Abstract
Bacterial pathogens colonize a host plant by growing between the cells by utilizing the nutrients present in apoplastic space. While successful pathogens manipulate the plant cell membrane to retrieve more nutrients from the cell, the counteracting plant defense mechanism against nonhost pathogens to restrict the nutrient efflux into the apoplast is not clear. To identify the genes involved in nonhost resistance against bacterial pathogens, we developed a virus-induced gene-silencing-based fast-forward genetics screen in Nicotiana benthamiana. Silencing of N. benthamiana SQUALENE SYNTHASE, a key gene in phytosterol biosynthesis, not only compromised nonhost resistance to few pathovars of Pseudomonas syringae and Xanthomonas campestris, but also enhanced the growth of the host pathogen P. syringae pv tabaci by increasing nutrient efflux into the apoplast. An Arabidopsis (Arabidopsis thaliana) sterol methyltransferase mutant (sterol methyltransferase2) involved in sterol biosynthesis also compromised plant innate immunity against bacterial pathogens. The Arabidopsis cytochrome P450 CYP710A1, which encodes C22-sterol desaturase that converts β-sitosterol to stigmasterol, was dramatically induced upon inoculation with nonhost pathogens. An Arabidopsis Atcyp710A1 null mutant compromised both nonhost and basal resistance while overexpressors of AtCYP710A1 enhanced resistance to host pathogens. Our data implicate the involvement of sterols in plant innate immunity against bacterial infections by regulating nutrient efflux into the apoplast.
A given plant species is normally susceptible to only a few pathogens although they are exposed to a vast number of potential pathogens in nature. Plants have evolved intricate defense mechanisms referred to as innate immunity to recognize and defend themselves against a wide array of pathogens (Hofius et al., 2007). Plant immunity involves pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI; Chisholm et al., 2006; Hofius et al., 2007). The resistance shown by all members of a plant species to the majority of potentially pathogenic microbes is known as nonhost resistance (Heath, 2000). Nonhost resistance is the most common and durable form of plant disease resistance in nature and is of great interest to agriculture (Ellis, 2006). Investigations of the molecular basis of nonhost resistance against bacterial pathogens are just emerging (Mysore and Ryu, 2004; Niks and Marcel, 2009).
Nonhost disease resistance involves two major components (Mysore and Ryu, 2004; Ellis, 2006). One is preformed defenses wherein the microbes are not able to overcome preexisting obstacles such as the cytoskeleton, antimicrobial compounds, and secondary metabolites. Another one is inducible defense responses that can be triggered by general elicitors from invading pathogens including PAMPs, such as lipopolysaccharides, flagellin and oligomers, or specific elicitors/effectors (Nürnberger and Lipka, 2005). ETI is also proposed to be a part of nonhost resistance (Schulze-Lefert and Panstruga, 2011). Many plant defense responses to pathogen infections are shared by nonhost and R-gene-mediated resistance (Heath, 2000; Mysore and Ryu, 2004; Freialdenhoven et al., 2005). For example, hypersensitive response (HR) is a typical resistance phenomenon that is associated with both R-gene and some nonhost resistance. Similar global gene expression profiles (Tao et al., 2003) and other induced responses (Zimmerli et al., 2004; Aghnoum and Niks, 2010; Zellerhoff et al., 2010; Hiruma et al., 2011) have been observed during nonhost resistance against fungal and bacterial pathogens.
Virus-induced gene silencing (VIGS) has been a popular approach to identify genes involved in disease resistance (Lu et al., 2003; Liu et al., 2004; del Pozo et al., 2004). One of the advantages of VIGS is that it allows the researcher to assess the function of genes where a null mutation might be lethal in sexually propagated plants or of genes that have functionally redundant family members (Senthil-Kumar et al., 2008; Senthil-Kumar and Mysore, 2011). In this study, we used VIGS-mediated fast-forward genetic screens (Lu et al., 2003; Anand et al., 2007) and identified several plant genes that play a role in nonhost disease resistance. We show squalene synthase (SQS), a key enzyme catalyzing the first enzymatic step in sterol biosynthesis, as a new player in nonhost disease resistance. Few studies have related the sterol metabolism to plant-microbe interaction (Devarenne et al., 1998, 2002; Griebel and Zeier, 2010). Previously, increased contents of stigmasterol in leaves through stimulation of sterol C22 desaturation after Pseudumonas syringae infection have been shown to attenuate pathogen-induced expression of the defense regulator flavin-dependent monooxygenase-1 (Griebel and Zeier, 2010). However, changes in membrane properties, due to alteration in sterols, leading to alteration in control of the efflux of cytoplasmic nutrients and solutes into apoplastic space where bacterial pathogens grow have not been explored (Sattelmacher, 2001; Bernsdorff and Winter, 2003; Hodzic et al., 2008; Melotto et al., 2008). Pathogens have mechanisms to derive nutrients from host plants, and it is speculated that there is a relationship between apoplastic nutrient levels and bacterial growth (Rico and Preston, 2008), but the mechanism is largely not studied. We systematically studied the relevance of membrane leakage and pathogen growth in two different plant species. Our results show that sterol content plays an important role in plant innate immunity against bacterial infections by regulating nutrient efflux into the apoplast.
RESULTS
Identification of Genes That Play a Role in Nonhost Resistance
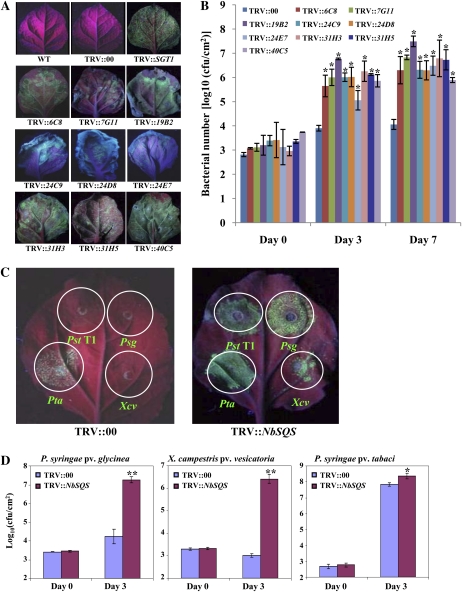
A normalized Nicotiana benthamiana cDNA library (made from mixed-elicitor-treated leaf tissues; Anand et al., 2007) in tobacco rattle virus (pTRV2)-VIGS vector (Liu et al., 2002) was used in a forward-genetics screen to identify plant genes that play a role in nonhost resistance. Two leaves of each silenced plant were inoculated with a nonhost pathogen, P. syringae pv tomato T1 (Pst T1), expressing GFPuv (Wang et al., 2007) at a concentration of 1 × 105 colony forming units (cfu)/mL. Since the innate immunity limits the growth of nonhost pathogens, little or no green fluorescence emitted from GFPuv-expressing Pst T1 was observed in the wild-type and mock (TRV::00)-inoculated plant leaves under long-wavelength UV light (Fig. 1A). Of the 3,840 clones silenced, after three rounds of screening, nine of them showed varying levels of increased nonhost bacterial growth visualized by the intensity of the green fluorescence in the inoculated plant leaves (Fig. 1A).
Figure 1.
Identification of NbSQS-gene-silenced plants that compromise resistance to both host and nonhost pathogens. A, N. benthamiana plants (3 weeks old) were infiltrated with a mixture of Agrobacterium tumefaciens GV2260 carrying TRV1 and TRV2 containing NbSGT1 (TRV::NbSGT1; positive control) or preselected TRV2 clones from the cDNA library or TRV2 empty vector (TRV::00; control). After 3 weeks, top leaves were syringe inoculated (in multiple spots) with a GFPuv-expressing nonhost pathogen, Pst T1, at a concentration of approximately 1 × 105 cfu/mL. Photographs were taken under long-wavelength UV light 4 dpi. B, Inoculation of N. benthamiana plants silenced by different cDNA clones was conducted as described above. Bacterial population was quantified by plating serial dilutions of leaf extracts. Asterisks indicate a significant difference from the control (Student’s t test; *P < 0.01). Data were pooled from two independent experiments representing four biological replicates. C, The NbSQS-silenced (TRV::NbSQS) and control (TRV::00) leaves were syringe inoculated with GFPuv-expressing nonhost pathogens Pst T1, Psg, and Xcv, and a host pathogen, Pta, at a concentration of 1 × 105 cfu/mL. Photographs were taken under long-wavelength UV light at 4 dpi. D, NbSQS-silenced and control plants were inoculated by vacuum infiltration with host and nonhost pathogens at a concentration of 1 × 105 cfu/mL. Bacterial population was quantified by plating the serial dilutions of leaf extracts at 0 and 3 dpi. Asterisks indicate a significant difference from the control (Student’s t test; *P < 0.05 and **P < 0.01). Data were pooled from two independent experiments representing four biological replicates. See also Supplemental Figures S1 and S2.
To confirm whether the increase in green fluorescence correlated with the bacterial population, we conducted bacterial growth assays on N. benthamiana control (TRV::00) and gene-silenced plants that were vacuum infiltrated with Pst T1 (5 × 104 cfu/mL). As shown in Figure 1B, silenced plants accumulated 100- to 1,000-fold more bacteria by 7 d post inoculation (dpi) compared to the control. These results demonstrated the use of GFPuv-expressing bacteria in combination with VIGS-mediated forward genetics as a powerful approach to identify silenced plants that are compromised in nonhost disease resistance.
SQS Is Required for Plant Innate Immunity
The clone 6C8, that when silenced compromised nonhost resistance, was selected for further analyses. The insert in the 6C8 cDNA clone was sequenced, and this information was used to clone the full-length gene by RACE. The sequence was identical to a N. benthamiana gene (GenBank no. U46000) encoding SQS (NbSQS). Using real-time quantitative reverse transcription RT-PCR (qRT-PCR), we showed that NbSQS transcripts were significantly down-regulated in N. benthamiana plants silenced with the cDNA clone 6C8 (Supplemental Fig. S1A). The SQS gene is known to be involved in the biosynthesis of sterols in tobacco (Nicotiana tabacum; Devarenne et al., 2002). Consistent with its function, NbSQS-silenced plants showed a reduction in stigmasterol when compared to the control (TRV:00; Supplemental Fig. S1B).
We examined whether NbSQS is required for resistance against other nonhost pathogens and a host pathogen. Bacterial strains Pst T1, P. syringae pv glycinea (Psg), Xanthomonas campestris pv vesicatoria (Xcv), and P. syringae pv tabaci (Pta) expressing GFPuv were used to infiltrate NbSQS-silenced N. benthamiana and control plants using a needleless syringe. Strong green fluorescent spots produced by increased growth of Pst T1, Psg, and Xcv were observed in the NbSQS-silenced plant leaves, but not in the control plants, indicating that silencing of NbSQS compromised nonhost resistance to these bacteria (Fig. 1C). Silencing of this gene also resulted in the delayed occurrence of HR induced by nonhost pathogen Pst T1 (Supplemental Fig. S1C). Furthermore, the NbSQS-silenced leaf infiltrated with host pathogen Pta produced stronger green fluorescence than the control leaf (Fig. 1C). Estimation of bacterial number in planta by bacterial growth assay confirmed that there were more nonhost and host bacteria in the NbSQS-silenced plants than in the control plants (Fig. 1D). The silenced plants inoculated with Psg and Xcv also showed disease symptoms (Supplemental Fig. S1D). These results indicated that silencing of NbSQS not only compromised nonhost resistance, but also partially compromised basal resistance.
Silencing of NbSQS in N. benthamiana Results in Electrolyte Leakage and Nutrient Efflux
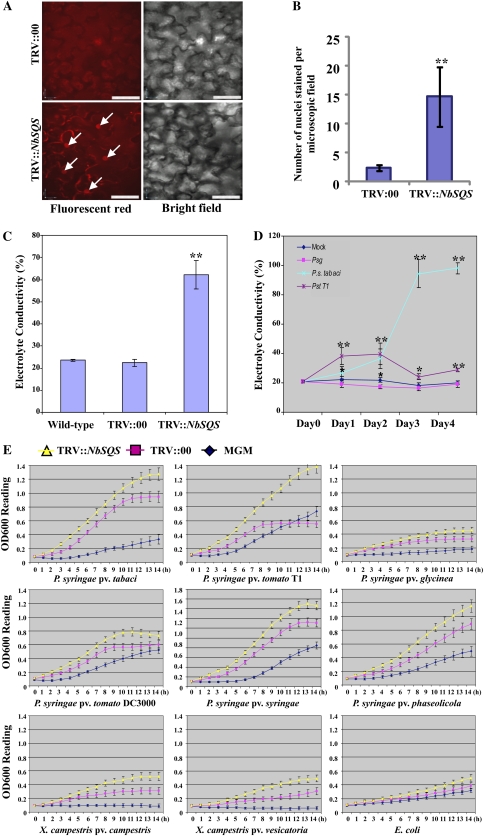
SQS is a key enzyme catalyzing the first enzymatic step in the biosynthesis of phytosterols, which are one of the major components of the cell membrane (Supplemental Fig. S2, A and B; Devarenne et al., 2002). Therefore, we tested through microscopy whether NbSQS-silenced plants have highly permeable membranes. Propidium iodide (PI), which binds to and stains cellular DNA, can only enter and stain cells with a permeable plasma membrane (Harrison and Vickers, 1990). In this study, PI was used to assess membrane permeability of control and NbSQS-silenced N. benthamiana cells. Leaf segments were stained with PI, and the entry of PI into plant cells was monitored under a confocal microscope. The NbSQS-silenced plants showed a significantly higher number of stained nuclei (red fluorescence) than the control plant cells (Fig. 2, A and B). Interestingly, the NbSQS-gene-silenced plants had larger cell size (Supplemental Fig. S2C). As a positive control, we showed that PI can enter and stain the cell nucleus much faster in heat-stressed tissues when compared to the control (Supplemental Fig. S2, D and E); this can be attributed to disruption of the plasma membrane due to heat stress (Wahid et al., 2007; Wang et al., 2011). In addition, we showed that the entry of FM1-43 dye was higher in the silenced cells (Supplemental Fig. S2F). These results indicate that the cell membrane of NbSQS-silenced plants is highly permeable.
Figure 2.
(Legend appears on following page.) Figure 2. Silencing of NbSQS leads to leaky cell membrane and the apoplastic fluid of silenced plants supports more bacterial multiplication in vitro. A and B, Leaf tissues were collected from TRV::00 (control) and NbSQS-silenced plants, and incubated in a solution containing PI for 30 min. Sections were made and observed under a confocal microscope (Perkin Elmer ultra view ERS spinning disk confocal) for PI-stained fluorescent nuclei (left). Corresponding bright-field images were also taken at the same focal area (right). Scale bar in each image represent 50 micron. The number of stained nuclei were counted and represented in the graph. An asterisk indicates a significant difference from the control (Student’s t test; P < 0.01). Data were pooled from two independent experiments representing four biological replicates. C, Three weeks after silencing, two leaf discs (0.5-cm2 each) of the NbSQS-silenced N. benthamiana plants were immersed into deionized water and electrolyte conductivity was measured. Electrolyte leakage was calculated as a percentage of the conductivity before autoclaving to the conductivity after autoclaving. Asterisks indicate a significant difference from the control (Student’s t test; P < 0.01). Data were pooled from two independent experiments representing four biological replicates. D, The wild-type N. benthamiana plant leaves were inoculated with Pst T1, Psg, and Pta at a concentration of approximately 5 × 104 cfu/mL. Electrolyte leakage was measured every 24 h after inoculation. Asterisks indicate a significant difference from the control (*P < 0.05 and **P < 0.01). Data were pooled from two independent experiments representing four biological replicates. E, Apoplastic fluid was extracted from TRV::00 and TRV::NbSQS-silenced N. benthamiana leaves by the vacuum-centrifuge method. The filter-sterilized apoplastic fluid was added to MGM to make 5% apoplastic fluid. The bacterial population was quantified by measuring the optical density at 600 nm (OD600) of bacterial cultures every hour. Diamond: MGM alone; square: MGM containing 5% apoplast extract from control plants; triangle: MGM containing 5% apoplast extract from NbSQS-silenced N. benthamiana. x axis represents time in hours. Error bars represent sds of three technical replicates. Results were similar for two independent experiments. See also Supplemental Table S2.
We further examined whether silencing of NbSQS resulted in altered cell membrane permeability by measuring the electrolyte conductivity. NbSQS-silenced N. benthamiana leaves showed more than 50% ion leakage, while the control leaves had only approximately 20% ion leakage (Fig. 2C). This result indicates that silencing of NbSQS in N. benthamiana plants releases more ions from the cytoplasm into the apoplast. This is analogous to the situation wherein the host pathogen Pta manipulates the host cells to release more electrolytes in N. benthamiana and multiplies in planta, unlike nonhost pathogens (Fig. 2D). Pst T1 is known to induce HR cell death at early stages of infection and, therefore, slightly higher leakage can be observed in leaves inoculated with this nonhost pathogen (Fig. 2D).
To check whether the increased electrolyte leakage in NbSQS-silenced plants is associated with organic compounds, we performed metabolite profiling of apoplastic fluids from NbSQS-silenced and control plant leaves using gas chromatography-mass spectrometry (GC-MS; Broeckling et al., 2005). Interestingly, 420 ± 11 components were detected by GC-MS analyses in the apoplast from NbSQS-silenced N. benthamiana, while only 261 ± 5 were detected from the control plants. Among the commonly identified compounds, 35 compounds were at least 2-fold higher in the apoplast from NbSQS-silenced N. benthamiana than in the control plants (Supplemental Table S1). Out of these 35 compounds, 21 were sugars. In fact, we observed that Pta causes more efflux of nutrients into the apoplast of N. benthamiana when compared to nonhost pathogens that were similar to the mock treatment (Supplemental Table S2). These results indicate that, much like a virulent pathogen infection led to nutrient efflux, silencing of NbSQS in N. benthamiana leads to nutrient efflux from the plant cell into the apoplastic space that could serve as nutrients for multiplication of host and nonhost pathogens.
Apoplastic Extracts from NbSQS-Silenced N. benthamiana Enhances Bacterial Multiplication in Vitro
To examine whether the nutrient-rich apoplastic fluid from the NbSQS-silenced N. benthamiana plants supports faster bacterial multiplication than the apoplastic fluids from control plants, we measured the multiplication rate of different bacterial strains including host, nonhost, and nonpathogenic bacteria using minimal growth medium (MGM) supplemented with 5% of apoplastic extracts from NbSQS-silenced and control plants. All tested bacteria, including nonpathogenic bacterium Escherichia coli, multiplied faster in MGM containing 5% apoplast from the NbSQS-silenced N. benthamiana plants than in MGM containing 5% apoplast from the control plants (Fig. 2E).
Silencing of AtSQS Compromises Plant Innate Immunity in Arabidopsis
Arabidopsis (Arabidopsis thaliana) contains two SQS-annotated genomic sequences: At4g34640 (AtSQS1) and At4g34650 (AtSQS2), which are located in tandem array in chromosome 4 and share 80.7% identity in amino acid sequences. The amino acid sequence of NbSQS shows 73.9% identity to AtSQS1 and 72.8% to AtSQS2. Several potential Arabidopsis individual T-DNA knockout lines for AtSQS1 and AtSQS2 genes were obtained from the SALK mutant collection, but no homozygous mutants for either AtSQS1 or AtSQS2 knockouts could be isolated. Therefore, to examine the function of Arabidopsis SQS in disease resistance, we developed Arabidopsis SQS RNA interference (RNAi) lines. A recent study has demonstrated that Arabidopsis contains only a single gene (AtSQS1) encoding SQS since the expressed protein encoded by AtSQS2 is unable to synthesize squalene from farnesyl diphosphate (Busquets et al., 2008). Therefore, the nucleotide sequence of AtSQS1 was used to make a construct for development of AtSQS1 RNAi lines. AtSQS1 RNAi homozygous lines showed an elongated petiole compared to wild type (Supplemental Fig. S3A). Examination of the AtSQS1 transcript in the RNAi lines by qRT-PCR indicated that four out of five transgenic lines tested showed a reduction in the expression of AtSQS1, but not AtSQS2, compared to the vector control (Supplemental Fig. S3B).
AtSQS1 RNAi lines were challenged with a nonhost pathogen, Pta, by leaf infiltration at a concentration of 1 × 106 cfu/mL. The disease symptoms were observed only in the transgenic AtSQS1 RNAi lines of Arabidopsis that showed reduced transcription of AtSQS1, but not in the empty vector control line and an RNAi line (SSi1e) that did not reduce AtSQS1 transcripts (Supplemental Fig. S3C). Correspondingly, bacterial growth assays indicated that the transgenic AtSQS1 RNAi lines of Arabidopsis were susceptible to the nonhost pathogen Pta when compared to control plants (Supplemental Fig. S3D). These results demonstrated that the AtSQS1 was required for plant nonhost resistance and is a functional analog of NbSQS.
Arabidopsis sterol methyltransferase2 Compromises Plant Innate Immunity
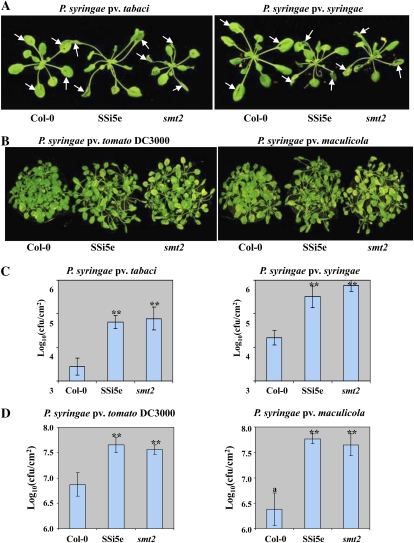
Silencing of AtSQS1 could affect multiple pathways involved in the biosynthesis of brassicasterol, brassinolide, and stigmasterol. Therefore, it is important to elucidate which phytosterol-specific pathway contributes to plant nonhost resistance. Sterol methyltransferase2 (SMT2), catalyzing the second methyl transfer reaction in the biosynthesis of phytosterols, serves as a branch point between in vivo synthesis of β-sitosterol/stigmasterol and brassinosteroids (Supplemental Fig. S2A). Thus, we tested the susceptibility of an Arabidopsis smt2 mutant, previously identified as cotyledon vascular pattern1 (cvp1; Carland et al., 2002), to nonhost pathogens. Upon challenging with nonhost pathogens Pta and P. syringae pv syringae (Pss), disease symptoms were developed on AtSQS1 RNAi line and smt2 mutant (cvp1-4) plant leaves, but not on the wild-type Columbia-0 (Col-0; Fig. 3A).
Figure 3.
Silencing of the AtSQS1 gene and smt2 mutation in Arabidopsis leads to disease susceptibility. A and C, Four-week-old Col-0, smt2 mutant, and SQS1 RNAi (SSi5e) plant leaves were syringe infiltrated with nonhost pathogens Pss or Pta at a concentration of 1 × 107 cfu/mL or dip inoculated with host pathogens Pst DC3000 or Psm at a concentration of 1 × 108 cfu/mL. B and D, Four-week-old plant leaves were syringe inoculated with Pta or Pss at a concentration of 1 × 106 cfu/mL or dip inoculated with Pst DC3000 or Psm at a concentration of approximately 1 × 108 cfu/mL. Bacterial population was quantified by plating serial dilutions of plant leaf extracts at 3 dpi. Asterisks indicate a significant difference from the wild type (Student’s t test; P < 0.01). Data were pooled from two independent experiments representing four biological replicates. Photographs were taken at 4 dpi. See also Supplemental Figure S3.
To test whether an Arabidopsis AtSQS1 RNAi line (SSi5e) and smt2 mutant compromise basal resistance against virulent pathogens, 4-week-old seedlings were inoculated with Pst DC3000 and P. syringae pv maculicola (Psm). All plants were inoculated by dipping in 1 × 108 cfu/mL of bacteria. The SSi5e RNAi line and smt2 mutant plants showed enhanced chlorotic and necrotic disease symptoms at 4 dpi, compared to wild-type Col-0 plants infected with the same pathogens (Fig. 3B). Bacterial growth assay confirmed that both smt2 mutant and AtSQS1 RNAi line supported higher levels of bacterial multiplication of Pta and Pss than the wild-type Col-0 plants in repeated experiments (Fig. 3C). Similarly, SSi5e and smt2 mutant plants supported more Pst DC3000 and Psm bacterial multiplication than Col-0 plants (Fig. 3D). These results indicated that smt2 plants were susceptible, to a similar degree as the AtSQS1 RNAi line, to host and nonhost pathogens tested, suggesting that the SMT2-mediated phytosterol biosynthetic pathway plays a key role in plant innate immunity.
AtCYP710A1 Gene Positively Regulates Plant Innate Immunity
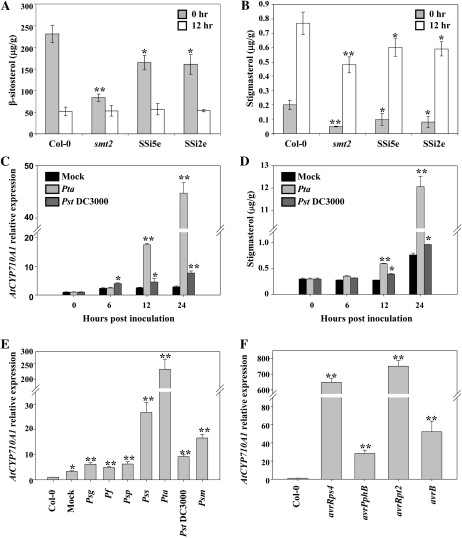
SQS catalyzes the first step in the biosynthesis of various phytosterols, while sterol methyltransferase is a key branching-point enzyme leading to biosynthesis of β-sitosterol and stigmasterol. Therefore, Arabidopsis smt2 mutant and AtSQS1 RNAi lines may contain altered levels of β-sitosterol and stigmasterol. Maintaining the correct balance of these phytosterols in plants may be related to plant defense because phytosterols can modulate permeability and fluidity of membrane (Schuler et al., 1991). To test this hypothesis, we first examined the content of β-sitosterol and stigmasterol in a smt2 mutant and AtSQS1 RNAi lines by GC-MS. As expected, β-sitosterol and stigmasterol in the smt2 mutant and AtSQS1 RNAi lines were reduced compared to the wild-type Arabidopsis (Fig. 4, A and B). To examine whether the levels of these sterols are affected when plants are infected with nonhost pathogens, we inoculated the plants by vacuum infiltration with a nonhost pathogen, Pta, at a concentration of 1 × 105 cfu/mL. The content of β-sitosterol was reduced in all tested lines, while stigmasterol was dramatically induced 12 h post inoculation (hpi; Fig. 4, A and B).
Figure 4.
Differential expression of the AtCYP710A1 gene and accumulation of stigmasterol due to plant defense. A and B, Four-week-old Arabidopsis wild-type (Col-0), smt2 mutant, and SQS1 RNAi (SSi5e and SSi2e) plants were vacuum infiltrated with a nonhost pathogen, Pta, at a concentration of 1 × 105 cfu/mL. Sterols were extracted from the leaves of inoculated plants and quantified at 12 hpi by GC-MS. C, Col-0 plants were vacuum infiltrated with 10 mm MgCl2 (mock), Pta, and Pst DC3000 at a concentration of 1 × 105 cfu/mL. Transcript levels of AtCYP710A1 were measured by qRT-PCR and the expression levels relative to 0 h (1.0) are shown. D, Stigmasterol level in the plant leaves was determined as described in B. E, Col-0 leaves were syringe infiltrated with various bacterial pathogens at a concentration of 1 × 106 cfu/mL. The relative expression of AtCYP710A1 to uninoculated Col-0 (1.0) was determined at 24 hpi by qRT-PCR. Col-0: Uninoculated; mock: 10 mm MgCl2; Pf: P. fluorescens; Psp: P. syringae pv phaseolicola. F, Col-0 plants were syringe inoculated with Pst DC3000 containing different avirulence genes (avrRps4, avrPphB, avrRpt2, and avrB) at a concentration of 1 × 105 cfu/mL. The relative expression of AtCYP710A1 to uninoculated Col-0 (1.0) was determined. Asterisks indicate a significant difference from the wild type (Student’s t test; *P < 0.05 and **P < 0.01). Data were pooled from two independent experiments representing four biological replicates. See also Supplemental Figures S4 and S5.
Since Arabidopsis C22-sterol desaturase encoded by the gene AtCYP710A1 (At2g34500) specifically converts β-sitosterol to stigmasterol (Morikawa et al., 2006), we examined the expression levels of AtCYP710A1 in wild-type Col-0 by qRT-PCR in addition to the measurement of stigmasterol content at different time intervals after inoculation with different pathogens, including a nonhost pathogen, Pta, and a host pathogen, Pst DC3000. The transcript level of AtCYP710A1 was dramatically increased 12 hpi with Pta and increased over 45-fold more than the control at 24 hpi (Fig. 4C). Correspondingly, the levels of stigmasterol were more than 12-fold higher at 24 hpi with Pta compared to the controls (Fig. 4D). An increase of AtCYP710A1 expression and stigmasterol was also observed at 12 hpi with the host pathogen Pst DC3000 when compared to mock treatment (Fig. 4, C and D).
We also observed a dosage-dependent induction of stigmasterol that correlates with the amount of pathogen inoculums. We inoculated the wild-type Arabidopsis Col-0 with a nonhost pathogen, Pta, at different concentrations. As shown in Supplemental Figure S4, at 12 hpi the transcripts of AtCYP710A1 gene were increased 115- or 15-fold when inoculated at 1 × 106 cfu/mL or 1 × 105 cfu/mL, respectively. Similarly, at 24 hpi the AtCYP710A1 gene was induced 272-fold or 45-fold at 1 × 106 cfu/mL or 1 × 105 cfu/mL, respectively.
AtCYP710A1 Gene Is Induced during Both PTI and ETI Responses
We examined whether AtCYP710A1 could be induced upon inoculation with other host and nonhost pathogens. Four-week-old Arabidopsis (Col-0) seedlings were individually inoculated with different pathovars of P. syringae and nonpathogenic Pseudomonas fluorescens at a concentration of 1 × 106 cfu/mL by leaf infiltration. qRT-PCR analysis indicated that AtCYP710A1 was increased by 24 hpi with most bacteria tested, compared to the mock control (Fig. 4E). These data demonstrated that AtCYP710A1 is commonly induced during both basal and nonhost defense responses with variation in the degree of induction.
Since stigmasterol is a component of cell membranes, we speculated that the AtCYP710A1 gene might also play a role in ETI. Pst DC3000 expressing AvrB, AvrPphB, AvrRps4, or AvrRpt2 was used to inoculate the wild-type Arabidopsis Col-0 (has the corresponding R genes) individually by leaf infiltration at a concentration of 1 × 105 cfu/mL. AvrRps4 and AvrRpt2 expressing Pst DC3000 induced AtCYP710A1 by approximately 600-fold, while AvrB and AvrPphB expressing Pst DC3000 induced AtCYP710A1 by approximately 30-fold at 24 hpi when compared to mock inoculation (Fig. 4F). These data suggested that AtCYP710A1 is induced during ETI. To check the induction of AtCYP710A1 during PTI, we examined the expression of AtCYP710A1 in Arabidopsis leaves inoculated with various elicitors in the publicly available gene expression database, GENEVESTIGATOR (www.genevestigator.com). Induction of AtCYP710A1 was seen with PAMP elicitor FLG22 (Supplemental Fig. S5A). Taken together, our data and already-published results demonstrated that AtCYP710A1 is induced during both PTI and ETI.
Arabidopsis Atcyp710A1 Mutant Compromises Plant Innate Immunity
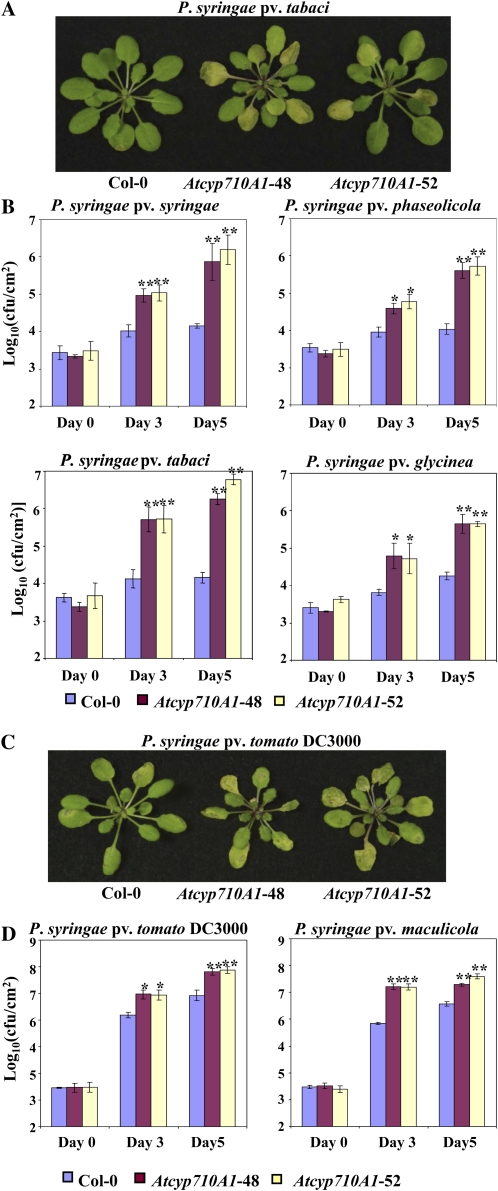
To further investigate the function of the AtCYP710A1 gene in plant innate immunity, Arabidopsis Atcyp710A1 T-DNA insertion mutants were obtained from SALK (SALK_112491 and SALK_014626) and the European Arabidopsis Stock Center (GK-325E09). In both the SALK lines, the T-DNA insertion was in 5′ untranslated region and did not have a complete loss of transcripts and therefore was not characterized further. In the GK-325E09 mutant, the T-DNA was inserted in the coding region (i.e. 98 nucleotides after the start codon) of AtCYP710A1 gene and was a null mutant (Supplemental Fig. S5, B and C). Two homozygous plants (Atcyp710A1-48 and Atcyp710A1-52) were selected for characterization. The mutant plants had no obvious developmental abnormalities (Supplemental Fig. S5D). Atcyp710A1 mutant plants along with wild-type Col-0 were inoculated (by leaf infiltration using needleless syringe) with nonhost pathogens Pta, Pss, Psp, and Psg, as well as host pathogens Psm and Pst DC3000. The results showed that all tested nonhost pathogens multiplied more, and some even produced disease symptoms in the Atcyp710A1 mutant plant leaves compared to Col-0 at 3 and 5 dpi (Fig. 5, A and B). The virulent pathogens Psm and Pst DC3000 also grew up to 15-fold more in the Atcyp710A1 mutant and produced enhanced disease symptoms compared to Col-0 plants (Fig. 5, C and D; Supplemental Fig. S5E). Silencing of the AtCYP710A1 gene by VIGS in wild-type N. benthamiana and tomato (Solanum lycopersicum) plants also leads to susceptibility upon inoculation with nonhost pathogens Pst T1 and Pta, respectively (Supplemental Fig. S6, A and B).
Figure 5.
Arabidopsis Atcyp710A1 mutant compromises disease resistance. A and C, Arabidopsis wild type (Col-0) and Atcyp710A1 mutants (48 and 52) were syringe inoculated with a nonhost pathogen, Pta, or a host pathogen, Pst DC3000, at a concentration of 1 × 106 cfu/mL. Photographs were taken at 3 dpi. B and D, Plant leaves were syringe infiltrated with various host and nonhost pathogens at a concentration of approximately 1 × 106 cfu/mL. Bacterial population was quantified at 0, 3, and 5 dpi. Asterisks indicate a significant difference from the wild type (Student’s t test; *P < 0.05 and **P < 0.01). Data were pooled from two independent experiments representing four biological replicates. See also Supplemental Figures S5 to S7.
Apoplastic Extracts from Arabidopsis Atcyp710A1 Mutant Plants Support Higher Bacterial Multiplication
To investigate whether the Arabidopsis Atcyp710A1 mutant also has increased nutrient efflux into the apoplast, similar to N. benthamiana SQS-silenced plants, both virulent and avirulent pathogens were grown in vitro in MGM containing 5% apoplastic fluid extracted from wild-type (Col-0) and mutant (Atcyp710A1-48 and Atcyp710A1-52) leaves. The results indicated that the apoplastic fluid from the mutant leaves supported more multiplication of virulent and avirulent pathogens compared to apoplastic fluid from Col-0 leaves (Supplemental Fig. S7A). Consistent with this data, higher membrane permeability, as reflected by higher entry of PI, was also seen in Atcyp710A1 mutant cells (Supplemental Fig. S7, B–D). These results suggest that the AtCYP710A1 gene plays a crucial role in controlling nutrient efflux into the apoplast by maintaining membrane integrity.
Overexpression of the AtCYP710A1 Gene Confers Disease Resistance in Arabidopsis
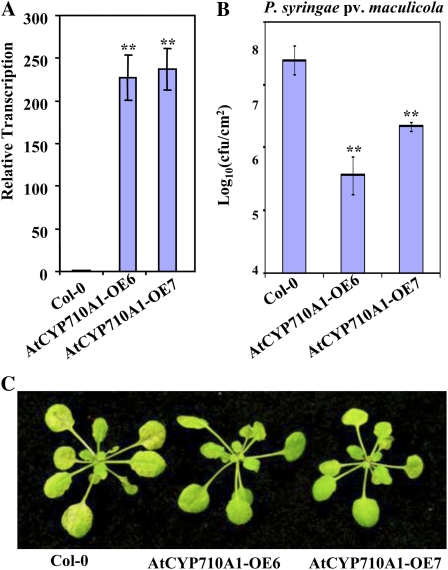
Since the expression of the AtCYP710A1 gene increased during both compatible and incompatible interaction with pathogens, we reasoned that AtCYP710A1 is a plant defense gene and its overexpression might confer disease resistance to host pathogens. High levels of AtCYP710A1 gene transcripts were confirmed by qRT-PCR in Arabidopsis AtCYP710A1 gene overexpression lines (Fig. 6A; Morikawa et al., 2006; Arnqvist et al., 2008). We challenged these plants through dip inoculation of host pathogen Psm (1 × 108 cfu/mL). As speculated, disease symptoms and bacterial multiplication were suppressed in AtCYP710A1 gene overexpression lines when compared to wild-type Col-0 (Fig. 6, B and C). AtCYP710A1 gene overexpression plants also showed a mild decrease in host-pathogen-induced ion leakage at 12 hpi (Supplemental Fig. S8). These results further suggested that the AtCYP710A1 gene positively mediates plant innate immunity.
Figure 6.
Overexpression of the AtCYP710A1 gene in Arabidopsis confers disease resistance. A, Transcripts of AtCYP710A1 in two independent AtCYP710A1 overexpressor lines (AtCYP710A1OE-6 and AtCYP710A1OE-7) were determined by qRT-PCR. Expression of AtCYP710A1 in the overexpressors relative to expression in wild type (Col-0; 1.0) is shown. Values for individual plants from each line are shown here. B, Arabidopsis Col-0 and AtCYP710A1 gene overexpressors were inoculated with a host pathogen, Psm, by spraying at a concentration of 1 × 108 cfu/mL. Bacterial population in plants shown in B was quantified at 3 dpi. C, Photographs of disease symptoms were taken at 3 dpi. Asterisks indicate a significant difference from the wild type (Student’s t test; *P < 0.05 and **P < 0.01). Data were pooled from two independent experiments representing four biological replicates. See also Supplemental Figure S8.
DISCUSSION
Unlike R-gene-mediated resistance, which is specific for a particular bacterial pathovar, nonhost resistance is governed by a broad defense mechanism that can defend against a wide range of bacterial pathovars (Heath, 2000; Mysore and Ryu, 2004; Ellis, 2006). Nonhost resistance involves many aspects of plant defense mechanisms such as production of antimicrobial compounds and HR (Nürnberger and Lipka, 2005). In addition, many of the subsequent plant defenses that determine the success or failure of pathogenic bacteria take place in the apoplast, the final battlefield of plant-bacterial interactions (Hoefle and Hückelhoven, 2008). The leaf apoplast constantly exchanges components with the cytosol and contains minimal nutrients, mostly transferred from the cell due to membrane permeability (Sattelmacher, 2001). Biotrophic and hemibiotrophic bacterial pathogens colonize a host by growing in the apoplast and absorbing nutrients from the cytoplasm (Melotto et al., 2008). A recent report indicated that to get nutrients from cells, pathogen virulence factors directly bind promoters of sugar transporters like SWEET11 and SWEET14 in Arabidopsis, and efflux sugars from the cytosol (Chen et al., 2010). Therefore, restricting the availability of nutrients to pathogens can be a potential plant defense mechanism; such a strategy can be exploited for broad spectrum disease resistance.
We developed a VIGS-based novel approach to identify plant genes that play a role in nonhost disease resistance. Based on the multiplication of bacteria directly placed in the apoplast, we identified the role of SQS in limiting the multiplication of nonhost bacteria. Our results showed that SQS-silenced N. benthamiana plants leaked more nutrients into the apoplast, thus supporting increased bacterial multiplication. We demonstrated that other sterol biosynthetic pathway genes like AtSMT2 and AtCYP710A1 are also involved in plant defense by regulating nutrient efflux into the apoplast. Arabidopsis mutant lines of these genes were compromised in nonhost and basal disease resistance. After a pathogen manages to overcome preformed constitutive and inducible barriers, it is subjected to recognition at the plasma membrane (Alfano and Collmer, 1996; Hoefle and Hückelhoven, 2008). At this point, the PAMP can be recognized and plants induce PTI (Hofius et al., 2007). A recent study (Griebel and Zeier, 2010) showed that AtCYP710A1 gene-mediated biosynthesis of stigmasterol is induced upon inoculation with a virulent pathogen, Psm, avirulent pathogen Psm-avrRpm1, nonhost pathogen, Psg, and PAMPs (FLG22). Similarly, data from the publicly available gene expression database GENEVESTIGATOR showed induction of this gene upon FLG22 treatment. Therefore, plants have a strategy to manipulate sterol synthesis to prevent nutrient efflux into the apoplast. β-Sitosterol and stigmasterol are the predominant sterols in plant membranes (Hodzic et al., 2008). Since plant sterols are structurally related to cholesterol and control membrane mechanical properties (Hodzic et al., 2008), changes in sterol content and composition can affect membrane permeability, hydrocarbon chain order, condensation efficacy, and elastic behavior (Bernsdorff and Winter, 2003).
The fact that both SQS-gene-silenced N. benthamiana and the Arabidopsis Atcyp710A1 mutant increased nutrient efflux into the apoplast indicates the relevance of sterols to prevent nutrient efflux into the apoplast. Our results also suggest that plant cells can induce defense mechanisms to deprive potential pathogens of the nutrients available in the apoplast. The SQS-gene-silenced plants were susceptible to a wide range of pathogens and consistently showed more nutrients in the apoplast compared to wild-type plants. This is mainly due to excessive nutrient efflux into the apoplast. Although specific transporters are known to be involved in efflux of nutrients into the apoplast upon pathogen infection (Chen et al., 2010), our data suggest that membrane permeability also plays an important role in nutrient efflux into the apoplast. To defend against pathogens effectively, we speculate that plants have to reduce excess nutrient efflux immediately after pathogen infection and well before the start of disease.
Stigmasterol levels in Arabidopsis transgenic lines with AtCYP710A1 gene overexpression were increased up to 32-fold (Morikawa et al., 2006). We believe that Arabidopsis AtCYP710A1 gene-overexpressing plants have a less-permeable membrane and hence resisted pathogen growth while the Atcyp710A1 mutant compromised both host and nonhost resistance. Recent studies also showed the relevance of stigmasterol in plant-pathogen interaction (Griebel and Zeier, 2010; Wang et al., 2012). Consistent with our results, resistance of 3-hydroxy-3-methylglutaryl-CoA synthase overexpressed Arabidopsis plants to Botrytis cinerea was shown to be due to up-regulation of AtCYP710A1 gene expression and stigmasterol content (Wang et al., 2012). However, contrary to our results, study by Griebel and Zeier (2010) indicated less host bacterial growth in different Arabidopsis Atcyp710A1 mutants, Salk_112491, and Salk_014626. In the Salk_112491 line, T-DNA is inserted nearly 780-bp upstream of the start codon of the AtCYP710A1 gene. The Salk_014626 line has two T-DNA insertions where one of the T-DNA is inserted in the potential promoter region (480-bp upstream of the start codon) of the AtCYP710A1 gene and another T-DNA is inserted in the exon of a gene that encodes isopentenyl diphosphate isomerase 2. The Atcyp710A1 mutant used in this study has T-DNA insertion in the exon and is a null mutant. At this point, we are unable to explain the exact reason for the discrepancy in our results. It is possible that the reduced AtCYP710A1 expression in the SALK lines may have other unknown pleiotropic effects on the plant, contrary to the null mutant.
Our results showed that the AtCYP710A1 gene also plays a role during R-gene-mediated plant disease resistance. The Arabidopsis Col-0 ecotype encodes many resistance proteins that can recognize their cognate avirulence effectors, such as AvrB, AvrPphB, AvrRps4, and AvrRPt2, leading to HR. The AtCYP710A1 gene was slightly induced upon infection with the host pathogen Pst DC3000, and the induction was up to 600-fold more when the Pst DC3000 expressed one of the corresponding avirulence genes, suggesting that AtCYP710A1 is also induced during ETI.
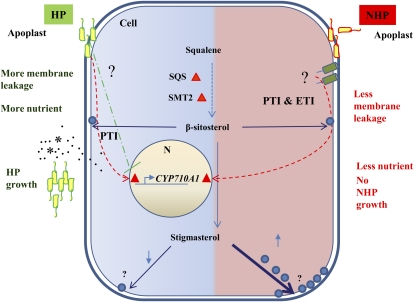
Based on our data showing the up-regulation of sterol biosynthetic genes during both PTI and ETI, we propose a model shown in Figure 7 for the role of sterols (especially stigmasterol) in plant defense. During an incompatible interaction (nonhost or R-gene-mediated resistance), either PTI or ETI, or both together, induces sterol pathway genes like AtCYP710A1 and prevents nutrient efflux to the apoplast. During a compatible interaction, stigmasterol biosynthesis is initially induced by PTI, but suppressed by pathogens. We speculate that the virulence proteins (effectors) injected by the host pathogens into the plant cell will interfere with the induction of stigmasterol biosynthesis to combat plant defense mechanisms and favor nutrient efflux into the apoplast. A host pathogen is also known to manipulate alkalinization of the apoplast to favor nutrient efflux (Atkinson and Baker, 1987).
Figure 7.
Sterols play a role in nonhost resistance and basal resistance by regulating nutrient efflux into the apoplast. Successful plant pathogenic bacteria multiply in the apoplast by drawing nutrients from cytoplasm. During attack by nonhost bacteria, the plant cell initiates signaling events leading to enhanced expression of genes involved in sterol biosynthesis. An unknown mechanism makes the cell reduce nutrient efflux, thereby depriving nutrient availability in the apoplast, thus arresting nonhost pathogen growth. When pathogenic bacteria attack their host plants, bacterial PAMPs trigger a general plant defense response, leading to induction of stigmasterol. However, unknown virulence factors of host pathogenic bacteria might suppress the production of stigmasterol by inhibiting the expression of the AtCYP710A1 gene, thus allowing nutrient efflux from cytoplasm to the apoplastic space. HP, Host pathogen; NHP, nonhost pathogen. [See online article for color version of this figure.]
In conclusion, our results show that plants alter sterol biosynthesis upon infection by a potential bacterial pathogen through PTI and/or ETI. This restricts nutrient transfer from cytosol to the apoplast. Overexpression of the AtCYP710A1 gene improved plant immunity against pathogens. Our results show promise for a novel strategy to engineer plants for broad spectrum disease resistance by manipulating sterol levels. The exact role of stigmasterol or β-sitosterol in membrane permeability and the relevance of sterols as signaling molecules warrant further investigation.
MATERIALS AND METHODS
Plant Materials, Bacterial Pathogens, and Growth Conditions
Nicotiana benthamiana seeds were germinated as previously described (Anand et al., 2007). Arabidopsis (Arabidopsis thaliana) Atsqs1 T-DNA mutants (Salk_034266, 087515, 034431, and 077057), Atsqs2 T-DNA mutants (Salk_018690 and 018698), and Atcyp710A1 mutants (SALK_112491 and SALK_014626) were obtained from the Arabidopsis Biological Resource Center. Atcyp710A1 (locus: At2G34500) T-DNA mutant (GK-325E09; Nottingham Arabidopsis Stock Centre ID N376345-62) was obtained from Nottingham Arabidopsis Stock Centre. Arabidopsis AtCYP710A1 overexpression lines (Morikawa et al., 2006) were provided by Dr. Daisaku Ohta, Osaka Prefecture University, Osaka, Japan. Atsmt2 mutant (Carland et al., 2002; cvp1-3) was obtained from Dr. Timothy Nelson, Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT. Arabidopsis seeds were cold treated for 3 to 4 d at 4°C and germinated in Professional Blend soil in a growth chamber at 20°C to 22°C under short-day conditions (8-h light). Agrobacterium tumefaciens GV2260 was grown in Luria-Bertani medium at 28°C. Pseudomonas syringae strains were grown in King’s B medium at 28°C. Bacterial growth was monitored as described (Wang et al., 2007).
VIGS in N. benthamiana
For screening cDNA library, VIGS was carried out as described earlier (Mysore and Ryu, 2004; Ryu et al., 2004; Anand et al., 2007) with slight modifications. See Supplemental Materials and Methods S1 for more details.
Plant Inoculation, Disease Assay, and Quantification of Bacteria
Bacterial cells of P. syringae strains were collected by centrifugation of overnight culture at 5,000 rpm for 10 min, washed twice, and resuspended to the desired concentration in 10 mm MgCl2 with 0.01% (v/v) Silwet L-77 (Osi Specialties). The bacterial suspension was used to inoculate fully expanded leaves of gene-silenced N. benthamiana plants 2 weeks after TRV infection or 4-week-old Arabidopsis plants by dipping or leaf infiltration using a needleless syringe or vacuum, and incubated in growth chambers at 20°C to 22°C. Two leaf discs (0.5-cm2 each) of each plant were homogenized and serial diluted in 10 mm MgCl2 and plated on King’s B agar medium for quantification of in planta bacterial growth. Bacterial population was calculated as cfu per square centimeter of leaf area. Experiments were performed with at least four replicates.
Extraction of Apoplastic Fluid and Metabolite Profiling
N. benthamiana leaves were submerged in a beaker containing 1,000 mL milliQ water and a vacuum was applied for 1 min to remove the air pockets in the apoplast. Vacuum-infiltrated leaves were then blotted dry, cut into two pieces along the midrib to remove major leaf veins, carefully rolled, and placed into a 15-mL tube with a hole punched at the bottom leading to a 1.5-mL collection tube. This setup was then inserted into a 50-mL conical tube (Corning) for spinning. Apoplast fluid was extracted by centrifugation at 500g for 10 min at 4°C. The fraction collected in the 1.5-mL tube was transferred to a fresh tube and centrifuged for 10 min at 3,000g at 4°C. The supernatant was then filter sterilized (to remove bacterial contamination) and was either used for further analyses immediately or stored at −80°C. Apoplastic fluid from Arabidopsis was extracted similarly except centrifugation was at 2,000g for 30 min at 4°C. Metabolite profiling of apoplastic fluid was performed as previously described (Broeckling et al., 2005) with minor modifications. In brief, 300 μL of apoplastic fluid was transferred to a 4-mL glass vial and lyophilized in a freeze drier (VirTis) overnight. HPLC grade water (1.5 mL) containing 25 μg/mL ribitol (internal standard) and 1.5 mL of chloroform was added, vortexed, and incubated for 45 min at 50°C. The samples were then centrifuged and the polar phase was transferred (1 mL) to an autosampler vial and dried by vacuum centrifugation at room temperature. The dried extracts were methoximated in 50 μL of pyridine with 15 mg/mL of methoxyamine hydrochloride, sonicated, and incubated at 50°C until the residue was resuspended. The samples were then derivatized with 50 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide + 1% trimethylchlorosilane for 1 h at 50°C. One microliter of the solution was analyzed by GC-MS.
Phytosterol Extraction and Analysis
Phytosterols were extracted by chloroform/methanol as previously described (Morikawa et al., 2006) with minor modifications. Leaf tissues were frozen in liquid nitrogen and lyophilized in a freeze drier (VirTis) for 3 d. The homogenized dry tissue (20 mg) was used to extract the total sterols in 5-mL chloroform/methanol (1:2, v/v) with gentle shaking at room temperature as described in detail in Supplemental Materials and Methods S1.
Statistical Analysis
Student’s t test was conducted for statistical analysis of the data presented in the figures. Error bars indicate the se values of replicate means. Asterisks indicate a significant difference from the wild type (*P < 0.05 and **P < 0.01).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Endogenous NbSQS gene transcript levels, sterol levels, occurrence of HR, and disease symptoms in NbSQS-gene-silenced plants.
Supplemental Figure S2. Simplified biosynthetic pathway for plant sterols and structure of β-sitosterol and stigmasterol.
Supplemental Figure S3. Silencing of the AtSQS1 gene and smt2 mutation in Arabidopsis leads to disease susceptibility.
Supplemental Figure S4. Dosage-dependent expression of the AtCYP710A1 gene upon nonhost pathogen inoculation.
Supplemental Figure S5. AtCYP710A1 gene expression upon elicitor treatment and details of Arabidopsis Atcyp710A1 mutants.
Supplemental Figure S6. Silencing of the AtCYP710A1 gene by VIGS supports more multiplication of nonhost bacteria in N. benthamiana and tomato.
Supplemental Figure S7. Analysis of pathogen growth in apoplastic fluid and membrane permeability characteristics.
Supplemental Figure S8. Membrane leakage in AtCYP710A1 overexpression plants upon host pathogen infection.
Supplemental Table S1. Increased compounds in apoplast extracted from the SQS-silenced N. benthamiana compared to the control.
Supplemental Table S2. Metabolite profiling of apoplast.
Supplemental Materials and Methods S1. Additional details for identification and characterization of SQS, SMT2, and CYP710A1 genes in N. benthamiana and Arabidopsis.
Supplementary Material
Acknowledgments
We thank Ms. Janie Gallaway (Noble Foundation [NF]) and Colleen Elles (NF) for excellent plant care, Drs. Elison Blancaflor (NF) and Rao Uppalapati (NF) for critical reading of the manuscript, Mrs. Jackie Kelley (NF) for editorial corrections, Mr. David Huhman at the analytical chemistry core facility at NF for help with quantification of sterols, Dr. Daisaku Ohta (Osaka Prefecture University) for seeds of AtCYP710A1 overexpression lines, and Dr. Timothy Nelson (Yale University, New Haven, CT) for seeds of the Atsmt2 mutant. We also thank Drs. Elison Blancaflor and Jin Nakashima at the imaging facility at NF for help with microscopy.
References
- Aghnoum R, Niks RE. (2010) Specificity and levels of nonhost resistance to nonadapted Blumeria graminis forms in barley. New Phytol 185: 275–284 [DOI] [PubMed] [Google Scholar]
- Alfano JR, Collmer A. (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell 8: 1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Vaghchhipawala Z, Ryu C-M, Kang L, Wang K, del-Pozo O, Martin GB, Mysore KS. (2007) Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol Plant Microbe Interact 20: 41–52 [DOI] [PubMed] [Google Scholar]
- Arnqvist L, Persson M, Jonsson L, Dutta PC, Sitbon F. (2008) Overexpression of CYP710A1 and CYP710A4 in transgenic Arabidopsis plants increases the level of stigmasterol at the expense of sitosterol. Planta 227: 309–317 [DOI] [PubMed] [Google Scholar]
- Atkinson MM, Baker CJ. (1987) Alteration of plasmalemma sucrose transport in Phaseolus vulgaris by Pseudomonas syringae pv. syringae and its association with K+/H+ exchange. Phytopathology 77: 1573–1578 [Google Scholar]
- Bernsdorff C, Winter R. (2003) Differential properties of the sterols cholesterol, ergosterol, sitosterol, trans-7-dehydrocholesterol, stigmasterol and lanosterol on DPPC bilayer order. J Phys Chem 107: 10658–10664 [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW. (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Busquets A, Keim V, Closa M, del Arco A, Boronat A, Arró M, Ferrer A. (2008) Arabidopsis thaliana contains a single gene encoding squalene synthase. Plant Mol Biol 67: 25–36 [DOI] [PubMed] [Google Scholar]
- Carland FM, Fujioka S, Takatsuto S, Yoshida S, Nelson T. (2002) The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Q, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB. (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarenne TP, Ghosh A, Chappell J. (2002) Regulation of squalene synthase, a key enzyme of sterol biosynthesis, in tobacco. Plant Physiol 129: 1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarenne TP, Shin DH, Back K, Yin S, Chappell J. (1998) Molecular characterization of tobacco squalene synthase and regulation in response to fungal elicitor. Arch Biochem Biophys 349: 205–215 [DOI] [PubMed] [Google Scholar]
- Ellis J. (2006) Insights into nonhost disease resistance: can they assist disease control in agriculture? Plant Cell 18: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freialdenhoven A, Orme J, Lahaye T, Schulze-Lefert P. (2005) Barley Rom1 reveals a potential link between race-specific and nonhost resistance responses to powdery mildew fungi. Mol Plant Microbe Interact 18: 291–299 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J. (2010) A role for beta-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J 63: 254–268 [DOI] [PubMed] [Google Scholar]
- Harrison RAP, Vickers SE. (1990) Use of fluorescent probes to assess membrane integrity in mammalian spermatozoa. J Reprod Fertil 88: 343–352 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. (2011) Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J 67: 980–992 [DOI] [PubMed] [Google Scholar]
- Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G. (2008) Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys J 94: 3935–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Hückelhoven R. (2008) Enemy at the gates: traffic at the plant cell pathogen interface. Cell Microbiol 10: 2400–2407 [DOI] [PubMed] [Google Scholar]
- Hofius D, Tsitsigiannis DI, Jones JDG, Mundy J. (2007) Inducible cell death in plant immunity. Semin Cancer Biol 17: 166–187 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 38: 800–809 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu A-J, Rathjen JP, Bendahmane A, Day L, Baulcombe DC. (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, He SY. (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T, Mizutani M, Aoki N, Watanabe B, Saga H, Saito S, Oikawa A, Suzuki H, Sakurai N, Shibata D, et al. (2006) Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell 18: 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Ryu C-M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Niks RE, Marcel TC. (2009) Nonhost and basal resistance: how to explain specificity? New Phytol 182: 817–828 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Lipka V. (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6: 335–345 [DOI] [PubMed] [Google Scholar]
- Rico A, Preston GM. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact 21: 269–282 [DOI] [PubMed] [Google Scholar]
- Ryu C-M, Anand A, Kang L, Mysore KS. (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J 40: 322–331 [DOI] [PubMed] [Google Scholar]
- Sattelmacher B. (2001) The apoplast and its significance for plant mineral nutrition. New Phytol 149: 167–192 [DOI] [PubMed] [Google Scholar]
- Schuler I, Milon A, Nakatani Y, Ourisson G, Albrecht AM, Benveniste P, Hartman MA. (1991) Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc Natl Acad Sci USA 88: 6926–6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R. (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Ajith A, Uppalapati SR, Mysore KS. (2008) Virus-induced gene silencing and its applications. CAB Rev Perspect Agric Vet Sci Nutr Nat Res 3: 1–18 [Google Scholar]
- Senthil-Kumar M, Mysore KS. (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang H-S, Han B, Zhu T, Zou G, Katagiri F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR. (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61: 199–223 [Google Scholar]
- Wang H, Nagegowda DA, Rawat R, Bouvier-Navé P, Guo D, Bach TJ, Chye M-L. (2012) Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol J 10: 31–42 [DOI] [PubMed] [Google Scholar]
- Wang K, Kang L, Anand A, Lazarovits G, Mysore KS. (2007) Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol 174: 212–223 [DOI] [PubMed] [Google Scholar]
- Wang L-C, Tsai M-C, Chang K-Y, Fan Y-S, Yeh C-H, Wu S-J. (2011) Involvement of the Arabidopsis HIT1/AtVPS53 tethering protein homologue in the acclimation of the plasma membrane to heat stress. J Exp Bot 62: 3609–3620 [DOI] [PubMed] [Google Scholar]
- Zellerhoff N, Himmelbach A, Dong W, Bieri S, Schaffrath U, Schweizer P. (2010) Nonhost resistance of barley to different fungal pathogens is associated with largely distinct, quantitative transcriptional responses. Plant Physiol 152: 2053–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40: 633–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.