (Arabidopsis thaliana). In contrast, the histidine-containing phosphotransfer proteins (OsHPs) and response regulators (OsRRs) display a higher degree of lineage-specific expansion. The intracellular localizations of several OsHPs and OsRRs were examined in rice and generally found to correspond to the localizations of their dicot counterparts. The functionality of rice type-B OsRRs was tested in Arabidopsis; one from a clade composed of both monocot and dicot type-B OsRRs complemented an Arabidopsis type-B response regulator mutant, but a type-B OsRR from a monocot-specific subfamily generally did not. The expression of genes encoding two-component elements and proteins involved in cytokinin biosynthesis and degradation was analyzed in rice roots and shoots and in response to phytohormones. Nearly all type-A OsRRs and OsHK4 were up-regulated in response to cytokinin, but other cytokinin signaling elements were not appreciably affected. Furthermore, multiple cytokinin oxidase (OsCKX) genes were up-regulated by cytokinin. Abscisic acid treatment decreased the expression of several genes involved in cytokinin biosynthesis and degradation. Auxin affected the expression of a few genes; brassinosteroid and gibberellin had only modest effects. Our results support a shared role for two-component elements in mediating cytokinin signaling in monocots and dicots and reveal how phytohormones can impact cytokinin function through modulating gene expression.
Plants make use of “two-component systems” for signal transduction, and these are involved in important cellular processes, such as the responses to cytokinin, ethylene, red light, and osmosensing (Mizuno, 2005; Schaller et al., 2008). Two-component systems were originally identified in bacteria, and in their simplest form they involve a receptor kinase that autophosphorylates on a conserved His residue in response to an environmental stimulus (Stock et al., 2000; Gao and Stock, 2009). This phosphate is then transferred to a conserved Asp residue within the receiver domain of a response regulator. Phosphorylation of the response regulator modulates its ability to mediate downstream signaling in the pathway. Of particular relevance to plants is a permutation on the two-component system known as the multistep phosphorelay (Appleby et al., 1996; Goulian, 2010; Wuichet et al., 2010). The multistep phosphorelay typically makes use of three components: a “hybrid” receptor kinase that contains both His kinase and receiver domains in a single protein; a histidine-containing phosphotransfer (HPt) protein; and a separate response regulator (RR). In these multistep phosphorelays, the phosphate is transferred from amino acid to amino acid in sequence His to Asp to His to Asp.
Plants, as exemplified by the dicot Arabidopsis (Arabidopsis thaliana) and the monocot rice (Oryza sativa), contain all the elements of a multistep phosphorelay (Mizuno, 2005; Jain et al., 2006; Pareek et al., 2006; Schaller et al., 2007, 2008; Pils and Heyl, 2009). Separate His kinase families have been shown to function as cytokinin receptors and ethylene receptors, pointing toward key roles in mediating phytohormone signaling (Schaller and Bleecker, 1995; Inoue et al., 2001; Kakimoto, 2003; Yau et al., 2004; Ito and Kurata, 2006; Du et al., 2007). The response regulators can be classified into three distinct groups based on domain structure and sequence: type A, type B, and type C (Schaller et al., 2008). The type-A response regulators are relatively small, containing the receiver domain common to response regulators along with short N- and C-terminal extensions, and are transcriptionally up-regulated by cytokinin (Brandstatter and Kieber, 1998; D’Agostino et al., 2000; Jain et al., 2006). The type-B response regulators each have a large C-terminal extension following the receiver domain and act as transcriptional regulators (Hosoda et al., 2002; Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008b). The type-C response regulators are similar to the type-A response regulators in domain architecture but are clearly distinct based on phylogenetic analysis (Schaller et al., 2008; Pils and Heyl, 2009), and they are not transcriptionally regulated by cytokinin. Two-component signaling elements are involved in several signaling pathways in plants, most notably cytokinin signaling (To and Kieber, 2008). The initial steps of cytokinin signaling are mediated by a multistep phosphorelay involving cytokinin receptors, phosphotransfer proteins, and type-B ARRs (Hwang and Sheen, 2001; Inoue et al., 2001; Suzuki et al., 2001; Higuchi et al., 2004; Nishimura et al., 2004; Mason et al., 2005; Hutchison et al., 2006; Argyros et al., 2008; Ishida et al., 2008b). These relay the cytokinin signal from membrane to nucleus, where the type-B ARRs regulate gene expression, including transcriptional induction of the type-A response regulator genes (Mason et al., 2005; Argyros et al., 2008; Ishida et al., 2008a). The type-A response regulators mediate downstream responses to cytokinin and act as negative regulators of the initial signal transduction pathway (To et al., 2004; Hirose et al., 2007; Cheng et al., 2010).
In addition to the two-component signaling elements that have all the conserved residues required for activity, plants also contain diverged two-component elements that lack residues essential for phosphotransfer function. Phytochromes and a subset of the ethylene receptor family are diverged His kinases (Moussatche and Klee, 2004; Rockwell et al., 2006; Schaller et al., 2008). A diverged phosphotransfer protein, lacking the conserved His for phosphorylation, acts as a negative regulator of cytokinin signaling in Arabidopsis (Mähönen et al., 2006b), and multiple genes encoding such pseudo-HPt proteins are found in rice (Hutchison and Kieber, 2007). Diverged response regulators, referred to as pseudo-response regulators, lack the conserved Asp for phosphorylation, but in many cases the Asp residue is replaced by a Glu that may mimic the phosphorylated form (Makino et al., 2000). The best characterized of the pseudo-response regulators contain a distinctive, plant-specific CCT motif in their C-terminal extensions and function in the regulation of circadian rhythms (Más, 2008; McClung, 2010).
Only limited characterization has been performed on the rice two-component signaling elements compared with those of Arabidopsis (Jain et al., 2006; Pareek et al., 2006; Du et al., 2007; Hirose et al., 2007; Jain et al., 2008; Karan et al., 2009; Pils and Heyl, 2009; Cheng et al., 2010). Here, we characterize the rice signaling elements based on several complementary approaches, with a particular focus on their role in cytokinin signaling. We describe the subcellular localization of a subset of these elements, the ability of the rice type-B response regulators to functionally substitute for Arabidopsis type-B response regulators, and the transcriptional responsiveness to cytokinin and other phytohormones of these genes and other genes involved in cytokinin function.
RESULTS
Phylogenetic Relationship of Two-Component Signaling Elements from Rice and Arabidopsis
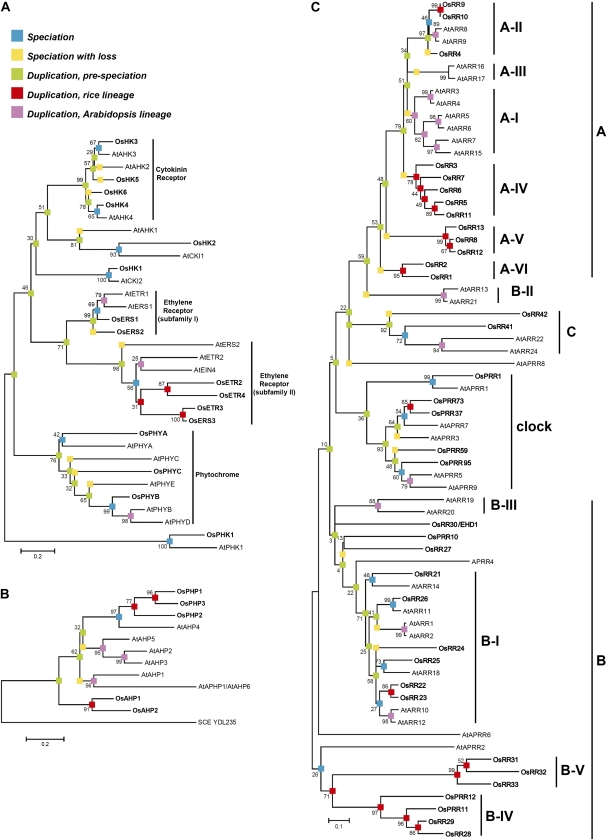
We performed a phylogenetic analysis to elucidate the conserved and unique features of the two-component signaling systems in the monocot rice as compared with the dicot Arabidopsis (Fig. 1). For this purpose, we used the complete repertoire of currently known genes with characteristic protein domains found in two-component genes, including the diverged ones that lack essential residues required for activity (e.g. pseudo-response regulators; Schaller et al., 2007, 2008; Supplemental Table S1). Based on this analysis, Arabidopsis and rice have a similar repertoire of His kinases (Fig. 1A), which would function as receptors for signal input into the two-component signaling system, although we were unable to identify a rice ortholog of AtAHK1, suggesting a potential gene loss in the rice lineage. Arabidopsis contains five genes encoding His-containing phosphotransfer proteins, which contain the conserved His that serves as the site of phosphorylation (AtAHP1–AtAHP5; Schaller et al., 2008), whereas rice contains two (OsAHP1 and OsAHP2; Fig. 1B). Both Arabidopsis and rice contain representatives for type-A, type-B, and type-C response regulators (Imamura et al., 1999). No additional families of response regulators were found unique to rice or Arabidopsis. These results confirm and expand on previous analyses (Pareek et al., 2006; Pils and Heyl, 2009; Mochida et al., 2010), with minor differences likely due to our analyses being performed on a larger complement of signaling elements and/or being based on the conserved domains rather than the entire sequence.
Figure 1.
Phylogenetic relationship of two-component signaling elements from rice and Arabidopsis. Alignments were based on His kinase domains (A), HPt domains (B), or receiver domains (C). Sequences from rice are in boldface and designated by the prefix Os; sequences from Arabidopsis are designated with the prefix At. Bootstrap supports for individual branches are given as percentages based on 1,000 bootstrap trials. For analysis of the HPt proteins, the yeast HPt protein YDL235C was included as an outgroup. Colored boxes indicate points of speciation and gene duplication. The nomenclature of rice elements is based on Schaller et al. (2007), and that of Arabidopsis elements on Schaller et al. (2008).
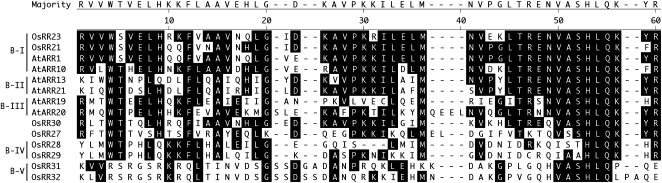
Although all three types of response regulators are present in rice and Arabidopsis, substantial lineage-specific expansions of the type-A and type-B response regulators have occurred, such that there are few 1:1 orthologous relationships, suggesting a higher rate of duplicate retention compared with that of His kinases (Fig. 1C). Potentially as a result of lineage-specific expansion, there is little interdigitation in the phylogenies of the monocot and dicot type-A response regulators, indicative of differential gene losses in lineages leading to these two species. For example, in subfamily C, OsRR41 is in a fairly well-supported clade as AtARR22 and AtARR24. Meanwhile, OsRR42 is sister to the clade with both rice and Arabidopsis genes, which suggests the loss of an Arabidopsis response regulator after rice and Arabidopsis diverged. There is only one distinct subfamily of type-A response regulators that contains members from both rice and Arabidopsis (subfamily A-II). The type-A ARRs in subfamilies A-I and A-II have been found to negatively regulate cytokinin signaling in Arabidopsis (To et al., 2004). Evidence for lineage-specific expansion can also be seen with the type-B response regulators of rice and Arabidopsis (a polyphyletic group with clades that do not share a most recent common ancestor), of which only subfamily B-I contains members from both species (Fig. 1C). Members of subfamily B-I have been shown in Arabidopsis to participate as positive regulators in cytokinin signaling (Hwang and Sheen, 2001; Sakai et al., 2001; Mason et al., 2005). The potential for the regulation of different gene sets by the type-B response regulators is emphasized by the divergence within their Myb domains (Fig. 2; Sakai et al., 2000; Hosoda et al., 2002).
Figure 2.
Amino acid alignment of the Myb domains from type-B response regulators. Representative sequences from various subfamilies of rice and Arabidopsis type-B response regulators were aligned by ClustalW. Residues identical to that of the consensus are boxed in black.
Subcellular Localization of Rice Phosphorelay Proteins
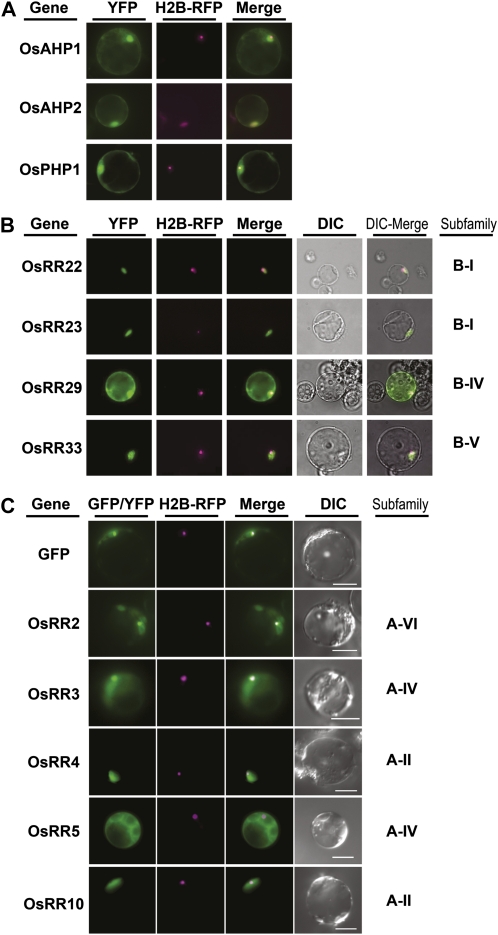
Recent evidence from monocots and dicots supports the localization of cytokinin receptors to the endoplasmic reticulum (Caesar et al., 2011; Lomin et al., 2011; Wulfetange et al., 2011); however, information on the subcellular localization of downstream two-component elements from monocots is lacking. To determine the subcellular localization of rice two-component proteins, we transiently transformed rice protoplasts with plasmids designed to express yellow fluorescent protein (YFP) or GFP fusions to representative OsHPs, type-B OsRRs, and type-A OsRRs, emphasizing members from rice subfamilies most likely to participate in cytokinin signaling based on their relationship to functionally characterized elements of Arabidopsis (Fig. 1). The reporter plasmids were cotransformed with a plasmid encoding a Histone-2B fusion to red fluorescent protein, which localizes to the nucleolus. The two phosphotransfer protein (OsAHP1 and OsAHP2) fusions localized to the nucleus and the cytosol (Fig. 3A). The same localization pattern was seen with the pseudo-phosphotransfer protein OsPHP1 (Fig. 3A), suggesting that the phosphate-receiving His is not required to establish the localization pattern. Treatment with the cytokinin zeatin did not result in any apparent change in the localization pattern of the phosphotransfer proteins (data not shown), consistent with recent data from Arabidopsis (Punwani et al., 2010; Punwani and Kieber, 2010).
Figure 3.
Subcellular distribution of two-component signaling elements in rice protoplasts. Representative images of fluorescence signals are shown from rice protoplasts transfected with plasmids encoding YFP/GFP fusions to phosphotransfer proteins (A), type-B response regulators (B), and type-A response regulators (C). Protoplasts were cotransfected with a plasmid encoding a Histone-2B fusion to red fluorescent protein (H2B-RFP) for identification of the nucleus. DIC, Differential interference contrast. Bars = 10 μm.
The rice type-B response regulators showed variability in their subcellular localization patterns. The subfamily-I response regulators OsRR22 and OsRR23 as well as the subfamily-V response regulator OsRR33 exhibited tight nuclear localization, with no fluorescence in other parts of the cell (Fig. 3B). This nuclear localization pattern matches that of the type-B response regulators of Arabidopsis (Sakai et al., 2000; Hwang and Sheen, 2001; Imamura et al., 2001; Lohrmann et al., 2001; Hosoda et al., 2002; Mason et al., 2004; Dortay et al., 2008). However, the subfamily-IV response regulator OsRR29 localized to both the nucleus and the cytosol (Fig. 3B), a localization pattern not previously observed with subfamily-I, -II, or -III type-B response regulators of Arabidopsis.
The rice type-A response regulators also showed variability in their subcellular localization (Fig. 3C). Two subfamily-II type-A proteins (OsRR4 and OsRR10) both exhibited a tight nuclear localization pattern (Fig. 3C). However, two type-A subfamily-IV proteins (OsRR3 and OsRR5) were present in both the nucleus and the cytosol (Fig. 3C). Likewise, a type-VI protein (OsRR2) was also present in both the nucleus and the cytosol. The intracellular localization of the type-A OsRR proteins was not affected by treatment with exogenous cytokinin (data not shown). The type-A response regulators of Arabidopsis also show variability in their subcellular localization, with some showing only nuclear localization and some being present in both the nucleus and the cytosol (Hwang and Sheen, 2001; Hwang et al., 2002; Kiba et al., 2002; Dortay et al., 2008).
Functional Complementation of an Arabidopsis Type-B Response Regulator Mutant
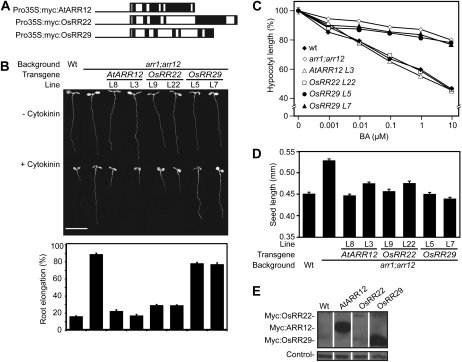
To test the conservation of function between the monocot and dicot type-B response regulators, genomic clones of OsRR22 and OsRR29 driven by the cauliflower mosaic virus (CaMV) 35S promoter were stably transformed into the Arabidopsis arr1-3;arr12-1 double mutant background (Fig. 4). The Arabidopsis arr1-3; arr12-1 double mutant line exhibits decreased sensitivity to cytokinin due to loss-of-function mutations in two of its subfamily-I type-B response regulators (Mason et al., 2005). OsRR22 was chosen for characterization because, like AtARR1 and AtARR12, it is a member of the subfamily-I type-B response regulators (Fig. 1). OsRR29 was chosen for characterization because it is a member the monocot-specific subfamily IV (Fig. 1) and exhibits substantial differences from the subfamily-I type-B response regulators in its Myb-like DNA-binding motif (Fig. 2). As a positive control, we used AtARR12, which should complement the arr12-1 loss-of-function mutation and restore cytokinin sensitivity to arr1-3;arr12-1. The constructs also encoded a Myc epitope tag fused to the N terminus of the response regulator to allow for comparison of protein levels.
Figure 4.
Functional complementation analysis of the Arabidopsis arr1;arr12 mutant with OsRR22 and OsRR29. A, Schematic of the constructs used in this study. Line, CaMV 35S promoter; light gray boxes, Myc sequence; black boxes, exons; white boxes, introns. B, Root growth inhibition by 1 μm BA. The top panel shows representative 7-d-old seedlings grown in the presence or absence of 1 μm BA. Bar = 1 cm. The bottom panel shows the root lengths of seedlings grown on medium containing 1 μm BA expressed as a percentage of the lengths of siblings grown on dimethyl sulfoxide control medium. Error bars represent se. C, Hypocotyl elongation response to BA of 4-d-old dark-grown seedlings. The hypocotyl length of each line receiving cytokinin treatment is expressed as a percentage of its dimethyl sulfoxide control. Error bars indicate se (n ≥ 8). D, Mean length of seeds from the wild type (Wt), arr1-3;arr12-1, and transgenic lines. Error bars indicate se (n = 80). E, Protein levels of selected response regulator transgenes based on immunoblot analysis using the Myc epitope tag. Hsp70 protein was immunologically detected as a loading control.
The wild type, the arr1-3;arr12-1 mutant, and the transgenic arr1-3;arr12-1 lines containing AtARR12, OsRR22, and OsRR29 were assayed for the effect of cytokinin on root and hypocotyl elongation as well as their effect on seed size, as this is affected in cytokinin signaling mutants. Root elongation of wild-type seedlings is strongly inhibited by cytokinin (1 μm 6-benzyladenine [BA]), but the roots of the arr1-3;arr12-1 mutant are largely insensitive (Fig. 4B). As predicted, transgenic expression of AtARR12 restored cytokinin sensitivity of arr1-3;arr12-1 to wild-type levels. Transgenic expression of the subfamily-I member OsRR22 also restored cytokinin sensitivity of arr1-3;arr12-1 to wild-type levels, indicating that OsRR22 can functionally complement the missing subfamily-I members of Arabidopsis. In contrast, the subfamily-IV member OsRR29 was unable to complement the arr1-3;arr12-1 mutant (Fig. 4B). We observed a similar effect of the transgenes when we examined their ability to rescue the cytokinin insensitivity of arr1-3;arr12-1 in a hypocotyl elongation assay. Hypocotyl elongation of wild-type seedlings grown in the dark is inhibited by BA, the arr1-3;arr12-1 mutant being largely insensitive (Fig. 4C; Argyros et al., 2008). AtARR12 and OsRR22 rescued the mutant phenotype, but OsRR29 did not. The cytokinin-insensitive mutant arr1-3;arr12-1 also has larger seeds than the wild type (Argyros et al., 2008); interestingly, we found that all three transgenes (AtARR12, OsRR22, and OsRR29) could rescue the mutant seed phenotype (Fig. 4D). Immunoblot analysis confirmed protein expression from all three transgenes (Fig. 4E). OsRR29 exhibited the highest protein level, indicating that its inability to complement arr1-3;arr12-1 in the root and hypocotyl elongation assays was not due to a problem with expression or degradation. The ability of OsRR29 to rescue the seed size phenotype suggests that the gene is functional but within a more limited developmental context.
Expression Analysis of Genes Involved in Cytokinin Function from Rice
To address the regulation of cytokinin function in monocots, we examined the expression of genes involved in cytokinin biosynthesis, degradation, and signaling in roots and shoots of rice and in response to treatment with cytokinin and other phytohormones. To this end, we used the NanoString nCounter system, which is a relatively high-throughput and extremely sensitive method to quantify RNA transcript levels (Geiss et al., 2008; Malkov et al., 2009). The NanoString system allows for the assessment of transcript levels with similar sensitivity to what is found with quantitative reverse transcription (qRT)-PCR, but with the added benefit that many genes can be analyzed in tandem. Furthermore, NanoString analysis does not require enzymatic reactions (e.g. RT) or amplification of targets and thus eliminates the potential biases introduced by these steps. We generated probes for rice two-component signaling elements, representing four cytokinin OsHK receptors, two OsAHPs (authentic HPts), three OsPHPs (pseudo-HPts), 13 type-B OsRRs, and 10 type-A OsRRs (the high sequence similarity of OsRR9 to OsRR10, and of OsRR13 to OsRR8 and OsRR12, precluded the generation of probes specific to these genes; Table I; Supplemental Table S2). Six non-cytokinin-regulated genes were included as controls in the probe set.
Table I. Contents of the NanoString nCounter code set list.
| Family | No. of Genes |
| OsIPT | 10 |
| CYP735A | 2 |
| LOG/LOGL | 11 |
| OsCKX | 11 |
| OsAHP/OsPHP | 5 |
| OsHK | 5 |
| Type-A OsRR | 10 |
| Type-B OsRR | 13 |
| Control | 6 |
Within the same NanoString probe set, we also included probes for genes involved in cytokinin biosynthesis and metabolism (Table I; Supplemental Table S2). Probes for genes involved in cytokinin biosynthesis were targeted against 10 isopentenyltransferases (OsIPTs), 11 LONELY GUY/LOG LIKE phosphoribohydrolases (LOG/LOGLs), and two cytochrome P450 cytokinin hydroxylases (CYP735As). Of the 10 IPTs encoded in the rice genome, eight (OsIPT1–OsIPT8) are likely to function in cytokinin biosynthesis (Sakamoto et al., 2006), with the remaining two (OsIPT9 and OsIPT10) likely acting as tRNA IPTs. The two CYP735A orthologs, CYP735A3 and CYP735A4, catalyze the hydroxylation of the side chain of isopentenyladenine to make trans-zeatin (Takei et al., 2004). The LOG family of phosphoribohydrolases converts inactive cytokinin nucleotides to the active, free-base forms (Kurakawa et al., 2007; Kuroha et al., 2009). Probes were also targeted against 11 OsCKX cytokinin oxidases, which degrade cytokinin.
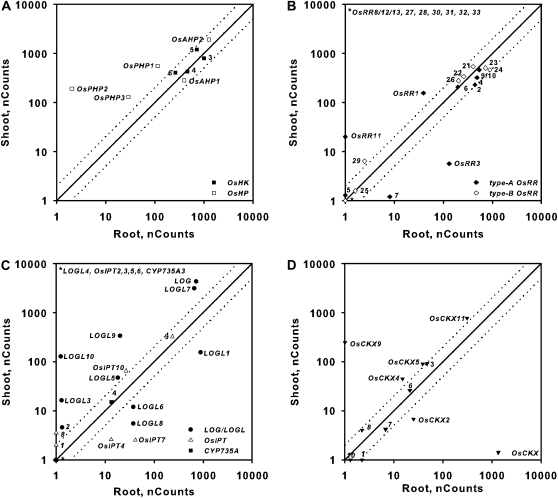
To establish the basal levels of expression for these genes, 12-d-old rice seedlings were grown hydroponically, total RNA was prepared from isolated roots and shoots, and transcript levels were analyzed using the NanoString nCounter system. The data were normalized to positive and negative controls and to the six control genes (see “Materials and Methods”). The six control genes (Supplemental Fig. S1) showed similar expression levels in tissues from cytokinin- and vehicle control-treated plants (Supplemental Fig. S1). The OsHK and OsHP genes were all fairly highly expressed in both roots and shoots (Fig. 5). In contrast, there was a wide variation in the relative expression of individual genes within the OsRR, LOG, OsIPT, and OsCKX gene families in both roots and shoots (Fig. 5). Of the 66 genes involved in cytokinin function that were analyzed, 42 were present above background levels (less than 10 counts) in roots and/or shoots, including five of 13 type-B OsRRs, seven of 10 type-A OsRRs, four of 10 OsIPTs, nine of 12 LOGLs, 7 of 11 OsCKXs, and all OsHK and OsHP genes. (Supplemental Tables S3 and S4). Most of the type-B OsRRs were expressed at very low levels; indeed, only five of 13 type-B OsRRs (OsRR21, OsRR22, OsRR23, OsRR24, and OsRR26) were expressed at clearly detectable levels in roots and shoots. Although several of the type-A OsRRs were expressed below background levels, the cytokinin induction of the type-A OsRRs allowed for the detection of most family members, as described in the next section. Of the eight IPTs implicated in cytokinin biosynthesis, only OsIPT4 and OsIPT7 were present substantially above background and only in roots.
Figure 5.
Expression of genes involved in cytokinin signaling or metabolism in roots and shoots of rice. Comparison plots show the expression in roots versus shoots of genes encoding cytokinin receptors (OsHKs) and HPts (OsHPs; A), response regulators (OsRRs; B), cytokinin biosynthetic enzymes (LOG/LOGL, OsIPT, and CYP35A; C), and cytokinin oxidases (OsCKX; D). Expression from 12-d-old hydroponically grown rice seedlings was quantified using the NanoString nCounter system (see “Materials and Methods”). The normalized counts for each transcript in the roots or shoots as indicated on each axis are shown; note that the counts are plotted on a log10 scale. Dotted lines represent variation greater than 2-fold in expression levels between roots and shoots. The symbols for the different classes of genes are shown at the bottom right of each graph, and the gene number (or whole gene name) for each gene within each class is indicated next to its symbol. Note that one of the probe sets targets two type-A response regulators (OsRR9 and OsRR10) and a second probe set targets three closely related type-A response regulators (OsRR8, OsRR12, and OsRR13). The counts represent means of three biological replicates.
Many genes displayed tissue-specific expression and/or had greater than 2-fold differences in expression in shoots or roots (Fig. 5), although no clear pattern emerged linking either tissue to a particular cytokinin function. Putative elements of the primary cytokinin signal transduction pathway (OsHKs, OsAHPs, and type-B OsRRs) tended to be similarly expressed in both root and shoot. Some type-A OsRRs showed different expression patterns, most notably OsRR11, which was essentially shoot specific. All three OsPHPs, which function as negative regulators of cytokinin signaling in Arabidopsis, were more highly expressed in the shoot than in the root. Many of the genes involved in cytokinin biosynthesis or degradation showed differing expression. For example, the transcripts of five of the LOG/LOGL genes (LOG, LOGL5, LOGL7, LOGL9, and LOGL10) were more abundant in shoots relative to roots, and the transcripts of the remaining LOGLs (LOGL1, LOGL6, and LOGL8) had higher expression in roots (Fig. 5). In addition, the expression of OsCKX9 was essentially shoot specific.
Response of Genes to Exogenous Cytokinin
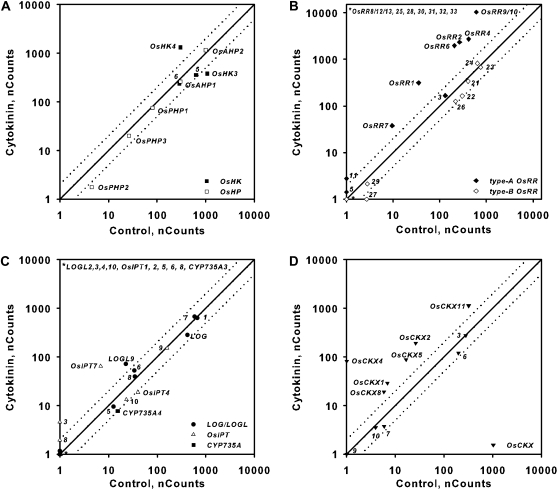
To examine the effect of cytokinin on the expression of genes controlling its signal transduction, synthesis, and degradation, rice seedlings were treated with 5 μm BA for 30, 60, and 120 min. The four putative cytokinin receptors in rice were previously reported to not be cytokinin responsive, based on studies using the Affymetrix GeneChip rice genome array (Hirose et al., 2007). However, the NanoString analysis demonstrates that OsHK4 is elevated in response to cytokinin in roots (Fig. 6A), and OsHK6 is elevated in shoots (Fig. 7A). Interestingly, AtAHK4, which is likely orthologous to OsHK4 (Fig. 1A), is also up-regulated in response to cytokinin in Arabidopsis (Rashotte et al., 2003). The rice genome encodes two authentic His-containing phosphotransfer proteins (OsAHPs) and three pseudo-phosphotransfer proteins lacking the conserved His (OsPHPs; Hutchison and Kieber, 2007; Fig. 1). While OsAHP levels were not appreciably altered by cytokinin, the OsPHP transcripts were modestly down-regulated in shoots 2 h after BA treatment (Fig. 7). The expression of the type-B OsRR genes was not altered by cytokinin treatment (Figs. 6 and 7), which is consistent with what has been observed in Arabidopsis and what has been reported previously in rice (Hirose et al., 2007).
Figure 6.
Expression of genes involved in cytokinin signaling or metabolism in response to exogenous cytokinin treatment in roots. Comparison plots show expression in roots of cytokinin receptors and HPts (A), response regulators (B), cytokinin biosynthetic enzymes (C), and cytokinin oxidases (D). Expression from 12-d-old hydroponically grown rice seedlings was quantified using the NanoString nCounter system (see “Materials and Methods”). The normalized counts for each transcript from tissue derived from seedlings treated for 2 h with 5 μm BA or a vehicle control as indicated on each axis are shown. Dotted lines represent variation greater than 2-fold in expression levels between control and BA-treated samples. The counts represent means of three biological replicates.
Figure 7.
Expression of genes involved in cytokinin signaling or metabolism in response to exogenous cytokinin treatment in shoots. Comparison plots show expression in shoots of cytokinin receptors and HPts (A), response regulators (B), cytokinin biosynthetic enzymes (C), and cytokinin oxidases (D). Expression from 12-d-old hydroponically grown rice seedlings was quantified using the NanoString nCounter system (see “Materials and Methods”). The normalized counts for each transcript from tissue derived from seedlings treated for 2 h with 5 μm BA or a vehicle control as indicated on each axis are shown. Dotted lines represent variation greater than 2-fold in expression levels between control and BA-treated samples. The counts represent means of three biological replicates.
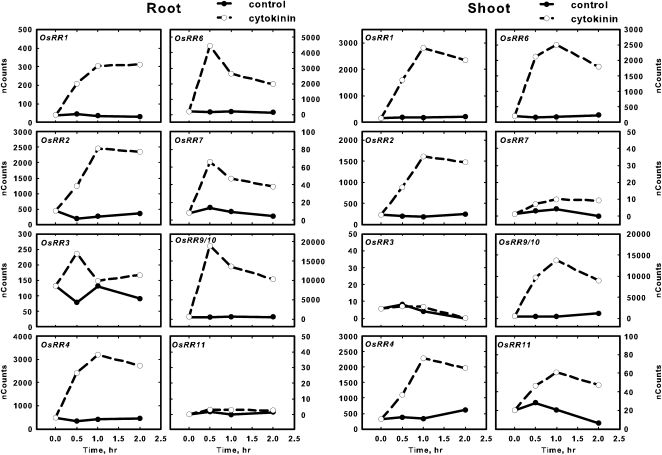
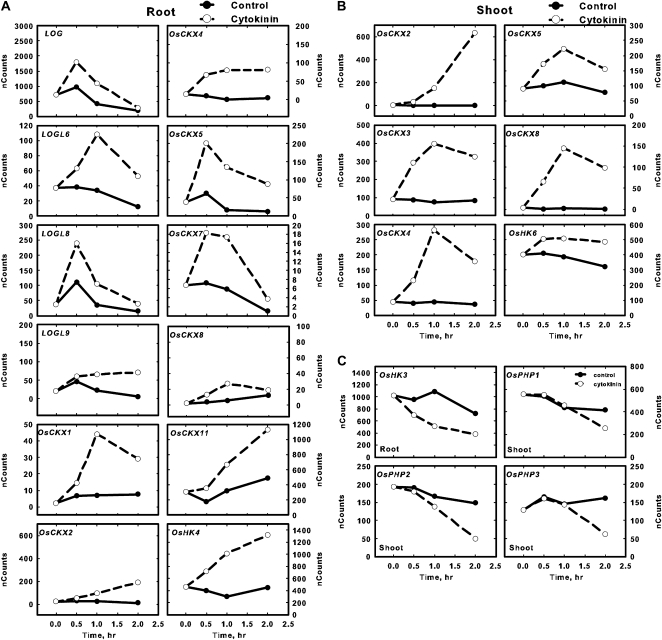
The type-A response regulators have been shown to be rapidly and specifically induced by cytokinin treatment in monocots and dicots (Brandstatter and Kieber, 1998; D’Agostino et al., 2000; Jain et al., 2006; Hirose et al., 2007). Consistent with this, most of the 10 type-A OsRR genes were up-regulated 2 h after BA treatment in the roots and shoots (Figs. 6B and 7B), which is consistent with a previous report (Jain et al., 2006). In contrast, only three of the type-A OsRR genes were determined to be up-regulated in response to cytokinin from an analysis using the Affymetrix GeneChip rice genome array (Hirose et al., 2007). The only type-A OsRRs that could not be detected in either roots or shoots after BA treatment were OsRR8/12/13 (the probe used could not distinguish these three genes; Figs. 6B and 7B), which is consistent with publicly available expression profiling of rice that indicates that these three type-A OsRR genes are expressed mostly in the axillary and vegetative shoot apical meristems (http://bioinformatics.med.yale.edu/riceatlas/overview.jspx). OsRR5 had a very low basal expression in both roots and shoots and was only robustly detectable in shoots treated for 120 min with cytokinin (Figs. 6B and 7B). The induction of the type-A OsRRs by cytokinin generally peaked at 30 to 60 min and then declined by 2 h in both roots and shoots (Fig. 8), which is similar to the kinetics observed in Arabidopsis (D’Agostino et al., 2000).
Figure 8.
Induction kinetics of type-A OsRR gene expression in response to cytokinin treatment. Twelve-day-old hydroponically grown rice seedlings were treated with cytokinin for various times by addition of 5 μm BA to the hydroponic medium. Total RNA was isolated, and the transcript levels for the various type-A OsRR genes in the roots and shoots as indicated were quantified using NanoString nCounter analysis. The normalized counts for each gene are plotted on the y axis. Solid and dotted lines represent control and BA treatment, respectively. Note that the 120-min time points show the same data shown in Figures 6 and 7. Genes with counts greater than 11 at all points in roots and shoots are not shown. The default scale was set at 50; for genes with higher expression, plots were scaled appropriately. Values are means derived from three biological replicates.
Cytokinin-dependent changes were also observed in genes involved in cytokinin biosynthesis and degradation. Transcripts for the two detectable IPTs involved in cytokinin biosynthesis both demonstrate a cytokinin response: OsIPT7 transcript levels were elevated and OsIPT4 levels were slightly reduced in the roots 2 h after cytokinin treatment (Fig. 6C). Only one CYP735A family member (CYP735A4) was detectable, and its expression is repressed by cytokinin, most notably in the shoot (Fig. 7C). Several of the LOG/LOGL genes are induced by cytokinin in roots (Fig. 6C), especially at 30 and 60 min after cytokinin treatment (Fig. 9A). The expression of genes encoding cytokinin oxidases is particularly sensitive to cytokinin. Multiple OsCKX genes are induced in response to cytokinin in roots and/or in shoots, and one (OsCKX6) is down-regulated specifically in the shoot (Figs. 6D and 7D). In some cases, this induction is rapid and transient (e.g. OsCKX7, in roots), in others, it is rapid and sustained (e.g. OsCKX4, in roots), and in still others, the transcripts continue to increase throughout the 120 min of cytokinin treatment (e.g. OsCKX2, in shoots; Fig. 9). OsCKX1, OsCKX5, and OsCKX11 were up-regulated by BA only in roots, and OsCKX3 only in shoots. The differing kinetics and tissue specificity of the cytokinin responses of the OsCKX genes suggest that these cytokinin oxidases each have distinct roles in regulating cytokinin levels in rice. The up-regulation of OsCKXs by cytokinin is consistent with the cytokinin-inducible expression of multiple Arabidopsis CKX genes (AtCKX3, AtCKX4, AtCKX5, and AtCKX6) and maize (Zea mays) Ckx1 (Brugière et al., 2003; Hoth et al., 2003; Rashotte et al., 2003; Werner et al., 2006).
Figure 9.
Expression kinetics of changes of various genes regulated by cytokinin. Twelve-day-old hydroponically grown rice seedlings were treated with cytokinin for various times by addition of 5 μm BA to the hydroponic medium. Total RNA was isolated, and the transcript levels for the indicated genes were quantified using NanoString nCounter analysis. The genes depicted represent genes up-regulated by cytokinin in the root (A), up-regulated by cytokinin in the shoot (B), or down-regulated by cytokinin in the root or shoot (C). The normalized counts for each gene are plotted on the y axis. Solid and dotted lines represent control and BA treatment, respectively. Note that the 120-min time points show the same data shown in Figures 6 and 7. Values are means derived from three biological replicates.
To validate the results from the NanoString assay, we performed qRT-PCR with the same RNA samples and analyzed representative high-abundance (i.e. OsRR9/10), medium-abundance (i.e. OsRR1, OsAHP2), and low-abundance (i.e. OsRR7) genes. Consistent with the results from the NanoString analysis, the OsRR1, OsRR7, and OsRR9/10 transcripts were found to be elevated in response to cytokinin by qRT-PCR analysis (Table II). Likewise, similar to what was determined by the NanoString method, the OsAHP2 transcript was not cytokinin responsive when analyzed by qRT-PCR.
Table II. Validation of NanoString nCounter analysis using qRT-PCR.
| NanoString |
||||
| Gene | Control | Cytokinin | Change | qRT-PCR Change |
| nCounts | fold change | |||
| 30-min root | ||||
| OsAHP2 | 1,284 | 1,344 | 1.05 | 1.04 |
| OsRR1 | 45 | 208 | 4.62 | 2.93 |
| OsRR7 | 14 | 66 | 4.64 | 12.43 |
| OsRR9/10 | 487 | 18,848 | 38.70 | 26.89 |
| 2-h root | ||||
| OsAHP2 | 955 | 1,151 | 1.21 | 1.03 |
| OsRR1 | 30 | 311 | 10.32 | 6.32 |
| OsRR7 | 5 | 38 | 8.12 | 16.60 |
| OsRR9/10 | 517 | 10,299 | 19.92 | 8.08 |
Response of Cytokinin Genes to Other Phytohormones
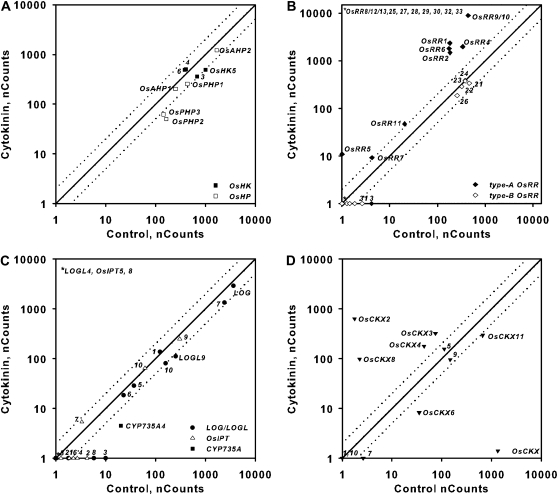
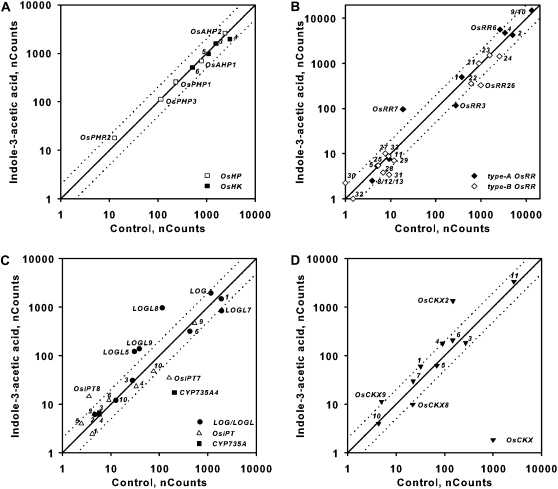
Phytohormones often alter the synthesis, degradation, or perception of each other to regulate growth and development in an integrated manner. To determine if other phytohormones influence cytokinin in rice, we examined the transcript abundance of the genes involved in cytokinin function in response to treatment with auxin, GA, brassinosteroid, and abscisic acid (ABA). Auxin treatment has been shown to induce and repress two type-A ARRs in Arabidopsis roots and shoots, respectively (Müller and Sheen, 2008; Zhao et al., 2010). Treatment of rice with 10 μm of the auxin indole-3-acetic acid (IAA) had little or no effect on the expression of OsHKs, OsAHPs, and type-B OsRRs (Fig. 10; Supplemental Fig. S2). Among the type-A OsRRs, OsRR7 was induced by IAA treatment in both roots and shoots (Fig. 10B; Supplemental Fig. S2). In the roots, in addition to the induction of OsRR7, the OsRR6 gene was slightly elevated and the OsRR3 gene was slightly repressed by auxin (Fig. 10B). In the shoots, only OsRR7 was elevated, and several type-A OsRRs were slightly decreased by auxin treatment (Supplemental Fig. S2B). IAA treatment caused a reduction of OsIPT7 in roots (opposite to the effect of cytokinin on this gene) and induced OsIPT4 (and perhaps OsIPT7) in shoots. CYP735A4 displayed reduced transcript abundance after IAA treatment, which is consistent with what has been observed in Arabidopsis (Takei et al., 2004). Multiple LOGL genes were up-regulated after IAA treatment in roots and shoots (Fig. 10C; Supplemental Fig. S2C). OsCKX2, a regulator of rice grain production (Ashikari et al., 2005), was up-regulated by IAA treatment in both roots and shoots (Fig. 10D; Supplemental Fig. S2D). In the shoot, in addition to OsCKX2, auxin up-regulated OsCKX1 and OsCKX11 and caused a decrease in the level of OsCKX8 (Supplemental Fig. S2). Auxin may have a more pronounced effect if one examines more discrete tissues, such as meristems. These results suggest that auxin has a modest effect on cytokinin two-component signaling genes, primarily the type-A OsRRs, and that it alters cytokinin levels in a complex manner.
Figure 10.
Effect of auxin treatment on the expression of genes involved in cytokinin function in rice roots. Comparison plots show expression in roots of genes encoding cytokinin receptors and HPts (A), response regulators (B), cytokinin biosynthetic enzymes (C), and cytokinin oxidases (D). Twelve-day-old hydroponically grown rice seedlings were treated with auxin for 2 h by addition of 10 μm IAA to the hydroponic medium. Total RNA was isolated from the roots, and the transcript levels for the indicated genes were quantified using NanoString nCounter analysis. The normalized counts for each transcript from roots derived from seedlings treated with 10 μm IAA or a vehicle control as indicated on each axis are shown. Dotted lines represent variation greater than 2-fold in expression levels between control and BA-treated samples. The counts represent means of three biological replicates.
We also examined the expression of genes involved in cytokinin function in roots treated for 2 h with GA, brassinosteroid, and ABA. There was a slight increase in the expression of OsCKX2 and LOGL10, and a slight decrease in the expression of LOGL9 and OsCKX10, in response to GA3, but otherwise GA had little or no effect on the expression of the genes examined (Supplemental Fig. S3). In response to brassinosteroid, the transcript levels of OsIPT2 and OsIPT4 decreased slightly, and those of LOGL2 and LOGL10 increased slightly, but otherwise there was little effect on the expression of these genes (Supplemental Fig. S4). Treatment with ABA for 2 h had a more substantial effect on expression (Supplemental Fig. S5). In the signaling pathway, OsHK3 and OsRR4 (a type-A response regulator) were slightly elevated in response to ABA. The expression of OsIPT2, OsIPT4, and OsIPT7, the CYP735A genes, and several OsCKX genes was down-regulated in response to ABA. Multiple LOGL genes were altered both positively and negatively in response to ABA treatment. These results suggest that in rice roots, brassinosteroid and GA have relatively minor effects on the transcription of genes involved in cytokinin function in these conditions; ABA has a modest effect on the level of transcripts of genes encoding cytokinin signaling elements but a substantial effect on genes encoding proteins involved in cytokinin synthesis and degradation.
DISCUSSION
Phylogenetic analysis reveals a similar cohort of His kinases and phosphotransfer proteins in rice and Arabidopsis. But, although similar numbers of type-A, type-B, and type-C response regulators exist in both rice and Arabidopsis, substantial lineage-specific expansions of the type-A and type-B response regulators have occurred, such that there are few 1:1 orthologous relationships. This is likely due in part to recent genome duplications and retention, which can account for how the Arabidopsis type-A response regulators are found as five closely related pairs (Vision et al., 2000; To et al., 2004). There is, however, lineage-specific expansion beyond that of the recent genome duplications. The finding of tandem copies of several subfamilies of type-B response regulators on the same chromosome (subfamily B-IV and B-V) is consistent with the expansion of some individual subfamilies of response regulators through repeated unequal crossovers. In addition, the general lack of pairwise relationships among the rice regulators suggests frequent gene loss events in the response regulator family. It remains unclear whether such expansion and frequent contraction is due to random events indicative of neutral evolution or to adaptive evolution that resulted in the initial retention and subsequent loss due to the absence of a selective agent. The expansions found in rice and Arabidopsis result in distinct and sometimes novel subfamilies of the type-A and type-B response regulators, which likely lead to the acquisition of lineage-specific roles in monocots and dicots. Thus, based on phylogenetic analysis, one would predict that rice and Arabidopsis (1) share similar signal inputs (His kinases); (2) transmit these signals to the response regulators by a similar mechanism (phosphotransfer proteins); (3) share some overlapping signal outputs (e.g. subfamily-I of the type-B response regulators); and (4) also have lineage-specific signal outputs (e.g. unique subfamilies of type-B response regulators).
These data suggest a shared mechanism of cytokinin signaling between monocots and dicots, based on the presence of similar cohorts of two-component genes, the similar intracellular localization of two-component signaling elements, the similar induction and induction kinetics of type-A response regulators in response to cytokinin, and the ability of a rice type-B response regulator to complement an Arabidopsis type-B mutant. Consistent with this, overexpression of the type-A response regulators OsRR3, OsRR5, or OsRR6 in rice reduced cytokinin sensitivity (Hirose et al., 2007; Cheng et al., 2010), suggesting that these type-A OsRRs are negative regulators of cytokinin signaling, as is the case in Arabidopsis. However, both phylogenetic as well as functional analyses of a subset of rice type-B OsRRs suggest that some functions likely have diverged in rice and Arabidopsis response regulators. For example, the ability of the subfamily B-IV OsRR29 to rescue a seed size phenotype but not the effects of cytokinin on root and hypocotyl elongation in the subfamily B-I Arabidopsis mutant arr1-3;arr12-1 suggests that the rice gene is functional but within a more limited developmental context. Consistent with this, two response regulator mutants in monocots have phenotypes (Doi et al., 2004; Giulini et al., 2004) not reported for Arabidopsis response regulators.
To explore how cytokinin and other phytohormones affect cytokinin function in rice roots and shoots, we examined the expression of genes encoding two-component proteins implicated in cytokinin signaling as well as genes encoding enzymes implicated in cytokinin biosynthesis and degradation. Of course, as with any expression study focused on transcript levels, it is possible that posttranscriptional controls also modulate the levels of the proteins encoded by these genes in addition to the effects observed on the RNA levels. The rationale for exploring roots and shoots separately is that cytokinin has been determined to exert opposite effects on growth in these tissues, stimulating meristem activity in the shoot but inhibiting meristem activity in the root (Werner et al., 2001). Various studies, primarily in Arabidopsis, have demonstrated that subsets of these signaling, biosynthesis, and degradation genes are regulated by cytokinin itself as well as by other plant hormones, most notably auxin and ABA (Sakakibara, 2005). The effect of cytokinin on the expression of these genes generally leads to a negative feedback on cytokinin responsiveness and levels. This feedback is also observed in rice, as type-A OsRRs (negative regulators of the response pathway) and cytokinin oxidase genes (which degrade cytokinin) are the most prevalent transcripts induced by exogenous cytokinin. Exogenous cytokinin thus induces its own degradation and reduces the sensitivity of the cell to itself. Such negative feedback regulation is a common theme observed in many cell signaling systems in plants and animals. Furthermore, in Arabidopsis, cytokinin treatment represses the expression of at least four IPT genes (Miyawaki et al., 2004), although this does not appear to be the case in rice. The differences in the regulation of IPT by cytokinin could reflect differences in treatment times (2 versus 4 h), or it may reflect differences between rice and Arabidopsis in the circuitry of the feedback regulation. The induction of the OsHK4 receptor by cytokinin, which is similar to the induction of AtAHK4 in Arabidopsis (Rashotte et al., 2003), appears to run counter to the trend of the induction of negative components. However, AtAHK4 has been demonstrated to act both as an HPt kinase and phosphatase, depending on whether cytokinin is bound to the CHASE domain or not, and thus it can act as either a positive element in the presence of cytokinin or it can inhibit cytokinin signaling when not bound to cytokinin (Mähönen et al., 2006b). The induction of OsHK4 (and AtAHK4) in response to cytokinin could serve to promote the rapid shutoff of the signaling pathway upon cytokinin removal.
The pseudo-Hpt family is larger in rice as compared with Arabidopsis (three versus one genes). The Arabidopsis AtPHP1/AtAHP6 pseudo-Hpt acts as a negative regulator of cytokinin responsiveness, and its expression is down-regulated in response to cytokinin (Mähönen et al., 2006a). Likewise, in rice, the OsPHPs are down-regulated by exogenous cytokinin. As the OsPHPs are likely negative regulators as well, this down-regulation of these pseudo-HPts may reflect a positive feedback loop in cytokinin signaling.
In other systems, auxin has been shown to affect both cytokinin biosynthesis and responsiveness. In terms of cytokinin signaling, auxin regulates two type-A AtARRs in Arabidopsis: in the root apical meristem, auxin induces AtARR7 and AtARR15 expression, and in the shoot, auxin represses the expression of these genes (Müller and Sheen, 2008; Zhao et al., 2010). In rice, the OsRR7 type-A response regulator is induced by auxin in both roots and shoots, and there are modest effects of auxin on the expression of a few other type-A OsRRs. It may be that one needs to examine specific rice tissues, such as the apical meristem, in order to see the opposing effects of auxin on type-A gene expression observed in Arabidopsis. Alternatively, as the type-A response regulator gene families have substantially diverged in rice and Arabidopsis, and there is no clear ortholog of the AtARR7/AtARR15 pair in rice, the transcriptional regulation of the members of this gene family may have diverged.
Using an in vivo labeling/mass spectrometry approach, it was found that auxin caused a rapid (approximately 6 h) down-regulation of zeatin biosynthesis in Arabidopsis (Nordström et al., 2004). Consistent with this, auxin represses IPT expression in lateral buds of pea (Pisum sativum; Tanaka et al., 2006), although in Arabidopsis, auxin treatment resulted in the up-regulation of two IPT genes (Miyawaki et al., 2004). The transcripts of most of the OsIPTs involved in cytokinin synthesis are present at low abundance in the tissues examined here, which is similar to the expression levels of IPT genes in Arabidopsis. Auxin appears to affect a few members of the OsIPT gene family in rice, up-regulating at least one in the root (OsIPT8) and decreasing another (OsIPT7), but in general, the level of expression of most of these genes was too low to detect robustly. The expression of CYP735A genes is down-regulated by auxin in roots in Arabidopsis (Takei et al., 2004), and a similar repression of CYP735A gene expression by auxin is seen in rice. Auxin had a substantial effect on the expression of multiple LOG/LOGL genes in rice, mostly increasing the level of transcripts. Thus, in general, auxin appears to have a complex effect on the expression of genes involved in cytokinin biosynthesis in rice: up-regulating several LOG/LOGL genes, down-regulating CYP735A genes, and differentially affecting OsIPT gene expression. Auxin also has a complex effect on the genes encoding cytokinin-degrading enzymes, up-regulating several OsCKX genes in the shoot (and one in the root), which may be analogous to the up-regulation of cytokinin oxidase genes in the axillary bud of pea (Tanaka et al., 2006; Shimizu-Sato et al., 2009), although it also decreases the expression of OsCKX8 in the shoot.
ABA has been shown to affect various aspects of cytokinin function in other systems. In Arabidopsis, ABA treatment down-regulates multiple IPT, CKX, and CYP735A genes (Takei et al., 2004; Nishiyama et al., 2011). In maize leaf discs, multiple CKX genes are up-regulated by ABA (Brugière et al., 2003), although the effect was examined more than 16 h after ABA application. Here, we find that multiple OsCKX genes are down-regulated within 2 h of ABA treatment, consistent with the Arabidopsis results. The difference in maize could be the result of the extended time of ABA treatment. We also find that CYP735A and two OsIPT genes are down-regulated, as observed in Arabidopsis. In addition, we find a complex effect of ABA on LOG/LOGL gene expression, with some genes being repressed and some being elevated in response to ABA. These results suggest that ABA has a substantial effect on cytokinin levels in the plant, although only a few (and perhaps only one significantly) type-A response regulator is altered. It may be that one needs to examine more discrete tissue types, or longer ABA treatments, to discern a more dramatic effect on cytokinin responses in the plant.
Several other studies have examined the expression of two-component genes in rice. Hirose et al. (2007) examined the rice transcriptome in response to exogenous cytokinin and in OsRR6-overexpressing lines using microarrays. Many of the low-abundance genes detected here by NanoString were not detectable by microarray analysis, reflecting the high sensitivity of the NanoString assay. For example, of the 11 cytokinin oxidase genes, only OsCKX11 expression was reliably detected using Affymetrix GeneChip arrays (Hirose et al., 2007), as compared with the eight OsCKX genes robustly detected here using NanoString. In any case, consistent with the results presented here, the microarray analysis indicated that OsCKX11 was induced by cytokinin (Hirose et al., 2007). Furthermore, the microarray analysis indicated that cytokinin had no effect on the expression of the OsHPs or the type-B OsRRs, consistent with the NanoString analysis, although the microarray study did not detect the induction of OsHK4 (Hirose et al., 2007), as was found here. Several studies have examined the expression of the type-A OsRRs in response to cytokinin (Ito and Kurata, 2006; Jain et al., 2006; Du et al., 2007; Hirose et al., 2007), and consistent with this NanoString analysis, most found that the majority are up-regulated by exogenous cytokinin, although some differences can be found. For example, only three of the type-A OsRR genes were determined to be up-regulated in response to cytokinin using microarrays (Hirose et al., 2007). OsRR3 and OsRR8/12/13 were found to be not affected by cytokinin using a qRT-PCR approach (Jain et al., 2006), although in a second study both of these genes were reported to be up-regulated (Du et al., 2007). We found that OsRR8/12/13 was expressed below the detection limit of NanoString, and OsRR3 was induced by cytokinin specifically in the roots, and only transiently, 30 min after cytokinin application. Jain et al. (2006) also examined the expression of type-A OsRRs using a qRT-PCR approach. They found no significant effects of IAA, GA3, 1-aminocyclopropane-1-carboxylic acid, ABA, or brassinolide on type-A OsRR expression. The discrepancies among these studies could reflect differences in growth conditions, tissues examined, or the details of the hormonal treatments (concentration, duration, or species used). Alternatively, they could reflect variations due to the variability of the qPCR assay resulting from the required enzymatic manipulation and amplification of the samples, which are not necessary for the NanoString approach.
In conclusion, we have shown spatial and temporal control of rice cytokinin signaling gene expression in response to different phytohormones by using NanoString analysis. The localization of rice two-component elements reveals similar and unique characteristics compared with dicot plants. Additional studies will be required to dissect the regulation mechanism of these genes and their roles in the phytohormone signaling network.
MATERIALS AND METHODS
Phylogenetic Analysis
Several complementary approaches were taken to identify the cohort of rice (Oryza sativa) two-component signaling elements, the results from this analysis being published previously (Schaller et al., 2007). First, candidate rice sequences were identified by performing BLAST searches (Altschul et al., 1997) with the known complement of Arabidopsis sequences (Schaller et al., 2008). Second, hidden Markov models and alignment seed sequences for the His kinase catalytic domain (HATPase_c) and acceptor domain (HisKA), the His phosphotransfer domain (Hpt), and the receiver domain (RR) were obtained from Pfam (Sonnhammer et al., 1998). These hidden Markov models were used to search against the protein sequences of the rice genome using HMMER (Eddy, 1998). The HisKA domain proved the most discriminating for the identification of His kinases, with most sequences recovered containing a HATPase-c domain in addition to the HisKA domain, as would be predicted for an authentic His kinase. Instead of using the expect value for defining cutoff threshold, any entry with a score greater than 1 was regarded as a candidate containing the domain in question. Third, as part of an iterative approach, the rice sequences identified using the above strategies were used in turn for additional BLAST searches against the rice genome. For phylogenetic analysis, the domain protein sequences of rice and Arabidopsis (Arabidopsis thaliana) were aligned using ClustalW (Higgins et al., 1996). The phylogenetic trees were generated with MEGA3.1 (Kumar et al., 2001) with 1,000 bootstrap replicates using the neighbor-joining algorithm (Saitou and Nei, 1987). Gaps and missing data were dealt with by pairwise deletion. Poisson correction distance was used to correct for multiple substitutions.
Plant Material and Growth Conditions
For gene expression analysis, japonica rice (cv Nipponbare) seeds were surface sterilized with 2.5% sodium hypochlorite for 15 min, washed with distilled water, and germinated on Whatman No. 1 filter paper containing distilled water at 37°C for 1 d. The uniformly germinated rice seeds were grown for 11 d in a hydroponic culture system with Kimura B nutrient medium (Ma et al., 2001) and a photoperiod of 14 h of light (30°C)/10 h of dark (25°C) in a growth chamber. Twelve-day-old rice seedlings were treated with 5 μm BA or 0.05 mn NaOH in nutrient medium as a control. Roots and shoots were harvested at 0, 30, 60, or 120 min. For other hormone treatments, rice seedlings were treated with 10 μm IAA, 50 μm ABA, 1 μm brassinolide, 10 μm GA3, or 0.05% ethanol as a control. Three biological replicates were performed in each experiment, except for the IAA, ABA, BR, and GA3, treatments.
The Columbia ecotype was used for all Arabidopsis experiments. Isolation of the arr1-3 and arr12-1 mutant alleles and creation of the arr1-3;arr12-1 double mutant were described previously (Mason et al., 2005). Arabidopsis root elongation was measured as described (Mason et al., 2005) using Arabidopsis seedlings grown on vertical plates containing either BA at the indicated concentration or 0.1% dimethyl sulfoxide as a vehicle control. Hypocotyl and seed length measurements were made as described (Argyros et al., 2008).
Preparation of DNA Constructs
Rice genomic DNA was isolated using the International Rice Research Institute isolation protocol (Zheng et al., 1995). Genomic sequences encoding the two-component signaling elements used for subcellular localization or functional complementation were amplified from genomic DNA using PrimeSTAR HS DNA Polymerase (Takara) according to the manufacturer. Genomic sequences were amplified from within 50 bp of the beginning of the 5′ untranslated region and ending before the stop codon for subcellular localization constructs, and beginning with the start codon and ending with the stop codon for functional complementation constructs. Primers used are given in Supplemental Table S5. The Gateway cloning system (Invitrogen) was used for the preparation of plasmid constructs. For this purpose, amplified genes were cloned into the pCR8-TOPO entry vector (Invitrogen) according to the manufacturer. Inserts for all entry clones were confirmed by sequencing.
For determination of the subcellular localization of OsHPs, type-B OsRR, and type-A OsRRs (OsRR4, OsRR5, and OsRR10), the 35S::GW:YFP:HA expression cassette from the pEarleygate101 vector (Earley et al., 2006) was cloned into the EcoRI site of the pBluescript II KS+ vector (Stratagene). The OsAHP and OsRR genes were cloned into the Gateway site using LR Clonase II according to the manufacturer (Invitrogen) and propagated in TOP10 Escherichia coli (Invitrogen). For determination of the subcellular localization of OsRR2 and OsRR3, the target genes were fused to GFP driven by the 35S promoter in the pUC18 vector. Plasmids were isolated using the PureYield Plasmid Midiprep Kit (Promega) according to the manufacturer, precipitated with 2-propanol, and resuspended in water at a concentration of 1 to 2 μg μL−1 for transformation into protoplasts.
For functional complementation experiments in Arabidopsis, the type-B OsRR sequences were cloned into the pEarleygate203 vector (Earley et al., 2006) using LR Clonase II (Invitrogen) according to the manufacturer. The pEarleygate203 vector results in the fusion of a Myc sequence tag to the insert and drives its expression from the CaMV 35S promoter. Constructs were electroporated into the Agrobacterium tumefaciens GV1301 strain and transformed into the arr1-3;arr12-1 double mutant (Mason et al., 2005) using the floral dip method (Clough and Bent, 1998).
Subcellular Localization in Rice Protoplasts
Japonica rice cv Kitaake (OsHp and type-B OsRRs) or cv Nipponbare (all the type-A OsRRs) was used for analysis of rice protoplasts. Plants were grown on soil under an AcroDome (Acro Plastics) for 11 d on a 10-h-light, 28°C/14-h-dark, 22°C regimen. The AcroDome was covered with an opaque plastic bag after 4 d to block light. Protoplast isolation and transfection were performed using a protocol adapted from published procedures (Bart et al., 2006; Chen et al., 2006; Yoo et al., 2007). Briefly, rice coleoptile and leaf tissue was cut into 1-mm-thick cross-sections and shaken for 5 h at 40 rpm in 1.5% Cellulase RS (Yakult), 0.3% Macerozyme R10 (Yakult), 0.6 m mannitol, 10 mm KCl, 20 mm MES, pH 5.7, 0.05% (w/v) bovine serum albumin, and 5 mm CaCl2. Shaking speed was increased to 100 rpm for 2 min at the end of the digestion to release protoplasts. An equal volume of W5 wash buffer (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES, pH 5.7) was added to the digestion mixture, and protoplasts then were filtered through a 35-μm nylon mesh filter. The mixture was centrifuged 5 min at 300g in a swinging-bucket rotor to pellet protoplasts. Protoplasts were washed in W5 buffer and then resuspended in MMg solution (0.32 m mannitol, 15 mm MgCl2, and 4 mm MES, pH 5.7) to a final concentration of 106 protoplasts mL−1.
For DNA transfection of protoplasts, 10 μg of DNA was mixed with 200 μL of protoplasts, to which was then added 110 μL of polyethylene glycol-Ca2+ solution (40% [w/w] polyethylene glycol-4000, 100 mm CaCl2, and 200 mm mannitol). Samples were incubated 10 min at 22°C and then diluted with 1 mL of W5 buffer. Samples were centrifuged 3 min at 100g in a swinging-bucket rotor to pellet protoplasts, washed twice with W5 buffer, and then resuspended in 1 mL of WI incubation buffer (0.4 m mannitol, 4 mm MES, pH 5.7, and 20 mm KCl). Samples were incubated in the dark at 22°C for 16 h before being visualized with a Zeiss Axioplan 2 microscope using a 40× objective. For cytokinin treatment, 2 μm trans-zeatin was added for various times (45–120 min).
Protein Isolation and Immunoblotting
For protein isolation from transgenic Arabidopsis plants, seedlings were ground in liquid nitrogen, and the powder was resuspended in isolation buffer containing 50 nm Tris-HCl, pH 7.5, 50 mm NaCl, and 0.1% (v/v) Nonidet P-40. Samples were centrifuged at 16,000g for 1.5 min, and the supernatant was retained for further analysis. Protein concentration was determined by use of the bicinchoninic acid reagent (Pierce) according to the manufacturer’s instructions after first adding 0.2 mL of 0.5% (w/v) SDS to the samples, with bovine serum albumin as a standard. Samples were heated above 65°C in gel-loading buffer for 5 min, and SDS-PAGE and immunoblotting were performed as described previously (Gao et al., 2008) using 8% (w/v) polyacrylamide gels (Laemmli, 1970). Myc-tagged proteins were detected with a monoclonal anti-Myc antibody conjugated to horseradish peroxidase (monoclonal 9E-10; Santa Cruz Biotechnology). Hsp70 protein was used as a loading control and detected with a monoclonal anti-Hsc70 antibody (monoclonal N27F34; StressGen) and goat anti-mouse IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology).
RNA Preparation, Quantitative Real-Time PCR, and NanoString nCounter Assay
Total RNA was prepared from roots or shoots using Trizol (Invitrogen) and RNeasy (Qiagen) followed by treatment with TURBO DNase (Ambion) as described by the manufacturer. One microgram of total RNA was reverse transcribed using SuperScript III (Invitrogen) with oligo(dT)12-18 primers. Quantitative real-time PCR was performed using SYBR Premix Ex Taq polymerase (Takara) on a DNA Engine OPTICON 2 system (MJ Research). Primers used for real-time PCR analysis are shown in Supplemental Table S5.
NanoString nCounter analysis was performed by the University of North Carolina Genomics and Bioinformatics Core Facility. Total RNA (160 ng) was directly hybridized with gene-specific color-coded probes, and data collection was carried out in the nCounter Digital Analyzer as described by the manufacturer (NanoString Technologies). The NanoString Codeset (Supplemental Table S2) was designed and synthesized by NanoString Technologies, which including six reference genes: LOC_Os01g22490 (40S ribosomal protein S27a), LOC_Os01g64630 (actin 7), LOC_Os03g08010 (elongation factor Tu), LOC_Os03g13170 (ubiquitin fusion protein), LOC_Os03g50885 (actin 1), and LOC_Os11g06390 (actin 2). In addition, six positive-control and eight negative-control probes were added to each reaction to produce a standard curve for normalization. All the reaction counts were within the linear dynamic range of the standard curve. For each gene analyzed, the average plus 2× sd of the negative controls was subtracted from the raw data and then normalized to the standard curve within each reaction and six reference genes. For comparison plots, all counts below 1 were defined as 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of control genes in roots treated with 5 μm BA for the indicated times as determined by NanoString analysis.
Supplemental Figure S2. Effect of auxin treatment on the expression of cytokinin function genes in the shoot.
Supplemental Figure S3. Effect of GA3 treatment on the expression of cytokinin function genes in the root.
Supplemental Figure S4. Effect of brassinosteroid treatment on the expression of cytokinin function genes in the root.
Supplemental Figure S5. Effect of ABA treatment on the expression of cytokinin function genes in the root.
Supplemental Table S1. Nomenclature for two-component elements of rice.
Supplemental Table S2. Sequences for the code set for the NanoString probes used in this study.
Supplemental Table S3. Normalized nCounts for transcript levels at various times following cytokinin treatment for all the genes in roots analyzed in this study.
Supplemental Table S4. Normalized nCounts for transcript levels at various times following cytokinin treatment for all the genes in shoots analyzed in this study.
Supplemental Table S5. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Young Hu at the University of North Carolina Genomics and Bioinformatics Core Facility for technical help with the NanoString analysis, and Christopher Cahn for assistance in protoplast localization.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby JL, Parkinson JS, Bourret RB. (1996) Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86: 845–848 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ. (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière N, Jiao S, Hantke S, Zinselmeier C, Roessler JA, Niu X, Jones RJ, Habben JE. (2003) Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol 132: 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar K, Thamm AMK, Witthöft J, Elgass K, Huppenberger P, Grefen C, Horak J, Harter K. (2011) Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J Exp Bot 62: 5571–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cheng X, Jiang H, Zhang J, Qian Y, Zhu S, Cheng B. (2010) Overexpression of type-A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet Mol Res 9: 348–359 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. (2008) Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res 7: 3649–3660 [DOI] [PubMed] [Google Scholar]
- Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. (2007) The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89: 697–707 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. (2009) Biological insights from structures of two-component proteins. Annu Rev Microbiol 63: 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wen C-K, Binder BM, Chen Y-F, Chang J, Chiang Y-H, Kerris RJ, III, Chang C, Schaller GE. (2008) Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem 283: 23801–23810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D. (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Goulian M. (2010) Two-component signaling circuit structure and properties. Curr Opin Microbiol 13: 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Ikeda Y, Morgante M, Wang X, Zuo J, Hanafey MK, Gaasterland T, Tingey SV, Chua NH. (2003) Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett 554: 373–380 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ. (2007) Signaling via histidine-containing phosphotransfer proteins in Arabidopsis. Plant Signal Behav 2: 287–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen H-C, Sheen J. (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T. (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Imamura A, Yoshino Y, Mizuno T. (2001) Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci Biotechnol Biochem 65: 2113–2117 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Mizuno T. (2008a) Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 3025–3029 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. (2008b) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kurata N. (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2008) Differential gene expression of rice two-component signaling elements during reproductive development and regulation by abiotic stress. Funct Integr Genomics 8: 175–180 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Karan R, Singla-Pareek SL, Pareek A. (2009) Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct Integr Genomics 9: 411–417 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T. (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schäfer E, Kudla J, Harter K. (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear-encoded mitochondrial complex I genes. Mol Gen Genet 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. (2011) Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot 62: 5149–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Goto S, Tamai K, Ichii M. (2001) Role of root hairs and lateral roots in silicon uptake by rice. Plant Physiol 127: 1773–1780 [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006a) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. (2006b) Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41: 791–803 [DOI] [PubMed] [Google Scholar]
- Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, Fare T. (2009) Multiplexed measurements of gene signatures in different analytes using the NanoString nCounter assay system. BMC Res Notes 2: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P. (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18: 273–281 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2010) A modern circadian clock in the common angiosperm ancestor of monocots and eudicots. BMC Biol 8: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Mizuno T. (2005) Two-component phosphorelay signal transduction systems in plants: from hormone responses to circadian rhythms. Biosci Biotechnol Biochem 69: 2263–2276 [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2010) Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res 17: 303–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ. (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279: 48734–48741 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G. (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. (2006) Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol 142: 380–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils B, Heyl A. (2009) Unraveling the evolution of cytokinin signaling. Plant Physiol 151: 782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. (2010) The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J 62: 473–482 [DOI] [PubMed] [Google Scholar]
- Punwani JA, Kieber JJ. (2010) Localization of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin. Plant Signal Behav 5: 896–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ. (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su Y-S, Lagarias JC. (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2005) Cytokinin biosynthesis and regulation. Vitam Horm 72: 271–287 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M. (2006) Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu S-H, Wu P, et al. (2007) Nomenclature for two-component signaling elements of rice. Plant Physiol 143: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Kieber JJ, Shiu S-H. (2008) Two-component signaling elements and histidyl-aspartyl phosphorelays. The Arabidopsis Book 6: e0112, doi/10.1199/tab.0112 [DOI] [PMC free article] [PubMed]
- Shimizu-Sato S, Tanaka M, Mori H. (2009) Auxin-cytokinin interactions in the control of shoot branching. Plant Mol Biol 69: 429–435 [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26: 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai K, Ueguchi C, Mizuno T. (2001) Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol 42: 37–45 [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. (2004) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J Biol Chem 279: 41866–41872 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ. (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Cantwell BJ, Zhulin IB. (2010) Evolution and phyletic distribution of two-component signal transduction systems. Curr Opin Microbiol 13: 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T. (2011) The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol 156: 1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau CP, Wang L, Yu M, Zee SY, Yip WK. (2004) Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J Exp Bot 55: 547–556 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]
- Zheng K, Subudhi PK, Domingo J, Magpantay G, Huang N. (1995) Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genet Newsl 12: 225–258 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.