Abstract
Sugar is essential for all cellular activities, but at high levels it inhibits growth and development. How plants balance the tradeoffs between the need for sugars and their growth inhibitory effects is poorly understood. SHORT-ROOT (SHR) and SCARECROW (SCR) are key regulators of stem cell renewal and radial patterning in the root of Arabidopsis (Arabidopsis thaliana). Recently, we identified direct targets of SHR at the genome scale. Intriguingly, among the top-ranked list, we found a number of genes that are involved in stress responses. By chromatin immunoprecipitation-polymerase chain reaction (PCR), we showed that SHR and SCR regulate a similar but not identical set of stress response genes. Consistent with this, scr and shr were found to be hypersensitive to abscisic acid (ABA). We further showed that both mutants were hypersensitive to high levels of glucose (Glc) but responded normally to high salinity and osmoticum. The endogenous levels of sucrose, Glc, and fructose were also elevated in shr and scr. Intriguingly, although shr had sugar content and developmental defects similar to those of scr, it was much less sensitive to Glc. Chromatin immunoprecipitation-PCR and reverse transcription-PCR assays as well as transgenic studies with an ABA-INSENSITIVE2 (ABI4)-β-glucuronidase reporter construct revealed that in root, SCR, but not SHR, repressed ABI4 and ABI5 directly and specifically in the apical meristem. When combined with abi4, scr became much more tolerant of high Glc. Finally, transgenic plants expressing ABI4 under the control of the SCR promoter manifested a short-root phenotype. These results together suggest that SCR has a SHR-independent role in mitigating the sugar response and that this role of SCR is important for root growth.
In both animals and plants, sugar is critically important for all cellular activities, but at high concentrations, it becomes inhibitory to growth and development (Rolland et al., 2006). Although soluble sugar concentration may be low on average in plants, it can reach an inhibitory level in some organs or cell types, particularly when the rate of photosynthesis is high. Sugar is made in photosynthetic organs, fully expanded leaves in particular, and transported to nonphotosynthetic organs such as root, shoot apical meristem, and developing embryo. In most plant species, Suc is the form that is transported over long distances, but Glc is the sugar that cells can directly utilize. At the site of sugar production, Glc is converted to Suc, which is in turn loaded into the phloem for long-distance transport. Phloem companion cells are known to facilitate this process by concentrating Suc through an active mechanism, but the bundle sheath cells also play a role in sugar loading (Leegood, 2008). In organs that import sugar, Suc is converted to Glc by locally expressed Suc synthase or invertase (Koch, 2004). In root, for example, Suc invertase and synthase are expressed preferentially in the meristem and early elongation zone (Duke et al., 1991; Barratt et al., 2009). Consequently, sugar concentration is high in cells that are involved in uploading and downloading. To ensure optimal growth, plants must monitor and regulate the sugar level closely and suppress the harmful effects of exposure to high sugar.
How plants balance the tradeoffs between the need for sugars and their growth inhibitory effects is poorly understood, but presumably, this involves regulation of the sugar-signaling pathways. Sugar signaling is a conserved mechanism in eukaryotic organisms (Rolland et al., 2006). In yeast, sugar availability is sensed by HEXOKINASE2 (HXK2) and the signal is relayed to SUCROSE NONFERMENTATION1 (SNF1), which in turn alters the expression of sugar-responsive genes (Rolland et al., 2006). In plants, HXK1 and SNF1-RELATED PROTEIN KINASE1 (SnRK1) are the major players in sugar signaling (Moore et al., 2003), with ABA-INSENSITIVE4 (ABI4) acting downstream in the regulation of nuclear genes involved in the sugar response (Arenas-Huertero et al., 2000; Koussevitzky et al., 2007). In addition to HXK1 and HXK2, which are localized in the mitochondria and nucleus (Cho et al., 2006), plants possess a HXK in the chloroplasts that also acts as a Glc sensor (Zhang et al., 2010). Evidence suggest that other sugar-signaling pathways exist (Xiao et al., 2000), although little is known about these HXK-independent pathways.
A potential mechanism for plants to orchestrate the sugar response and development is to regulate both processes with the same factors. Several genes have been identified that appear to play such a role. PLEIOTROPIC REGULATORY LOCUS1 (PRL1), for example, is known to negatively regulate Glc signaling by interacting with SnRK1 kinase (Bhalerao et al., 1999). The prl1 mutant is hypersensitive to high levels of Glc, but only under light (Németh et al., 1998), suggesting that PRL1 plays an important role in coordinating light and sugar signaling. STIMPY/WOX9, on the other hand, seems to be a positive coordinator between sugar signaling and plant growth, as the stip mutant is compromised in shoot apical meristem development but this defect can be rescued by exogenous Suc (Wu et al., 2005). Other proteins that play dual roles in sugar signaling and plant development include HYPERSENESCENCE1, which was initially found to be a player in the defense response (Aki et al., 2007); LOW-LEVEL BETA-AMYLASE1, a UPF1 RNA helicase (Yoine et al., 2006); and HIGH SUGAR RESPONSE8, which is involved in Ara biosynthesis (Li et al., 2007b).
SHORT-ROOT (SHR) and SCARECROW (SCR) are key regulators of root growth and development (Di Laurenzio et al., 1996; Helariutta et al., 2000). They are both essential for the maintenance of the stem cell niche, which is a group of pluripotent cells surrounding the quiescent center (QC) cells, with the QC acting as the organizing center (Supplemental Fig. S1). In shr and scr, the stem cell niche is depleted early, resulting in plants with substantially shorter roots than the wild type (Benfey et al., 1993). SHR and SCR also play an important role in radial patterning. In wild-type primary root, the ground tissue consists of two cell layers, the cortex and endodermis, which lie between the epidermis and the central vasculature or stele (Supplemental Fig. S1). The cortex and endodermis are derived from a common progenitor cell called the cortex/endodermis initial daughter cell (CEID) through a longitudinal asymmetric cell division (Supplemental Fig. S1). In shr and scr mutants, the asymmetric cell division does not occur, resulting in a ground tissue consisting of only a single cell layer.
Although SHR and SCR play a similar role in ground tissue patterning, they are expressed in different domains. SCR is expressed in the QC and endodermis and is required for the longitudinal asymmetric cell division in the CEID that gives rise to the cortex and endodermis lineages (Di Laurenzio et al., 1996; Cui et al., 2007; Cui and Benfey, 2009). In contrast, SHR is expressed in the stele, but the protein moves to the adjacent cell layer including the endodermis, CEID, and QC, where it physically interacts with SCR to activate a positive feedback loop for SCR transcription and a feed-forward loop for other factors that appear also to play a role in radial patterning (Helariutta et al., 2000; Cui et al., 2007). Independent of SCR, SHR activates genes that are involved in endodermis cell fate specification. A role for SHR in stele development has also been identified (Levesque et al., 2006; Carlsbecker et al., 2010; Yu et al., 2010). SCR expression in root apical meristem is largely dependent on SHR, placing SHR upstream in the mechanism that determines radial patterning. However, there is evidence that SCR expression is also regulated by a SHR-independent mechanism. SCR is still expressed in shr, albeit at a reduced level, and its expression pattern is similar to that in the wild type (Cui et al., 2007). Thus, it appears that SHR and SCR control root development through both common and distinct mechanisms.
SHR and SCR belong to the GRAS family of transcriptional regulators (Pysh et al., 1999), and we have shown that they regulate downstream genes directly (Levesque et al., 2006; Cui et al., 2007; Sozzani et al., 2010). To better understand how they control root growth and development, recently we have determined the genome-wide locations of SHR binding sites using chromatin immunoprecipitation ChIP followed by analysis on microarrays (ChIP-chip; Sozzani et al., 2010). In further analysis of the ChIP-chip data, we uncovered a role for SHR in vascular tissue patterning through the regulation of cytokinin homeostasis (Cui et al., 2011). Intriguingly, among the top-ranked list of SHR direct targets, we noticed a number of genes that are associated with stress responses in addition to those that presumably play a role in development. We showed that both SHR and SCR directly regulate these stress response genes. This preliminary observation has led us to the findings that SCR, but not SHR, suppresses the sugar response through direct repression of ABI4 in the root apical meristem and that this role appears to be important for normal root growth.
RESULTS
SHR and SCR Control Genes Involved in Both Development and Stress Responses
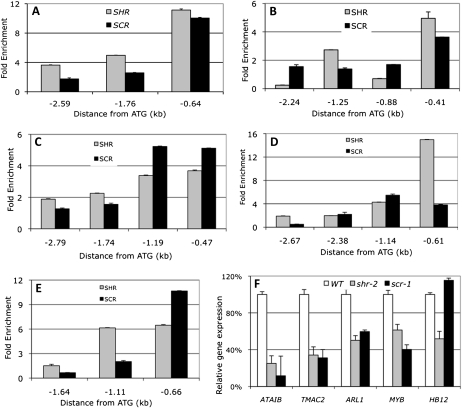
Listed in Supplemental Table S1 are the 25 top-ranked SHR targets that we recently have identified by ChIP-chip (Cui et al., 2011). In addition to genes that are known to play a role in development, such as MAGPIE (Welch et al., 2007), NUTCRACKER (Welch et al., 2007), SCARECROW-LIKE3 (Heo et al., 2011), and microRNA166B (Carlsbecker et al., 2010), we noticed that some of these genes are associated with stress responses, such as ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR (ATAIB; Li et al., 2007a), TWO OR MORE ABRES-CONTAINING GENE2 (TMAC2; Huang and Wu, 2007), and ARABIDOPSIS THALIANA HOMEOBOX12 (ATHB12; Olsson et al., 2004). To determine whether these genes are true SHR targets, we performed ChIP-PCR and confirmed binding by SHR (Fig. 1, A–E). We also confirmed binding by SHR to the promoters of At1g68670, a MYB domain transcription factor, and At1g24120, which appear to be involved in stress responses as well on the basis of our analysis of T-DNA knockout mutants (Supplemental Fig. S2). We next compared their transcript levels in shr and the wild type by reverse transcription (RT)-PCR and found that all these stress-associated genes were reduced in expression in the shr-2 mutant (Fig. 1F).
Figure 1.
SHR and SCR directly control stress response genes. A to E, ChIP-PCR assay showing binding of SHR and SCR to the promoters of ATAIB (A), TMAC2 (B), ATHB12 (C), At1g68670, a MYB transcription factor (D), and ARL1 (At1g24120; E). The x axis indicates the distance (kb) from the translation start site. F, Real-time RT-PCR assay showing the transcript level for the above genes in wild-type (WT), shr-2, and scr-1 root. Error bars represent sd from technical replicates. Note the different y axis scales.
Previously, we demonstrated that SHR and SCR work as a heterodimer in radial patterning (Cui et al., 2007), so SCR probably also directly regulates these stress-associated genes. ChIP-PCR and RT-PCR assays showed that, indeed, SCR binds to the promoters of the same set of genes (Fig. 1, A–E) and that their transcript levels decreased in the scr mutant (Fig. 1F), except for ATHB12, whose expression does not seem to be altered.
SCR Plays a Role in the Sugar Response
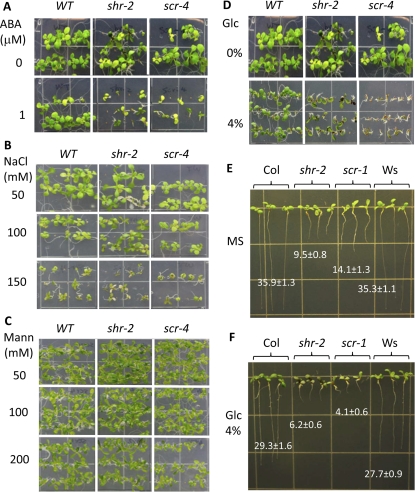
The observation that SHR and SCR directly regulate stress-associated genes suggests that these two proteins are involved in stress responses. To test this hypothesis, we treated shr and scr mutants with the plant hormone abscisic acid (ABA), because ABA accumulates in response to various stresses, and we reasoned that exogenous application would mimic stress. The shr-2 and scr-4 alleles were used in this test. Although shr-2 and scr-4 have significantly shorter roots than the wild type, when grown on normal growth (Murashige and Skoog [MS]) medium their shoots are of similar size during early stages of seedling development (Fig. 2A, top row). In contrast, on MS medium containing ABA, the shoots of both mutants were much smaller than when grown on MS medium alone, and root growth was severely retarded (Fig. 2A, bottom row). These results extend our recent finding that SCR mediates developmental processes and stress responses (Iyer-Pascuzzi et al., 2011) but also suggest that SHR plays a role in stress responses too.
Figure 2.
shr and scr are hypersensitive to high Glc but not salt and mannitol. A, Wild-type (WT), shr-2, and scr-4 seedlings on MS medium (top row) or MS medium containing 1 μm ABA (bottom row), 10 d after germination. B, The same three seedling types grown on MS medium containing 50, 100, and 150 mm NaCl, 2 weeks after germination. C, The same three seedling types grown on MS medium containing 50, 100, and 200 mm mannitol (Mann), 2 weeks after germination. D, The same three seedling types grown on MS medium (top row) or MS medium containing 4% Glc (bottom row), 10 d after germination. E and F, shr-2 and scr-1 as well as wild-type Col and Ws seedlings growing on MS medium with (F) or without (E) 4% Glc, 10 d after germination. The numbers represent root length in millimeters (means ± sd) from measurements of 15 seedlings. In A to D, only Col is shown as the wild type, as Col and Ws behaved similarly under the conditions tested.
ABA signaling is involved in numerous stress responses. To identify the specific response in which SHR and SCR are involved, we challenged shr-2, scr-4, and the wild type with a number of stresses. When grown under conditions of high salinity (NaCl) or high osmoticum (mannitol), the shr-2 and scr-4 mutants responded similarly to the wild type (Fig. 2, B and C). We also tested their response to Glc, as GBF5, a key component in the Glc-signaling pathway (Rolland et al., 2006), is also among the top-ranked putative SHR direct targets. Interestingly, we found that both mutants were hypersensitive to Glc at a concentration of 4% and above, as indicated by pigment accumulation and retarded growth, but scr-4 was apparently much more sensitive (Fig. 2D). Because the two mutants (scr-4 and shr-2) are in different ecotype backgrounds (Wassilewskija [Ws] and Columbia [Col], respectively), we next tested the response of different scr and shr alleles to Glc (shr-2, shr-3, shr-5, and scr-3 are in Col; scr-1, scr-2, and scr-4 are in Ws). All shr and scr alleles examined showed Glc hypersensitivity (Supplemental Fig. S3). Regardless of their genetic background, all scr alleles consistently showed a much more severe Glc hypersensitivity phenotype than the shr alleles: scr mutant plants were very small and their leaves were bleached, whereas shr seedlings were much larger and the leaves were largely green despite the accumulation of purple pigments. Consistent with this result, scr root growth was also inhibited to a greater extent than shr when they were grown vertically (Fig. 2, E and F). Because shr and scr have similar defects in root growth and ground tissue patterning, the sugar hypersensitivity of scr is unlikely to be a secondary effect from its developmental defect. These results suggest that SCR plays a major role in the sugar response.
Sugar Levels Are Elevated in scr
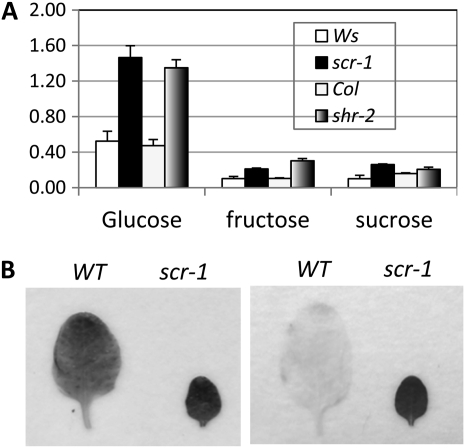
The scr mutant should become hypersensitive to high Glc if sugar metabolism or signaling is altered. To investigate this possibility, we measured Suc, Glc, and Fru content in scr and wild-type (Ws) plants. The concentrations of all three water-soluble sugars were significantly higher in both the root and shoot of scr than in the wild type when plants were grown on MS medium (Supplemental Table S2). An even larger difference in sugar content was apparent when the plants were grown in soil (Fig. 3A). To determine whether the increase in sugar content in scr results from defects in starch biosynthesis, we next compared starch content in scr and the wild type by Lugol staining. As shown in Figure 3B (left panel), at the end of the illumination period, starch accumulated to similar levels in scr and the wild type, indicating normal starch biosynthesis in scr. At the end of the dark period, all starch had been consumed in the wild type, but its level was still high in scr (Fig. 3B, right panel). These results suggest that the Glc hypersensitivity of scr is due to a defect in sugar homeostasis.
Figure 3.
Sugar homeostasis is compromised in scr and shr. A, Glc, Fru, and Suc contents (mg g−1 fresh weight) in leaves of 10-d-old wild-type (Ws and Col), scr-1 and shr-2 plants, measured by HPLC. B, Starch content of wild-type (WT) and scr-1 leaves, as revealed by Lugol staining. Left, samples taken in the evening; right, samples taken in the morning.
ABI4 Is Dramatically Up-Regulated in scr but Not in shr
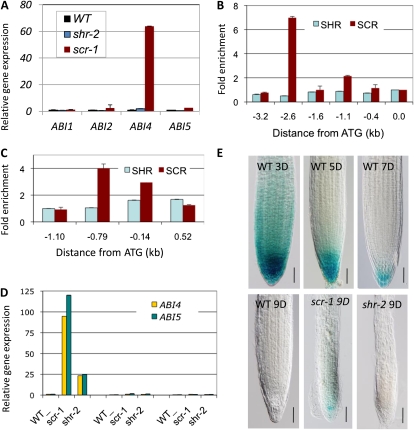
Because shr was much less sensitive to high Glc than scr, we reasoned that shr should have lower sugar levels. Surprisingly, we found that the concentrations of Suc, Glc, and Fru in shr were as high as those in scr (Fig. 3A; Supplemental Table S2). One explanation for this difference is that other stress responses were also altered in scr and that this would cause sugar hypersensitivity as a consequence of cross talk between different signaling pathways. An alternative explanation is that the sugar response is somehow suppressed in shr. To determine whether a second stress response is altered in scr but not in shr, we examined the transcript level of some key ABA signaling regulators in root, including ABI3, ABI4, and ABI5 in shr-2 and scr-1, as these ABI genes are involved in different stress responses (Arroyo et al., 2003). For example, ABI3 is primarily involved in responses to cold (Tamminen et al., 2001), desiccation (Tamminen et al., 2001), and nutrient deficiency (Walker and Connolly, 2008), whereas ABI4 plays a major role in sugar signaling and plastid-derived retrograde signaling (Arenas-Huertero et al., 2000; Koussevitzky et al., 2007). ABI5 is required for seedling growth, but it also plays a role in sugar signaling (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001). ABI1 and ABI2 were also included in this comparison. As shown in Figure 4A, the scr mutation caused a dramatic increase in the transcript level of ABI4 and some increase in ABI5 but had no apparent effect on RNA levels of ABI1 and ABI2. The transcript level of ABI3 was too low to be determined by quantitative RT-PCR. In contrast, the ABI4 transcript level in the shr mutant remained almost unchanged, and the ABI5 transcript level was only moderately increased. Because SCR and SHR are also expressed in the shoot apical meristem, leaf primordium, and young leaves (Wysocka-Diller et al., 2000; Dhondt et al., 2010; Gardiner et al., 2010) and shr and scr leaves showed obvious differences in sugar hypersensitivity, we also analyzed these ABI genes in the shoot. A similar trend was observed for ABI4 and ABI5 (Supplemental Fig. S4). ABI3 transcript can be reliably quantified, but no difference in its level was detected between the wild type, scr, and shr. Because ABI4 is a key regulator of Glc-signaling pathways (Rolland et al., 2006), these results suggest that an elevated level of ABI4 in scr is a major cause of its sugar hypersensitivity.
Figure 4.
SCR, but not SHR, regulates ABI4 and ABI5 directly and specifically in the root apical meristem. A, Real-time RT-PCR assay of the transcript level of ABI genes in the root of shr-2, scr-4, and wild-type Col (WT). B, ChIP-PCR assay for SHR and SCR binding to the ABI4 promoter, with primer pairs that tile the promoter sequence. C, ChIP-PCR assay of SHR and SCR binding to the ABI5 promoter. D, RT-PCR assay of ABI4 and ABI5 transcripts in the root meristem, the elongation zone, and the maturation zone in wild-type and scr-1 roots. Error bars represent sd from technical replicates. E, pABI4::GUS expression in the root of the wild type at different times after germination and in scr-1 and shr-2 at 9 d after germination. Bars = 50 μm.
SCR Modulates the Sugar Response Directly and Specifically in the Root Apical Meristem through ABI4
The results described above also suggest that, in contrast to the case of radial patterning, SHR and SCR do not act as a heterodimer in the regulation of the sugar response. To determine whether SCR regulates ABI4 and ABI5 independently of SHR, we examined SHR and SCR binding to the ABI4 and ABI5 promoters by ChIP-PCR. Interestingly, both the ABI4 and ABI5 promoters were bound by SCR but not by SHR (Fig. 4, B and C). This result demonstrated that SCR has a SHR-independent role in transcriptional regulation but also provides evidence that SCR modulates the sugar response directly.
Along the longitudinal axis of a root, three developmental stages with distinct mitotic activity and morphology can be recognized: the apical meristem, the elongation zone, and the maturation zone (Ishikawa and Evans, 1995). To determine the developmental stage when SCR is required to regulate ABI4 and ABI5, we examined their transcript levels in the three zones in scr and wild-type roots by quantitative RT-PCR. As shown in Figure 4D, ABI4 and ABI5 transcripts were very low in all developmental stages but were much higher in the meristematic zone in scr. Although ABI4 and ABI5 transcript levels were also elevated in the meristem zone in shr-2, this was to a much lesser extent (Fig. 4D). To validate this result, we next generated transgenic plants expressing the GUS reporter gene under the control of a 2.6-kb sequence upstream of the ABI4 coding region and examined its expression in the wild type, scr, and shr. As reported previously (Arroyo et al., 2003), in the wild type the pABI4::GUS construct was expressed in the root tip during early postembryonic growth (less than 3 d after germination), but its expression level decreased rapidly (Fig. 4E, top panel). Seven days after germination, only weak GUS staining was visible in some of the root cap cells, and by day 9, GUS activity was no longer detectable even after prolonged staining (Fig. 4E). In scr, however, pABI4::GUS was still expressed strongly in the apical meristem, with the maximum in the QC at 7 d after germination (Supplemental Fig. S5), and remained expressed in the QC at 9 d after germination (Fig. 4E, bottom row). In contrast, in shr, no GUS activity was visible 9 d after germination, similar to the case in the wild type. Together, these results suggest that in root, SCR suppresses the sugar response specifically in the apical meristem.
To determine whether ABI4 misexpression indeed causes sugar hypersensitivity in scr, we generated the scr-4 abi4-104 double mutant and compared its response to 4% Glc with that of the scr-4 and abi4-104 single mutants. Because scr-4 and abi4-104 are in different ecotype backgrounds (Ws and Col, respectively), to minimize ecotype-related variability, we analyzed seeds derived from the same cross but with different genotypes. As shown in Figure 5A, the scr-4 abi4-104 double mutant was much more tolerant to high Glc than the scr-4 single mutant, although it was still slightly more sensitive to sugar than the abi4-104 mutant, probably because of the presence of a higher level of ABI5. This result supports the notion that SCR suppresses the sugar response mainly by repressing ABI4.
Figure 5.
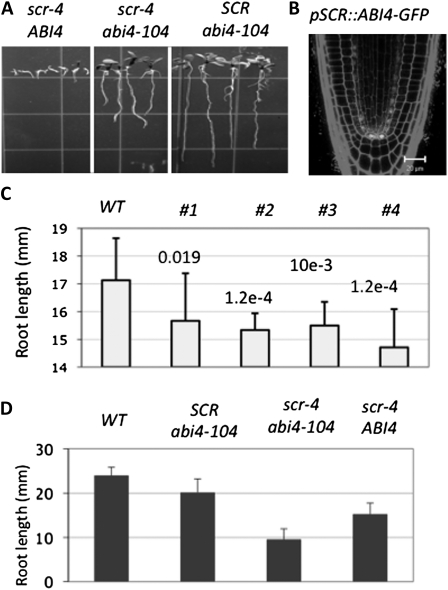
ABI4 regulates root growth under high-sugar and normal growth conditions. A, scr-4 ABI4, scr-4 abi4-104, and SCR abi4-104 seedlings grown on MS medium containing 4% Glc, 10 d after germination, showing that mutation in ABI4 confers scr tolerance to high Glc. B, Confocal microscopy image showing the expression of the ABI4-GFP fusion protein in transgenic plants harboring the pSCR::ABI4-GFP construct. Bar = 20 μm. C, Root length of wild-type Col (WT) and pSCR::ABI4-GFP transgenic plants on MS medium, 7 d after germination. Numbers 1 to 4 indicate independent lines, and the numbers above each bar are the P values for pairwise comparison between the wild type and transgenic lines (t test, n = 20). D, Root length of wild-type Col, SCR abi4-104, scr-4 abi4-104, and scr-4 ABI4 seedlings grown on MS medium, 7 d after germination. Error bars represent sd from technical replicates.
ABI4 Has a Dual Role in Root Growth
The elevated level of ABI4 transcript in scr could also be a cause of its root growth defect under normal growth conditions. To test this hypothesis, we expressed ABI4 as a fusion protein to GFP in the SCR expression domain using the SCR promoter. This strategy not only permits ABI4 overexpression in a cell type-specific manner but also allows us to monitor ABI4 expression. As shown in Figure 5B, the ABI4-GFP fusion protein was clearly expressed in the nucleus. As shown in Figure 5C, all transgenic plants that showed detectable levels of ABI4-GFP expression had significantly shorter roots than the wild type. To examine further the function of ABI4 in root development, we compared root lengths of scr, abi4, and the scr abi4 double mutant that are derived from the same cross. Interestingly, on MS medium, the scr-4 abi41-104 double mutants had even shorter roots than the scr-4 mutant (Fig. 5D; P < 0.001, n = 30, t test). Compared with the wild type, the abi4-104 single mutant also had shorter roots. These results suggest that ABI4 also has a positive role in root growth. Because ABI4 is mainly expressed in the meristem zone in primary roots, we reasoned that at low levels, ABI4 probably promotes meristematic activity. Therefore, we measured the size of the apical meristem in the wild type, scr-4, abi4-104, and the scr-4 abi4-104 double mutant. As expected, the abi4 mutation reduced the size of the root apical meristem in both the wild type and scr backgrounds (128 versus 109 μm in the wild type and abi4-104, respectively; 108 versus 90 μm in scr-4 and scr-4 abi4-104, respectively; P < 0.05, n = 12, t test). These results together suggest that ABI4 regulates root growth positively or negatively, depending on its level of expression; they further suggest that ABI4 levels must be strictly regulated to ensure root growth and that SCR plays this important role.
DISCUSSION
Plants are sessile; therefore, to ensure optimal growth and development in an ever-changing environment, they must be able not only to respond appropriately to various stimuli but also to overcome the deleterious effects from stress responses. Although recent studies have shed significant light on the mechanisms by which SHR and SCR control radial patterning, how they regulate root meristematic activity remains unclear. Our findings that SCR modulates the sugar response in the root apical meristem through direct repression of ABI4 and ABI5 provides an example of a mechanism that plants use to coordinate developmental programs with stress response mechanisms. Because high levels of ABI4 inhibit root growth, we suggest that SCR promotes root growth by suppressing the ABI4-mediated sugar response. Although SHR and SCR act in the same pathway regulating root stem cell renewal and radial patterning, we showed that SHR and SCR function differently in the sugar response. This study also revealed a dual role for ABI4 in regulating root growth in response to environmental cues.
SCR Modulates the Sugar Response Directly
Evidence for a role of SCR in the sugar response comes from the observations that scr was hypersensitive to high Glc and that endogenous levels of Suc and Glc in scr were elevated. The higher sugar content in scr does not seem to be the sole cause of sugar hypersensitivity, however, because shr also had higher levels of water-soluble sugars but was much less sensitive to high Glc. The similarity of the developmental defects in shr and scr argues against the possibility that the sugar hypersensitivity in scr is a secondary effect. The sugar hypersensitivity in scr is not due to a general stress response either, because scr responded normally to high concentrations of NaCl and mannitol. By performing the sugar-sensitivity assay on MS medium in a closed environment (petri dish), we also minimized the effect of the root growth defects in shr and scr on nutrient uptake, which is evidenced by the observation that the shoots of scr and shr were similar in size to those of wild-type plants. In further studies, we found that SCR directly regulates ABI4 and ABI5. Because ABI4 is a key component in sugar-signaling pathways and ABI5 also has a role in the sugar response, these results lend strong support to the conclusion that SCR modulates the sugar response directly. In line with this argument, we showed that mutation in ABI4, which positively regulates sugar signaling, alleviated the sugar hypersensitivity phenotype in scr.
The Role of SCR in the Sugar Response Is SHR Independent
Although SCR transcription is largely dependent on the presence of SHR, the role of SCR in the sugar response does not appear to require SHR. First, although both shr and scr were hypersensitive to high Glc, the scr mutant had a much more severe phenotype. Second, by ChIP-PCR, we showed that SHR did not bind to the promoters of ABI4 and ABI5. Consistent with this result, by RT-PCR we found that the transcript level of ABI4 and ABI5 was dramatically elevated in scr but remained almost unchanged in shr, except in the meristem zone, where the basal level of SCR is lowest (Cui et al., 2007). The distinct roles for SHR and SCR in the regulation of ABI4 expression was further confirmed by transgenic analysis with the pABI4::GUS reporter construct.
The SHR-independent role of SCR in the sugar response is not unexpected, because we have previously shown that, in addition to an overlapping set of targets, SHR and SCR control other genes differently (Cui et al., 2007; Cui and Benfey, 2009; Sozzani et al., 2010). Due to a positive feedback loop for SCR transcription, which is dependent on the SHR/SCR complex (Cui et al., 2007), SCR would accumulate to a level much higher than that of SHR, and consequently, some SCR protein is predicted not to form a complex with SHR. Our hypothesis is that at least two distinct pools of SCR are present, one in a complex with SHR and the other acting alone or interacting with other proteins to regulate a set of genes different from those controlled by the SHR/SCR complex. ABI4 and ABI5 are among the genes that are regulated only by SCR. Down-regulation of ABI4 and ABI5 by SCR is an example of SCR’s action as a transcriptional repressor. In the shr mutant background, SCR is still expressed because of a SHR-independent mechanism, which could explain the much less severe Glc hypersensitivity phenotype in shr.
The QC-specific regulation of ABI4 by SCR is somewhat surprising, however, because SCR is expressed not only in the QC but also in the endodermis, and mutation in SCR is expected to relieve the repression on ABI4 in both the QC and endodermis. Intriguingly, a cell type- or developmental stage-specific expression pattern has been reported for other SCR targets. WOX7, for example, is expressed specifically in the cortex/endodermis initial cell and its daughter cell (Cui et al., 2011), and several genes, including MAGPIE and NUTCRACKER, are not expressed even in the QC, although they are expressed in the meristem zone (Cui et al., 2007; Welch et al., 2007). These data suggest that the regulatory mechanism by SCR is complex. Although SCR is a transcriptional regulator, it does not appear to bind to DNA directly (Ogasawara et al., 2011). It is conceivable, therefore, that the expression pattern of SCR targets is actually determined by DNA-binding factors that are expressed in a cell type- or developmental stage-specific manner and recruit SCR to target promoters through protein-protein interaction, which warrants further investigation.
SCR Is Probably Also Involved in Other Stress Responses
The sugar response is unlikely to be the only stress condition in which SCR is involved, as most of its direct targets confirmed in this study do not appear to play a role in the sugar response. For example, several of its top-ranked direct targets, including ATAIB, TMAC2, and HB12, are known to be involved in drought tolerance (Olsson et al., 2004; Huang and Wu, 2007; Li et al., 2007a), suggesting that SCR also plays a role in drought tolerance. Because these stress-associated genes are also directly regulated by SHR, the role of SCR in other stress responses presumably depends on the SHR/SCR complex. The observation that both shr and scr were sensitive to the stress hormone ABA supports this possibility. Further elucidation of the mechanism by which SCR regulates plant growth and development will require the identification of additional stress conditions in which SCR is involved.
SCR May Have a Similar Function in the Shoot
SCR is expressed not only in root but also in shoot organs such as the apical meristem, leaf primordium, and bundle sheath cells in fully expanded leaves (Wysocka-Diller et al., 2000), but its function in these tissues and cell types remains unclear. scr has been shown, though, to have growth defects in the shoot (smaller leaves and shoot), but these are generally attributed to its root growth defects. By means of grafting experiments, a recent study demonstrated that SCR regulates leaf growth independently of its function in the root (Dhondt et al., 2010). This study showed that the cell cycle in scr leaf primordia is prolonged, entailing less frequent cell divisions and ultimately a smaller number of cells in mature leaves (Dhondt et al., 2010), but the restricted expression of SCR in only a subset of leaf cells suggests that SCR does not regulate the mitotic cycle in the whole leaf directly. Although sugar accumulation in scr leaves may result from low mitotic activity and therefore reduced sugar utilization, it is equally likely to be the cause of the leaf growth defect in scr. Further studies will be needed to determine whether SCR also promotes meristematic activity in the shoot by suppressing the sugar response.
ABI4 Regulates Root Growth in a Dosage-Dependent Manner
An essential role for ABI4 in sugar signaling and seed development has long been established, but recent studies show that ABI4 is also involved in other processes. Shkolnik-Inbar and Bar-Zvi (2010) showed that the abi4 mutant produced more lateral roots than the wild type at a similar developmental stage, suggesting an inhibitory role for ABI4 in lateral root formation. Consistent with this observation, they showed that ABI4 is expressed in the maturation zone of the root. Surprisingly, they found that ABI4 expression is regulated not only by ABA but also by cytokinin and auxin and that ABI4 also modulates cytokinin and auxin responses. Consistent with this, lateral root formation in abi4 is no longer affected by exogenous cytokinin. ABI4 appears to be also required for lipid biosynthesis in seedlings during nitrogen deficiency (Yang et al., 2011). In our study here, we showed that ABI4 plays an important role in regulating root growth. In agreement with other reports (Dekkers et al., 2008; Shkolnik-Inbar and Bar-Zvi, 2010), we showed that ectopic expression of ABI4 causes retardation in root growth. Our study, however, also reveals a positive role for ABI4 in root growth under normal conditions, as the abi4 mutant had a shorter root and a smaller apical meristem than the wild type. How ABI4 promotes root growth is currently unknown, but probably this involves a complex interaction between ABI4 and several hormone-signaling pathways. Because ABI4 is stress inducible and it inhibits root growth at elevated levels but is also required for root growth under normal growth conditions, we suggest that ABI4 is an important component in the mechanism that orchestrates plant growth with stress responses.
MATERIALS AND METHODS
Plant Materials and Treatments
All the Arabidopsis (Arabidopsis thaliana) seedlings used in this study were grown on MS agar plates containing 1% Suc (w/v), which were positioned vertically in a Percival growth chamber at 22°C with 16 h of daily illumination. One percent Suc was added because without it, the shr and scr mutants do not grow well. For Glc-sensitivity assays, Glc was added to the standard MS medium to a final concentration of 4% to 6%. Concentrations for other chemicals tested were 0.5, 1, and 2 μm ABA; 50, 100, and 150 mm NaCl; and 50, 100, and 200 mm mannitol.
All the shr and scr mutant alleles have been described previously: scr-1 and scr-2 by Di Laurenzio et al. (1996), scr-3 and scr-4 by Fukaki et al. (1998), shr-2 and shr-3 by Helariutta et al. (2000), and shr-5 by Gallagher et al. (2004). The aba2-1 and abi4-104 mutants were obtained from the Arabidopsis Stock Center. T-DNA insertion lines (FLAG_587G11 for ARL1 (AT1G24120) and FLAG_532D03 for MYB [AT1G68670]) were obtained from the INRA (http://www-ijpb.versailles.inra.fr/en/sgap/equipes/fichiers/FST_information.htm).
Soluble Sugar Measurement
Plants used for sugar measurement were 1-week-old seedlings grown in MS medium or 3-week-old plants grown in soil. Water-soluble sugars were extracted by grinding of fresh tissues in water (3 mL g−1 fresh tissue) on ice. The mixture was centrifuged at 12,000 rpm, 4°C, for 10 min. The supernatant was further cleared by filtering through a Microcon centrifugal filter column with a molecular weight cutoff of 10,000 (Millipore). Suc, Glc, and Fru were then separated by HPLC on a Hamilton RCX10 ion-exchange chromatography column with 150 mm NaOH for elution and quantified according to sugar standards.
Other Methods
ChIP-PCR and RT-PCR assays were performed as described previously (Cui et al., 2007), except that a StepOnePlus real-time PCR system (Applied Biosystems) was used. For cloning of the pSCR::ABI4 construct, RT was first performed with root total RNA; the ABI4 cDNA was then amplified with the high-fidelity Phusion DNA polymerase (New England Biolabs) and cloned into the pDONR221 vector. The pSCR::ABI4 construct was obtained by subcloning the ABI4 cDNA clone and the SCR promoter clone into a destination vector (dpGreenBarT) with the Multi-Site Gateway system. The pABI4::GUS construct was cloned similarly with the same Gateway system. Briefly, a 2.6-kb fragment upstream of the start codon of the ABI4 gene was first amplified using the primer pair attB4_pABI4_FW and attB1_pABI4_RV and was then cloned into the pDONR-P4-P1R vector. The pEnter-pABI4 entry clone was then subcloned together with the pEnter-GUS entry clone into dpGreenBarT, yielding the pABI4::GUS construct. After confirmation by restriction analysis, the final constructs were transformed into wild-type Arabidopsis (ecotype Col) by the Agrobacterium tumefaciens-mediated method. The pSCR::ABI4-GFP and pABI4::GUS transgenic lines were selected by epifluorescence microscopy and GUS staining, respectively.
The primers used for the ChIP-PCR and RT-PCR assays as well as for cloning and genotyping are listed in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic of the radial pattern in wild type and shr and scr roots.
Supplemental Figure S2. ARL1 and the MYB factor, at1g68670, are probably involved in stress response too.
Supplemental Figure S3. The sugar hypersensitivity in scr and shr is independent of their genetic backgrounds.
Supplemental Figure S4. ABI4 transcript level is dramatically elevated in the shoot of scr, but not that of shr.
Supplemental Figure S5. pABI4::GUS expression pattern in wild type and scr-1 root, 7 d after germination.
Supplemental Table S1. Top-ranked direct SHR targets identified by ChIP-chip.
Supplemental Table S2. Soluble sugar content in wild-type, scr-1, and shr-2 seedlings grown on MS medium.
Supplemental Table S3. Primers used for ChIP-PCR, RT-PCR, genotyping, and cloning.
Supplementary Material
Acknowledgments
We thank Dr. George Bates (Florida State University) and Dr. Hironaka Tsukagoshi, Dr. Jaimie Van Norman, and Natalie Breakfield (Duke University) for helpful comments and critiques. We also thank Dr. Anne B. Thistle (Florida State University) for careful editing of the manuscript.
References
- Aki T, Konishi M, Kikuchi T, Fujimori T, Yoneyama T, Yanagisawa S. (2007) Distinct modulations of the hexokinase1-mediated glucose response and hexokinase1-independent processes by HYS1/CPR5 in Arabidopsis. J Exp Bot 58: 3239–3248 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, León P. (2003) Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol 133: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106: 13124–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Okrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee J-Y, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Cui H, Benfey PN. (2009) Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 58: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Hao Y, Kovtun M, Stolc V, Deng XW, Sakakibara H, Kojima M. (2011) Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol 157: 1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC. (2008) Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol Biol 67: 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt S, Coppens F, De Winter F, Swarup K, Merks RM, Inzé D, Bennett MJ, Beemster GT. (2010) SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiol 154: 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Duke ER, McCarty DR, Koch KE. (1991) Organ-specific invertase deficiency in the primary root of an inbred maize line. Plant Physiol 97: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Donner TJ, Scarpella E. (2010) Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev Dyn 240: 261–270 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, Lee MM, Lim J. (2011) Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA 108: 2166–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Wu WL. (2007) Overexpression of TMAC2, a novel negative regulator of abscisic acid and salinity responses, has pleiotropic effects in Arabidopsis thaliana. Plant Mol Biol 63: 557–569 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. (1995) Specialized zones of development in roots. Plant Physiol 109: 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Leegood RC. (2008) Roles of the bundle sheath cells in leaves of C3 plants. J Exp Bot 59: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sun J, Xu Y, Jiang H, Wu X, Li C. (2007a) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65: 655–665 [DOI] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW. (2007b) Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19: 2500–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Okrész L, Stabel S, et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H, Kaimi R, Colasanti J, Kozaki A. (2011) Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol Biol 77: 489–499 [DOI] [PubMed] [Google Scholar]
- Olsson AS, Engström P, Söderman E. (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen I, Mäkelä P, Heino P, Palva ET. (2001) Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J 25: 1–8 [DOI] [PubMed] [Google Scholar]
- Walker EL, Connolly EL. (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D. (2005) Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15: 436–440 [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC. (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Yang Y, Yu X, Song L, An C. (2011) ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol 156: 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 47: 49–62 [DOI] [PubMed] [Google Scholar]
- Yu NI, Lee SA, Lee MH, Heo JO, Chang KS, Lim J. (2010) Characterization of SHORT-ROOT function in the Arabidopsis root vascular system. Mol Cells 30: 113–119 [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Yuan S, Xu F, Yang H, Zhang NH, Cheng J, Lin HH. (2010) The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis. FEBS Lett 584: 3573–3579 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.