Abstract
5′-Methylthioadenosine (MTA) is the common by-product of polyamine (PA), nicotianamine (NA), and ethylene biosynthesis in Arabidopsis (Arabidopsis thaliana). The methylthiol moiety of MTA is salvaged by 5′-methylthioadenosine nucleosidase (MTN) in a reaction producing methylthioribose (MTR) and adenine. The MTN double mutant, mtn1-1mtn2-1, retains approximately 14% of the MTN enzyme activity present in the wild type and displays a pleiotropic phenotype that includes altered vasculature and impaired fertility. These abnormal traits were associated with increased MTA levels, altered PA profiles, and reduced NA content. Exogenous feeding of PAs partially recovered fertility, whereas NA supplementation improved fertility and also reversed interveinal chlorosis. The analysis of PA synthase crystal structures containing bound MTA suggests that the corresponding enzyme activities are sensitive to available MTA. Mutant plants that expressed either MTN or human methylthioadenosine phosphorylase (which metabolizes MTA without producing MTR) appeared wild type, proving that the abnormal traits of the mutant are due to MTA accumulation rather than reduced MTR. Based on our results, we propose that the key targets affected by increased MTA content are thermospermine synthase activity and spermidine-dependent posttranslational modification of eukaryotic initiation factor 5A.
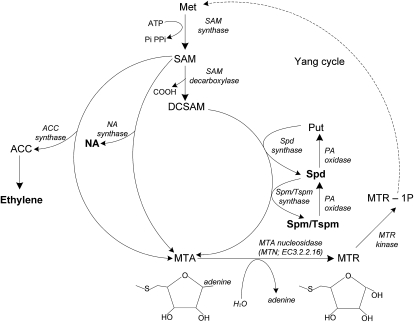
The Met recycling pathway (or Yang cycle) is present in all types of organisms, where its primary role is to recycle sulfur-containing metabolites (Albers, 2009). In most plants, bacteria, and protozoa, 5′-methylthioadenosine nucleosidase (MTN; EC 3.2.2.16) irreversibly hydrolyses 5′-methylthioadenosine (MTA) to methylthioribose (MTR) and adenine (Fig. 1); the resulting MTR is phosphorylated to 5′-methylthioribose 1-phosphate (MTR-1P) by methylthioribose kinase (MTK; EC 2.7.1.100). Recently, two novel enzymes that catalyze intermediate steps in generating Met from MTR-1P were identified and characterized: 5′-methylthioribose-1-phosphate isomerase and dehydratase-enolase-phosphate complex 1 (Pommerrenig et al., 2011).
Figure 1.
Overview of enzymatic reactions that generate MTA. MTA is generated as a by-product of PA, NA, and ethylene biosynthesis. MTN (EC 3.2.2.16) hydrolyzes MTA to MTR and adenine. The MTR is converted to Met through several steps, the first of which, MTR kinase (EC 2.7.1.100), catalyzes the production of MTR-1P. Met adenosyltransferase (EC 2.5.1.6) condenses Met and ATP to form S-adenosyl-Met (SAM). SAM decarboxylase (EC 4.1.1.50) removes the carboxyl group from SAM to generate decarboxylated SAM. An aminopropyl group is transferred from decarboxylated SAM when Put is converted to Spd via Spd synthase (EC 2.5.1.16). Similarly, another aminopropyl group is transferred from decarboxylated SAM when Spd is converted to Spm or its isomer Tspm via Spm synthase (EC 2.5.1.22) or Tspm synthase (ACL5; EC 2.5.1.79), respectively. NA is produced from the condensation of three SAM molecules by NA synthase (EC 2.5.1.43). 1-Aminocyclopropane-1-carboxylic acid (ACC) is also derived from SAM by ACC synthase (EC 4.4.1.14) in the rate-limiting step in the biosynthesis of ethylene. The dotted line indicates the additional steps involved in converting MTR into Met. The compounds in boldface letters are the key compounds of interest in this study. Pi, Inorganic phosphate or orthophosphate; PPi, inorganic pyrophosphate.
Several of the enzyme activities involved in Met recycling in eukaryotes such as humans differ from those in plants. For example, MTA is converted to MTR-1P by methylthioadenosine phosphorylase (MTAP; EC 2.4.2.28). Continuous MTA metabolism is extremely important in mammals, as loss of MTAP activity is associated with cancer (Bertino et al., 2011) and accumulation of MTA has been linked to tumor progression (Stevens et al., 2009).
MTA metabolism may be equally important to plants, as it is the by-product of polyamine (PA), nicotianamine (NA), and ethylene biosynthesis (Fig. 1). PAs such as putrescine (Put), spermidine (Spd), spermine (Spm), and thermospermine (Tspm) are cationic organic molecules that are essential for plant development and stress responses. Some of the documented roles of PAs include vascular differentiation, embryogenesis, cell division, and responses to abiotic stresses such as salt, osmotic, drought, and wounding (Takahashi and Kakehi, 2010; Vera-Sirera et al., 2010). For example, recent evidence suggests roles for both Tspm and hydrogen peroxide (H2O2) produced from PA catabolism in the control of xylem differentiation (Muñiz et al., 2008; Tisi et al., 2011). PAs have also been implicated in RNA processing, chromatin remodeling, membrane fluidity, and protein activation (Baron and Stasolla, 2008; Takahashi and Kakehi, 2010). NA, a product of another MTA-generating reaction, is a metal chelator involved in long-distance transport of ions. NA ion complexes ultimately support gametogenesis and embryo development (Curie et al., 2009; Lan et al., 2011). Finally, ethylene is a phytohormone that impacts numerous processes, including seed germination, seedling growth, fruit ripening, flower development, and abscission (Yang and Hoffman, 1984).
The Arabidopsis (Arabidopsis thaliana) genome has two MTN-encoding genes (At4g38800 and At4g34840) that are annotated as AtMTN1 (MTN1) and AtMTN2 (MTN2), respectively (Rzewuski et al., 2007). Using northern-blot analysis, Oh et al. (2008) showed that MTN1 is expressed in roots, stems, flowers, and cauline and rosette leaves. Public microarray data reveal that MTN2 transcripts are about 10 times less abundant than those of MTN1 in these organs (Winter et al., 2007). When compared on a tissue level, MTN1 is expressed preferentially in the cortex of roots and xylem of stems. On the other hand, MTN2 transcripts are most abundant in apical-basal cells of embryos and in developing pollen, stigma, ovules, and leaf guard cells (Winter et al., 2007). Recently, it has also been shown that both MTNs are abundant in the phloem tissue (Pommerrenig et al., 2011). Although MTN1 and MTN2 polypeptides share 64% amino acid sequence identity, they have distinct substrate specificities and pH optima (Siu et al., 2008). Thus, it was initially inferred that these two enzymes may have distinct roles in plant metabolism: in vitro, MTN1 accepts only MTA as a substrate, while MTN2 can also accept S-adenosylhomocysteine to a limited extent (Siu et al., 2008). More recent crystallography and protein dynamic analyses revealed that MTN1 binds to S-adenosylhomocysteine but is incapable of hydrolyzing it (Siu et al., 2011).
Recently we described the physiology of several single T-DNA insertion mutants in MTN1 and MTN2: mtn1-1 (T-DNA insertion in the third intron), mtn1-2 (T-DNA insertion in the sixth exon), mtn2-1 (T-DNA insertion in the fourth exon), and mtn2-2 (T-DNA insertion in the fourth exon; Bürstenbinder et al., 2010). Based on biochemical analyses, we established that MTN1 is responsible for 80% of the MTN activity in crude extracts of 4-d-old seedlings and rosette leaves of 3-week-old plants grown on medium (Bürstenbinder et al., 2010). When grown on soil or germinated on sulfur-sufficient medium containing 500 μm MgSO4, seedlings of single mutants were phenotypically indistinguishable from the wild type. However, when the sulfur source was 500 μm MTA, both mtn1-1 and mtn1-2 mutants had impaired seedling and root growth (Bürstenbinder et al., 2010). These MTN-deficient seedlings also have altered PA profiles, with increased Put and Spm compared with seedlings grown on medium containing 500 μm MgSO4. Interestingly, no significant changes in either NA content or ethylene production were detected in either mtn1-1 or mtn2-1. In an attempt to lower MTN levels further, the two homozygous mutant lines were crossed and a double mutant was isolated. The resulting mtn1-1mtn2-1 plants exhibited a pleiotropic phenotype with developmental abnormalities (Bürstenbinder et al., 2010). The details and physiological basis for these pleiotropic traits, in terms of MTN’s enzyme activity and MTA regulation, was unclear.
Here, we report on the effect of MTN deficiency based on a comprehensive examination of the development and physiology of the homozygous mtn1-1mtn2-1 double mutant. In addition to measuring its residual MTN enzyme activity and profiling relevant metabolites, we analyzed the effect of MTA accumulation based on the predicted structures of Spd and Tspm synthases cocrystallized with this ligand. The effect of exogenous supplementation of relevant compounds on the mutant phenotype was tested to reveal abnormalities in the cellular metabolism of the MTN-deficient plants. The results of these analyses indicate the critical importance of MTA metabolism in maintaining the biosynthesis of NA and PAs that are essential for normal plant vascular development and reproduction.
RESULTS
MTN Activity in mtn1-1mtn2-1 Mutants Reflects Decreased Transcript and Protein Abundance
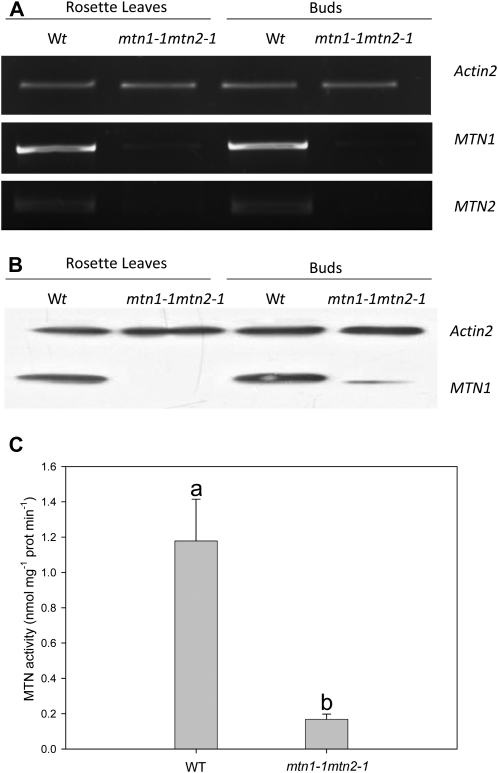
Reverse transcription (RT)-PCR was performed on cDNA isolated from the buds and rosette leaves of wild-type and mtn1-1mtn2-1 plants. In both organs, MTN1 transcript levels were higher than MTN2 in the wild-type background (Fig. 2A). A comparison of transcript abundance in mtn1-1mtn2-1 revealed substantially decreased MTN1 and undetectable levels of MTN2 in both leaves and buds. Immunoblot analysis (Fig. 2B) showed a similar abundance of MTN1 protein in wild-type leaves and buds but reduced levels in mtn1-1mtn2-1. Interestingly, MTN-deficient buds had higher amounts of MTN1 in their buds than in their leaves. As documented previously by Bürstenbinder et al. (2010), MTN2 protein cannot be detected in either wild-type or mtn1-1mtn2-1 tissues by immunoblotting with an MTN2-specific polyclonal antibody.
Figure 2.
MTN transcript and protein abundance with corresponding enzyme activity. Results are averages ± sd of three to 10 independent biological replicates. A, RT-PCR showed that MTN1 and MTN2 transcript abundance was reduced in mtn1-1mtn2-1. Actin was used as the reference gene to ensure equal loading of the cDNA (n = 3). B, Immunoblot analysis indicated that MTN1 was present in buds and leaves of wild-type plants. In mtn1-1mtn2-1, MTN1 was considerably lower in buds and undetectable in leaves. Detection of actin protein confirmed equal protein loading (n = 3). C, Specific enzyme activity of MTN1 was measured in buds harvested from plants 2 weeks after bolting. Statistically significant variables are indicated by different letters (P < 0.05; n = 10). Wt/WT, Wild type.
Wild-type and mutant bud extracts were assayed for MTN enzyme activity to determine whether this correlated with MTN protein abundance and phenotype. Floral buds were chosen for the assay based on preliminary studies indicating that this organ had the highest MTN-specific activity (Supplemental Fig. S1). Compared with the 7% to 8% retained MTN activity observed in mtn1-1 seedlings grown on MTA (Bürstenbinder et al., 2010), mtn1-1mtn2-1 buds had 14% residual MTN activity (1.21 ± 0.27 nmol mg−1 protein min−1 in the wild type; Fig. 2C). Thus, our results show that mtn1-1mtn2-1 is a knockdown mutant, with MTN1 providing the residual MTN activity. Attempts to create a mutant completely lacking MTN activity by crossing homozygous knockout lines, mtn1-2 and mtn2-2, failed due to embryo lethality.
MTN Deficiency Affects Both Vegetative and Reproductive Development
The mtn1-1mtn2-1 mutants developed no obvious defects until the formation of the first true leaves. Subsequently, an array of abnormalities developed upon activation of the floral meristem. Here, we focus on the changes in vascular development and reproduction.
Vegetative Phase Phenotypes
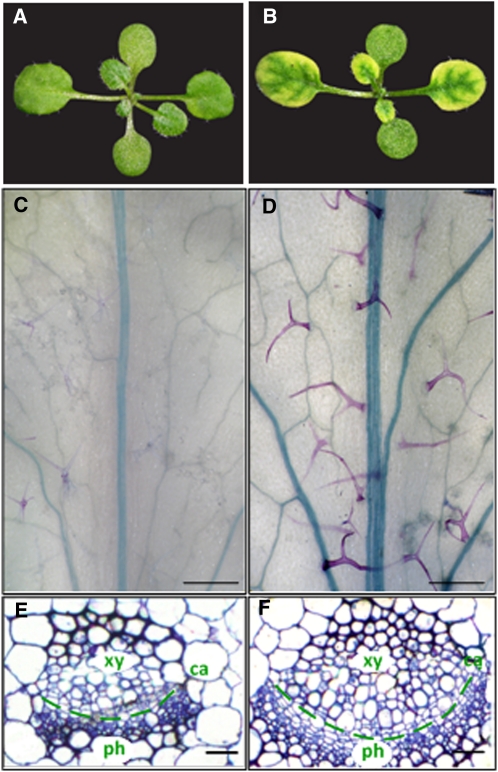
Ten days after germination, the first true leaf of wild-type seedlings is greater than 1 mm in length (stage 1.02 as described by Boyes et al. [2001]; Fig. 3A). In comparison with the wild type, mtn1-1mtn2-1 seedlings did not reach this stage until 12 d after sowing. In addition to their delayed development, the first true leaves of mtn1-1mtn2-1 exhibited interveinal chlorosis (Fig. 3B), a condition where the veins of the leaf remain green while the area between the veins becomes yellow. This chlorosis was most notable in emerging young leaves when seedlings were grown on half-strength Murashige and Skoog medium (1/2 MS; Murashige and Skoog, 1962) as opposed to soil. Upon maturation of the leaves or transfer of the plants to soil, the interveinal chlorosis gradually disappeared.
Figure 3.
Vegetative phenotypes of mtn1-1mtn2-1 mutants compared with the wild type. A and B, Wild-type and mtn1-1mtn2-1 seedlings at the 1.02 development stage, when the first true leaves are more than 1 mm. Wild-type seedlings had green true leaves (A) compared with mutant seedlings, which exhibited interveinal chlorosis (B). C and D, Developmentally matched adult leaves (equal number of secondary veins arising from the mid veins) of the wild type (C) and mtn1-1mtn2-1 (D) were cleared and stained with toluidine blue O to reveal the increased vein thickness in mtn1-1mtn2-1. Trichomes of the mutant were larger and the leaves tended to acquire more stain compared with the wild type. Bars = 1.5 mm. E and F, Transverse sections through the mid veins of developmentally matched adult leaves of wild-type (E) and mtn1-1mtn2-1 (F) plants. mtn1-1mtn2-1 exhibited increased numbers of xylem (xy), phloem (ph), and cambial (ca) cells. Bars = 15 μm.
As the mtn1-1mtn2-1 mutants matured, abnormalities of the rosette leaves became apparent. Clearing and observation of the sixth leaf revealed an increase in venation, particularly adjacent to the leaf margins (Supplemental Fig. S2, A and B). Leaves of mtn1-1mtn2-1 leaves also had thicker mid veins (Fig. 3, C and D). Further examination of embedded leaf sections showed that the thicker veins in mtn1-1mtn2-1 resulted from an increase in xylem, phloem, and cambial cells (Fig. 3, E and F). Irrespective of the larger leaves, the leaf vasculature of mtn1-1mtn2-1 was similar to that of the Tspm synthase mutant, thick vein (tkv; Clay and Nelson, 2005), which is allelic to acaulis5 (acl5; Hanzawa et al., 1997; Kakehi et al., 2008). ACL5 is specifically expressed in xylem vessel elements, and both acl5 and tkv have an overproliferation of xylem vessels in their stems. This increased xylem is proposed to result from reduced polar auxin transport (PAT; Clay and Nelson, 2005). Thus, based on similarities to tkv and acl5, we assayed auxin transport and followed vascular patterning in the stems of mtn1-1mtn2-1.
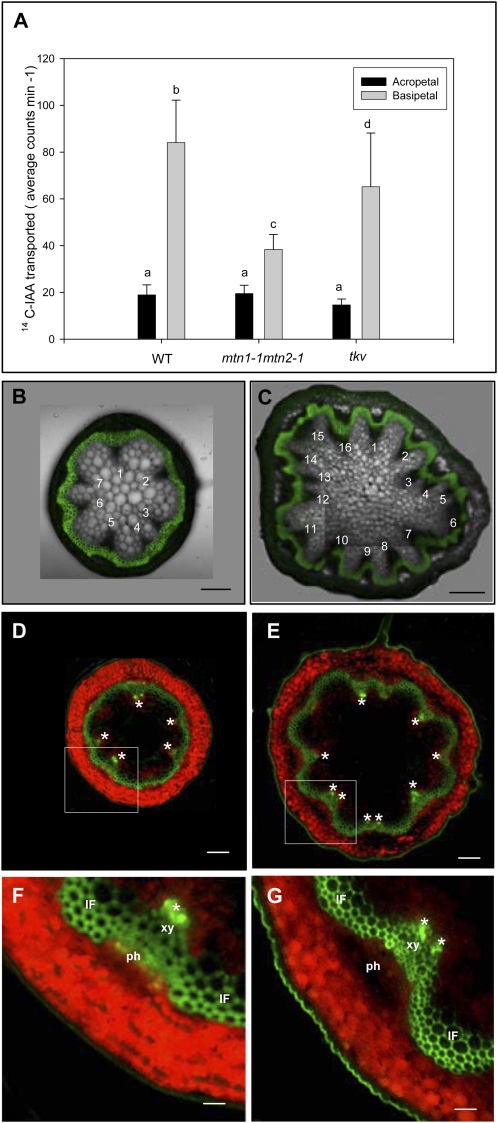
Auxin transport was measured in wild-type and mutant inflorescence stem segments using 14C-labeled indole acetic acid (IAA). The basipetal transport of [14C]IAA in the mtn1-1mtn2-1 mutant was 46% of that detected in wild-type stems, while the acropetal transport in the stem segments was normal. IAA transport in tkv, measured as a control, was 64% that of the wild type (Fig. 4A), which is consistent with the initial description of this mutant by Clay and Nelson (2005).
Figure 4.
PAT and stem vascular arrangement. A, Reduced PAT of [14C]IAA in excised inflorescence stem segments of mtn1-1mtn2-1 mutants placed basipetally (inverted orientation) in [14C]IAA. Three independent experiments were conducted using segments of 32 plants for each genotype. Error bars indicate se. Statistically significant values (P < 0.05) are indicated by different letters. WT, Wild type. B and C, Free-hand cross-sections of mtn1-1mtn2-1 and the wild type taken at the basal end of the primary inflorescence stem. Composite images of confocal laser scanning images and pseudo-differential interference contrast images are shown. The pseudo-differential interference contrast images show the increased number of vascular bundles of mtn1-1mtn2-1 (C) compared with the wild type (B). Confocal laser images show the lignified interfascicular and intrafascicular fibers stained with berberine. Bars = 50 μm. D to G, Auxin maxima (asterisks) visualized using DR5rev::GFP mostly corresponded to the number of vascular bundles in cross-sections taken from the basal end of the primary inflorescence stem (D and E) of wild-type (D and F) and mtn1-1mtn2-1 (E and G) backgrounds. Higher magnification images reveal auxin maxima primarily localized to the xylem (xy) tissue of the vascular bundles (F and G). Note that the green fluorescence observed on lignified xylem of the interfascicular and intrafascicular fibers (IF) was autofluorescence. ph, Phloem. Bars = 50 μm (D and E) and 150 μm (F and G).
We next examined stem cross-sections of the MTN double mutant in more detail. Wild-type stems contained six to eight symmetrically arranged vascular bundles approximately 2 to 3 cm above the base of the primary inflorescence (Fig. 4B), whereas corresponding sections of mtn1-1mtn2-1 stems usually had 10 or more vascular bundles. These mutants also had increased numbers of small cells in all stem tissues (Fig. 4C). Furthermore, the symmetrical arrangement of the vascular bundles in the wild type was disrupted in the mtn1-1mtn2-1 mutant, leading it to have an irregular stem circumference.
Auxin maxima resulting from differences in auxin distribution in the shoot apical meristem determine the positions of leaf primordia and the formation of leaf traces (Dengler, 2006). The leaf traces arise from vascular bundles or vascular sympodia in the stem (Dengler, 2006). Thus, we considered that the increased number of vascular bundles in mtn1-1mtn2-1 mutants might reflect changes in auxin distribution. To examine this, we monitored GFP expression from a synthetic auxin-responsive promoter, DR5rev::GFP (Friml et al., 2003), in this mutant background. The resulting images showed that mtn1-1mtn2-1 stem cross-sections always had a greater number of auxin maxima than the wild type, corresponding to the mutant’s increased number of vascular bundles (Fig. 4, D–G).
Reproductive Phase Phenotypes
The mtn1-1mtn2-1 siliques arrested when they were 2 to 3 mm (stage 16 of the developmental milestones described by Smyth et al. [1990]), and very few siliques produced viable seeds. To determine the basis for this observed sterility, reciprocal crosses were performed: wild-type pollen was applied to mtn1-1mtn2-1 pistils, and mtn1-1mtn2-1 pollen grains were transferred to stigma of the male-sterile mutant apt1-3. The resulting fertilization was documented by recording the number of crosses producing viable seeds (Table I). Both mtn1-1mtn2-1 and apt1-3 pistils were receptive to wild-type pollen, and mtn1-1mtn2-1 pollen was able to germinate on wild-type and apt1-3 pistils, resulting in successful seed production in all cases. Thus, both mtn1-1mtn2-1 male and female gametes were capable of producing viable heterozygous progeny, although with reduced efficiency relative to the wild type.
Table I. Viable seeds obtained from reciprocal crosses between the wild type and mutants (apt1-3 and mtn1-1mtn2-1).
σσ, Pollen donor; ϕ, stigma.
| Cross | Percentage of Crosses with Viable Seeds | No. of Crosses from 10 Plants |
| apt1-3 (φ) × wild type (σ) | 84 | 25 |
| mtn1-1mtn2-1 (φ) × wild type (σ) | 14 | 50 |
| apt1-3 (φ) × mtn1-1mtn2-1 (σ) | 28 | 50 |
Further examination revealed several factors that contribute to decreased fertility in mtn1-1mtn2-1. Anthers of stage 14 mtn1-1mtn2-1 flowers were indehiscent (Fig. 5A). In order to observe pollen, anthers of mtn1-1mtn2-1 needed to be opened manually (Fig. 5D). These anthers contained abnormally formed pollen grains (Fig. 5, E and F). Specifically, most of the mtn1-1mtn2-1 grains were round and lacked the groove present in wild-type pollen (Fig. 5, B and C). To assess the viability of the abnormal pollen grains, anthers of stage 14 flowers were stained with fluorescein diacetate, a stain that is taken up only by viable cells. Almost all of the wild-type pollen was able to take up the fluorescein diacetate (n = 45), whereas mtn1-1mtn2-1 had more unstained than stained pollen: 37% of MTN-deficient anthers had no viable pollen, 20% had less than five viable pollen grains, almost half (40%) had five to 10 viable pollen grains, and only 3% had an excess of 10 viable pollen grains (n = 65).
Figure 5.
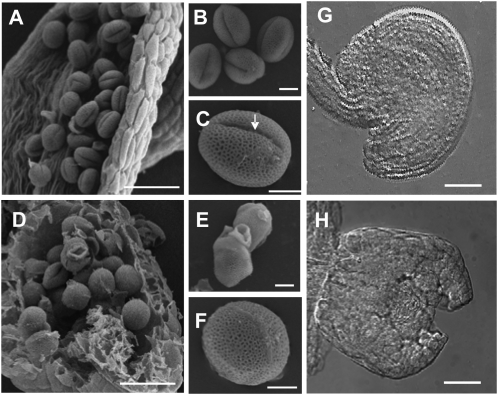
Reproductive abnormalities of mtn1-1mtn2-1. A to F, Scanning electron micrographs of wild-type (A) and manually opened (D) anther sacs showing abnormal pollen development in the mutant. The mtn1-1mtn2-1 pollen grains were irregularly shaped (F) and lacked the characteristic furrow (indicated by the arrow) seen in the wild type (B and C). At times, the mutant pollen appeared clumped with each other (E) compared with the wild type (B). Bars = 45 μm (A and D) and 5 μm (B, C, E, and F). G and H, Differential interference contrast images of stage 14 flower pistils. The wild type had mature ovules in the post fertilization stage (G), while the mtn1-1mtn2-1 ovules (H) corresponded to those normally observed in stage 12 wild-type flowers: outer integuments were not fully extended over the nucellus. Bars = 25 μm.
Next, in vivo pollen tube formation was assessed: pistils were fixed 24 h after pollination and visualized with aniline blue (Supplemental Fig. S2, C–H). Reciprocal crosses performed between mtn1-1mtn2-1 and wild-type pollen proved the mutant stigma to be receptive to pollen, with 90% of the successfully pollinated pistils containing pollen tubes. Attempts to manually pollinate apt1-3 or mtn1-1mtn2-1 with mtn1-1mtn2-1 pollen were not successful, as no pollen tubes were observed for either cross 24 h later. Collection of stage 14 mtn1-1mtn2-1 flowers and staining with aniline blue showed that 22% had self-pollinated, and of those pollinated, 48% had pollen tubes. Thus, mtn1-1mtn2-1 pollen are capable of forming pollen tubes in vivo but apparently need longer than 24 h to extend.
Carpel and ovule development was also aberrant in stage 14 mtn1-1mtn2-1 flowers: carpels had wide or duplicated stigmas, while ovules of the pistils had unextended integuments when compared with the wild type (Fig. 5, G and H). About 10% of mtn1-1mtn2-1 flowers examined contained one to three fully developed ovules that appeared to be wild type.
The mtn1-1mtn2-1 Phenotype Complemented by MTN1 Overexpression
In the Yang cycle, MTA is converted to MTR-1P via two steps: first, MTA is hydrolyzed by MTN to form MTR and adenine, and the resulting MTR is subsequently metabolized by MTK. Unlike mtn1-1mtn2-1 seedlings, MTK-deficient seedlings are not impaired in their growth and development when provided with sulfur-sufficient conditions (Sauter et al., 2004), suggesting that MTN is the more crucial of the two enzyme activities. In order to verify that the complex phenotype of mtn1-1mtn2-1 was solely due to MTN deficiency, the MTN1 gene was reintroduced to the double mutant. The pleiotropic phenotype of mtn1-1mtn2-1 was fully complemented by a 2.4-kb genomic fragment that contained the full-length coding sequence of MTN1 along with 356 bp of the upstream region (Fig. 6A). These results were further confirmed by the ectopic expression of MTN1 cDNA under the control of the constitutive Polyubiquitin10 (UBQ10) promoter in the mtn1-1mtn2-1 background. Therefore, we concluded that the pleiotropic phenotype of mtn1-1mtn2-1 was the result of MTN deficiency and that replenishing MTN1 alone was sufficient to compensate for the lack of MTN2.
Figure 6.
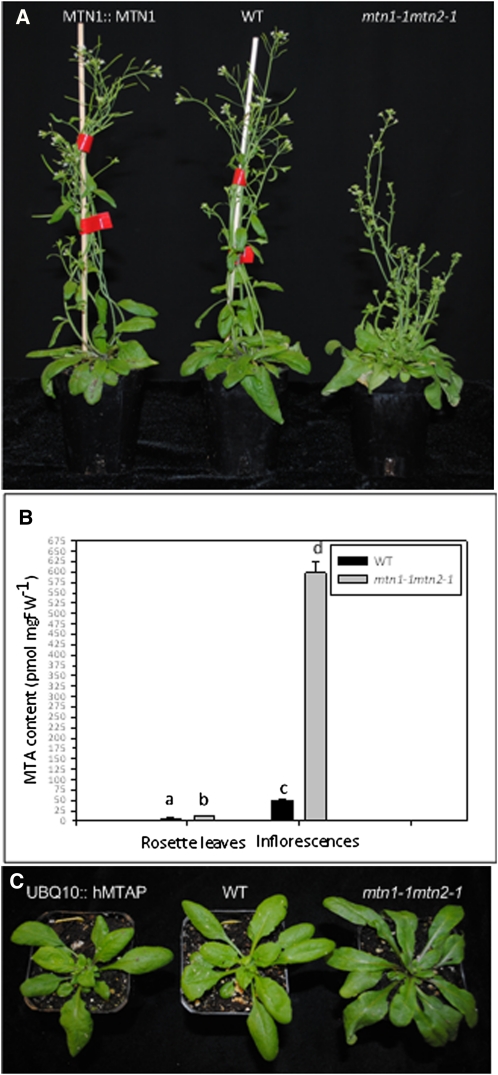
Complementation of the mtn1-1mtn2-1 phenotype by MTN1 expression, MTA content, and complementation of the mtn1-1mtn2-1 phenotype by hMTAP. A, Complementation of mtn1-1mtn2-1 with MTN1::MTN1 3 weeks after bolting. B, Steady-state levels of MTA in inflorescences and rosette leaves. Mean values of five replicates were plotted, with error bars indicating sd. Statistically significant values (P < 0.05) are indicated by different letters. C, Complementation of mtn1-1mtn2-1 with UBQ10::hMTAP shown at bolting. FW, Fresh weight; WT, wild type.
The Basis of the mtn1-1mtn2-1 Phenotype Is the Accumulation of MTA, Not the Loss of MTR
Although the observed vascular and reproductive defects of mtn1-1mtn2-1 were restored by MTN transgene expression, we reasoned that these abnormal traits could be due to either a lack of MTA metabolism or decreased MTR. Targeted profiling of Met metabolites in inflorescences and rosette leaves revealed statistically significant increases of MTA in mtn1-1mtn2-1 (P < 0.05). MTA was elevated 2-fold in rosette leaves (Fig. 6B) and 5-fold in the mutant inflorescences, leading us to conclude that MTN deficiency leads to increased MTA content. However, based on these data, it could not be excluded that the lack of MTR might also contribute to the complex phenotype of the mutant. To test this, we introduced a cDNA encoding human MTAP activity (UBQ10::hMTAP) into the mutant. This enzyme phosphorylates MTA directly into MTR-1P without producing MTR. The phenotype of the double mutant was fully complemented by constitutive expression of hMTAP (Fig. 6C).
Modeling of Spd Synthase and Tspm Synthase Complexes with MTA
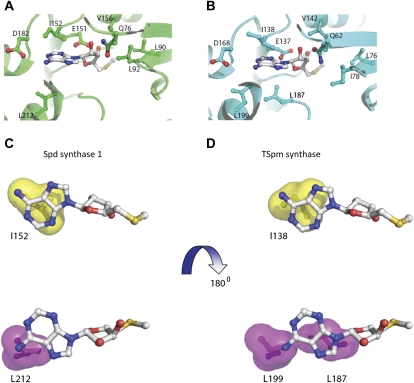
MTA is a potent inhibitor of both human Spm synthase (approximate Ki = 0.3 μm) and Spd synthase (Ki = 2–10 μm; Wu et al., 2008). Analysis of crystal structures containing bound ligands of these enzymes with MTA explains the difference in affinity: the hydrophobic binding site of Spm synthase is larger, allowing it to have more extensive interactions with the adenine residue of MTA than does Spd synthase. The conserved structural folds of the human and plant PA enzymes allowed us to model MTA binding to these targets. Three-dimensional models were developed for Arabidopsis Spd synthase1 and Tspm synthase, and then their MTA complexes were visualized as for human Spd synthase1 and Spm synthase (Wu et al., 2008). Analysis of these three-dimensional models revealed that the MTA-binding pockets of Spd synthase1 and Tspm synthase are very similar (Fig. 7, A and B): the CH3S moiety of MTA makes van der Waals contact with Leu-90, Leu-92, and Val-156 of Spd synthase1 or Leu-76, Ile-78, and Val-142 of Tspm synthase. The Rib ring of MTA is predicted to establish hydrogen bonds with the side chains of Gln-76 and Glu-151 of Spd synthase 1 or the side chains of Gln-62 and Glu-137 of Tspm synthase. N-6 of MTA adenine forms hydrogen bonds with the side chain of Asp-182 of Spd synthase1 or Asp-168 of Tspm synthase. The adenine ring of MTA is placed between Ile-137 on one side and Leu-187 and Leu-199 on the other side in the Tspm synthase-MTA complex. In the Spd synthase1-MTA complex, the adenine ring of MTA is sandwiched between Ile-151 and Leu-212. The residues Ile-151 and Leu-212 of Spd synthase1 and the residues Ile-137 and Leu-199 of Tspm synthase are structurally conserved, while the Leu-187 residue of Tspm synthase is substituted by Ser (Ser-202) in Spd synthase1 (Hanzawa et al., 2000). Therefore, the hydrophobic surfaces involved in interaction with the adenine ring of MTA are predicted to be very different. The hydrophobic surface (in yellow) made by Ile-152 in Spd synthase1 is similar to the corresponding one in Tspm synthase (Ile-138; Fig. 7, C and D). On the other side of the adenine ring of MTA, the hydrophobic surface (in magenta) of Spd synthase1 made by Leu-212 is shifted and partially covers the adenine ring of MTA, whereas in the Tspm synthase, the hydrophobic surfaces made by Leu-187 and Leu-199 are large and positioned to cover the adenine ring completely. Although MTA binds to all three PA synthases, these modeling experiments predict that, similar to human Spm synthase, Tspm synthase may be more sensitive to inhibition by MTA than Spd synthase1, due to extensive hydrophobic interactions between Tspm synthase and the adenine moiety of MTA.
Figure 7.
Structural basis for the inhibition of Tspm synthase and Spd synthase1 enzymes by MTA. A and B, Closeup views of the MTA-binding pocket for Spd synthase1 and Tspm synthase, respectively. MTA and the residues interacting with MTA in both structures are shown in stick-and-ball representation; carbon atoms for residues are in the same color as the corresponding structure, whereas carbon atoms of MTA are in gray. C and D, Hydrophobic surfaces interacting with the adenine ring in MTA. The two surfaces stacking with the adenine ring are in magenta and yellow.
mtn1-1mtn2-1 Plants Exhibit Altered PA Profiles
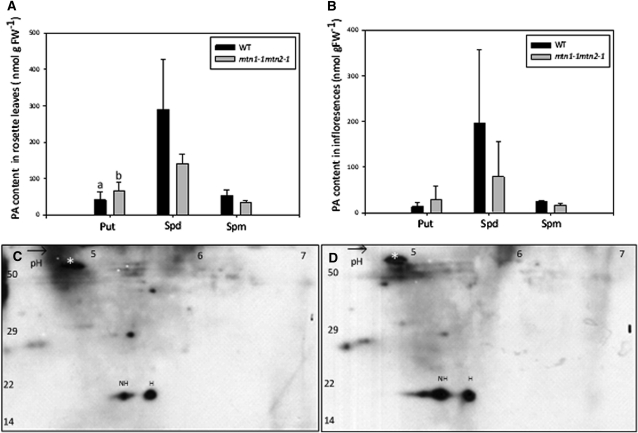
We next investigated whether increased MTA abundance in the mutant was associated with changes in free PA content. To do this, we monitored both leaves and inflorescences for Put, Spd, and Spm using HPLC analysis. Unfortunately, Tspm abundance was so low that it was not readily detected in wild-type samples (Naka et al., 2010) and, as a result, could not be measured. For those PAs that could be measured, differences were observed in both mtn1-1mtn2-1 leaves and inflorescences: Put levels increased in mtn1-1mtn2-1, whereas Spd and Spm levels decreased (Fig. 8, A and B). Of these, a statistically significant difference was only observed in the Put content of inflorescences.
Figure 8.
PA profiles and two-dimensional gels showing eIF5A. A and B, Altered PA profiles of inflorescences (A) and rosette leaves (B) of mtn1-1mtn2-1 and the wild type (n = 4). Error bars represent sd. Statistically significant values (P < 0.05) are indicated by different letters. FW, Fresh weight; WT, wild type. C and D, Hypusinated (H) and nonhypusinated (NH) eIF5A in rosette leaves of mtn1-1mtn2-1 (D) compared with the wild type (C). Images are representations of three replicates. The Rubisco large subunit polypeptide was used as the reference for quantification (white asterisks). n = 3. Molecular mass standards (in kD) are indicated to the left.
We reasoned that if these changes were physiologically relevant, reactions dependent on PAs would be affected. Spd is a cofactor for the posttranslational activation of the eukaryotic translation initiation factor eIF5A. The butylamine moiety that is provided solely by Spd is transferred to eIF5A in a two-step reaction, causing it to become hypusinated on a specific Lys residue (Pegg and Casero, 2011). Thus, we tested the degree of hypusination of eIF5A in mtn1-1mtn2-1 leaf extracts by two-dimensional gel electrophoresis followed by immunoblotting with eIF5A-specific antibodies as an indicator of intracellular Spd availability. The amount of hypusinated eIF5A was 2-fold lower in mtn1-1mtn2-1, indicating a clear reduction in hypusination of elF5A in comparison with wild-type leaf extracts (Fig. 8, C and D).
MTN Deficiency Results in Altered NA Levels
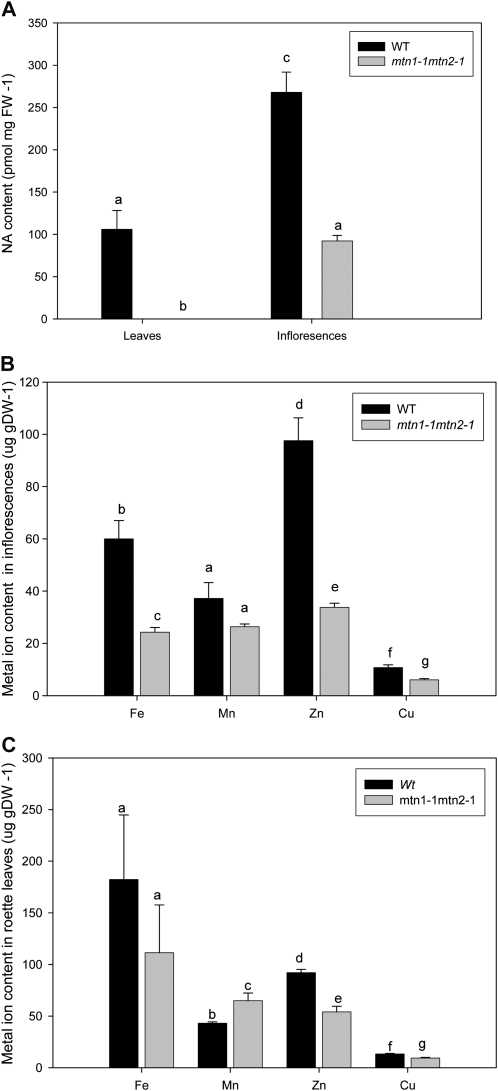
Upon recognizing that the free PA profiles changed in response to increased MTA abundance, we next analyzed NA levels of mtn1-1mtn2-1. We determined that NA was significantly reduced in inflorescences and mature rosette leaves of mtn1-1mtn2-1 compared with the wild type (P < 0.05; Fig. 9A). In wild-type plants, the highest NA content (268 ± 58 pmol mg−1 fresh weight) was detected in inflorescences, with leaves having 60% less NA (106 ± 50 pmol mg−1 fresh weight), whereas the NA content of mtn1-1mtn2-1 inflorescences was 34% lower than in the wild type and undetectable in rosette leaves.
Figure 9.
NA content and metal profiles of leaves and buds. NA could not be detected in the rosette leaves (A; n = 5) compared with the wild type and was lower in mtn1-1mtn2-1 inflorescences (n = 3). In addition to NA, Fe, Mn, Zn, and Cu levels were also reduced in mtn1-1mtn2-1 inflorescences (B) and leaves (C; n = 5) compared with the wild type. Each value represents a pool of three biological replicates. Error bars indicate se. Statistically significant values (P < 0.05) are indicated by different letters, while insignificant values are indicated by same letters. DW, Dry weight; FW, fresh weight; WT, wild type.
Since NA is known to play a key role in ion homeostasis and long-distance transport, we investigated whether the ion content (copper [Cu], iron [Fe], zinc [Zn], and manganese [Mn]) of mtn1-1mtn2-1 was different from that in the wild type, using inductively coupled plasma atomic emission spectroscopy (Gadapati and Macfie, 2006). In the mutant inflorescences, contents of Cu, Fe, and Zn were significantly lower, with Fe and Zn showing the greatest reduction compared with the wild type (P < 0.05; Fig. 9B). The ion profiles of mutant and wild-type rosette leaves were similar, with reduced Zn and Cu abundance, except the Mn content in the mutant was significantly higher than in the wild type (P < 0.05; Fig. 9C). Interestingly, mtn1-1mtn2-1 had significantly decreased Fe levels in the inflorescences compared with the wild type, while these levels were not significantly different in rosette leaves (P < 0.05). Confirming these results, interveinal chlorosis was absent in the reproductive leaves of mature mtn1-1mtn2-1 plants.
Exogenous Feeding of PA and NA Partially Restored the Fertility of mtn1-1mtn2-1
Chemical Rescue of Fertility
Since both free PA and NA levels were affected in the mtn1-1mtn2-1 mutant, we postulated that MTA-producing reactions were being product inhibited. We then hypothesized that the exogenous application of target compounds would reverse abnormal phenotypes. To test this, we chemically complemented mtn1-1mtn2-1 mutants with exogenous application of Spd, Spm, Tspm, or NA (NA was extracted from NA-overproducing transgenic Arabidopsis plants [Pianelli et al., 2005]).
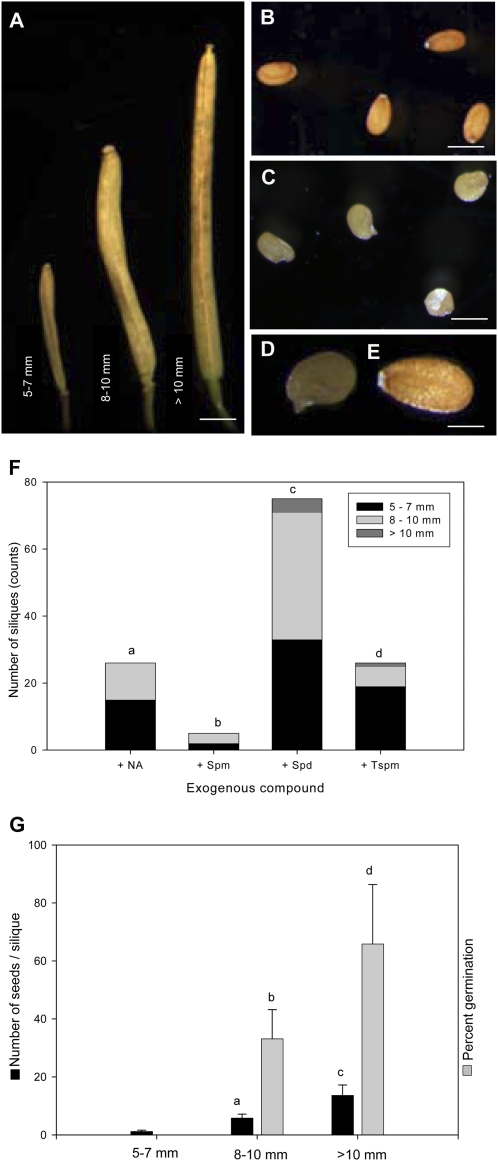
To treat MTN-deficient plants, F2 seeds of MTN1-1/mtn1-1mtn2-1/mtn2-1 plants were grown on medium supplemented with Spm, Spd, Tspm, or NA. After 2 to 3 weeks on medium, seedlings were transplanted to soil and genotyped via PCR. With the exception of Tspm, drops of the corresponding compound were then applied daily to shoot apices of mtn1-1mtn2-1 plants until the plants were 8 weeks old. Examination of the treated plants showed that mtn1-1mtn2-1 fertility was partially recovered using any of three compounds (Spm, Spd, or NA): partial recovery of fertility was indicated by the presence of extended siliques (Fig. 10A) with fertile seed (Fig. 10, B–E) on individual branches. Surprisingly, partial fertility was also recovered in a similar manner when the seeds were germinated on the indicated compounds for 14 d or more and transferred to soil without being provided with further supplement (Fig. 10F).
Figure 10.
Exogenous NA and PA restored the fertility phenotype of mtn1-1mtn2-1. A to E, Silique and seed morphology of restored plants. The restored seeds of mtn1-1mtn2-1 were categorized in groups based on their lengths; most of the seeds in siliques of more than 10 mm had normal wild-type-looking seeds (B), while the other two categories had seeds with abnormal color and shape (C). Closer observation of these abnormal seeds (D) showed that they had developed seed coats similar to the wild type (E). Bars = 1.5 mm (A ), 300 μm (B and C), and 100 μm (D and E ). F, Number of siliques that developed when mtn1-1mtn2-1 was grown in 1/2 MS medium supplemented with 20 μm NA, 100 μm Spm or Spd, or 50 μm Tspm. The restored siliques were of various lengths and were categorized into three groups based on those lengths (n = > 20 siliques per treatment). Statistically significant values (P < 0.05) are indicated by different letters. G, Number of seeds per silique and percentage of seeds germinated from mtn1-1mtn2-1 plants grown on 1/2 MS supplemented with 100 μm Spd. Seed counts were averages of 60 siliques from six plants for 5 to 7 mm and 8 to 10 mm but 36 siliques from six plants for 10 mm. Percentage germination was assessed only on seeds collected for the latter two categories (n = 6 siliques). Bars represent sd. Statistically significant values (P < 0.05) are indicated by different letters, while insignificant values are indicated by the same letters.
Generally, siliques with unfertilized ovules fail to extend beyond 3.5 mm, while siliques containing 60 seeds reach a maximal length of 15 mm (Meinke, 1994). Thus, the fertility of the plants without further supplement was scored by categorizing the siliques based on their length into three groups: (1) 5 to 7 mm; (2) 8 to 10 mm; and (3) more than 10 mm (Fig. 10A). Most of the siliques produced were in the first two categories, while only Spd and Tspm gave rise to siliques that were more than 10 mm long. Of the four treatments, the most effective was 100 μm Spd (78 ± 5 siliques per plant) and the least effective was 100 μm Spm (Fig. 10F). Both NA and Tspm treatments produced 25 to 35 siliques per plant.
The number of seeds per silique in each group of the Spd experiment was further determined: the 5- to 7-cm group had 1 ± 0.4 seeds per silique, while the 8- to 10-mm and greater than 10-mm groups had 6 ± 1 and 14 ± 4 seeds per silique, respectively. Some seeds in the 5- to 7-mm and 8- to 10-mm categories showed abnormal shape and color (Fig. 10B) when compared with the wild type (Fig. 10C). To determine the viability of these abnormal seeds, approximately 100 seeds from six siliques were germinated on medium and scored: 8- to 10-mm and more than 10-mm siliques had 33% ± 10% and 66% ± 20% germination, respectively (Fig. 10G).
Chemical Rescue of Interveinal Chlorosis by NA
In addition to testing for recovered fertility, rescue of interveinal chlorosis with the application of exogenous PAs (Put, Spd, Spm, or Tspm) or NA was also investigated. Of the compounds tested, only medium supplemented with NA completely eliminated the interveinal chlorosis normally observed at 10 d after germination in mtn1-1mtn2-1 seedlings. Mutant seedlings that exhibited interveinal chlorosis on 1/2 MS developed green leaves 2 d after being transferred to 1/2 MS medium supplemented with NA. Moreover, hydroponically grown mtn1-1mtn2-1 supplemented with 20 μm NA lacked interveinal chlorosis (n = 8; Supplemental Fig. S3).
DISCUSSION
Our detailed characterization of the mtn1-1mtn2-1 mutant has established a correlation between MTN deficiency, abnormal vasculature, and low reproductive efficiency. A similar phenotype has been recovered in mtn1-1 plants expressing an artificial microRNA specific for MTN2, further substantiating the link between MTN deficiency and these traits (data not shown). The first distinct phenotype of mtn1-1mtn2-1 coincides with the development of true leaves. As recently reported, in Arabidopsis and Plantago major, the Yang cycle (i.e. MTA recycling) mainly occurs in leaf veins (Pommerrenig et al., 2011). Thus, it is likely that the flux of MTA synthesis first increases substantially in leaves as vein development proceeds.
Molecular characterization of the mtn1-1mtn2-1 mutant showed a reduction in MTN transcript levels consistent with the presence of a T-DNA insertion within the transcribed region of each gene. Protein abundance and enzyme activity reflected transcript levels, with the mtn1-1mtn2-1 mutant having approximately 14% residual MTN activity, indicating its substantially reduced capacity for MTA hydrolysis. The MTN assay results were consistent with our previous analysis of mtn1-1 and mtn1-2 single mutant seedlings grown on sulfur-deficient medium supplemented with MTA (Bürstenbinder et al., 2010). These seedlings have less than 20% of the MTN activity of the wild type and display severe growth retardation. Interestingly, they also have increased MTN activity when grown in the presence of MTA. Since MTN transcript levels did not change substantially under these conditions, the activation of MTN must occur at the protein level (Bürstenbinder et al., 2010). While the molecular basis of this activation remains unknown, we assume that the increased MTA content in the double mutant activates the residual MTN enzyme, leading to cellular variations in enzyme activity.
MTA Accumulation Is the Basis for the mtn1-1mtn2-1 Phenotype
While our data pointed to the lack of MTA hydrolysis as being the cause of the abnormalities in mtn1-1mtn2-1 plants, the contribution of reduced MTR content was also considered. This concept arose from comparing the completely normal phenotype of MTR kinase mutants (Sauter et al., 2004) with the complex phenotype of MTN-deficient plants. Both mutants are unable to salvage Met, yet MTR kinase mutants have MTR and are normal, whereas the MTN-deficient plants lack MTR and are abnormal. Thus, we tested the involvement of MTR deficiency in the phenotype of mtn1-1mtn2-1 by introducing constitutive expression of MTN1 and hMTAP transgenes into mtn1-1mtn2-1 plants. MTAP metabolizes MTA to MTR-1P without producing MTR. Complementation of the mutant phenotype by either transgene confirms that the abnormal development of MTN-deficient plants is due to their increased MTA content rather than a lack of MTR or Met salvage itself.
MTA accumulates in cells of mtn1-1mtn2-1 that are actively synthesizing PA or NA; this MTA accumulation is obviously both tissue specific and developmentally dependent. When mtn1-1 seedlings are grown on sulfur-deficient medium supplemented with MTA, presumably all their cells are exposed to MTA, resulting in retarded shoot and root growth (Bürstenbinder et al., 2010). We propose that these phenotypes are also due to MTA inhibition of NA and PA synthases, based on the similarity of their metabolite profiles with those of mtn1-1mtn2-1 plants.
The MTA abundance in the leaves of mtn1-1mtn2-1 plants was 2-fold higher than in the wild type: 1.7 ± 0.2 pmol mg−1 fresh weight as compared with 0.8 ± 0.1 pmol mg−1 fresh weight in the wild type. This change was in the same range as that of mtn1-1 grown on MTA: a 2-fold increase in 4-d-old seedlings and a 1.5-fold increase in 3-week-old plants (Bürstenbinder et al., 2010). Interestingly, the MTA accumulation in the double mutant was greater in flowers than in leaves. The MTN level of the mtn1-1mtn2-1 inflorescences was 24 ± 2 pmol mg−1 fresh weight compared with 4.6 ± 0.2 pmol mg−1 fresh weight in the wild type. Thus, in mtn1-1mtn2-1 plants, the degree of MTA accumulation is tissue specific. In accordance with our MTA abundance values, Tassoni et al. (2000) and Naka et al. (2010) found that all PAs (Put, Spd, Spm, and Tspm) are higher in wild-type flowers compared with mature leaves. Thus, flowers must have an increased requirement for MTN activity to hydrolyze the MTA produced during PA biosynthesis.
MTA Binds NA Synthase, Spd Synthase, Spm Synthase, and Tspm Synthase
MTA is known to inhibit PA synthetic enzyme activities while increasing S-adenosyl-Met decarboxylase activity (Albers, 2009). For example, mammalian Spm synthase and Spd synthase activities are MTA sensitive (Pegg et al., 1981; Wu et al., 2008). Our in silico modeling leads us to propose that MTA inhibits Arabidopsis Spd synthase and Tspm synthase, with the latter being the most sensitive. This inhibition is likely quite dynamic, since the stimulation of S-adenosyl-Met decarboxylase by MTA theoretically increases flux to PA synthesis and further enhances MTA production.
Barley (Hordeum vulgare) NA synthase is product inhibited by MTA (Ki = 5 μm; Herbik, 1997). We suspect that the Arabidopsis enzyme is similarly affected, given its amino acid sequence identity (46%) and similar domains (Herbik et al., 1999). Moreover, the mutant plants have chlorotic leaves that are reversed by NA supplementation. Thus, we conclude that MTA inhibits PA and NA synthases in a spatially and temporally dependent manner, reflecting the flux through the Yang cycle and the cellular abundance of the target enzyme activities.
Molecular Basis of the Altered Vascular Development of mtn1-1mtn2-1
The increased venation of mtn1-1mtn2-1 rosette leaves likely contributes to the variation in their free PA content, as PA biosynthesis predominantly occurs in leaf vasculature (Pommerrenig et al., 2011). The increased vein thickness of the MTN mutant rosette leaves is similar to that reported for tkv (Clay and Nelson, 2005). Since our modeling predicted that Tspm synthase activity is inhibited by MTA, and Tspm is required for normal xylem differentiation (Muñiz et al., 2008), it was of interest to examine Tspm content. Unfortunately, Tspm quantification is a challenge due to its low abundance; Tspm was not detected in our system. Despite this observation, our results lead us to believe that mtn1-1mtn2-1 plants have reduced Tspm.
Aside from Tspm effects, PA catabolism may also contribute to the changes in vascular development in mtn1-1mtn2-1. The altered PA content of the mutant (e.g. increased Put) may lead to increased PA oxidase activity and H2O2, a documented signal for increased vascular tissue differentiation (Tisi et al., 2011). However, we did not detect increased H2O2 in 14-d-old mtn1-1mtn2-1 seedlings using a histochemical stain (Supplemental Fig. S4). Similarly, semiquantitative RT-PCR of mature leaves of mtn1-1mtn2-1 did not show significant changes in the transcript levels of three representative PA oxidases (PAO1, PAO2, and PAO5) compared with the wild type (Supplemental Fig. S4). However, more detailed analysis is necessary to determine if PA oxidase activities change in the mature mutant plants or as a result of Spd treatments.
The vascular abnormalities of mtn1-1mtn2-1 appear to be a direct effect of changes in PAT. We base this conclusion on the similarities between the mtn1-1mtn2-1 vasculature and the effects of reducing PAT by naphthylphthalamic acid treatment (Mattsson et al., 1999); both sets of plants have thick veins in their rosette leaves and an increased number of vascular bundles in their stems. Ibañes et al. (2009) created a computational model for vascular bundle development based on DR5::GUS expression of naphthylphthalamic acid-treated plants and anatomical analysis of brassinosteroid (BR) signaling mutants. Their model predicts that PAT-related auxin maxima determine the spacing between vascular bundles while BR controls bundle number. If this is the case, then our results suggest that BR signaling changes may also contribute to the vascular phenotype of mtn1-1mtn2-1.
The question arises, then, why do acl5 and tkv, which also have reduced PAT (Clay and Nelson, 2005; Vera-Sirera et al., 2010), lack the increase in vascular bundles characteristic of mtn1-1mtn2-1 plants? We propose that the differing phenotypes of these Tspm-deficient plants reflect the degree to which PAT is inhibited. mtn1-1mtn2-1 plants have less residual auxin transport as compared with tkv (46% versus 64%, respectively). The greater reduction in PAT in mtn1-1mtn2-1 plants may lead to an increase in their BR content or sensitivity, causing them to have an increased number of bundles. The modest PAT changes in tkv (and acl5) may not be sufficient to trigger the cross talk with BR. This reasoning is consistent with the proposal that the PAT changes in acl5 are secondary to its vascular defects (Vera-Sirera et al., 2010). Direct investigation of BR content and signaling in Tspm-deficient mutants is needed to clarify BR involvement in these phenotypes.
NA Deficiency of mtn1-1mtn2-1 Causes Altered Ion Homeostasis Leading to Interveinal Chlorosis and Reproductive Abnormalities
In addition to free PAs, NA levels were significantly affected in mtn1-1mtn2-1 tissues: NA was undetectable in double mutant rosette leaves, while inflorescences exhibited a 3.7-fold reduction relative to the wild type. This NA deficiency is similar to that of the chloronerva tomato (Solanum lycopersicum) mutant (Böhme and Scholz, 1960), quadruple NA synthase Arabidopsis mutants (nas4x-2 and nas4x-1; Klatte et al., 2009), and transgenic naat tobacco (Nicotiana tabacum) lines overexpressing NA aminotransferase (Takahashi et al., 2003). All these plants have interveinal chlorosis and reduced fertility along with reduced NA content. In all cases, this deficiency is associated with altered profiles of Fe, Zn, Mn, and Cu. Similar to the previously published reports describing mutants with decreased NA, mtn1-1mtn2-1 plants had lower levels of all these ions in both inflorescences and rosette leaves, with the exception of Mn, which increased in leaves.
Restoration of Seed Set in mtn1-1mtn2-1
The reduced presence of double mutants in the segregating progeny of MTN1/mtn1-1mtn2-1mtn2-1 plants is indicative of a reproduction problem. The double mutant was commonly recovered from these segregating populations at a frequency of 1:7 to 1:10, in contrast to the expected 1:4. At least two factors may contribute to the reduction of the double mutant in these F2 populations. (1) mtn1-1mtn2-1 embryos may have insufficient Spd to support their development. This hypothesis is based on the observation that the Spd content of mtnt1-1mtn2-1 inflorescences is lower than in the wild type. If the MTN-deficient embryos are similarly affected, this would cause them to delay or arrest their development, as reported for the Spd synthase double mutant spds1-1spds2-1 that lacks Spd (Imai et al., 2004). The biochemical basis for the embryo lethality of Spd mutants is not known but may relate to the Spd-dependent modification of eIF5a; deoxyhypusine synthase mutants that lack the rate-limiting enzyme activity required for eIF5a hypusination arrest during embryo sac development (Pagnussat et al., 2005). (2) Alternatively, the abnormal growth of mtn1-mtn2-1 pollen tubes may contribute to the reduced recovery of the homozygous double mutant seed. Although we do not know the basis of the pollen tube defect in mtn1-1mtn2-1, this may be associated with their altered polyamine metabolism, since mutants with reduced PAO3 activity also have abnormal pollen tube growth (Wu et al., 2010).
Exogenous supply of 20 μm NA, 100 μm Spd, 100 μm Spm, or 50 μm Tspm improved the fertility of mtn1-1mtn2-1 to a limited extent. When each of these chemicals is supplied exogenously, the plants presumably have a reduced need to synthesize them. As a result, the amount of MTA generated in vivo by these biosynthetic pathways is reduced and the product inhibition of PA or NA synthases is alleviated, thereby improving fertility. Among the compounds tested, 100 μm Spd was the most effective, Spm was the least, and Tspm and NA had intermediate effects. Application of 100 μm Put, 10 or 100 μm IAA, or 5 μm 1-aminocyclopropane-1-carboxylic acid did not produce observable differences. The simplest explanation for these results is that exogenous Spd supplies a limiting compound that is required for fertility or proper embryo development. For example, the Spd supplement may provide an increased precursor pool for Tspm synthesis. Tspm may induce more normal xylem differentiation and restore sufficient transport of compounds essential for seed development, including phytohormones and metal cofactors. Interestingly, a recent report by Tisi et al. (2011) shows that increased xylem differentiation and secondary wall deposition are induced in maize (Zea mays) roots by exogenous Spd feeding. This is apparently induced by H2O2 arising from PA catabolism. Similar H2O2 signaling may contribute to the Spd effect on mtn1-1mtn2-1 plants reported here. It is also possible that the Spd acts by another route aside from simply being the precursor for Tspm or affecting PA catabolism, such as via eIF5A modification. A recent study of pumpkin (Cucurbita maxima) phloem has shown that eIF5A modification occurs in phloem sap (Ma et al., 2010), consistent with the localization of MTN enzymes in companion cells (Pommerrenig et al., 2011). A comprehensive developmental and metabolite analysis of Spd-treated mtn1-1mtn2-1 plants will be required to elucidate the molecular mechanism underlying the effects of both MTN deficiency and Spd feeding. A deeper understanding of this process will provide important insight into the long recognized link between PA and plant development.
CONCLUSION
Although MTN-deficient plants have a complex phenotype, each abnormal trait can be traced back to an effect of MTA accumulation. Given the different enzyme activities inhibited by MTA and their broad range of contributions to plant metabolism, increases in MTA content have a broad impact on both cellular and developmental scales.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds for mtn1-1 (SALK_085385) and mtn2-1 (SALK_071127) single T-DNA insertion lines of Arabidopsis (Arabidopsis thaliana) used to generate mtn1-1mtn2-1 were obtained from the Arabidopsis Biological Resource Center. The male-sterile mutant apt1-3 (Moffatt and Somerville, 1988) was used for reciprocal crosses. All Arabidopsis seeds were surface sterilized with chlorine gas before sowing on plates with 1/2 MS medium solidified with 0.8% (w/v) agarose (Murashige and Skoog, 1962); these plates were incubated in the dark at 4°C for 48 h for seed stratification. On day 3 after sowing, plates were transferred to a tissue culture chamber (TC7; Conviron) under continuous light: 24 h at 21°C with 80 μmol m−2 s−1 photosynthetically active radiation for 10 d. Seedlings were then transplanted to individual pots (25 cm2) containing a 1:1 soil mixture of Sunshine LC1 mix and Sunshine LG3 germination mix (SunGro Horticultural) and maintained in a growth chamber (Conviron) under long-day conditions: 16 h at 22°C with 150 μmol m−2 s−1 photosynthetically active radiation. The plants were watered every 2 d and fertilized with 20:20:20 (nitrogen:phosphorus:potassium) fertilizer mix (Plant Products Co.) once per week until maturity. Hydroponic experiments were set up as described by Tocquin et al. (2003) and maintained in the same growth chambers.
To document the first obvious developmental defect of mtn1-1mtn2-1 compared with the wild type, plate-based and soil-based growth assays were conducted as detailed by Boyes et al. (2001). In the plate-based assay, the time taken to develop the first true leaves of more than 1 mm along with daily lengths of roots were measured over a duration of 11 d. The reproductive defects were evaluated in stage 14 flowers as described by Smyth et al. (1990).
Buds and rosette leaf samples were obtained from wild-type and mtn1-1mtn2-1 plants 2 weeks after bolting unless otherwise stated. When sampled, wild-type plants were 5 weeks old whereas mtn1-1mtn2-1 plants were approximately 7 weeks old, in order for similar developmental stages to be evaluated. Samples were collected before noon for all the experiments.
Microscopy
Bright-Light Microscopy
Developmentally similar (i.e. having the same number of first-order veins arising from the mid vein) wild-type and mtn1-1mtn2-1 rosette leaves and stage 14 pistils (Smyth et al., 1990) were first incubated in clearing solution (3:1, glacial acetic acid:ethanol) for 2 h. Upon clearing, the tissue was rehydrated by moving it from 70% ethanol to water in a gradient series of 15-min intervals each and softened by incubating overnight in 8 m NaOH. For visualization, leaf samples were then stained with 0.1% (w/v) toluidine blue O (pH 6.8) for 15 s and observed with a Zeiss Axiovert 200M microscope (Carl Zeiss). The ovules of the cleared pistils were observed with differential interference contrast optics on a Zeiss Axiovert 200M microscope.
The same stage leaves of both genotypes were fixed in solution (50 mm sodium cacodylate, 2% [w/v] paraformaldehyde [pH 7], and 0.1% glutaraldehyde) for 1 h under vacuum at room temperature. The samples were then transferred to fresh fixative and incubated overnight at 4°C. After the appropriate amount of fixation, the samples were dehydrated in a series of ethanol under vacuum at room temperature, going from 10% to 70% ethanol in 15% intervals for 1 h each. The samples were left in 70% ethanol and 0.2% eosin overnight at 4°C. Once fully dehydrated, the tissue was slowly infiltrated with LR White plastic resin (hard grade; Canemco) in a series of ethanol:resin volumes (3:1, 1:1, and 1:3 for 2 h each), then in 100% LR White for 1 h, and then in fresh LR White overnight. Once the samples were fully infiltrated with resin, the LR White was refreshed and the samples were polymerized in a 60°C to 65°C oven for 2 to 24 h. Once the resin had hardened, tissue was sectioned at 1 μm thickness using an ultramicrotome (UltracutE; Reichert) and stained with toluidine blue O for observation with a Zeiss Axiovert 200M microscope.
Scanning Electron Microscopy
Anthers of stage 14 flowers from the wild type and mtn1-1mtn2-1 were placed in fixative (4% [w/v] paraformaldehyde [pH 7] and 2% [w/v] glutaraldehyde dissolved in potassium-buffered saline for 1 h) under vacuum at room temperature. Upon replacing the fixative solution, the samples were left to incubate overnight at 4°C. The tissue was then dehydrated in a 10% ethanol gradient (50%–95%) at 30-min intervals under vacuum. Upon reaching 95% ethanol concentration, samples were left overnight at 4°C.The samples were then moved to HPLC-grade acetone for 2 h at room temperature. The samples were dried using a mass critical point dryer, mounted on 2-inch stubs, and sputter coated with gold particles. The tissue was visualized using a Hitachi S570 scanning electron microscope.
Laser Scanning Confocal Microscopy
The expression patterns of DR5rev::GFP stem sections of both wild-type and mtn1-1mtn2-1 backgrounds were observed using a Zeiss LSM 510 confocal microscope. The GFP signals were excited with an argon ion laser at 488 nm, and emissions were captured with a 505/530-nm band-pass filter. Chlorophyll autofluorescence was also excited at 488 nm and was a captured with 650-nm long-pass filter.
Chemical Complementation Experiments
Seeds were placed on 1/2 MS medium supplemented with one of the following chemicals: 20 μm NA (crude K1 extract; see below), 20 μm wild-type extract, 100 μm Spd, 100 μm Spm, and 50 μm Tspm. After 2 d of cold incubation, these plates were transferred to continuous light for 21 d. The seedlings were then transplanted to soil or hydroponic medium and grown under long-day conditions. In the case of soil-grown plants, chemicals were supplied daily as drops (approximately 20 μL) in 1/2 MS to each vegetative and reproductive meristem over a period of 3 to 4 weeks. However, Tspm was not applied as daily drops due to its limited availability.
Extraction of NA from K1 Plants
NA-overproducing Arabidopsis transgenic lines (line K1 described by Pianelli et al., 2005) were grown until the full rosette stage. NA was extracted from the rosette leaves ground in 80°C deionized water; corresponding extracts from wild-type leaves were used as controls. The K1 extract contained 50 pmol mg−1 fresh weight NA compared with the wild type, and neither K1 nor wild-type extracts contained detectable PAs, indicating that the K1 is a source of NA but not PAs.
Complementation of mtn1-1mtn2-1
For complementation, pMTN1::MTN1, UBQ10::MTN1 coding sequences (CDS), and UBQ10::hMTAP CDS were introduced into mtn1-1mtn2-1 mutants. The UBQ10 (AT4G05320) promoter (693 bp) was amplified using UBQ10pF and UBQp blunt R primers (Supplemental Table S1) and cloned into pSAT5.nosP.RNAi.nosT (EUO49865) with AgeI and HincII to generate pSAT5.UB10p.nosT. This vector was then used as a recipient for MTN1 and hMTAP CDS. Binary vectors pGreenII 0229 (www.pgreen.ac.uk) and pPZP-RCS2-BAR (CD3-1057; Arabidopsis Biological Resource Center) were used for Agrobacterium tumefaciens-mediated transformation.
Genomic MTN1 was amplified using the following primers: gMTN1-F and gMTN1-R (Supplemental Table S1). The 1.9-kb genomic MTN1 fragment with 162 bp of promoter was then digested with SpeI and BamHI and cloned directly into binary vector pGreenII 0229. The MTN1 cDNA was amplified with the following primers: MTN1CDS-F and MTN1CDS-R (Supplemental Table S1). The resulting 0.8-kb fragment was digested with XmaI and BamHI and cloned first into pSAT5.UB10p.nosT. After sequencing, the UB10p::CDS cassette was transferred to PZP-RCS2-BAR. Similarly, the 0.8-kb hMTAP cDNA fragment was amplified from hMTAP cDNA with hMTAP-F and hMTAP-R primers (Supplemental Table S1) and cloned into the SmaI site of pSAT5-UBQ10p-nosT, and the resulting UBQ10::hMTAP cassette was inserted into pPZP-RCS2-BAR.
MTN1 constructs were introduced into Agrobacterium strain GV3101Soup, while UBQ10::MTN1CDS and UBQ10::hMTAP were placed in Agrobacterium strain GV3101. Homozygous mtn1-1 plants were transformed with their respective Agrobacterium strains using the floral dipping method (Clough and Bent, 1998). Transgenic T1 individuals were selected for by spraying 10-d-old soil-grown seedlings with 40 mg L−1 Basta (glufosinate ammonium; Wilson Laboratories). The expression of MTN1 and hMTAP in the surviving T1 plants was confirmed by immunoblot, and successful transformants were crossed with mtn1-1/mtn1-1MTN2-1/mtn2-1plants. The resulting F1 progeny were germinated on medium containing Basta and genotyped by PCR to identify mtn1-1mtn2-1 F2 individuals. F2 seeds of three lines were selected and screened on Basta-supplemented MS medium and genotyped to identify mtn1-1mtn2-1 seedlings.
RT-PCR
Total RNA was extracted from leaves and buds of wild-type and mtn1-1mtn2-1 plants using Tri Pure Isolation Reagent (Roche). Two micrograms of total RNA was used for first-strand synthesis with SuperScript reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. PCR was performed as described by Bürstenbinder et al. (2007) using AtMTN1-F3, AtMTN1-R3, AtMTN2-F1, AtMTN2-R1, Actin 8F, and Actin 8R primers (Supplemental Table S1) to determine the transcript abundance of MTN1 (851 bp), MTN2 (306 bp), and actin (700 bp).
MTN Enzyme Assay
Bud protein extracts were prepared from developmentally matched flowering plants in 50 mm potassium phosphate buffer (pH 7; Bürstenbinder et al., 2010). Twenty micrograms of soluble protein from buds was used to determine MTN-specific activity using a xanthine oxidase-coupled enzyme assay (Lee et al., 2005). Absorbance was measured every 10 min over 1 h with a DU530 spectrophotometer (Beckman Coulter) at 470 nm modified from the protocol outlined by Bürstenbinder et al. (2010).
PAT Assay
PAT in primary inflorescence stem segments of wild-type, mtn1-1mtn2-1, and tkv plants was measured using a modification of the procedure provided by Okada et al. (1991) as described by Clay and Nelson (2005). The inflorescence stem segments were immersed in 14C-labeled IAA (Sigma) in either an acropetal or basipetal orientation. After 19 h of incubation, the uppermost 5 mm of the stem segments was removed and incubated overnight in liquid scintillation cocktail (Cytoscint; MP Biomedicals). The released radioactivity was counted using a scintillation counter (model LS 1701; Beckman).
Immunoblot Analysis
Ten micrograms of total soluble protein was extracted as described above, separated on a 12.5% SDS-polyacrylamide gel (Laemmli, 1970), and transferred onto a polyvinylidene difluoride membrane using a semidry electroblotting system (Bio-Rad Laboratories). The membrane blot was cut near the 34-kD marker so the bottom portion could be incubated for 2 h with anti-MTN1 (1:2,500) antibody, while the top portion was incubated for the same time with monoclonal antibody for actin (1:3,000; MP Biomedicals). Bound antibodies were detected following incubation with horseradish peroxidase-conjugated secondary antibodies using the ECL Plus Western Blotting Detection System (GE Healthcare) and Amersham Hyperfilm ECL.
Two-Dimensional Electrophoresis
Protein was extracted using phenol followed by ammonium acetate-methanol precipitation, as described by Zheng et al. (2007). Protein was quantified using the RC/DC Protein Assay Kit (Bio-Rad Laboratories). These proteins were then separated based on isoelectric focusing on a Multiphore II system (GE Healthcare) using Immobiline dry strip gels with nonlinear pH gradients (pH 4–7). These strips were placed on 12.5% SDS-polyacrylamide gels as detailed by Zheng et al. (2007). The separated proteins were then blotted as described for immunoblot analysis above. The membrane blot was incubated overnight at 4°C with eIF5A-specific primary antibody (1:3,000). The blots were detected as described above. Digital images of these films were analyzed using ImageQuant TL analysis software (GE Healthcare).
Determination of PA Content
Rosette leaves and inflorescences of wild-type and mtn1-1mtn2-1 plants were mixed with 5% (v/v) perchloric acid at a ratio of 1:4 (w/v) and frozen at –20οC. Samples were further processed, and Put, Spd, and Spm were quantified by HPLC as described Minocha et al. (1990, 1994).
Determination of NA Content
NA was extracted from leaves and inflorescences of wild-type and mtn1-1mtn2-1 plants and quantified after labeling with 9-fluorenyl methyl chloroformate, as described in the supplemental material of Klatte et al. (2009).
Determination of MTA Content
Leaves and inflorescences of wild-type and mtn1-1mtn2-1 plants (0.1 g) were ground in liquid nitrogen to a fine powder, which was extracted with 1 mL of 0.1 m HCl for 15 min at 4°C. Cell debris were removed by centrifugation twice at 28,000g for 15 min at 4°C. MTA was determined as described by Rzewuski et al. (2007) using the same HPLC system.
Detection of Metal Contents
Samples were dried for constant weight at 60°C, subjected to acid digestion with Omni-Trace1 nitric acid (EMI Scientific), and then heated to 85°C to 90°C. Nitric acid blanks were processed to ensure that metals were not added during sample preparation. The standards tomato (Solanum lycopersicum; National Institute of Standards and Technology standard no. 1573a) and corn (Zea mays; National Institute of Standards and Technology standard no. 1573a) leaves were also processed without sample to quantify the recovery of Zn, Cu, Mn, and Fe during the extraction procedure. Filtered samples were prepared and analyzed for metals as described by Gadapati and Macfie (2006) by inductively coupled plasma atomic emission spectroscopy.
Modeling of MTA Inhibition for Tspm Synthase and Spd Synthase1
A three-dimensional model of ACL5 was built by the Swiss-model suite (Arnold et al., 2006) using the crystal structure of PA aminopropyl transferase from Thermus thermophilus (Protein Data Bank identifier 1UIR; Ohnuma et al., 2011) as a template (39% identity). The crystal structure of Arabidopsis Spd synthase1 is available in the Protein Data Bank (identifier 1XJ5). To obtain Tspm synthase-MTA and Spd synthase1-MTA complexes, MTA was docked using DOCK 4.0 (Ewinget al., 2001). The programs COOT (Emsley and Cowtan, 2004) and PyMOL (DeLano Scientific) were used for structure visualization and figure preparation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MTN activity of various organs harvested from 6-week-old wild-type plants.
Supplemental Figure S2. Leaf vein pattern and pollen tube growth of mtn1-1mtn2-1 compared with the wild type.
Supplemental Figure S3. Rescue of interveinal chlorosis.
Supplemental Figure S4. Relative transcript abundance of polyamine oxidases and H2O2 production.
Supplemental Table S1. Primer sequences.
Supplemental Materials and Methods S1. Leaf clearing, aniline blue staining, 3,3′-diaminobenzidine staining, and RT-PCR of polyamine oxidases.
Supplementary Material
Acknowledgments
We thank Steven Chatfield (University of Guelph) for helpful early discussions of the mutant; Prof. Shiela Macfee (University of Western Ontario) for generous guidance of the sample preparation for ionomic analysis; Yong Li (University of Waterloo) for constructing the pSAT5.UB10p.nosT vector and help with designing the cloning strategy for complementation experiments, cloning of UBQ10::hMTAP, and conducting PA oxidase RT-PCR; and Jaideep Mathur (University of Guelph) for help with imaging and Tzann-Wei Wang (University of Waterloo), Qifa Zheng (University of Waterloo), and Lilian Lee (University of Waterloo) for technical assistance. Seeds of the auxin reporter line DR5rev::GFP, the NA-overproducing Arabidopsis line, and tkv were kind gifts from Jiri Friml (Ghent University), Pierre Czernic (Université Montpellier), and Timothy Nelson (Yale University), respectively. T-DNA-tagged lines were supplied by the Arabidopsis Biological Resource Center. The hMTAP cDNA was a gift of W.D. Kruger (Fox Chase Cancer Center), and M. Niitsu of the Faculty of Pharmaceutical Sciences, Josai University, generously provided Tspm.
References
- Albers E. (2009) Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life 61: 1132–1142 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Baron K, Stasolla C. (2008) The role of polyamines during in vivo and in vitro development. In Vitro Cell Dev Biol 44: 384–395 [Google Scholar]
- Bertino JR, Waud WR, Parker WB, Lubin M. (2011) Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies. Cancer Biol Ther 11: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme H, Scholz G. (1960) Versuchezur Normalisierung des Phänotyps Mutante Chloronerva von Lycopersicon esculentum Mill. Kulturpflanze 8: 93–109 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M. (2007) The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J 49: 238–249 [DOI] [PubMed] [Google Scholar]
- Bürstenbinder K, Waduwara I, Schoor S, Moffatt BA, Wirtz M, Minocha SC, Oppermann Y, Bouchereau A, Hell R, Sauter M. (2010) Inhibition of 5′-methylthioadenosine metabolism in the Yang cycle alters polyamine levels, and impairs seedling growth and reproduction in Arabidopsis. Plant J 62: 977–988 [DOI] [PubMed] [Google Scholar]
- Clay NK, Nelson T. (2005) Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol 138: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S. (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot (Lond) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler NG. (2006) The shoot apical meristem and development of vascular architecture. Can J Bot 84: 1660–1671 [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Ewing TJA, Makino S, Skillman AG, Kuntz ID. (2001) DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 15: 411–428 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gadapati WR, Macfie SM. (2006) Phytochelatins are only partially correlated with Cd-stress in two species of Brassica. Plant Sci 170: 471–480 [Google Scholar]
- Hanzawa Y, Takahashi T, Komeda Y. (1997) ACL5: an Arabidopsis gene required for internodal elongation after flowering. Plant J 12: 863–874 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y. (2000) ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J 19: 4248–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A. (1997) Proteinchemische und molecular biologische Charakterisierung der Tomaten mutante chloronerva. PhD thesis. Humboldt University, Berlin [Google Scholar]
- Herbik A, Koch G, Mock HP, Dushkov D, Czihal A, Thielmann J, Stephan UW, Bäumlein H. (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley: a key enzyme for iron homeostasis in plants. Eur J Biochem 265: 231–239 [DOI] [PubMed] [Google Scholar]
- Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI. (2009) Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA 106: 13630–13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A, Akiyama T, Kato T, Sato S, Tabata S, Yamamoto KT, Takahashi T. (2004) Spermine is not essential for survival of Arabidopsis. FEBS Lett 556: 148–152 [DOI] [PubMed] [Google Scholar]
- Kakehi J, Kuwashiro Y, Niitsu M, Takahashi T. (2008) Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol 49: 1342–1349 [DOI] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P. (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W. (2011) iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Luong W, Huang DJT, Cornell KA, Riscoe MK, Howell PL. (2005) Mutational analysis of a nucleosidase involved in quorum-sensing autoinducer-2 biosynthesis. Biochemistry 44: 11049–11057 [DOI] [PubMed] [Google Scholar]
- Ma Y, Miura E, Ham BK, Cheng HW, Lee YJ, Lucas WJ. (2010) Pumpkin eIF5A isoforms interact with components of the translational machinery in the cucurbit sieve tube system. Plant J 64: 536–550 [DOI] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T. (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979–2991 [DOI] [PubMed] [Google Scholar]
- Meinke DW. (1994) Seed development in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 253–295 [Google Scholar]
- Minocha R, Shortle WC, Long SL, Minocha SC. (1994) A rapid and reliable procedure for extraction of cellular polyamines and inorganic ions from plant tissues. J Plant Growth Regul 13: 187–193 [Google Scholar]
- Minocha SC, Minocha R, Robie CA. (1990) High-performance liquid chromatographic method for the determination of dansyl-polyamines. J Chromatogr A 511: 177–183 [Google Scholar]
- Moffatt B, Somerville C. (1988) Positive selection for male-sterile mutants of Arabidopsis lacking adenine phosphoribosyl transferase activity. Plant Physiol 86: 1150–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz L, Minguet EG, Singh SK, Pesquet E, Vera-Sirera F, Moreau-Courtois CL, Carbonell J, Blázquez MA, Tuominen H. (2008) ACAULIS5 controls Arabidopsis xylem specification through the prevention of premature cell death. Development 135: 2573–2582 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Naka Y, Watanabe K, Sagor GH, Niitsu M, Pillai MA, Kusano T, Takahashi Y. (2010) Quantitative analysis of plant polyamines including thermospermine during growth and salinity stress. Plant Physiol Biochem 48: 527–533 [DOI] [PubMed] [Google Scholar]
- Oh SI, Park J, Yoon S, Kim Y, Park S, Ryu M, Nam MJ, Ok SH, Kim JK, Shin JS, et al. (2008) The Arabidopsis calcium sensor calcineurin B-like 3 inhibits the 5′-methylthioadenosine nucleosidase in a calcium-dependent manner. Plant Physiol 148: 1883–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma M, Ganbe T, Terui Y, Niitsu M, Sato T, Tanaka N, Tamakoshi M, Samejima K, Kumasaka T, Oshima T. (2011) Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus. J Mol Biol 408: 971–986 [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pegg AE, Casero RA., Jr (2011) Current status of the polyamine research field. Methods Mol Biol 720: 3–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE, Shuttleworth K, Hibasami H. (1981) Specificity of mammalian spermidine synthase and spermine synthase. Biochem J 197: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianelli K, Mari S, Marquès L, Lebrun M, Czernic P. (2005) Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Res 14: 739–748 [DOI] [PubMed] [Google Scholar]
- Pommerrenig B, Feussner K, Zierer W, Rabinovych V, Klebl F, Feussner I, Sauer N. (2011) Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23: 1904–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzewuski G, Cornell KA, Rooney L, Bürstenbinder K, Wirtz M, Hell R, Sauter M. (2007) OsMTN encodes a 5′-methylthioadenosine nucleosidase that is up-regulated during submergence-induced ethylene synthesis in rice (Oryza sativa L.). J Exp Bot 58: 1505–1514 [DOI] [PubMed] [Google Scholar]
- Sauter M, Cornell KA, Beszteri S, Rzewuski G. (2004) Functional analysis of methylthioribose kinase genes in plants. Plant Physiol 136: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KKW, Asmus K, Zhang AN, Horvatin C, Li S, Liu T, Moffatt B, Woods VL, Jr, Howell PL. (2011) Mechanism of substrate specificity in 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidases. J Struct Biol 173: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KKW, Lee JE, Sufrin JR, Moffatt BA, McMillan M, Cornell KA, Isom C, Howell PL. (2008) Molecular determinants of substrate specificity in plant 5′-methylthioadenosine nucleosidases. J Mol Biol 378: 112–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AP, Spangler B, Wallner S, Kreutz M, Dettmer K, Oefner PJ, Bosserhoff AK. (2009) Direct and tumor microenvironment mediated influences of 5′-deoxy-5′-(methylthio)adenosine on tumor progression of malignant melanoma. J Cell Biochem 106: 210–219 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK. (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15: 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kakehi JI. (2010) Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann Bot (Lond) 105: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassoni A, van Buuren M, Franceschetti M, Fornalè S, Bagni N. (2000) Polyamine content and metabolism in Arabidopsis thaliana and effect of spermidine on plant development. Plant Physiol Biochem 38: 383–393 [Google Scholar]
- Tisi A, Federico R, Moreno S, Lucretti S, Moschou PN, Roubelakis-Angelakis KA, Angelini R, Cona A. (2011) Perturbation of polyamine catabolism can strongly affect root development and xylem differentiation. Plant Physiol 157: 200–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Périlleux C. (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol 3: 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Sirera F, Minguet EG, Singh SK, Ljung K, Tuominen H, Blázquez MA, Carbonell J. (2010) Role of polyamines in plant vascular development. Plant Physiol Biochem 48: 534–539 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. (2008) Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J Biol Chem 283: 16135–16146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Shang Z, Wu J, Jiang X, Moschou PN, Sun W, Roubelakis-Angelakis KA, Zhang S. (2010) Spermidine oxidase-derived H2O2 regulates pollen plasma membrane hyperpolarization-activated Ca(2+)-permeable channels and pollen tube growth. Plant J 63: 1042–1053 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Zheng Q, Song J, Doncaster K, Rowland E, Byers DM. (2007) Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis. J Agric Food Chem 55: 1663–1673 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.