Abstract
Plant defense responses are tightly controlled by many positive and negative regulators to cope with attacks from various pathogens. Arabidopsis (Arabidopsis thaliana) ENHANCED DISEASE RESISTANCE2 (EDR2) is a negative regulator of powdery mildew resistance, and edr2 mutants display enhanced resistance to powdery mildew (Golovinomyces cichoracearum). To identify components acting in the EDR2 pathway, we screened for edr2 suppressors and identified a gain-of-function mutation in SIGNAL RESPONSIVE1 (SR1), which encodes a calmodulin-binding transcription activator. The sr1-4D gain-of-function mutation suppresses all edr2-associated phenotypes, including powdery mildew resistance, mildew-induced cell death, and ethylene-induced senescence. The sr1-4D single mutant is more susceptible to a Pseudomonas syringae pv tomato DC3000 virulent strain and to avirulent strains carrying avrRpt2 or avrRPS4 than the wild type. We show that SR1 directly binds to the promoter region of NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), a key component in RESISTANCE TO PSEUDOMONAS SYRINGAE2-mediated plant immunity. Also, the ndr1 mutation suppresses the sr1-1 null allele, which shows enhanced resistance to both P. syringae pv tomato DC3000 avrRpt2 and G. cichoracearum. In addition, we show that SR1 regulates ethylene-induced senescence by directly binding to the ETHYLENE INSENSITIVE3 (EIN3) promoter region in vivo. Enhanced ethylene-induced senescence in sr1-1 is suppressed by ein3. Our data indicate that SR1 plays an important role in plant immunity and ethylene signaling by directly regulating NDR1 and EIN3.
Plants encounter a wide variety of pathogens in the wild, and to counter this threat, they have evolved two layers of immune defenses: pathogen/microbe-associated molecular pattern-trigged immunity and effector-triggered immunity (Chisholm et al., 2006; Jones and Dangl, 2006). In effector-triggered immunity, pathogen effectors delivered into the plant cell are recognized by cognate cytoplasmic immune receptors traditionally called resistance (R) proteins, which subsequently trigger specific defense responses. In Arabidopsis (Arabidopsis thaliana), many R genes encode structurally related proteins containing nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains. Based on N-terminal sequences, the NBS-LRR proteins can be further divided into two subfamilies: proteins containing a coiled-coil domain (CC-NBS-LRR) and proteins containing a domain homologous to Toll and IL-1 receptors (TIR-NBS-LRR). In general, CC-NBS-LRR-mediated resistance requires NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), a plasma membrane-localized protein (Century et al., 1997; Coppinger et al., 2004), and TIR-NBS-LRR-mediated resistance requires ENHANCED DISEASE SUSCEPTIBLITY1 (EDS1), a protein with similarity to lipases (Aarts et al., 1998). For instance, RESISTANCE TO PSEUDOMONAS SYRINGAE2 (RPS2), RPM1-, and RPS5-mediated resistance is dependent on NDR1, but RESISTANCE TO HYALOPERONOSPORA PARASITICA2 (RPP2), RPP4-, and RPS4-mediated resistance is dependent on EDS1.
Based on their infection strategy, pathogens can be divided into two broad classes: the first class is biotrophic pathogens, such as the fungal pathogen powdery mildew (Golovinomyces cichoracearum); the second class is necrotrophic pathogens, such as Botrytis cinerea (Glazebrook, 2005). Biotrophic pathogens depend on living host cells for invasion and reproduction. Increasing evidence has shown that salicylic acid (SA) signaling usually is involved in the defense against biotrophic pathogens, while jasmonic acid and ethylene (ET) signaling are involved in the defense against necrotrophic pathogens. Powdery mildew pathogens are obligate biotrophs that infect a broad range of crop species, including barley (Hordeum vulgare), wheat (Triticum aestivum), and grape (Vitis vinifera), and cause large worldwide economic losses (Micali et al., 2008). In the study of the interactions between Arabidopsis and powdery mildew, three major types of mutants with altered responses to powdery mildew pathogens have been identified. The first class of Arabidopsis mutants show defects in nonhost penetration resistance to the barley powdery mildew pathogen Blumeria graminis f. sp. hordei; these mutants include penetration1 (pen1), pen2, and pen3 (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). The second class of mutants show increased powdery mildew resistance but without mildew-induced cell death; this class is represented by powdery mildew-resistant1 (pmr1) to pmr6 (Vogel and Somerville, 2000; Vogel et al., 2002, 2004; Nishimura et al., 2003). The third class of mutants is represented by enhanced disease resistance (edr) mutants, including edr1, edr2, and edr3, which show increased disease resistance to powdery mildew that is accompanied by mildew-induced cell death (Frye et al., 2001; Tang et al., 2005a, 2006). Like the edr1 and edr3 mutants, edr2-mediated resistance is dependent on an intact SA signaling pathway. The EDR2 protein contains a pleckstrin homology domain, a StAR transfer domain, and a plant-specific domain of unknown function (Tang et al., 2005a; Vorwerk et al., 2007). The pleckstrin homology domain binds to phosphatidylinositol 4-phosphate in vitro. The EDR2 protein localizes to the endoplasmic reticulum, plasma membrane, and endosomes (Vorwerk et al., 2007). However, the mechanism by which EDR2 regulates powdery mildew resistance is not clear.
The interaction between plants and powdery mildew pathogens is conserved among different plant species. For example, in barley, MILDEW LOCUS O (MLO), an integral membrane protein with seven transmembrane domains, acts as a negative regulator of powdery mildew resistance (Büschges et al., 1997; Kessler et al., 2010). In Arabidopsis, MLO2, the ortholog of barley MLO, plays a similar role (Consonni et al., 2006). Interestingly, REQUIRED FOR MLO-SPECIFIED RESISTANCE2, the barley ortholog of PEN1, is required for mlo-mediated penetration resistance in barley (Bhat et al., 2005). Also, calcium signaling is required for MLO signaling (Kim et al., 2002). Calcium is a second messenger in biotic and abiotic stress signaling; these stresses induce temporal changes in cytosolic free Ca2+, which is called the calcium signature (Reddy, 2001; Hepler, 2005; Kim et al., 2009). Calcium signatures are decoded by calcium sensors, a class of calcium-binding proteins (Dodd et al., 2010). The predominant sensor is calmodulin, which has four EF hands that bind to calcium and relay calcium signaling by binding to its target proteins. Several calmodulin-binding proteins have been shown to play important roles in plant innate immunity. For instance, MLO binds to calmodulin in vitro, and loss of calmodulin-binding activity affects MLO function (Kim et al., 2002). In addition, CAM-BINDING PROTEIN 60G (CBP60g), a member of the Arabidopsis CBP60 gene family, regulates microbe-associated molecular pattern signaling and SA accumulation through calcium-dependent calmodulin binding (Wang et al., 2009). Furthermore, SIGNAL RESPONSIVE1 (SR1), a calmodulin-binding transcription factor, contributes to plant defense responses by binding to the CGCG box in the promoter of its target genes to regulate their expression (Yang and Poovaiah, 2002). One of its targets is EDS1, a positive regulator of SA signaling. SR1 calmodulin-binding activity is essential for its function (Du et al., 2009).
To identify genes that are involved in plant defense responses, we screened for suppressors of edr2. Here, we show that a gain-of-function mutation in the calmodulin-binding motif of SR1 suppressed edr2-mediated resistance to powdery mildew and enhanced ET-induced senescence in edr2. We also show that SR1 regulates plant defense responses and senescence by directly binding to the promoter regions of NDR1 and ETHYLENE INSENSITIVE3 (EIN3).
RESULTS
The sr1-4D Mutation Suppressed edr2-Mediated Powdery Mildew Resistance and ET-Induced Senescence
Previously, edr2 has been shown to display enhanced disease resistance to the powdery mildew pathogen strain UCSC1 (Tang et al., 2005a; Vorwerk et al., 2007). To identify components that are involved in EDR2 signaling, we screened for mutants that suppressed the edr2 enhanced resistance phenotype. In this screen, we identified a number of suppressors, including mutations in PHYTOALEXIN DEFICIENT4 (PAD4), SALICYLIC ACID INDUCTION-DEFICIENT2 (SID2), NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), and AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1), indicating that the screen was highly efficient (Nie et al., 2011). Here, we describe one of these mutants, which we named sr1-4D based on our subsequent characterizations described below; other edr2 suppressor mutants will be described elsewhere.
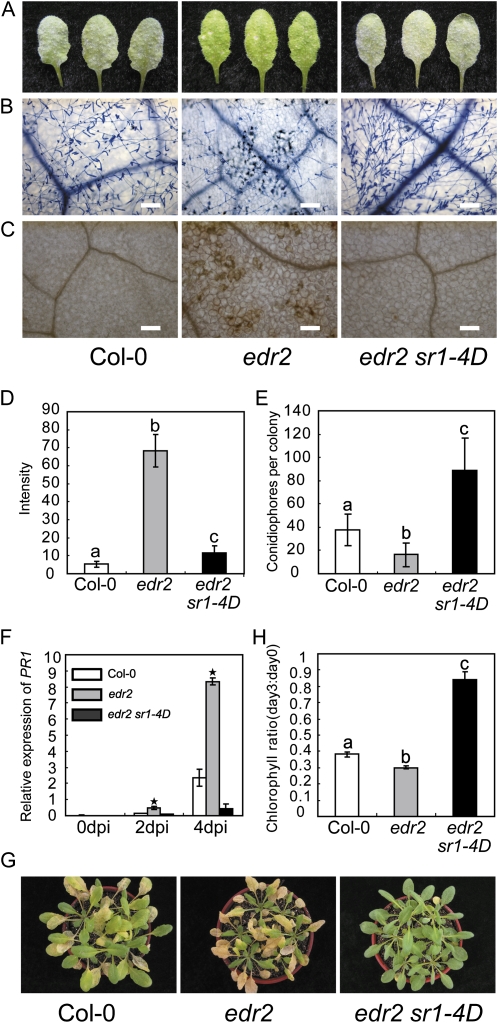
To characterize the powdery mildew resistance of edr2 sr1-4D, 4-week-old plants were inoculated with G. cichoracearum and the disease symptoms were scored at 8 d post infection (dpi). The wild-type plant was susceptible, and intensive sporulation was observed on the leaves, but the edr2 plant showed dramatic necrotic lesion formation upon mildew infection, and little powder was produced. The edr2 sr1-4D plants displayed a wild-type-like phenotype that supported the formation of a large number of conidia on the leaves, with no visible necrotic cell death observed at 8 dpi (Fig. 1A), indicating that sr1-4D suppressed the edr2 powdery mildew resistance phenotype. To further characterize the edr2 sr1-4D disease phenotype, we examined plant host cell death and fungal pathogen growth by staining the infected leaves with trypan blue at 8 dpi. As shown in Figure 1B, infected leaves of edr2 displayed massive necrotic cell death, with many fewer spores produced by the fungus compared with those of the wild type, but edr2 sr1-4D displayed extensive fungal growth with no obvious cell death, similar to the wild-type plants. Previously, it was shown that the edr2-mediated powdery mildew cell death was accompanied by the production of hydrogen peroxide (H2O2) in the cells that undergo cell death (Vorwerk et al., 2007). To examine whether H2O2 production was suppressed in edr2 sr1-4D, we monitored the H2O2 production in the wild type, edr2, and edr2 sr1-4D by staining infected leaves with 3,3′-diamino benzidine hydrochloride at 2 dpi. As shown in Figure 1, C and D, edr2 accumulated more H2O2 than the wild type, but edr2 sr1-4D accumulated H2O2 to a much lower level than edr2, indicating that the accumulation of H2O2 in edr2 was suppressed by the sr1-4D mutation. To further assess the effects of sr1-4D on the resistant phenotype of edr2, we monitored fungal growth by counting the conidiophores (asexual reproductive structures) per colony in wild-type, edr2, and edr2 sr1-4D leaves at 7 dpi. The edr2 mutant supported significantly fewer conidiophores than the wild type, but edr2 sr1-4D supported a much higher number of conidiophores than edr2 or the wild type (Fig. 1E).
Figure 1.
sr1-4D suppresses the edr2 phenotype of resistance to powdery mildew and ET-induced senescence. A, Four-week-old wild-type, edr2, and edr2 sr1-4D plants were infected with G. cichoracearum UCSC1, and representative leaves were removed and photographed at 8 dpi. The edr2 sr1-4D double mutant displayed a susceptible phenotype, showing visible powder and no necrosis, which was similar to the wild type. Thirty plants were evaluated for each genotype. B, Trypan blue staining to visualize plant cell death and fungal growth. Leaves were stained with trypan blue at 8 dpi. The edr2 mutant displayed massive cell death and very few conidia, while edr2 sr1-4D supported wild-type-like conidia formation. Bars = 100 μm. C, 3,3′-Diamino benzidine hydrochloride staining for H2O2 at 2 dpi. Note that edr2 accumulated more H2O2 than the wild type. Bars = 100 μm. Col, Ecotype Columbia. D, Accumulation of H2O2 was quantified as described previously (Wang et al., 2011). The bars represent means and sd of intensity per area from at least six leaves of three plants for each genotype. Lowercase letters indicate significant differences (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results. E, Quantification of fungal growth by counting the number of conidiophores per colony at 7 dpi. The bars represent means and sd of samples (n = 25). Lowercase letters indicate statistical significance (P < 0.01, one-way ANOVA). The experiment was repeated three times with similar results. F, Accumulation of PR1 mRNA in edr2 was suppressed by sr1-4D. Four-week-old plants were inoculated with G. cichoracearum. Accumulation of PR1 transcripts was examined by real-time PCR and normalized to ACT8 as an internal control. The bars represent means and sd from three biological replicates. Asterisks indicate significant differences from the wild type (P < 0.01, Student’s t test). G, ET-induced senescence. Four-week-old plants were treated with 100 μL L−1 ET for 3 d. H, Chlorophyll content of the fourth to sixth leaves at day 0 and day 3 after 100 μL L−1 ET treatment. The bars represent means and sd (n = 4). Statistical differences are indicated by lowercase letters (P < 0.01, one-way ANOVA). The experiment was repeated more than three times with similar results.
The disease resistance mediated by edr2 is correlated with activation of the SA signaling pathway. The defense-related gene PR1 is induced more quickly and to higher levels in edr2 than in the wild type (Tang et al., 2005a; Vorwerk et al., 2007). To investigate whether sr1-4D affects PR1 expression in edr2, we used quantitative reverse transcription-PCR to examine PR1 transcript levels in the wild type, edr2, and edr2 sr1-4D at different time points after powdery mildew infection. As shown in Figure 1F, the PR1 transcript level was very low in all plants in the absence of pathogen. However, at 4 dpi, the PR1 transcript level was much higher in edr2 than in the wild type, but it was much lower in edr2 sr1-4D than in edr2 and the wild type, indicating that sr1-4D fully suppressed the accumulation of PR1 transcripts upon powdery mildew infection in edr2.
In addition to powdery mildew resistance, edr2 also shows an enhanced ET-induced senescence phenotype (Tang et al., 2005a). To investigate whether sr1-4D suppressed the edr2 senescence phenotype, 4-week-old wild-type, edr2, and edr2 sr1-4D leaves were treated with 100 μL L−1 ET for 3 d. In the wild type, ET induced senescence in old leaves, but the edr2 mutant displayed more severe senescence phenotypes and the senescence occurred in much younger leaves (Fig. 1G). In contrast, the edr2 sr1-4D mutant displayed delayed senescence compared with edr2 and the wild type, indicating that sr1-4D also suppressed the edr2-mediated ET-induced senescence. To quantify this phenotype, we measured the senescence-associated decline in chlorophyll content and found that edr2 lost more chlorophyll than the wild type, but edr2 sr1-4D had significantly more chlorophyll than the wild type and edr2 after ET treatment (Fig. 1H). Taken together, these data indicated that sr1-4D fully suppressed all edr2-associated phenotypes and conferred enhanced disease susceptibility, further delaying ET-induced leaf senescence even in the edr2 background in comparison with the wild type (Fig. 1).
Identification of the sr1-4D Mutation
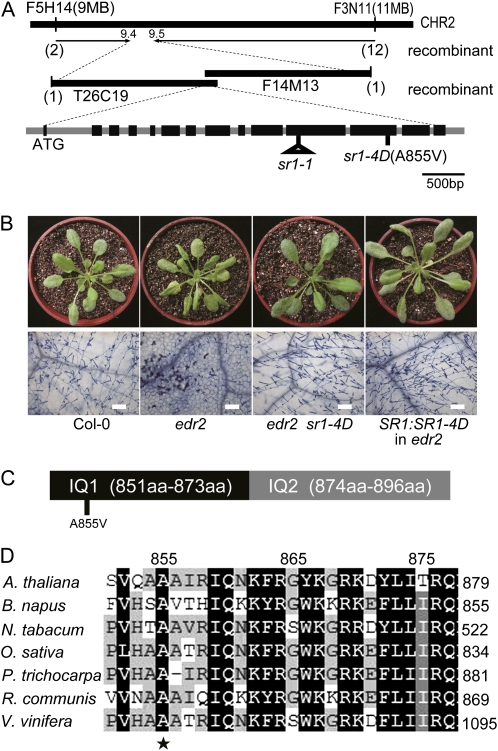
Genetic analysis showed that sr1-4D acts as a dominant mutation, as the original edr2 sr1-4D mutant segregated both edr2 and suppressed plants. Also, edr2/edr2 SR1/sr1-4D plants displayed the same phenotypes as edr2/edr2 sr1-4D/sr1-4D plants. To map the sr1-4D mutation, we crossed a homozygous edr2 sr1-4D plant with Landsberg erecta to generate a mapping population. Initially, we mapped the sr1-4D mutation to a region on chromosome 2 between markers T26C24 and F3N11. Using a large number of F3 plants, we narrowed the sr1-4D mutation to about 100 kb (Fig. 2A). We then sequenced the candidate genes in this region. A single-nucleotide (C-to-T) change was identified in At2g22300 at nucleotide 2,564 in the coding sequence; this change was predicted to produce an amino acid change (A855V; Fig. 2A).
Figure 2.
SR1 encodes a calmodulin-binding transcription factor. A, Positional cloning of SR1. A nucleotide change (C2564T) in the 12th exon in At2g22300 (SR1) was identified, which led to a substitution (A855V) in the SR1 protein. B, A genomic clone of SR1 from edr2 sr1-4D suppressed edr2-mediated powdery mildew resistance. Wild-type, edr2, edr2 sr1-4D, and edr2 plants transformed with the genomic clone of mutated SR1 (derived from the edr2 sr1-4D mutant) were inoculated with powdery mildew. The plants were photographed (top panel) and stained with trypan blue (bottom panel) at 8 dpi. Bars = 100 μm. Forty-nine independent T1 transgenic plants were evaluated, and 45 of them showed an sr1-4D-like susceptible phenotype. Col, Ecotype Columbia. C, The mutation site in SR1-4D is in the first IQ motif of SR1. aa, Amino acids. D, The mutation site of SR1-4D, Ala-855, is conserved in proteins homologous to SR1 in different organisms. The SR1 protein sequence was used to perform BLAST searches against the National Center for Biotechnology Information database. SR1 and its homologs identified in different organisms were aligned using Megalign software (DNASTAR), and the alignment was further edited with Genedoc software. A. thaliana, Arabidopsis thaliana SR1; B. napus, Brassica napus accession number AAM10969.1; N. tabacum, Nicotiana tabacum accession number AAG39222.1; O. sativa, Oryza sativa accession number EEC74662.1; P. trichocarpa, Populus trichocarpa accession number XP_002310562.1; R. communis, Ricinus communis accession number XP_002519355.1; V. vinifera, Vitis vinifera accession number CBI35638.3.
Because sr1-4D is a dominant mutation, it cannot be tested by traditional complementation. Instead, to confirm that At2g22300 is the gene responsible for the sr1-4D mutant phenotype, we tested whether introduction of the sr1-4D mutant genomic sequence could suppress edr2. To that end, we generated a genomic clone of At2g22300 by amplification of the genomic sequence from a homozygous edr2 sr1-4D mutant plant. This genomic clone contained the full-length At2g22300 gene, consisting of the coding sequence flanked by a 1.4-kb upstream promoter region and a 0.8-kb downstream sequence. We then transformed this genomic clone into the edr2 mutant, and the transgenic lines exhibited susceptibility to powdery mildew (Fig. 2B), indicating that this particular mutation in the At2g22300 gene suppressed the edr2 phenotype. Therefore, suppression of the edr2 phenotype in edr2 sr1-4D was caused by a mutation in the At2g22300 gene.
The At2g22300 gene was previously designated SR1 (also known as CALMODULIN BINDING TRANSCRIPTION ACTIVATOR3 [CAMTA3]); therefore, we designated the edr2 suppressor sr1-4D. SR1 is a transcription factor that contains two IQ motifs, which are known to be calmodulin-binding domains (Yang and Poovaiah, 2002). The sr1-4D mutation (A855V) is in the first IQ motif (Fig. 2C) in an amino acid that is highly conserved in the SR1 homologs in multiple plant species (Fig. 2D).
Responses of sr1-4D and sr1-1 to Bacterial and Fungal Pathogens
To investigate whether SR1 expression is induced by pathogens, we examined the SR1 transcript levels in plants inoculated with the bacterial pathogen Pseudomonas syringae pv tomato (Pto) DC3000 or the fungal pathogen G. cichoracearum. The levels of SR1 transcript were higher at 5 d post inoculation by G. cichoracearum (Supplemental Fig. S1A) and at 9 h post inoculation by Pto DC3000 (Supplemental Fig. S1B).
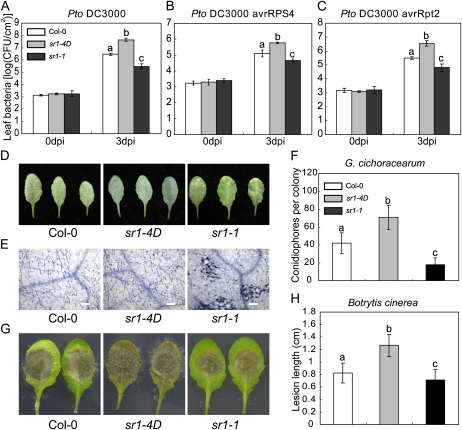
Previously, it was shown that the loss-of-function mutant sr1-1 displayed enhanced resistance to Pto DC3000 and B. cinerea (Galon et al., 2008; Du et al., 2009). To further investigate the role of SR1 in plant innate immunity, we tested the responses of both sr1-1 and sr1-4D mutants to virulent and avirulent strains of Pto DC3000 and to the fungal pathogens G. cichoracearum and B. cinerea. The sr1-1 mutant was more resistant to virulent Pto DC3000 and to the avirulent strains Pto DC3000 (avrRpt2) and Pto DC3000 (avrRPS4), which carry effectors that are recognized by the CC-NBS-LRR protein RPS2 and the TIR-NBS-LRR protein RPS4, respectively. In contrast, the sr1-4D mutant displayed enhanced susceptibility to these bacterial strains (Fig. 3, A–C).
Figure 3.
Response of the wild type, sr1-4D, and sr1-1 to other pathogens. A to C, Four-week-old plants were inoculated with virulent or avirulent strains of Pto DC3000. A, Pto DC3000. B, Pto DC3000 avrRPS4. C, Pto DC3000 avrRpt2. Ten plants were used for each genotype. The bars represent means and sd of three biological samples. Statistical differences are indicated by lowercase letters (P < 0.01, one-way ANOVA). The experiment was repeated more than three times with similar results. CFU, Colony-forming units. D, Four-week-old plants were infected with G. cichoracearum, and representative leaves were removed and photographed at 8 dpi. Thirty plants were evaluated for each genotype. Col, Ecotype Columbia. E, Leaves infected with G. cichoracearum at 8 dpi were stained with trypan blue to visualize fungal growth and plant cell death. Bars = 100 μm. F, The number of conidiophores per colony was counted at 7 dpi. The bars represent means and sd of 25 samples. Statistical differences are indicated by lowercase letters (P < 0.01, one-way ANOVA). The experiment was repeated three times with similar results. G, Leaves from 4-week-old plants were infected with B. cinerea and photographed at 3 dpi. Leaves from at least 30 plants were used for each genotype. H, Leaves were inoculated with B. cinerea. Lesion size was determined by measuring the major axis of the necrotic area. The bars represent means and sd of 30 samples. Statistical differences are indicated with lowercase letters (n = 30, P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
Similarly, for the fungal pathogen G. cichoracearum, sr1-1 displayed edr2-like powdery mildew resistance and mildew-induced necrotic cell death, but sr1-4D was highly susceptible and supported significantly more conidiophore formation than the wild type (Fig. 3, D–F). sr1-1 was also more resistant than the wild type to the necrotrophic pathogen B. cinerea (Galon et al., 2008; Fig. 3, G and H); by contrast, sr1-4D was more susceptible to this pathogen. Taken together, these data indicate that SR1 plays an important role in plant innate immunity by negatively regulating defense responses. Also, the loss-of-function mutant sr1-1 displayed opposite phenotypes to the sr1-4D mutant, suggesting that sr1-4D is a gain-of-function mutation.
Both edr1 and edr2 show enhanced disease resistance to powdery mildew, mildew-induced cell death, and ET-induced senescence. To examine whether the sr1-4D mutation can suppress the edr1 phenotypes, we infected the edr1 sr1-4D double mutant with G. cichoracearum and assessed the disease phenotype by staining the infected leaves at 8 dpi. The edr1 sr1-4D double mutant was susceptible to powdery mildew, supporting extensive fungal growth and showing no necrotic cell death at 8 dpi, indicating that the sr1-4D mutation also fully suppressed the edr1 mutant phenotype (Supplemental Fig. S2, A and B).
Previously, it was shown that the sr1-1 mutant accumulates high levels of SA and has a temperature-dependent growth phenotype (Du et al., 2009). To examine the growth phenotypes of sr1-4D, we grew wild-type, sr1-4D, and sr1-1 plants at lower (19°C–21°C) or higher (25°C–27°C) temperatures. At 25°C to 27°C, the growth of wild-type, sr1-4D, and sr1-1 plants was similar, and no difference between the wild-type and mutant plants was observed (Supplemental Fig. S3A). However, at 19°C to 21°C, the gain-of-function mutant sr1-4D was significantly larger than the wild type (Supplemental Fig. S3, B and C). Also, the relative expression of defense-related genes PR1, PR2, and PR5 was significantly lower in sr1-4D than in the wild type at 19°C to 21°C (Supplemental Fig. S3, D–F). To investigate whether sr1-4D has defects in SA accumulation, we measured the SA levels of 5-week-old wild-type, sr1-4D, and sr1-1 plants grown at 19°C to 21°C. Consistent with a previous finding, the sr1-1 mutant accumulated higher levels of SA (Du et al., 2009) while the sr1-4D mutant accumulated significantly lower levels of SA, compared with the wild type (Supplemental Fig. S4, A and B). Consistent with this observation, the relative expression of SID2, PAD4, EDS1, and EDS5 was significantly lower in sr1-4D than in the wild type (Supplemental Fig. S4, C–F).
SR1 Directly Binds to the NDR1 and EIN3 Promoters
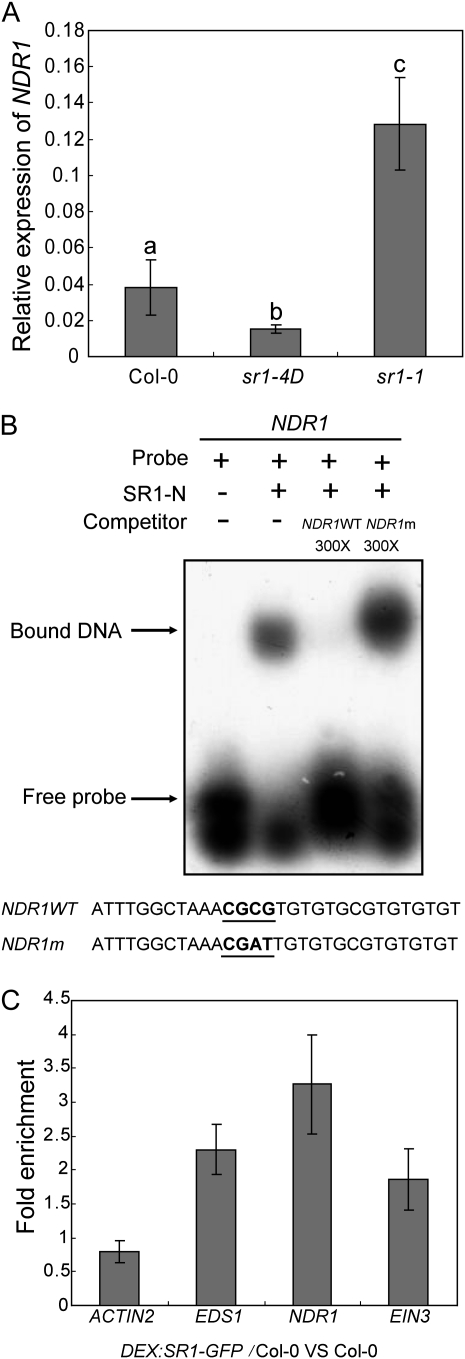
SR1 is a transcription factor that binds to promoters that contain a CGCG box (Yang and Poovaiah, 2002). Previously, it was shown that SR1 binds to the promoter of EDS1, a key regulator of plant defense responses, and represses EDS1 expression (Du et al., 2009). EDS1 is required by TIR-NBS-LRR-type R proteins, such as RPS4, which recognizes the bacterial effector avrRPS4. In contrast, NDR1, a membrane-associated protein, is required for several CC-NBS-LRR-type R proteins, including RPS2 (Century et al., 1995), which is responsible for resistance to Pto DC3000 carrying avrRpt2 (Aarts et al., 1998). Since the loss-of-function sr1-1 mutant displayed enhanced disease resistance and the gain-of-function sr1-4D mutant displayed enhanced disease susceptibility to Pto DC3000 (avrRpt2), we hypothesized that NDR1 may be another direct target of SR1. Consistent with this hypothesis, NDR1 is up-regulated in sr1-1 according to microarray data (Galon et al., 2008). In addition, analysis of the NDR1 promoter sequence revealed a CGCG box (Supplemental Fig. S5), which could be a potential SR1-binding site.
To investigate whether SR1 regulates NDR1, we first examined NDR1 expression levels in sr1-1 and sr1-4D. Interestingly, the level of NDR1 transcript was higher in sr1-1 but lower in sr1-4D compared with the wild type (Fig. 4A), indicating that mutations in SR1 do affect NDR1 expression. To examine whether SR1 directly binds to the NDR1 promoter, we expressed and purified recombinant SR1-N-terminal truncated protein (SR1-N; 1–146 amino acids), which contained the DNA-binding domain fused with a glutathione S-transferase (GST) tag, and performed DNA electrophoretic mobility-shift assays (EMSA). SR1-N was able to bind to the radiolabeled NDR1 promoter fragment in vitro, and the binding was blocked by the addition of an unlabeled NDR1 promoter fragment but not by an NDR1 promoter fragment with a mutation in the core binding sequence (CGCG box; Fig. 4B). To further confirm that SR1 binds to the NDR1 promoter, we performed chromatin immunoprecipitation (ChIP) assays. We first constructed transgenic plants that contained SR1-GFP with the dexamethasone (DEX)-inducible promoter. We then conducted ChIP assays with this transgenic line to examine whether SR1-GFP binds to the NDR1 promoter. The promoter of NDR1 was enriched in the chromatin-immunoprecipitated DNA with the anti-GFP antibody; as a control, an ACTIN2 promoter sequence was not enriched in the same assay (Fig. 4C), indicating that SR1-GFP binds to the promoter of NDR1 in vivo and, thus, that NDR1 is a direct target of SR1.
Figure 4.
SR1 directly binds to the promoter of NDR1 and EIN. A, Levels of NDR1 transcripts in 4-week-old wild-type, sr1-4D, and sr1-1 plants were examined by quantitative real-time PCR and normalized to ACT8 as an internal control. The bars represent means and sd from three independent biological replicates. The lowercase letters indicate significant differences (P < 0.01, one-way ANOVA). Col, Ecotype Columbia. B, EMSA for SR1 binding to the promoter fragment of NDR1 in vitro. GST-SR1-N (amino acids 1–146) was incubated with the radiolabeled NDR1 promoter fragment. The samples were loaded and separated on a polyacrylamide gel. The NDR1m sequence contained a mutated CGCG box (CGCG to CGAT). C, The promoter fragments of NDR1 and EIN3 were enriched in a ChIP assay. Chromatin from wild-type and DEX:SR1-GFP transgenic plants was immunoprecipitated by anti-GFP, and the enrichment of the fragments was determined by quantitative real-time PCR. The ACTIN2 promoter was used as a negative control, and the EDS1 promoter was used as a positive control. The bars represent means and sd of three samples. The experiment was repeated four times with similar results.
SR1 was first reported as an ET-induced gene (ETHYLENE INDUCED CALMODULIN BINDING PROTEIN1); also, SR1 was reported to bind to the promoter of EIN3, a key component of ET signaling, in vitro (Reddy et al., 2000; Yang and Poovaiah, 2002). However, to date, whether SR1 is involved in ET signaling has not been determined. To gain insight into the role of SR1 in ET signaling, we treated 4-week-old sr1-1 and sr1-4D plants with ET for 3 d and evaluated their leaf senescence phenotypes. We found that sr1-1 showed enhanced ET-induced senescence, but sr1-4D was insensitive to ET (Supplemental Fig. S6), indicating that SR1 may indeed regulate ET-induced senescence. To test whether SR1 binds to the EIN3 promoter, we performed ChIP assays, as described above. The EIN3 promoter was also enriched in the pool of sequences immunoprecipitated with the anti-GFP antibody (Fig. 4C), indicating that SR1 binds to the EIN3 promoter in vivo; thus, EIN3 is also a direct target of SR1.
To further investigate the regulation of EIN3 by SR1, We examined the relative expression of EIN3 in ET-treated or untreated wild-type, sr1-4D, and sr1-1 plants. As shown in Supplemental Figure S7, A and B, the relative expression of EIN3 is higher in sr1-1 but lower in sr1-4D than in the wild type, which is consistent with the negative role of SR1 in EIN3 expression.
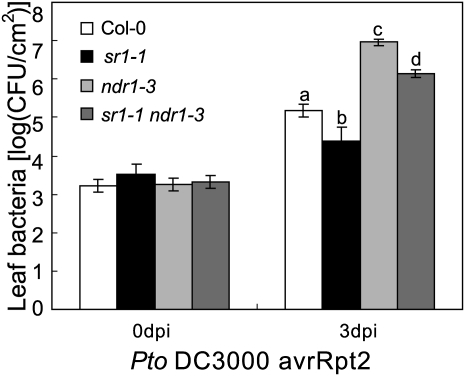
The ndr1 Mutation Suppresses sr1-1-Mediated Resistance to Pto DC3000 (avrRpt2) and G. cichoracearum
Since NDR1 is a direct target of SR1 and NDR1 expression increased in the loss-of-function sr1-1 mutant, SR1 likely regulates plant defense by repressing NDR1 expression. Therefore, the enhanced disease resistance phenotype of the sr1-1 mutant is at least partially due to the high expression of NDR1. To test this hypothesis, we examined whether the ndr1 mutation can suppress the sr1-1 phenotype of enhanced resistance to Pto DC3000 (avrRpt2). As shown in Figure 5, the ndr1-3 mutation suppressed the resistance phenotype of sr1-1 to Pto DC3000 (avrRpt2), indicating that NDR1 was required for sr1-1 resistance to Pto DC3000 (avrRpt2). This is consistent with our hypothesis that the responses of sr1-1 and sr1-4D to Pto DC3000 (avrRpt2) are due to higher and lower expression of NDR1, respectively. These observations are consistent with previous findings that overexpression of NDR1 enhances resistance to Pto DC3000 (avrRpt2) (Coppinger et al., 2004). However, the ndr1 sr1-1 double mutant was less susceptible than the ndr1-3 single mutant, suggesting that the modulation of Pto DC3000 (avrRpt2) resistance by SR1 is only partially dependent on NDR1 function.
Figure 5.
ndr1 suppresses the resistant phenotype of sr1-1 to Pto DC3000 carrying avrRpt2. Four-week-old plants were inoculated with Pto DC3000 avrRpt2. Ten plants were used for each genotype. The bars represent means and sd. Statistical differences are indicated with lowercase letters (n = 3, P < 0.01, one-way ANOVA). The experiment was repeated more than three times with similar results. CFU, Colony-forming units; Col, ecotype Columbia.
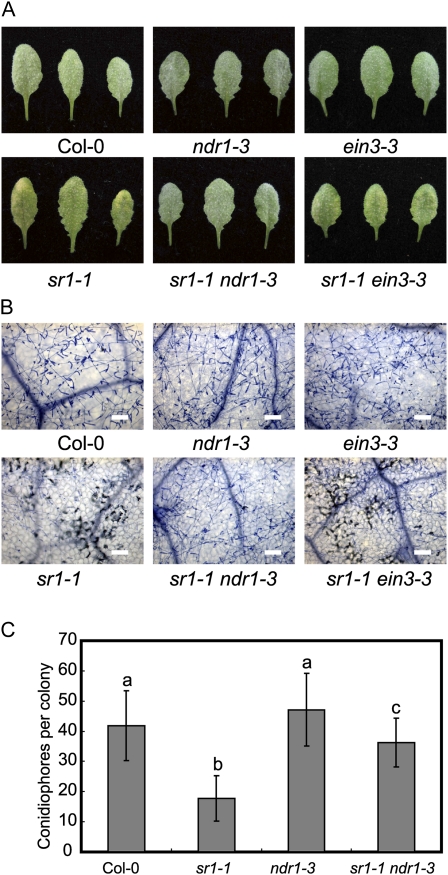
To further study the role of NDR1 in sr1-1-mediated powdery mildew resistance, we infected the wild type, sr1-1, ndr1-3, and the ndr1-3 sr1-1 double mutant with G. cichoracearum. The ndr1 mutant displayed a wild-type-like susceptible phenotype to powdery mildew and did not show enhanced susceptibility; however, ndr1 fully suppressed sr1-1-mediated mildew-induced cell death and partially suppressed powdery mildew resistance in sr1-1 (Fig. 6), indicating that NDR1 participated in sr1-1-mediated resistance to powdery mildew.
Figure 6.
ndr1, not ein3, suppresses the resistance of sr1-1 to powdery mildew. A, Four-week-old plants were inoculated with G. cichoracearum, and representative leaves were removed and photographed at 8 dpi. Thirty plants were evaluated for each genotype. B, Trypan blue staining of leaves inoculated with G. cichoracearum at 8 dpi. Bars = 100 μm. The fungal structures and dead plant cells were stained. C, The number of conidiophores per colony was counted at 7 dpi. The bars represent means and sd (n = 25, P < 0.01, one-way ANOVA). Different letters indicate significant differences between genotypes. The experiment was repeated three times with similar results. Col, Ecotype Columbia.
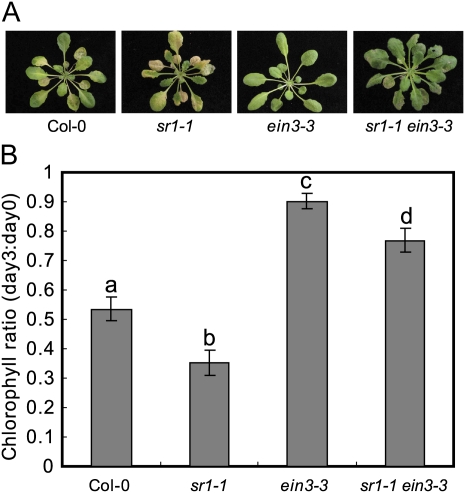
The ein3 Mutation Suppressed sr1-1-Mediated ET-Induced Senescence
Previously, it has been shown by EMSA that SR1 binds to the EIN3 promoter in vitro. Here, we show by ChIP that SR1 binds to the EIN3 promoter in vivo. However, the biological significance of SR1 binding to EIN3 has not yet been defined. Our observations that sr1-1 displayed enhanced ET-induced senescence, and that sr1-4D displayed delayed ET-induced senescence, may provide genetic evidence for the role of SR1 in ET signaling and in the regulation of EIN3 expression. In this scenario, the ET phenotypes of sr1-1 and sr1-4D might be due to the misregulation of EIN3 in these mutants. To test this hypothesis, we examined whether ein3 suppresses the enhanced ET-induced senescence in sr1-1. We treated the wild type, sr1-1, ein3-3, and ein3-3 sr1-1 with 100 μL L−1 ET for 3 d and found that the ein3-3 sr1-1 double mutant displayed insensitivity to ET, showing delayed senescence (Fig. 7), indicating that EIN3 is required for ET-induced senescence in sr1-1. However, ein3-3 had no effects on the sr1-1 resistance to powdery mildew (Fig. 6, A and B), indicating that defense responses and ET senescence are regulated by two distinct pathways.
Figure 7.
ein3 suppresses ET-induced senescence of sr1-1. A, Four-week-old plants were treated with 100 μL L−1 ET for 3 d. B, Decrease in chlorophyll content induced by ET treatment, measured by the ratio of chlorophyll content at day 3 to content at day 0, of the fourth to sixth leaves treated with 100 μL L−1 ET for 0 and 3 d. The bars represent means and sd (n = 4). Statistical differences are indicated by different lowercase letters (P < 0.01, one-way ANOVA). The experiment was repeated three times with similar results. Col, Ecotype Columbia.
To further investigate the role of SR1 in ET signaling, we tested the responses of sr1-1 and sr1-4D seedlings to 1-aminocyclopropane-1-carboxylic acid. Both sr1-1 and sr1-4D displayed the typical triple response, which was indistinguishable from the wild-type seedlings (Supplemental Fig. S6C), suggesting that ET-induced senescence is different from the classic ET signaling pathway.
To gain more insight into the function of SR1 in ET-induced senescence, we examined the relative expression of SR1 in response to ET treatment. As shown in Supplemental Figure S7C, the transcript accumulation of SR1 was increased after ET treatment. We then examined the relative expression of two senescence-associated genes, SENESCENCE-ASSOCIATED GENE12 (SAG12) and SAG24, in ET-treated wild-type, sr1-1, and sr1-4D plants. As shown in Supplemental Figure S7, D and E, the transcript accumulation of SAG12 and SAG24 was significantly higher in sr1-1 but much lower in sr1-4D than in the wild type. These data indicate that SR1 negatively regulates the expression of senescence-associated genes SAG12 and SAG24.
The Binding of SR1 and SR1-4D to Calmodulin Requires Calcium
SR1 is a calmodulin-binding transcription activator that contains a DNA-binding domain at the N terminus and two calmodulin-binding IQ motifs in the C-terminal 850 to 896 amino acids (Yang and Poovaiah, 2002). The sr1-4D mutation is located in the first IQ motif, which is the calmodulin-binding domain. To examine whether the SR1-4D mutation affected its binding to calmodulin, we expressed the SR1 calmodulin-binding domain with a GST tag in Escherichia coli and tested the calmodulin-binding activity of the wild-type and mutant versions of the SR1 calmodulin-binding domains. Both wild-type and mutated versions of SR1 proteins were able to bind to calmodulin in vitro. We then examined whether the binding between the SR1-4D protein and calmodulin requires calcium and found that both SR1 and SR1-4D bound to calmodulin in a calcium-dependent manner, as neither protein could bind to calmodulin in the absence of CaCl2 in vitro (Supplemental Fig. S8).
DISCUSSION
To search for components in the EDR2 signaling pathway, we performed a mutant screen and identified an edr2 suppressor mutation, sr1-4D, which affects a calmodulin-binding transcription factor. sr1-4D is a gain-of-function mutation that suppressed all edr2 phenotypes, including powdery mildew resistance and enhanced ET-induced senescence. In contrast, the loss-of-function sr1-1 mutant displayed increased disease resistance and enhanced ET-induced senescence. We show that SR1 negatively regulates plant immunity and leaf senescence by directly binding to the NDR1 and EIN3 promoters.
Although sr1-4D was identified in the edr2 suppressor screen, SR1 may not directly regulate the EDR2 signaling pathway, as the sr1-4D mutant showed enhanced susceptibility to multiple pathogens, including virulent and avirulent strains of the bacterial pathogen Pto DC3000, while edr2 did not show an altered response to these pathogens. Recently, Jing et al. (2011) identified an identical mutation (camta3-3D) in a screen for mutants that exhibit compromised systemic acquired resistance (SAR). In addition to defects in SAR, the camta3-3D mutant displays enhanced susceptibility to the virulent bacterium Pseudomonas syringae pv maculicola ES4326 and the oomycete pathogen Hyaloperonospora arabidopsidis Noco2 (Jing et al., 2011), which is consistent with our findings. Jing et al. (2011) also showed that the transgenic lines that express higher levels of SR1 have defects in basal defense and SAR. These data indicate that SR1 plays an important role in both SAR and basal defense; however, how SR1 regulates SAR is not clear.
Previously, it was shown that SR1 regulates EDS1 expression through binding to the EDS1 promoter (Du et al., 2009). Also, EDS1 is a positive regulator in basal defense and R gene-mediated responses. The eds1 mutation suppressed powdery mildew resistance mediated by edr1, atg2, and RPW8 (Frye et al., 2001; Xiao et al., 2005). As edr2-mediated resistance is dependent on SA signaling, one possibility is that sr1-4D suppressed the edr2-resistant phenotype to powdery mildew mainly through the repression of EDS1 expression by SR1-4D, which in turn leads to the inactivation of SA signaling.
sr1-1 displays resistance to Pto DC3000 and B. cinerea (Galon et al., 2008; Du et al., 2009). Also, microarray data showed that many disease resistance-related genes were up-regulated in sr1-1 (Galon et al., 2008). In this work, we show that sr1-1 is resistant to a virulent powdery mildew isolate and has further tightened resistance to avirulent strains of Pto DC3000 carrying avrRpt2 or avrRPS4. In contrast to sr1-1, the gain-of-function mutant sr1-4D displays susceptibility to each of these pathogens. Consistent with our finding, the gain-of-function mutant of SR1 shows enhanced disease susceptibility phenotypes to P. syringae pv maculicola ES4326 and H. arabidopsidis Noco2 (Jing et al., 2011). SR1 is a calcium-dependent calmodulin-binding transcription factor and binds to a CGCG box in the promoter of target genes to repress their expression (Yang and Poovaiah, 2002; Du et al., 2009). The mutation in sr1-4D likely leads to enhanced repression of the target genes. One possibility is that SR1-4D binds more tightly to calmodulin and thus constitutively binds to the target promoters, leading to reduced expression of the target genes. Alternatively, the threshold of calcium concentration required for the binding between calmodulin and SR1-4D may be lower than in the wild-type protein. Another possibility is that the SR1-4D protein accumulates to higher levels than wild-type SR1. Intriguingly, sr1-4D carries a C-to-T point mutation (A855V) that was exactly the same as that recently described for camta3-3D (Jing et al., 2011). These two mutants are identified from independent sources, suggesting that Ala-855 is the only or one of the few residues that play a critical role in the modulation of SR1 activity. The interactions between calcium signaling and plant defense responses are complicated, and further analysis is needed to determine why this particular mutation causes a gain-of-function phenotype.
Plants recognize pathogen effectors, directly or indirectly, by R proteins (Dangl and Jones, 2001). Many Arabidopsis R proteins contain an NB-LRR domain. According to the N-terminal structure, these R proteins can be divided into two classes, CC-NB-LRR and TIR-NB-LRR. In general, the resistance mediated by CC-NB-LRR proteins requires NDR1 function, but the resistance mediated by TIR-NB-LRR proteins requires EDS1 (Aarts et al., 1998; Feys and Parker, 2000). Although it has been well documented how NDR1 and EDS1 are involved in the defense response (Feys et al., 2001, 2005; Axtell and Staskawicz, 2003; Belkhadir et al., 2004; Day et al., 2006), it is not clear how NDR1 and EDS1 are regulated. Du et al. (2009) reported that SR1 directly binds to the EDS1 promoter and represses its expression, which revealed a mechanistic link between calcium signaling and SA-mediated disease resistance. Here, we report that NDR1 is also directly regulated by SR1. This finding provides new insights into the role of SR1 in plant immunity, providing a link between NDR1- and EDS1-mediated resistance pathways through the coregulator SR1.
The plant hormones SA and ET play important roles in plant defense responses. The cross talk between SA signaling and ET signaling in the defense response is complicated. In general, it is believed that SA signaling plays an important role in resistance to biotrophic pathogens and ET signaling plays a crucial role in resistance to necrotrophic pathogens (Glazebrook, 2005). However, there is evidence that these two pathways may be antagonistic or agonistic to each other. For instance, Chen et al. (2009) reported that EIN3 and EIN3 LIKE 1 (EIL1) bind to the SID2 promoter and repress SID2 expression. This is direct evidence of cross talk between SA and ET signaling, as EIN3 is one of the central components that positively regulates the ET signal transduction pathway (Chao et al., 1997). Also, SID2 is a key enzyme that is involved in SA synthesis, and mutations in SID2 compromise pathogen-induced SA accumulation. Consequently, loss-of-function mutants of ein2 and ein3 display enhanced disease resistance to bacterial Pto DC3000 (Bent et al., 1992; Chen et al., 2009), and overaccumulation of EIN3 protein leads to enhanced susceptibility to Pto DC3000 (Chen et al., 2009). Recently, it was reported that ein2 mutants are defective in all FLAGELLIN-SENSITIVE2 (FLS2)-mediated responses and that EIN3 and EIL1 directly bind to the receptor kinase FLS2 to mediate pathogen-associated molecular pattern signaling (Boutrot et al., 2010), which indicate a direct role of the ET pathway in plant immunity. SR1 may provide another link between SA and ET, as SR1 binds to the promoters of EDS1, a positive regulator of SA signaling, and EIN3, a positive regulator of ET signaling; SR1 also negatively regulates the expression of EDS1, NDR1, and EIN3. This indicates that plants can up-regulate or down-regulate both SA and ET signaling pathways by modulating SR1 function. These findings indicate that the relationship among SA signaling, ET signaling, and the immunity system is complicated. Negative regulation of both SA signaling and ET signaling by direct binding of SR1 to the promoter of EDS1, NDR1, and EIN3 may explain why sr1-1 is more resistant to both biotrophic and necrotrophic pathogens and why sr1-4D suppressed edr2-mediated resistance and ET-induced senescence.
The cross talk between defense responses and senescence has been discussed previously (Tang et al., 2005b; Consonni et al., 2006; Wang et al., 2011). Some mutants that display enhanced disease resistance show early senescence, such as edr1, atg2, and mlo2. However, the cross talk between defense responses and senescence appears to be complicated. For instance, edr1-mediated resistance is SA dependent, but senescence in edr1 is dependent on ET signaling; thus, resistance and senescence in edr1 are regulated by separate pathways (Tang et al., 2005b). However, the early senescence-like phenotype in mlo2 is suppressed by mutations in EDS5, NPR1, PAD4, and SID2 as well as by the NahG transgene, indicating that SA plays an important role in mlo2-associated senescence (Consonni et al., 2006). In addition, it was shown that SA levels are higher in senescent leaves in Arabidopsis (Morris et al., 2000). Because SR1 binds to the promoter of EDS1 and EIN3, plants may be able to control disease resistance and senescence by modulating SA signaling and ET signaling through their coregulation by SR1. Further analysis of global gene expression (e.g. RNA-seq) in the wild type, sr1-1, and sr1-4D may provide useful leads to identify connections between senescence and defense mediated by SR1.
In conclusion, Arabidopsis SR1 plays a critical role in plant immunity and ET-induced senescence. Our data support a model that SR1 fine-tunes plant immunity and senescence signaling by directly regulating the expression of NDR1, EDS1, and EIN3 (Supplemental Fig. S9). SR1 may represent another example of the complicated interactions between SA pathways, ET signaling, and plant immunity.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were sterilized in 10% bleach and sown on half-strength Murashige and Skoog (1/2 MS) medium containing 1% Suc. Plates were kept in 4°C for 3 d and then moved to the greenhouse (22°C–24°C, 9-h-light/15-h-dark photoperiod). Seedlings were transferred into soil after 7 d. Plants were grown in short-day conditions (9 h of light/15 h of dark) for phenotyping or in long-day conditions (16 h of light/8 h of dark) to set seeds as described previously (Nie et al., 2011), unless indicated otherwise. The sr1-1 mutant was from the Arabidopsis Biological Resource Center (SALK_001152). The ndr1-3 sr1-1 and ein3-3 sr1-1 double mutants were generated by standard crosses.
Pathogen Inoculation
Powdery mildew (Golovinomyces cichoracearum UCSC1) was kept on highly susceptible pad4-1 plants. Powdery mildew infection was performed with either high-density or low-density inoculation. High-density inoculation was used for mutant screening and mapping and was achieved by gently brushing the target leaves with infected leaves to pass the fungal spores (Adam and Somerville, 1996). To quantify the number of conidiophores per colony, low-density inoculation was used to achieve an even inoculation density as described previously (Wang et al., 2011). The number of conidiophores per colony was counted at 7 dpi (Consonni et al., 2006). Infections with Pseudomonas syringae pv tomato DC3000 virulent and avirulent strains were performed as described previously (Nie et al., 2011). Botrytis cinerea was grown on potato dextrose agar plates (Difco), and the leaves of 4-week-old plants were inoculated as described previously (Ferrari et al., 2003).
Staining and Microscopy
Fungal growth and host cell death were examined by staining infected leaves with trypan blue at 8 dpi for plants infected with powdery mildew (Frye and Innes, 1998). H2O2 was examined by staining infected leaves with 3,3′-diamino benzidine hydrochloride at 2 dpi (Xiao et al., 2003). Samples were observed and photographed using an Olympus BX60 microscope.
ET-Induced Senescence Assay
Four-week-old plants were kept in a sealed box with 100 μL L−1 ET for 3 d. Then, plants were photographed and chlorophyll was extracted using 100% ethanol. Chlorophyll content was measured with a Multiskan Spectrum spectrophotometer (Thermo Scientific) at 665- and 649-nm wavelengths (Tang et al., 2005a).
SA Measurement
SA extraction and measurement were performed as described previously (Gou et al., 2009).
Statistical Analyses
Statistical analyses were performed by Student’s t test for samples from two genotypes or one-way ANOVA for samples from multiple genotypes (Wang et al., 2011).
Mutant Screen and Mapping
The edr2 sr1-4D mutant was identified from an ethyl methanesulfonate-mutagenized population (Nie et al., 2011). To map the sr1-4D mutation, an edr2 sr1-4D plant was crossed with Landsberg erecta, and F2 homozygous edr2 plants were identified and used for rough mapping. For fine-mapping, a large number of F3 plants (derived from F2 plants that displayed the edr2 phenotype) were used; ultimately, the mutation was mapped to the region between markers T26C19 and F14M13. We then sequenced the candidate genes in this region. A nucleotide change (C2564T) in the 12th exon was found in At2g22300 (SR1); this mutation also leads to an amino acid change (A855V). Then, we amplified a 7-kb genomic DNA fragment from the edr2 sr1-4D mutant and cloned it into pEASY-blunting (TransGen Biotech). The genomic clone included 1.4 kb upstream of the ATG and 0.8 kb downstream of At2g22300. This genomic DNA was digested and inserted into binary vector pBINPLUS. The construct was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed to the edr2 plants by the floral dip method. The transformants were screened on 1/2 MS medium with 50 μg mL−1 kanamycin.
EMSA
The SR1 sequence encoding a truncated protein (amino acids 1–146) was constructed in pGEX4t, expressed in Escherichia coli BL21(DE3)pLysS (TransGen Biotech), and purified by GST beads (GE Healthcare). The probe was synthesized as forward and reverse strands and renatured to a double-stranded probe in 0.15 m NaCl under 70°C for 5 min. Then, the probe was labeled by [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) and purified by G-25 spin columns (GE Healthcare). The gel-shift assay was performed according to the Promega gel-shift assay system manual.
Calmodulin Binding
The SR1 calmodulin-binding domain (800–900 and 800–930 amino acids) was cloned into pGEX4t vector and expressed in E. coli BL21(DE3)pLysS (TransGen Biotech). An animal version of calmodulin was used in the experiments. The calmodulin-binding assay was performed using the AffinityH CBP Fusion Protein Detection Kit (Stratagene) according to the manufacturer’s instructions. For testing whether calmodulin binding is Ca2+ dependent, 1 mm CaCl2 or 5 mm EGTA was added to the reaction.
ChIP Assay
To produce an inducibly expressed, GFP-tagged SR1 (DEX:SR1-GFP), we cloned the full-length coding sequence of SR1 into pBAV150 and transformed this construct into wild-type ecotype Columbia. ChIP was performed as described previously with minor modifications (Bowler et al., 2004; Saleh et al., 2008). Briefly, wild-type and DEX:SR1-GFP transgenic seeds were grown on 1/2 MS plates for 8 to 10 d and then transferred to 20 μm DEX plates for 2 d. Roots were harvested and cross-linked by 1% formaldehyde for 15 min in vacuum and stopped by 0.125 m Gly. Roots were ground in liquid nitrogen, and nuclei were isolated. Chromatin was immunoprecipitated by anti-GFP (Roche) and protein G beads (Millipore). DNA was precipitated by isopropanol, washed by 70% ethanol, and dissolved in 30 μL of water with 20 μg mL−1 RNase. Gene-specific primers (NDR1-ChIP-F, NDR1-ChIP-R; EIN3-ChIP-F, EIN3-ChIP-R, EDS1-ChIP-F, EDS1-ChIP-R, ACTIN2-ChIP-F, ACTIN2-ChIP-R, SAG12-F, SAG12R; SAG24F, SAG24R) were used (Takara; sybgreen kit) to quantify the enrichment of each fragment. Primers used in this study are listed in Supplemental Table S1.
Gene Expression Analysis
RNA was extracted by TRIzol reagent (Invitrogen), and the first strand was synthesized using murine leukemia virus reverse transcriptase (Promega). Accumulation of transcripts was examined by real-time PCR using the sybgreen kit (Takara).
Primers used in this study are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SR1 was induced by powdery mildew and Pto DC3000.
Supplemental Figure S2. sr1-4D suppressed edr1-mediated powdery mildew resistance.
Supplemental Figure S3. Temperature-dependent growth phenotype of sr1-4D.
Supplemental Figure S4. SA accumulation of sr1-4D.
Supplemental Figure S5. The NDR1 promoter sequence contains a CGCG box.
Supplemental Figure S6. SR1 is involved in ET-induced senescence but is not involved in the 1-aminocyclopropane-1-carboxylic acid-induced triple response.
Supplemental Figure S7. Relative expression of several defense- and senescence-related genes.
Supplemental Figure S8. Calcium is needed for SR1-4D binding to the calmodulin in vitro.
Supplemental Figure S9. Model illustrating the role of SR1 in defense responses and senescence.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Joe Ecker for providing the ein3-3 seeds, Dr. Jane Parker for pad4-1 seeds, Dr. Roger Innes for ndr1-3 seeds, and the Arabidopsis Biological Resource Center for sr1-1 seeds. We thank Mr. Lu Gan for assistance with SA measurements.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam L, Somerville SC. (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J 9: 341–356 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. (2004) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ. (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, et al. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ. (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18: 2782–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16: 449–455 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Innes RW. (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW. (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G. (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Hepler PK. (2005) Calcium: a central regulator of plant growth and development. Plant Cell 17: 2142–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing B, Xu S, Xu M, Li Y, Li S, Ding J, Zhang Y. (2011) Brush and spray: a high throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol 157: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Kim MC, Chung WS, Yun DJ, Cho MJ. (2009) Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant 2: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Micali C, Göllner K, Humphry M, Consonni C, Panstruga R. (2008) The powdery mildew disease of Arabidopsis: a paradigm for the interaction between plants and biotrophic fungi. The Arabidopsis Book 6: e0115, doi/10.1199/tab.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Nie H, Wu Y, Yao C, Tang D. (2011) Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J Genet Genomics 38: 137–148 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Reddy AS. (2001) Calcium: silver bullet in signaling. Plant Sci 160: 381–404 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Reddy VS, Golovkin M. (2000) A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem Biophys Res Commun 279: 762–769 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW. (2005a) Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J 44: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW. (2006) A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J 47: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Christiansen KM, Innes RW. (2005b) Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol 138: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Somerville S. (2000) Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA 97: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC. (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, Vogel J, Somerville C, Somerville S. (2007) EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nishimura MT, Zhao T, Tang D. (2011) ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG. (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, Parker JE, Turner JG. (2005) The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J 42: 95–110 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.