Abstract
Amygdalin is a cyanogenic diglucoside and constitutes the bitter component in bitter almond (Prunus dulcis). Amygdalin concentration increases in the course of fruit formation. The monoglucoside prunasin is the precursor of amygdalin. Prunasin may be degraded to hydrogen cyanide, glucose, and benzaldehyde by the action of the β-glucosidase prunasin hydrolase (PH) and mandelonitirile lyase or be glucosylated to form amygdalin. The tissue and cellular localization of PHs was determined during fruit development in two sweet and two bitter almond cultivars using a specific antibody toward PHs. Confocal studies on sections of tegument, nucellus, endosperm, and embryo showed that the localization of the PH proteins is dependent on the stage of fruit development, shifting between apoplast and symplast in opposite patterns in sweet and bitter cultivars. Two different PH genes, Ph691 and Ph692, have been identified in a sweet and a bitter almond cultivar. Both cDNAs are 86% identical on the nucleotide level, and their encoded proteins are 79% identical to each other. In addition, Ph691 and Ph692 display 92% and 86% nucleotide identity to Ph1 from black cherry (Prunus serotina). Both proteins were predicted to contain an amino-terminal signal peptide, with the size of 26 amino acid residues for PH691 and 22 residues for PH692. The PH activity and the localization of the respective proteins in vivo differ between cultivars. This implies that there might be different concentrations of prunasin available in the seed for amygdalin synthesis and that these differences may determine whether the mature almond develops into bitter or sweet.
Prunasin is a cyanogenic monoglucoside present in almond (Prunus dulcis; syn. Prunus amygdalus) and is found in most tissue parts, including roots, shoots, and unripe fruits (Frehner et al., 1990; Arrázola, 2002; Dicenta et al., 2002; Wirthensohn et al., 2008). Prunasin is derived from the amino acid Phe (Mentzer and Favre-Bonvin, 1961; Ben-Yehoshua and Conn, 1964). The pathway for prunasin biosynthesis includes phenylacetaldoxime, phenylacetonitrile, and mandelonitrile as intermediates. In analogy with studies in other cyanogenic plant species, this part of the pathway is thought to be catalyzed by two multifunctional membrane-bound cytochrome P450 enzymes (Sánchez-Pérez et al., 2008), although these genes have not yet been characterized in almond. The last step in the pathway involves the conversion of mandelonitrile into prunasin and is catalyzed by a soluble UDP-glucosyltransferase UGT85A19 (Franks et al., 2008).
Biosynthetic studies in developing fruits based on the administration of radiolabeled precursors to excised tissues demonstrated prunasin synthesis to proceed in the tegument cells, with similar rates in sweet and bitter cultivars. In neither sweet nor bitter cultivars did the tegument tissue, also called integuments, testa, or seed coat (Hawker and Buttrose, 1980; Dourado et al., 2004), support amygdalin synthesis (Sánchez-Pérez et al., 2008). A transient accumulation of prunasin in the tegument of bitter almond cultivars was observed, whereas no such accumulation was seen in sweet cultivars (Sánchez-Pérez et al., 2008). Following the transient accumulation of prunasin in the tegument of the bitter cultivars, amygdalin was observed to gradually accumulate in the nucellus and endosperm in the mid developing stages. Once almonds are fully ripened, the only quantitative important site of accumulation is the embryo of the bitter cultivars. In comparison, the amounts of amygdalin detectable in sweet cultivars are minute (Sánchez-Pérez et al., 2008, Fig. 3).
Figure 3.
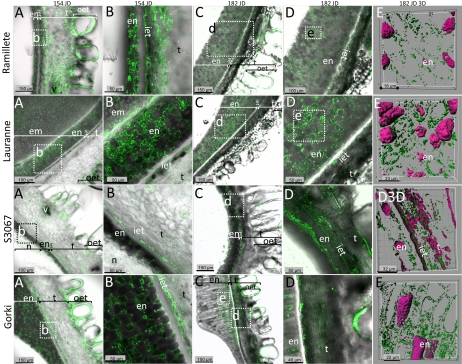
Immunolocalization studies of prunasin localization in four different almond cultivars in the course of fruit development. The cultivars Ramillete (sweet), Lauranne (sweet), S3067 (bitter), and Gorki (bitter) were analyzed for PH content at two different time points during mid season (154 and 182 JD) of fruit development using a polyclonal antibody against the sequence INKKGIEYY present in the PHs (green color) from almond. The first four columns represent overlay images of PH detected by the specific antibody (green) and translucent light micrographs to visualize cell walls (gray). The marked area in a panel represents the area shown at higher magnification in the panel to the right in the same row, except for Gorki, where the source of the images in columns d and e are derived from the marked segments in column c. The last column represents three-dimensional images obtained using sections containing mainly endosperm cells stained with propidium iodide in magenta color and green vesicles that represent the PHs. Abbreviations are as follows: endosperm (en), inner epidermis of the tegument (iet), tegument (t), sometimes with vascular bundles (v), and outer epidermis of the tegument (out). In a small number of the sections, the embryo (em) or the nucellus (n) remained attached to the rest of the tissue. In the later stage shown (182 JD), the tegument cells have collapsed. [See online article for color version of this figure.]
The conversion of prunasin to amygdalin requires the action of a yet unidentified UDP-glucosyltransferase (Frehner et al., 1990; Sánchez-Pérez et al., 2008; Wirthensohn et al., 2008). Biosynthetic studies based on the administration of radiolabeled UDPG in the presence of prunasin showed that the amygdalin-forming glucosyltransferase activity was essentially limited to the cotyledons, with residual activities detected in the nucellus and endosperm. Notably, the amygdalin-forming glucosyltransferase activity was at the same level in the sweet and bitter cultivars studied. Amygdalin accumulation is also observed in other Rosaceae seeds, such as in apples (Malus domestica), apricots (Prunus armenaica), black cherries (Prunus serotina), peaches (Prunus persica), and plums (Prunus domestica; McCarty et al., 1952; Conn, 1980; Frehner et al., 1990; Møller and Seigler, 1991; Swain et al., 1992; Poulton and Li, 1994).
Amygdalin constitutes the bitter principle in bitter almonds. The avoidance of bitterness in almond kernels is the main trait considered by almond breeders. Its single inheritance was discovered by Heppner (1923), who stated that the bitter phenotype is recessive. This single gene, called Sweet kernel (Sk), has been localized in linkage group 5 (out of eight) of two almond linkage maps (Joobeur et al., 1998; Sánchez-Pérez et al., 2007). Although molecular markers for bitterness have been added close to the Sk locus (Sánchez-Pérez et al., 2010), the sequence of this gene remains unknown.
Because biosynthetic studies had indicated that (1) the capacity to synthesize prunasin in the tegument tissue of sweet and bitter almond cultivars appeared similar and (2) the activity of the glucosyltransferase converting prunasin to amygdalin in the developing embryos was also similar in sweet and bitter almond cultivars, different rates of prunasin hydrolysis or turnover in the developing fruits may define whether their kernels are bitter or sweet. Prunasin may be hydrolyzed into mandelonitrile and Glc by the action of prunasin hydrolases (PHs), specific β-glucosidases belonging to family-1 β-glycosidases (EC 3.2.1.21). In a similar manner, amygdalin may be hydrolyzed by the action of amygdalin hydrolases into prunasin and Glc (EC 3.2.1.117). The mandelonitrile formed may dissociate into benzaldehyde and hydrogen cyanide (HCN) nonenzymatically or catalyzed by mandelonitrile lyase 1 (EC 4.1.2.10; Swain and Poulton, 1994a; Suelves and Puigdomènech, 1998). In a putative alternative pathway incorporating the action of heteromeric NIT4 nitrilases and additional, hitherto unidentified enzymes, prunasin may be degraded into benzoic acid, ammonia, and Glc and in this manner be redrawn into primary metabolism (Swain and Poulton, 1994b; Piotrowski et al., 2001; Piotrowski and Volmer, 2006; Jenrich et al., 2007; Kriechbaumer et al., 2007).
Recent studies of bitterness in almond showed a high total β-glucosidase activity in the inner epidermis of the tegument in sweet compared with bitter almond cultivars that might interfere with and decrease prunasin conversion into amygdalin (Sánchez-Pérez et al., 2008). Depending on the cellular localization of the PH activity (apoplastic, cell wall bound, vesicular, or cytosolic) and whether the route of transport of prunasin from the biosynthetic cells of the tegument to the nucellus, endosperm, or embryo takes place in the symplast or in the apoplast, the β-glucosidase activity might control the amount of prunasin available for amygdalin production (Sánchez-Pérez et al., 2008).
In this study, the localization and activity of PHs in different seed tissues were monitored in two sweet and two bitter almond cultivars during fruit development to investigate a possible correlation between the content of amygdalin in the almond kernel and the cellular localization of PHs.
RESULTS
PH Localization as Monitored Using the Sugar-Reducing Assay and Antibodies in Unripe Almond Seeds
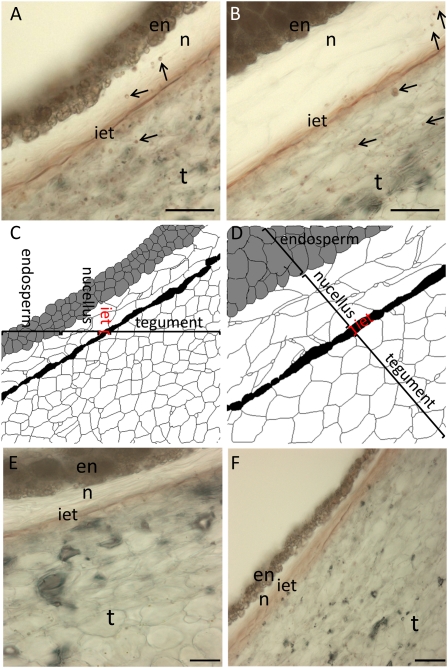
The localization of PH activity in thin sections of the sweet almond cultivar Lauranne and the bitter almond cultivar S3067 at 154 Julian days (JD; the number of days after January 1; Fig. 1) was monitored colorimetrically (red color formation) by the release of Glc following incubation with prunasin (Sánchez-Pérez et al., 2009). At this stage, the nucellus and endosperm were difficult to separate from the tegument; therefore, these are analyzed as a single combined sample. This also applies to the PH activity experiments (see below). The presence of PH was confined to small vesicles in both cultivars, as judged by the staining pattern observed (Fig. 1, A and B) when compared with control samples (Fig. 1, E and F). In both the bitter and sweet cultivars, vesicles in the tegument tissue layer stained strongly, while only a few vesicles in the nucella were found to react. In the endosperm, a slight reddish coloration was observed, reflecting the background reaction seen in this tissue. The strongest reaction was detected in the inner epidermis of the tegument and may represent the presence of a high amount of PH enzyme, the presence of PH with increased specific activity, or the presence of more than one or a different isoform in this tissue.
Figure 1.
Seeds from two almond cultivars, sweet (Lauranne; A and C) and bitter (S3067; B and D), at 154 JD were cross-sectioned to monitor the distribution of PH activity using the sugar-reducing assay after incubation with prunasin. A and B, PH was detected in the tegument (t), inner epidermis of the tegument (iet), nucellus (n), and endosperm (en), as observed by reddish vesicles indicated with arrows. C and D, Histological diagrams, defining the different type of tissues analyzed in the cross-sections. E and F, Background staining in the absence of added substrate in Lauranne (E) and S3067 (F). Bars = 50 μm. [See online article for color version of this figure.]
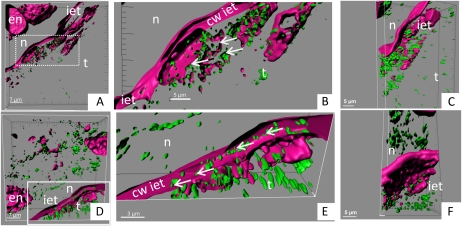
In order to directly monitor the localization of the PH protein, a parallel series of experiments was performed using an antibody known to specifically recognize PH. These studies were carried out using tissue sections at the same developmental stages as used for the activity stain-based experiments (Sánchez-Pérez et al., 2009; Fig. 2). PH was immunolocalized to specific vesicles as visualized by green fluorescence (Fig. 2), a localization being consistent with the vesicle-specific localization of the PH activity (Fig. 1). PH-containing vesicles were observed in cells of the endosperm, nucellus, and the tegument tissue layer in both cultivars (Fig. 2, A and D). A higher magnification of the inner epidermis of the tegument revealed that PH was exclusively localized in the symplast of the inner epidermis tissue layer in the sweet cultivar (Fig. 2, B and C). In the bitter almond cultivar, PH was observed in the apoplast (Fig. 2E). To study in more detail the possible differences in the localization of PH in sweet and bitter cultivars, the immunolocalization of PH was followed in the course of fruit development in the seed using two sweet and two bitter cultivars.
Figure 2.
Seeds from two almond cultivars, sweet (Lauranne; A–C) and bitter (S3067; D–F), at 154 JD were cross-sectioned to perform immunolocalization studies using a polyclonal antibody against the sequence INKKGIEYY present in the PHs from almond (green vesicles). PHs were detected in the tegument (t), inner epidermis of the tegument (iet), nucellus (n), and endosperm (en). Magenta color represents the cell wall (cw) and the DNA/RNA structures stained with propidium iodide in the confocal studies. Arrows indicate the PH vesicles in the cells belonging to the inner epidermis of the tegument. A and D show three-dimensional presentations of the z stack, whereas B, C, E, and F present magnifications of the cells in the inner tegument with the same view and different view angles. A, B, D, and E show views of open cells: in the sweet Lauranne cultivar (A and B), the cell wall of the inner epidermis of the tegument remains devoid of PH vesicles, whereas in the bitter S3067 cultivar (D and E), PH vesicles are observed within the cell wall. [See online article for color version of this figure.]
Immunolocalization of PH in Almond Seeds during Fruit Development in Sweet and Bitter Cultivars
The localization of PH protein in different tissues (tegument, nucellus, endosperm, and embryo) of seeds of two sweet (Ramillete and Lauranne) and two bitter (S3067 and Gorki) cultivars was monitored during fruit development using a specific antibody toward PHs (Figs. 3 and 4). Because of cultivar differences in flowering time and seed-ripening period, the thickness of the endosperm and the shape of the endosperm cells were not directly comparable when cultivars were analyzed at the same JD, but comparison of seeds of the same development stage was possible because of the sequential harvesting.
Figure 4.
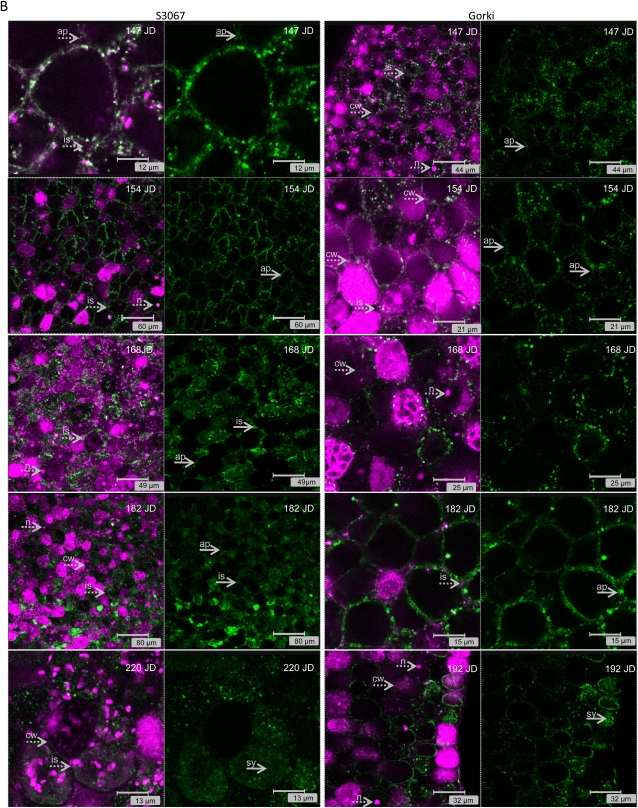
(Legend appears on following page.) Figure 4. A, Immunolocalization studies of PHs (green color) in the embryo of two sweet genotypes (Ramillete and Lauranne) at different time points during fruit development (Ramillete, 147, 154, 168, 182, and 220 JD; Lauranne, 147, 154, 168, 182, and 192 JD) using a polyclonal antibody against the sequence INKKGIEYY present in the PHs from almond. The presence of DNA/RNA structures (i.e. nucleus [n]) and cell wall (cw) is visualized by propidium iodide staining (magenta color) and indicated by dotted arrows. The localization of the PH changes during fruit development from being mainly in the symplast (sy; i.e. Ramillete at 147, 154, and 168 JD and Lauranne at 147, 154, and 168 JD) to being predominantly found in the apoplast (ap; i.e. Ramillete at 182 and 192 JD and Lauranne at 182 JD), as indicated by solid arrows, filling even the intercellular spaces (is). B, Immunolocalization studies of PHs (green color) in the cotyledon of two bitter genotypes (S3067 and Gorki) at different time points during fruit development (S3067, 147, 154, 168, 182, and 220 JD; Gorki, 147, 154, 168, 182, and 192 JD) using a polyclonal antibody against the sequence INKKGIEYY present in the PHs from almond. The presence of DNA/RNA structures (i.e. nucleus [n]) and cell wall (cw) is visualized by propidium iodide staining (magenta color) and indicated by dotted arrows. The localization of the PH changes during fruit development from being mainly in the apoplast (i.e. S3067 at 147,154, 168, and 182 JD and Gorki at 147, 154, 168, and 182 JD), with occasional presence in the intercellular spaces (is), to being found in the symplast (i.e. S3067 at 220 JD and Gorki at 192 JD). [See online article for color version of this figure.]
PH in the Tegument
PHs were present in vesicles in the vast majority of cells in the tegument and were most abundant in the inner epidermis of the tegument at 154 JD (Fig. 3). At the later stages of seed development (from 182 JD), the tegument collapses and dries out concomitant with a reduced content of PH. The inner epidermis of the tegument showed strong staining especially at the young stages, in a similar manner to that observed with the distribution of PH activity and the antibody staining patterns shown in Figures 1 and 2. This shows the presence of high concentrations of PH enzyme in this cell layer. The differences in localization between the cultivars are most clearly visualized at higher magnifications (Fig. 3, columns b, d, and e).
PH in the Endosperm
PH was not detected in all individual cells of the endosperm. In the two bitter cultivars, a distinct PH-staining zone was found in the endosperm cells adjacent to the inner epidermis of the tegument. In the two sweet cultivars, a more uniform presence of PH-containing cells was observed across the endosperm. In all four cultivars examined, PH was observed in the symplast of the cells, as shown by three-dimensional images (Fig. 3, last column, 182JD3D).
PHs in the Cotyledon of the Embryo
The localization of PH proteins in the developing cotyledons was followed from 105 to 220 JD (Fig. 4). The localization was found to vary over the course of seed development in a manner characteristic for each cultivar. Differences specific to sweet or bitter cultivars were also identified. In all cultivars tested, PH was found to be localized in large aggregates or in vesicles. In the sweet cultivar Ramillete (Fig. 4A), the localization of PH shifted from the symplast (i.e. 147, 154, and 168 JD) to the apoplast (182 and 220 JD) in the course of fruit development (Supplemental Videos S1 and S2). A parallel analysis of the sweet cultivar Lauranne at 147 JD showed a dual localization: PH was found in the apoplast surrounding some of the cotyledon cells but was found in the symplast of other cotyledon cells, as visualized by the overlay images reflecting the superposition of the green color representing immunostaining of PHs and the magenta color obtained by propidium iodide staining of cell walls and nuclei. At the subsequent stages of fruit development in Lauranne seeds, the PHs were found to be located in the symplast (154 and 168 JD), and in the final stages of fruit development, PH immunostaining was observed in the apoplast (182 and 192 JD). In the bitter cultivars (Fig. 4B), the pattern of PH immunostaining was more or less reversed compared with the observations with the sweet cultivars. In the early to middevelopmental stages of the embryo, PH immunostaining was observed in the apoplast in the cotyledons (S3067 at 147 JD and Gorki at 147 and 154 JD), even filling the intercellular spaces (i.e. S3067 at 168 JD). At the later developmental stages, the localization of the PHs shifted to the symplast (S3067 at 220 JD) and to some extent the apoplast (Gorki at 192 JD). Two videos are provided (Supplemental Videos S1 and S2) to more clearly document the symplastic or apoplastic localization of PH. When the localization of PH is in the symplast, a screen of the z-stack reveals green-staining zones surrounded by a nonstaining region representing the cell wall and apoplast space. At developmental stages where PH is localized in the apoplast, the stained patterns enclose nonstaining domains reflecting the internal cellular space.
Biochemical Measurements of PH Activity during Fruit Development in Almond Seeds
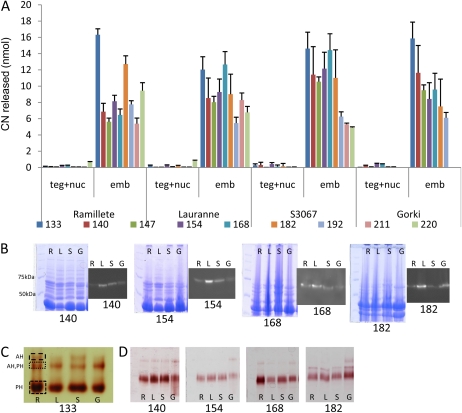
In parallel to the analysis of PH localization during seed development, the enzymatic PH activity was determined by measuring the ability of crude almond extracts to catalyze the release of HCN following incubation with prunasin. Measurements were carried out using dialyzed extracts from the tegument, from combined tissues of the nucellus and endosperm (as it was not possible to separate these tissues, especially in the mid season), and from the embryo composed of the two cotyledons and the small embryo axis. The cotyledons are the final tissue where amygdalin is stored (Frehner et al., 1990). Incubation of extracts from the tegument or from the combined nucellus and endosperm with 20 nmol of prunasin resulted in the release of minute amounts of HCN, reflecting very low PH activity. In contrast, assays with the cotyledon extract released more than 50-fold higher amounts of HCN using the same amount of total protein from each tissue (Fig. 5A). The ability of the cotyledon preparations to release HCN varied from one cultivar to the other, although similarities were observed between sweet and bitter phenotypes. In the sweet cultivars, the activity fluctuated significantly throughout the season, whereas in general, the activity in the bitter cultivars decreased as the fruit matured. These differences could be due to fruit developmental differences or caused by the presence of multiple PH isoforms. In control experiments, a crude commercially available emulsin preparation from almonds (Sigma) was added instead of an almond extract to liberate the maximum amount of HCN (20 nmol). A negative control, prepared with the reaction buffer instead of an almond extract, released about 0.5 nmol of HCN.
Figure 5.
A, HCN released (nmol) from the tegument and nucellus (teg+nuc) and embryo (emb) in two sweet cultivars (Ramillete [R] and Lauranne [L]) and two bitter cultivars (S3067 [S] and Gorki [G]) following incubation of crude protein extracts with 20 nmol of prunasin at different stages of fruit development (133–220 JD). B, Total protein extracts were analyzed by SDS-PAGE, stained with Coomassie Brilliant Blue, and PHs present were visualized by immunolocalization using a polyclonal antibody against the sequence INKKGIEYY present in the PHs from almond. C, Total β-glucosidase content was assayed using Fast Blue BB salt staining of SDS-PAGE following the application of crude protein extracts of cotyledons of the four different cultivars harvested at 133 JD and revealed the presence of different isoforms of PHs (PH) and the presence of amygdalin hydrolase (AH). D, The same assay was used to monitor the presence of different β-glucosidases during fruit development in the four cultivars. [See online article for color version of this figure.]
Detection of PH Isoforms in Almond Cotyledons by SDS-PAGE following Western Blotting and by Fast Blue BB Salt Staining
A single immunoreactive protein band with an apparent molecular mass of 63 kD was detected in all four cultivars following SDS-PAGE (Fig. 5B). β-Glucosidases retain their catalytic properties following SDS-PAGE when the enzyme-containing samples are not boiled before application. Protein bands containing β-glucosidase activity, therefore, can be detected following incubation with the Fast Blue BB salt and the 6-bromo-2-naphthyl-β-d-glucopyranoside, a general substrate for β-glucosidases. Several Fast Blue BB-generated reddish staining bands were obtained (Fig. 5C). The bands were excised from the gel and incubated with prunasin and amygdalin, as described previously, to analyze whether the β-glucosidase catalyzed HCN release and thus represented prunasin and amygdalin hydrolases, respectively. Two protein bands with apparent masses of 70 and 150 kD contained PH activity, while two bands with apparent masses of 150 and approximately 190 kD showed amygdalin hydrolysis activity. This suggests the presence of homodimeric or heterodimeric protein complexes of prunasin and amygdalin hydrolases and possible complexes with other proteins not denatured by the electrophoretic conditions chosen. The Fast Blue BB salt-staining method is more sensitive than western blotting, explaining why these additional bands were not observed on the immunoblot (Fig. 5B). When these assays were performed to follow the profile of β-glucosidase activity in the course of fruit development, all four cultivars showed a dominant immunostaining band with an apparent mass of 70 kD with PH activity and a number of weaker staining bands for which the intensity and mobility varied (Fig. 5D).
Isoforms of PH and Amygdalin Hydrolase in Almond Embryos as Analyzed by Liquid Chromatography-Tandem Mass Spectrometry
A liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach was undertaken to identify the presence of PH homologs in the two sweet and two bitter cultivars based on protein extracts from embryos composed of the two cotyledons and the small embryo axis isolated at 140 JD. At this developmental stage, the cotyledon tissues are low in lipids but rich in protein, amygdalin has started to accumulate in the cotyledons of the bitter cultivars, and minute amounts of prunasin are present in all four cultivars. Thus, this developmental stage would be expected to be suitable for isolating enzymes involved in the breakdown or turnover of cyanogenic glucosides in the sweet cultivars. The protein extracts of the four different cultivars were subjected to SDS-PAGE and the gel lanes stained with either Coomassie Brilliant Blue or with Fast Blue BB salt to identify proteins containing β-glucosidase activity. Prunasin and amygdalin hydrolases have expected masses of approximately 60 kD. Fast Blue BB staining confirmed that β-glucosidase activity was associated with proteins within this mass range. Gel blocks covering the mass range 50 to 75 kD were excised from the gels to be able to identify β-glucosidase isoforms by digestion with trypsin. Tryptic peptides were analyzed by LC-MS/MS, and protein identification was carried out by comparing the MS and MS/MS spectra with the protein sequences in the National Center for Biotechnology Information (NCBI) nonredundant protein sequence database. Preliminary ion-trap experiments identified several putative PH homologs to PHs of sweet cherry (Prunus avium) and black cherry. Sequencing of the tryptic fragments derived from the Coomassie Brilliant Blue-stained gel afforded 22 unique peptide sequences putatively derived from isoforms of five PHs and one isoform of amygdalin hydrolase, whereas analysis of the Fast Blue BB salt-stained gel afforded only nine unique peptide sequences, demonstrating that SDS-PAGE is more suitable for protein identification by LC-MS/MS (data not shown). To obtain additional sequence information for the putative PH homologs, embryo extracts of the four almond cultivars were subjected to SDS-PAGE, stained with Coomassie Brilliant Blue, and then processed as described above. The resulting peptides were subjected to LC-MS/MS by the more sensitive Orbitrap instrument. The same prunasin and amygdalin hydrolase homologs were identified as being present in all four cultivars. In most cases, the homologs were identified by the same matched tryptic peptides in all four cultivars, but in a few cases, a homolog was identified by a different peptide dependent on the cultivars (Table I). LC-MS/MS does not identify all peptides present in the eluate from the HPLC column because of the restriction in time points at which an MS/MS fragmentation may be recorded. Thus, peptides may be present in a sample even though they were not registered by a sequence. To test if this was indeed the case when PH homologs were identified by the detection of different peptides among the cultivars, ion chromatograms displaying the intensity of the differentially identified peptide mass-to-charge ratio (m/z) value were extracted. In all samples, the peptide mass in question (within ±2 ppm) eluted from the LC column at the same position of the gradient (±10 s). Thus, the identified homologs are represented by the same peptides in all four cultivars; consequently, we could not identify cultivar-specific PH isoforms (Table I). Clearly, more peptides were identified derived from PH homologs 1, 2, 4, and 5 from black cherry and β-glucosidase from sweet cherry than from amygdalin hydrolase I and PH 3 from black cherry. It is likely that the isoforms that afforded more peptide hits are more similar to abundant isoform(s) in almond.
Table I. Identified amygdalin hydrolase, PH, and β-glucosidase peptide sequences from Ramillete (R), Lauranne (L), S3067 (S), and Gorki (G) cotyledons.
Proteins migrating within the 50- to 75-kD mass region following SDS-PAGE were digested with trypsin and analyzed by LC-MS/MS. The resulting pairs of peptide mass data and fragment ion (sequence) data were searched against the NCBI nonredundant protein database. X, Sequence is identified (for the criteria for identification, see “Materials and Methods”). (X), Not fragmented but present in extracted ion chromatograms (m/z ±5 ppm) as a distinct chromatographic peak eluting at the same time as the others and with the same isotope (charge) distribution.
| Peptide Sequence | Genotype |
Isoform Assignment |
|||||||||
| Ra | La | Sa | Ga | AHIb | PH1b | PH2b | PH3b | PH4b | PH5b | Pa βGb | |
| AASDWLYVYPK | X | X | X | X | X | X | |||||
| AYADLCYK | X | X | X | X | X | ||||||
| DMGLDAYR | X | X | X | X | X | X | X | X | |||
| EKYNDPIMYITENGMDEFNNPK | X | X | X | X | X | ||||||
| FGINYIDYDNGLER | X | X | X | X | X | X | |||||
| FGINYVDYDNGLKR | X | X | X | X | X | X | X | ||||
| FSISWSR | X | X | X | X | X | X | X | X | X | X | |
| GDYPQSMR | X | X | X | X | X | X | |||||
| GLYDLVLYTK | X | X | X | X | X | X | X | X | |||
| HWTTLNEPYTISNHGYTIGIHAPGR | X | X | X | X | X | X | |||||
| ITDGSNGDVAIDQYHR | X | X | X | X | X | X | X | X | |||
| IVDDFK | X | X | X | X | X | X | |||||
| KFGDR | X | X | X | X | X | ||||||
| LPNFTEEQSK | (X) | (X) | (X) | X | X | X | X | X | X | ||
| LSTHWFK | X | X | X | X | X | X | X | X | |||
| SFLKK | X | X | X | X | X | ||||||
| SFLKR | X | X | X | X | X | ||||||
| SIVGER | X | X | X | X | X | ||||||
| SLSGSYDYIGVNYYSAR | X | X | X | X | X | ||||||
| THFDTLFPGFTFGAATAAYQLEGAANIDGR | (X) | (X) | X | X | X | X | |||||
| VKHWTTLNEPYTISNHGYTIGIHAPGR | (X) | X | X | (X) | X | X | |||||
| YASAYPK | X | X | X | X | X | ||||||
| YKEDVAIMK | X | X | X | X | X | X | X | X | X | ||
| YNDPIMYITENGMDEFNNPK | X | X | X | X | X | X | X | ||||
| Hits | 24 | 24 | 24 | 24 | 4 | 9 | 12 | 1 | 8 | 14 | 12 |
Peptide sequences obtained from Ramillete, Lauranne, S3067, and Gorki and identified by NCBI searches as part of amygdalin hydrolase, PH, or β-glucosidases.
The peptide sequences are part of the protein sequences according to NCBI (cDNA accessions): AHI (AF411130), amygdalin hydrolase from black cherry; PH, PH isoforms from black cherry as follows: PH1 (U50201), PH2 (AF221527), PH3 (AF221526), PH4 (AF411009), PH5 (AF411131); Pa βG, β-glucosidase from sweet cherry (AAA91166).
Identification of Two Genes, Each Encoding a PH in Almond Embryos
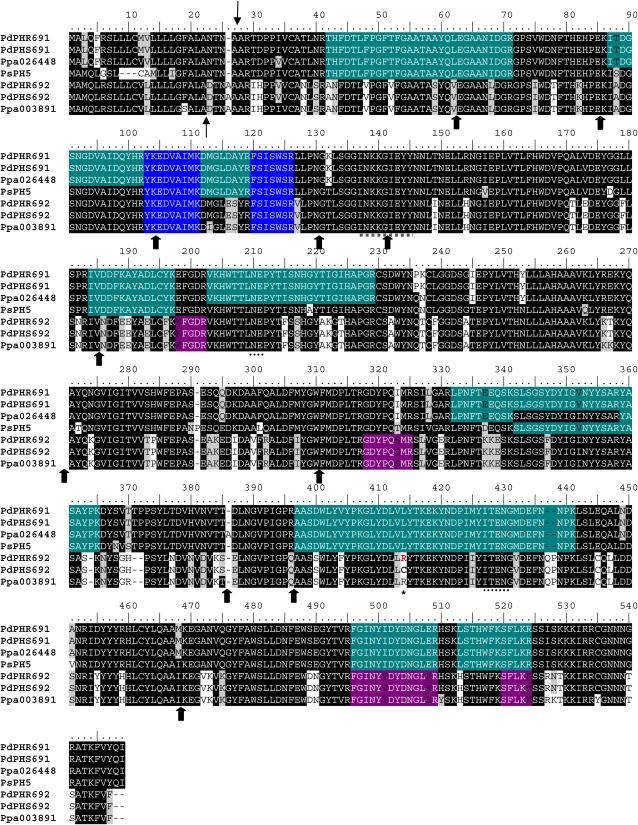
Encouraged by the identification of PH isoforms present in the embryo extracts of the four cultivars, ESTs with high similarity to PHs were searched for in the Genome Database of Rosaceae (http://www.rosaceae.org/). The search provided four almond accessions, BU572569, BU573272, BU574214, and BU574543, that displayed more than 80% nucleic acid identity with PHs from black cherry.
In order to isolate PH genes (Ph) from a sweet (Ramillete) and a bitter (S3067) almond cultivar, primers were designed to amplify these four database sequences by PCR (Table II). The amplified fragments obtained from two cultivars were cloned and sequenced to verify their identities. Interestingly, the fragments amplified from the four database accessions not only shared high similarity with PHs from black cherry but were identified as partial segments of a single Ph. This was confirmed by 5′ and 3′ RACE experiments, depending on the accessions. However, the RACE experiments provided two different 5′ untranslated regions. This indicated that there were at least two Ph genes encoding different isoforms in the two almond cultivars. The genes were assigned as Ph691 and Ph692. Both cDNAs shared 86% nucleotide identity, whereas the encoded protein sequences were 79% identical to each other. A BLAST analysis of the two cDNA sequences against the NCBI database showed that Ph691 showed 92% and Ph692 showed 86% nucleotide sequence identity to Ph1 from black cherry, with a 98% query coverage. When the protein sequences were compared with the five PHs from wild cherry, PH691 was slightly more similar to PH5 (94%) than to PH1 (93%), and for PH692, a similar identity (79%) with PH5 and PH1 was found (Table III). Based on the two cDNAs for the two isoforms, primers were designed to identify the genomic DNA encoding the two PHs by chromosome walking. Twelve introns were found in each Ph gene, with identical splice sites to those found in the Ph genes from black cherry (Zhou et al., 2002). The alignment of the sequences of the two PHs (PH691 and PH692) from the sweet (Ramillete) and the bitter (S3067) almond cultivars, of PH5 of black cherry, and of two peach accessions found in Phytozome (www.phytozome.org) is shown in Figure 6. The GenBank accession numbers in almond (Pd) for the genomics and the cDNAs for Ramillete (R) and S3067 (S) are as follows: PdPhR691 (JQ268617), PdPhS691 (JQ268618), PdPhR692 (JQ268619), PdPhS692 (JQ268620), PdPhR691 (JQ268621), PdPhS691 (JQ268622), PdPhR692 (JQ268623), PdPhS692 (JQ268624). The only sequence difference between the PH sequences from the sweet and the bitter cultivars was found in PH692 (Fig. 6). In the sweet cultivar as well as in a peach accession (ppa003891), the residue at position 414 was an Arg, whereas a Cys residue was found at this position in the bitter cultivar.
Table II. Designed primers for four almond ESTs and complete cDNA, annealing temperatures, and expected product size.
Arrows indicate the direction of the primers (→, forward; ←, reverse).
| Accession No. | Sequences (5′–3′) | Temperature | Expected Size |
| °C | bp | ||
| BU572569 | F: TAGCCATGGTTATGCAAAGG (→) | 60 | 525 |
| R: CTGTGGACCAATGGGAACTC (←) | |||
| BU573272 | F: CAACTTTGAATGGGACAATGG (→) | 60 | 600 |
| R: GGGGGCTGAACATTCATTTA (←) | |||
| BU574214 | F: GGCTTTGCTTTGGCAGATAC (←) | 60 | 568 |
| R: GGTCCAATGCTTCACTCGAT (→) | |||
| BU574543 | F: GGCTTTGCTTTGGCAGATAC (→) | 60 | 525 |
| R: CTGCGTATTCTTCAAAGTCATTG (←) | |||
| Ph691cDNA | CAP691F: GGGCACACCTAGCTCGATCATC (→) | 57 | 2,022 |
| 691polyAR: AGAAGAAATTGCATTTATAATTTAGCC (←) | |||
| Ph692cDNA | CAP146972F: GCTGCAAACTTGGACGGTAG (→) | 57 | 1,469 |
| 69LRd5R: GAAGGAGTTGTATTTTATTCATCACAT (←) |
Table III. Percentage of sequence identity of genes encoding PHs from almond (Ph691 and Ph692), cherry (Ph1–Ph5), and almond amygdalin hydrolase 1 (Ah1) BLASTed versus the peach genome (www.phytozome.org; ppa accessions).
Ph691 has the highest identity with the cherry Ph5 and with four peach accessions. Ph692 shows less than 80% identity with the cherry PHs and 98% with one of the peach accession (ppa08391).
| Gene or Accession | Ph691 | Ph692 | ppa026448 | ppa020839 | ppa021137 | ppa003831 | ppa003856 | ppa03891 |
| Ph691 | 100 | 79 | 99 | 98 | 99 | 98 | 87 | 79 |
| Ph692 | 79 | 100 | 80 | 79 | 79 | 80 | 78 | 98 |
| Ph1 | 93 | 79 | 93 | 92 | 93 | 93 | 88 | 78 |
| Ph2 | 87 | 78 | 87 | 86 | 87 | 87 | 90 | 77 |
| Ph3 | 73 | 71 | 72 | 73 | 74 | 72 | 73 | 71 |
| Ph4 | 89 | 78 | 89 | 88 | 88 | 89 | 85 | 77 |
| Ph5 | 94 | 79 | 93 | 95 | 93 | 93 | 87 | 78 |
| Ah1 | 70 | 71 | 69 | 70 | 69 | 69 | 68 | 71 |
Figure 6.
(Legend appears on following page.) Figure 6. Alignment of different PHs from the P. dulcis (Pd) isoforms 691 and 692 (PHR, Ramillete; PHS, S3067) identified in this study with black cherry isoform 5 (PsPH5) and peach (Ppa) 026448 and 003891. Thin arrows mark the predicted cleavage site of the signal peptide in the almond PHs. Thick arrows represent the splicing sites for the 12 introns for the almond sequences, characteristic for the PHs. ITENG and NEP motifs diagnostic for glycosyl hydrolase family 1 are marked by dotted lines. The peptide used to generate the polyclonal antibody in rabbit is underlined with a dotted gray line. The peptides identified by LC-MS/MS are highlighted in green (for the ones present in PH691), purple (PH692), and blue (present in all the sequences). The only difference between sequences from sweet and bitter cultivars was detected in PdPH692, at position 414 (asterisk). [See online article for color version of this figure.]
To determine whether the two almond PHs contained a signal peptide, PH691 and PH692 were analyzed using the SignalP 3.0 Server. Both proteins were predicted to possess an N-terminal signal peptide and a predicted cleavage site between amino acid residues 26 and 27 (TNA-AR) for PH691 (signal peptide probability, 0.996) and between amino acid residues 22 and 23 (ALA-DT) for PH692 (signal peptide probability, 0.999; Fig. 6). For protein subcellular localization prediction, the WoLF PSORT program predicted, based on nearest neighbor calculations (14 in total), that both PH691 and PH692 were most likely targeted to the vacuole (PH691 = vacuole: 7.0, chloroplast: 2.0, extracellular: 2.0, nucleus: 1.0, mitochondria: 1.0, Golgi: 1.0; PH692 = vacuole: 7.0, chloroplast: 2.0, endoplasmic reticulum: 2.0, nucleus: 1.0, extracellular: 1.0; Golgi: 1.0). In addition, when PH691 and PH692 were analyzed with TargetP 1.1, both proteins were predicted to contain signal peptides targeting the secretory pathway with a reliability class of 1 on a scale from 1 to 5, where 1 indicates the strongest prediction. These predictions are in agreement with the observed localization of the PH enzyme and activity in small vesicles that may change cellular location during fruit development and where the PH content of vesicles may end up being deposited in the apoplast.
To verify that the two cloned PH isoforms were also present in the protein extracts, the LC-MS/MS data sets obtained from the embryos of the four cultivars were searched against the two novel full-length almond PH sequences. Twelve tryptic peptide sequences were specifically assigned to PH691 and six to PH692. Thus, the cloned PH isoforms were present as proteins in the young developing embryos. These identified peptides are highlighted in the alignment shown in Figure 6. We did not observe any differences in the identified peptides from the four different cultivars, suggesting that these two PH isoforms are present in the embryos of all four cultivars. However, additional isoforms or alleles are present, because several of the peptides showing a clear match to PH sequences available from black cherry and sweet cherry are not contained within the two novel full-length sequences reported here from almond.
DISCUSSION
Sweetness or bitterness in almond kernels is determined by the genetic background of the sporophyte, which means that all almonds carried by a single tree will be either sweet or bitter, independent of the genotype of the pollen source (Kester and Asay, 1975; Dicenta et al., 2000; Arrázola, 2002). After fertilization, the fruit is composed of diploid mother tissues (exocarp, mesocarp, endocarp, tegument, and nucellus) and seed tissues containing one copy of the male gametophyte and two or a single copy of the female gametophyte (endosperm and embryo, respectively).
In some sweet almond cultivars, the concentration of prunasin in the vegetative parts of the plant is higher than the concentration in bitter cultivars (Dicenta et al., 2000). This illustrates that the concentration of prunasin in the vegetative parts is not correlated with the bitterness phenotype. This is in agreement with experiments demonstrating that the tegument is the first of the mother tissues present in the fruit in which prunasin is detected and prunasin is de novo synthesized.
In the tegument of the bitter cultivar S3067, prunasin starts to accumulate about 60 d after flowering, equivalent approximately to 101 JD, and proceeds until the tissue is dried out (199 JD or 160 d after flowering; Sánchez-Pérez et al., 2008). Prunasin does not accumulate in the tegument of the sweet cultivars. The dates for flowering and ripening are specific for each cultivar (Martínez-Gómez et al., 2008) and may be shifted from one year to another due to climatic differences and the quantitative nature of the flowering date (Tabuenca, 1972).
Although the biosynthesis of prunasin proceeds exclusively in the tegument, no amygdalin formation was observed in this tissue. Neither nucellus, endosperm, nor embryo tissues showed the ability to convert Phe into prunasin. Thus, prunasin has to be transported to other tissues for amygdalin synthesis to occur (Sánchez-Pérez et al., 2008), and this may include transport from the tegument to the nucellus, endosperm, and the nascent embryo. This transport would require the crossing of several “barriers” (e.g. within and across cells from the same or different tissues). The first would be related to the transport from the cytosolic surface of the endoplasmic reticulum, where the cytochrome P450-mediated synthesis of cyanogenic glucosides take place, to the vacuole or smaller vesicles, where cyanogenic glucosides are stored (Poulton, 1990; Selmar, 1994; Jones et al., 1999; Nielsen et al., 2008). In this context, it is interesting that the PHs are glycoproteins (Zhou et al., 2002). Glycoproteins are synthesized in the endoplasmic reticulum lumen or in the Golgi. This is in agreement with the results of this study demonstrating (Figs. 1–4) that PHs are localized in small vesicles that are supposedly targeted to the vacuole or to the secretory pathway (Fig. 6). Whether prunasin and PH at some stages during these transport processes gain contact is not known.
In the early stages of seed development, transport of prunasin from the tegument would proceed to the nucellus, which subsequently is displaced by the cellular endosperm. For such transport to proceed, the inner epidermis of the tegument has to be passed. These thick-walled cells are charged with PHs either inside or outside the cells in sweet or bitter cultivars, respectively (Figs. 1–3). If in the sweet cultivars, prunasin is transported across this layer via the symplast, the prunasin may be exposed to the PHs and hydrolyzed. The inner epidermis of the tegument may thus constitute a barrier and a major impediment for prunasin transport in the sweet cultivars. In the bitter cultivars, the PHs are mostly found in the apoplast (Fig. 2; Sánchez-Pérez et al., 2008). In the cotyledons of black cherry seeds, PHs have previously been shown to be localized in the vacuoles of the phloem parenchyma cells (Poulton and Li, 1994).
In a recent study carried out using sweet and bitter almond cultivars, it was reported that the UDPG-glucosyltransferase UGT85A19 catalyzing the conversion of mandelonitrile into prunasin showed a 3-fold higher activity in bitter compared with sweet cultivars and with the main activity localized in the tegument (Franks et al., 2008). This could suggest that a balance of the rates of biosynthesis and the degradation of prunasin may determine how much prunasin is available as a substrate for the production of amygdalin and thus whether a cultivar is sweet or bitter. The ability to store a secondary metabolite with a chemical structure larger than the precursor (i.e. amygdalin versus prunasin, raffinose versus Suc; McCaskill and Turgeon, 2007) is an option plants have developed to reduce back diffusion through plasmodesmata and thereby to achieve the accumulation of high concentrations of a product in a sink organ, such as the almond embryo or the phloem.
In a previous study, the inner epidermis of the tegument appeared to possess a higher total β-glucosidase activity in sweet compared with bitter cultivars (Sánchez-Pérez et al., 2008). To assess whether PH activity was part of the recorded total activity, and whether a different distribution was observed in teguments of bitter and sweet cultivars, a sugar-reducing assay with prunasin as substrate was performed (Sánchez-Pérez et al., 2009). This specific assay revealed PH activity in vesicles not only in the tegument but also in nucellus and endosperm, as evidenced by their reddish or even brownish staining (Fig. 1, A and B). Immunoassays were performed to localize specifically the protein responsible for prunasin hydrolysis. The immunohistological studies showed differences in the localization of PH in the cells of the inner epidermis of the tegument between the sweet and bitter cultivars. A symplastic localization was found in the sweet cultivar compared with a dual symplastic and apoplastic localization in the bitter cultivar (Fig. 2). Differences in the total amounts of PHs present in the sweet and bitter genotypes are very difficult to assess due to differences in the area sectioned, ripening state, and morphology between cultivars (Sánchez-Pérez et al., 2008).
PH was also detected in the embryo, especially in the cotyledons (Fig. 4). Localization appeared as a characteristic reticulate system, which, depending on the developmental stage analyzed, was more or less pronounced. A similar reticulate system has previously been reported for the glucosinolate-degrading myrosinases present in Arabidopsis (Arabidopsis thaliana) and Brassica napus, in which transmission electron microscopy analyses of mature embryos showed that the myrosinase was contained in several vacuoles within each cell, denoted as myrosin grains and referred to as constituting a “myrosin body” (Andréasson et al., 2001, Fig. 7B).
The cellular localization of the PHs changes through the development of the cotyledon. In the two sweet cultivars tested, PH is found in the symplast in the cotyledons at the early developmental stages, but the localization shifts to the apoplast at the later developmental stages (Supplemental Videos S1 and S2). In the two bitter cultivars examined, PH localization shifted from the apoplast to the symplast (Fig. 4B). The shifts in localization in the cotyledons may be the cause of the previously reported fluctuations in prunasin content during fruit development (Sánchez-Pérez et al., 2008). A change in cellular localization during fruit ripening was also detected by immunogold staining of a β-glucosidase present in the mesocarp of sweet cherry (Gerardi et al., 2001). At unripe developmental stages, the gold labeling was associated with the endoplasmic reticulum. In mid ripening stages, the labeling was localized in an endosome delimited by cytoplasmic membrane. At full ripening, the β-glucosidase was detected in the middle lamella of the cell wall.
The presence of PHs in the mesocarp has also been analyzed by confocal studies. In this almond tissue, the PH was surprisingly found to be localized within the chloroplasts (Sánchez-Pérez et al., 2009). In a recent study, a β-glucosidase localized to tobacco (Nicotiana tabacum) chloroplast was found to result in increased phytohormone levels by increasing the level of the active nonglucosylated phytohormones (zeatin and indole-3-acetic) and to confer a significant increase in biomass production as well as insect protection (Jin et al., 2011). Because tobacco does not contain cyanogenic glucosides, this study demonstrates that β-glucosidases involved in other processes may also be localized in the chloroplast.
The complex and developmentally dependent localization of PH prompted biochemical studies of sweet and bitter cultivars in the course of fruit ripening. When extracts containing the same amount of total protein were incubated with prunasin, the tegument and nucellus released about 50 times less HCN in comparison with the embryo (Fig. 5A). This result was observed with all four cultivars independent of the ripening stage, although the amount of HCN release from the embryo extracts was found to fluctuate within the ripening period. The cause of these fluctuations could be developmentally dependent changes in the expression profiles of different PH isoforms. In fact, comparison of the fluctuations of amygdalin concentration during ripening in S3067 embryos, as reported previously (Sánchez-Pérez et al., 2008, Fig. 3), and the HCN released in the same cultivar from our study here suggests that those increases and decreases of amygdalin content may be directly related to the fluctuations in PH activity.
In order to discover whether the fluctuations in PH activity and the shift in localization in the course of fruit development were due to the presence of different isoforms, western-blot analysis was carried out using a hapten antibody raised against a peptide sequence known to be conserved among all previously characterized PHs (Fig. 5B). The presence of a single immunoreactive protein band with an apparent molecular mass of 63 kD was observed throughout the entire ripening season. This mass is in accordance with the molecular mass of PHs previously characterized in black cherry (Zhou et al., 2002). The single band obtained by immunostaining may represent the presence of multiple comigrating PH isoforms with nearly identical molecular masses or may indicate that only a single isoform was indeed present. To further investigate these options, Fast Blue BB salt staining was performed to investigate the number of protein bands present carrying β-glucosidase activity (Fig. 5C) followed by excision of these protein bands and incubation with prunasin or amygdalin to specifically monitor prunasin and amygdalin hydrolase activity. The experiment was carried out using samples collected throughout the ripening season and demonstrated the continuous presence of a dominant β-glucosidase with a mass of less than 75 kD with PH activity and minor additional bands with prunasin and amygdalin hydrolase activities, most likely representing homodimeric or heterodimeric forms or aggregates with other proteins (Fig. 5D). Because these experiments were ambiguous with respect to the number of PH isoforms present, protein extracts from the four cultivars were fractionated by SDS-PAGE and proteins in the mass range 50 to 75 kD characterized by LC-MS/MS based on a search of known sequences of the Prunus taxon. When BLASTed versus the NCBI database, a total of 24 peptides were found to match sequences previously reported as present in five isoforms of PH, in one amygdalin hydrolase isoform from black cherry, and in a β-glucosidase from sweet cherry (Table I). All 24 peptides were found in the four cultivars analyzed, and some of them were present in only one of the isoforms, demonstrating the presence of different PHs in almond. In order to identify PHs in almond, we searched in the Rosaceae database for ESTs with high similarity to PHs from black cherry. Two different full-length cDNA sequences were obtained from almond encoding proteins designated PH691 and PH692. Whereas the amino acid sequence for PH691 was identical in the sweet and bitter almond cultivars, the PH692 sequence varied at a single position (residue 414), being an Arg residue in the sweet cultivar and a Cys residue in the bitter cultivar. Whether this change is genotype or phenotype specific is currently being studied. A sequence alignment of PH691 and PH692 versus PH5 from cherry and two accessions from peach is shown in Figure 6. Peptide sequences obtained by LC-MS/MS analyses are highlighted in Figure 6. A single peptide sequence (SIVGER) was not present in PH691 and PH692 and was assigned to amygdalin hydrolase 1.
Comparison of the almond and wild cherry PH sequences with the database hosting the peach genome version 1 sequences, released in 2010, showed six peach accessions with up to 99% sequence identity (Table III). For example, PH691 showed 99% sequence identity to ppa026448. The latter sequence showed 94% identity to PH5, strongly suggesting that Ph691 encodes a PH in almond. With respect to PH692, sequence identities were slightly lower than those observed with PH691. The sequence identity of PH692 to PH1 and PH5 was 79%. On the other hand, PH691 and PH3 showed only 73% identity. The high percentage of identities between the different isoforms prevents discrimination between the isoforms using in situ PCR or quantitative PCR. All the accessions and isoforms described in Table III contained the sequence INKKGIEYY used to identify PHs in almond. Accordingly, the antibody raised to detect PHs in almond would also be useful to identify PHs in other species, such as cherry, peach, and apricot. The INKKGIEYY sequence has not been found in any of the hitherto reported amygdalin hydrolases.
Allozymes have previously been reported as responsible for the multiplicity of cyanogenic glucoside-degrading enzymes (Zhou et al., 2002). However, these studies were performed without knowing the pedigree of the cultivars employed. Therefore, an inheritance study of these genes/alleles would help to resolve the precise genetic basis for the presence of multiple isoforms. The Almond Breeding Program from Centro de Edafología y Biología Aplicada del Segura-Consejo Superior de Investigaciones Científicas (CEBAS-CSIC; Spain) could provide the germplasm for these studies and serve to elucidate whether Ph691 represents a single locus with multiple alleles or whether additional PHs are encoded at different positions in the almond genome. Next-generation sequencing is being performed to help to elucidate this.
The genes encoding the two PHs identified in almond in this study each contain two AUG codons that could represent transcription start sites. In mammals, multiple transcripts derived from a single locus are frequently observed, and the gene products may differ from the use of different splice sites as well as from alternative sites of transcription initiation and termination (Shabalina et al., 2010). A positive correlation was demonstrated between alternative splicing within the coding sequence and alternative termination within the 3′ untranslated region. Therefore, depending on the site of transcription initiation, alteration of the splice site pattern or signal peptide may result in different localization of the protein or serve to modify its activity. Such mechanisms may add to and govern the complex and changing patterns of PH localization in the course of almond seed development in sweet and bitter almond cultivars.
MATERIALS AND METHODS
Plant Material
Almond (Prunus dulcis) fruits of the following four cultivars, Ramillete (SkSk; sweet), Lauranne (SkSk; sweet), Gorki (sksk; bitter), and S3067 (sksk; bitter), were provided by the Almond Breeding Program of CEBAS-CSIC. Almond fruits were collected every week from the beginning to the middle of the growing season and every second week from the middle to the end of the growing season (i.e. from fruit set in March to ripening in August) and examined at the Plant Biochemistry Laboratory, University of Copenhagen, or at CEBAS-CSIC, depending on the type of experiments. Fruits were dissected into tegument, nucellus including endosperm (as it was impossible to separate these two tissues, especially in the mid season), and embryo (embryonic axis plus cotyledon), depending on the developmental stage and experimental needs.
The four cultivars used in this work had different full blooming dates, JD (number of days after January 1), when 50% of flowers were opened: Ramillete (31 JD), Lauranne (60 JD), S3067 (51 JD), and Gorki (63 JD). The ripening dates of the four cultivars used were as follows: Ramillete (220 JD), Lauranne (211 JD), S3067 (220 JD), and Gorki (211 JD). The DAF for each cultivar at the different harvest days are shown in Table IV. From 85 to 105 JD, only the tegument and nucella were visible in the four cultivars. Cotyledon can be observed at 105 JD for Ramillete and Gorki, together with the endosperm. At 119 JD, the four cultivars contain tegument, nucella, endosperm, and cotyledon. In S3067, nucella is no longer observed after 154 JD. From 168 JD, the tegument starts to collapse in all the cultivars except S3067 and fully collapses in all cultivars in the period from 182 to 220 JD.
Table IV.
Relationship between the JD at which the samples were harvested and the DAF for the almond cultivars Ramillete, Lauranne, S3067, and Gorki
| JD | DAF |
|||
| Ramillete | Lauranne | S3067 | Gorki | |
| 85 | 54 | 25 | 34 | 22 |
| 91 | 60 | 31 | 40 | 28 |
| 98 | 67 | 38 | 47 | 35 |
| 105 | 74 | 45 | 54 | 42 |
| 112 | 81 | 52 | 61 | 49 |
| 119 | 88 | 59 | 68 | 56 |
| 126 | 95 | 66 | 75 | 63 |
| 133 | 102 | 73 | 82 | 70 |
| 140 | 109 | 80 | 89 | 77 |
| 147 | 116 | 87 | 96 | 84 |
| 154 | 123 | 94 | 103 | 91 |
| 168 | 137 | 108 | 117 | 105 |
| 182 | 151 | 122 | 131 | 119 |
| 192 | 161 | 132 | 141 | 129 |
| 211 | 180 | 151 | 160 | 148 |
| 220 | 189 | 160 | 169 | 157 |
Identification of Two PH-Encoding Genes in Almond
The sequences of four ESTs from almond with high similarity to PHs were obtained from the Genome Database of Rosaceae (http://www.rosaceae.org/): BU572569, BU573272, BU574214, and BU574543. For the amplification of these ESTs in almond, primers were designed using the Primer3 program (http://frodo.wi.mit.edu/; Table II), and RNA was extracted from Ramillete and S3067 using the RNeasy Plant Mini Kit (Qiagen). cDNA was produced using the iSCRIPT kit (Bio-Rad). The cDNA obtained was amplified using Phusion DNA Polymerase (Finnzymes) by the following PCR protocol: 94°C for 5 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and one cycle of 72°C for 5 min in an Eppendorf Thermocycler. Amplified products were analyzed on 2% agarose gels with the 1-kb Plus DNA Ladder (Invitrogen) and monitored using UV light. The authenticity of the reverse transcription-PCR amplicons was verified by DNA sequencing. Based on the verified sequences, full-length cDNAs were obtained by 5′ or 3′ RACE (Clontech). The corresponding genomic sequences harboring the 12 introns characteristic of PHs (Zhou et al., 2002) were obtained by chromosome walking (Michiels et al., 2003) using genomic DNA extracted using the Qiagen DNeasy Plant Mini kit.
Fast Blue BB Salt and Sugar-Reducing Assay and Immunolocalization of PH
From March and with 2-week intervals until ripening, fruit samples were collected and crude protein extracts prepared from seed tissues as tegument, nucellus plus endosperm (when present), and embryo from the four cultivars Ramillete, Lauranne, S3067, and Gorki as described previously (Sánchez-Pérez et al., 2009). Aliquots of the crude protein extracts were applied to SDS-PAGE and used for Fast Blue BB salt (30 μg of protein)-based total β-glucosidase staining assays using the β-glucosidase substrate, 6-bromo-2-naphthyl-β-d-glucopyranoside, for Coomassie Brilliant Blue staining (167 μg of protein) and for western-blot immunolocalization of PHs (167 μg) following the same protocol as described by Sánchez-Pérez et al. (2009).
Sugar-Reducing Assay
As has been described previously (Sánchez-Pérez et al., 2009), almond tissues from different cultivars (Lauranne and S3067) embedded in agarose and sectioned (70 μm thickness, more than seven replicates per sample) were incubated (more than 5 h, thermo block shaking at 150 rpm, 37°C, total volume of 1 mL) with 25 mm prunasin (Sigma-Aldrich) in 20 mm sodium citrate buffer (pH 6.0). At the end of the incubation period, the Glc released was oxidized by the addition of 0.1% triphenyl tetrazolium chloride in 0.5 n NaOH, resulting in the precipitation of red-colored reduced triphenyl tetrazolium chloride (1 min, 80°C). The sections were washed in 7.5% acetic acid followed by water. Sections incubated in the absence of prunasin were used as negative controls. Imaging for the sugar-reducing assay was obtained using a Leica DMR fluorescence microscope fitted with a Leica DC 300F camera.
Immunolocalization
PH was followed throughout seed development in tegument, nucellus, endosperm, and embryo (mainly cotyledons) using specific PH hapten antibodies obtained as described previously (Sánchez-Pérez et al., 2009). Before analysis with the confocal microscope, the sections were treated with propidium iodide (10 μg mL−1 water) to visualize DNA/RNA structures and the cell wall. Images were obtained using a Leica SPII confocal scanning microscope using 488-nm excitation for fluorescein isothiocyanate detection and 543 nm for propidium iodide; read detection emission was in the range of 500 to 540 nm and 590 to 680 nm, respectively. Stacked images were captured using 0.7-μm increments and deconvolved using Huygens Essential Confocal software 3.6 by Scientific Volume Imaging. Three-dimensional reconstruction of the deconvolved images was accomplished using the Imaris package 6.3.1 (Bitplane). The supplemental videos were made using confocal images and processed with Huygens (Scientific Volume Imaging). Videos were recorded by means of Imaris, at a speed of three frames per second, without compression.
Measurements of the Catabolic Activity of PH
The PH activity of the crude protein extracts obtained as above was determined using prunasin as a substrate. The reaction mixture (total volume of 200 μL) contained 20 nmol of prunasin, crude protein extract (4 μg of protein), and 16 mm citrate buffer, pH 6.0. Blank samples devoid of the crude protein extract were used as a control. Following incubation (10 min, 30°C, 300 rpm shaking) in a closed Eppendorf tube to avoid loss of volatile HCN formed, the enzyme reaction was stopped by freezing the samples in liquid nitrogen and adding NaOH (40 μL of 6 m) to the frozen sample. After thawing, the samples were left at room temperature (20 min), and a 60-μL aliquot was used to for colorimetric determination of HCN content (580- to 750-nm scan; UV/VIS spectrometer, Lambda 800 [Perkin-Elmer]; Halkier and Møller, 1989) by the addition of 100% glacial HOAc (12.5 μL) immediately followed by 50 μL of reagent A (50 mg of succinimide and 125 mg of N-chlorosuccinimide in 50 mL of water) and 50 μL of reagent B (3 g of barbituric acid and 15 mL of pyridine in 35 mL of water). After thorough mixing and incubation (5 min), the cyanide content was determined spectrophotometrically. A standard curve was created with the mixture of 0.2 mm KCN and 1 m NaOH in a total volume of 240 μL.
Identification of Almond Glucosidases by nanoLC-MS/MS Analysis and Database Searches
Protein extracts from the almond cultivars Ramillete, Lauranne, S3067, and Gorki were separated by 12% SDS-PAGE and stained with Coomassie Brilliant Blue. Two gel blocks of each lane spanning the 50- to 75-kD region were excised. The gel blocks were digested with trypsin after reduction and alkylation (Shevchenko et al., 2006). The peptides formed were eluted with 0.1% trifluoroacetic acid, desalted on C18 filters packed in the tip of a pipette tip (Rappsilber et al., 2003), and subjected to fractionation and characterization using a nanoreverse-phase HPLC online device attached to a tandem LTQ-Orbitrap XL mass spectrometer equipped with a nanoelectrospray ion source (ThermoFisher). Chromatographic separation was performed by a Proxeon EASY-nLC system (Proxeon Biosystems) with a 13-cm column of ReproSil-Pur 120 AQ-C18, 3 μm (Dr. Maisch), packed in 100 μm of fused silica fritted with a kasil plug and connected to a nanoelectrospray needle. The peptides were separated with a linear gradient (0%–30% B in 50 min, and 30%–100% B in 5 min at a flow rate of 300 nL min−1). Solvent A was composed of 0.1% formic acid in water and solvent B was composed of 95% acetonitrile, 0.1% formic acid, and 5% water.
Mass spectra were acquired in the positive ion mode. The electrospray voltage was kept at 2.3 kV with an ion-transfer temperature of 200°C without sheath gas flow. Data-dependent acquisition was employed for automated switching between MS mode in the Orbitrap and MS/MS mode in the LTQ. A total of 1,000,000 charges were accumulated in the LTQ prior to injection in the Orbitrap, where a parent ion scan from m/z 300 to 1,800 was performed with a target peak resolution of 60,000 at m/z = 400. The five most abundant ions with charge states above 1 and intensity above 15,000 counts were selected with an isolation width of 2.5 m/z units for MS/MS with collision-induced dissociation in the LTQ. A total of 30,000 charges were accumulated, and the normalized collision energy was set to 35% with activation q = 0.25 and activation time of 30 ms. The m/z values ±10 ppm of precursor ions that were selected for MS/MS were subjected to a dynamic exclusion list for 45 s.
Peak lists in the MGF format were generated by Proteome Discoverer version 1.2 (Thermo Scientific) and searched against all protein sequences of the Prunus taxon in the NCBI nonredundant database. The in-house MASCOT server version 2.3.02 (Matrix Sciences) was employed for searching. The parameters were set as follows: enzyme, trypsin, allowing one missed trypsin cleavage site; variable modifications, oxidation of Met and deamidation of Asp and Glu; fixed modification, carbamidomethyl on Cys. The MS and MS/MS tolerances were set to 5 ppm and 0.8 D, respectively. Only proteins with at least two unique peptides with a significance threshold of P < 0.003 were reported for Orbitrap data. These thresholds led to a false discovery rate of less than 1% as estimated by decoy database searching as integrated in MASCOT.
Signal Peptide Prediction
For the signal peptide prediction, three Web servers were used: SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/), WoLF PSORT (http://wolfpsort.org/), and Target P 1.1 (http://www.cbs.dtu.dk/services/TargetP/). The default conditions described in each of the Web servers were used.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JQ268617, JQ268618, JQ268619, JQ268620, JQ268621, JQ268622, JQ268623, and JQ268624
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Video S1. Immunolocalization studies of PHs (green color) in the embryo of the sweet cultivar Ramillete at 168 JD.
Supplemental Video S2. Immunolocalization studies of PHs (green color) in the embryo of the sweet cultivar Ramillete at 182 JD.
Supplementary Material
Acknowledgments
We thank Teresa Cremades Rosado for technical help and Dr. M. Acosta Echevarría for help in the discussion of this work. Imaging data were achieved at the Center for Advanced Bioimaging (University of Copenhagen).
References
- Andréasson E, Bolt Jørgensen L, Höglund A-S, Rask L, Meijer J. (2001) Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol 127: 1750–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrázola G. (2002) Análisis de glucósidos cianogénicos en variedades de almendro: implicaciones en la mejora genética. PhD thesis. Universidad de Alicante, Alicante, Spain [Google Scholar]
- Ben-Yehoshua S, Conn EE. (1964) Biosynthesis of prunasin, the cyanogenic glucoside of peach. Plant Physiol 39: 331–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn EE. (1980) Cyanogenic compounds. Annu Rev Plant Physiol 31: 433–451 [Google Scholar]
- Dicenta F, Martínez-Gómez P, Grané N, Martín ML, León A, Cánovas JA, Berenguer V. (2002) Relationship between cyanogenic compounds in kernels, leaves, and roots of sweet and bitter kernelled almonds. J Agric Food Chem 50: 2149–2152 [DOI] [PubMed] [Google Scholar]
- Dicenta F, Martínez-Gómez P, Ortega E. (2000) Cultivar pollinizer does not affect almond flavor. HortScience 35: 1153–1154 [Google Scholar]
- Dourado F, Barros A, Mota M, Coimbra MA, Gama FM. (2004) Anatomy and cell wall polysaccharides of almond (Prunus dulcis D. A. Webb) seeds. J Agric Food Chem 52: 1364–1370 [DOI] [PubMed] [Google Scholar]
- Franks TK, Yadollahi A, Wirthensohn MG, Guerin JR, Kaiser BN, Sedgley M, Ford CM. (2008) A seed coat cyanohydrin glucosyltransferase is associated with bitterness in almond (Prunus dulcis) kernels. Funct Plant Biol 35: 236–246 [DOI] [PubMed] [Google Scholar]
- Frehner M, Scalet M, Conn EE. (1990) Pattern of the cyanide-potential in developing fruits. Plant Physiol 94: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardi C, Blando F, Santino A, Zacheo G. (2001) Purification and characterisation of a β-glucosidase abundantly expressed in ripe sweet cherry (Prunus avium L.) fruit. Plant Sci 160: 795–805 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Møller BL. (1989) Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol 90: 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker JS, Buttrose MS. (1980) Development of the almond nut (Prunus dulcis (Mill.) D.A. Webb): anatomy and chemical composition of fruit parts from anthesis to maturity. Ann Bot (Lond) 46: 313–321 [Google Scholar]
- Heppner MJ. (1923) The factor for bitterness in the sweet almond. Genetics 8: 390–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenrich R, Trompetter I, Bak S, Olsen CE, Møller BL, Piotrowski M. (2007) Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc Natl Acad Sci USA 104: 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Kanagaraj A, Verma D, Lange T, Daniell H. (2011) Release of hormones from conjugates: chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol 155: 222–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PR, Møller BL, Hoj PB. (1999) The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor: isolation, cloning, heterologous expression, and substrate specificity. J Biol Chem 274: 35483–35491 [DOI] [PubMed] [Google Scholar]
- Joobeur T, Viruel MA, de Vicente MC, Jáuregui B, Ballester J, Dettori MT, Verde I, Truco MJ, Messeguer R, Battle I, et al. (1998) Construction of a saturated linkage map for Prunus using an almond × peach F2 progeny. Theor Appl Genet 97: 1034–1041 [Google Scholar]
- Kester DE, Asay RN. (1975) Almonds. In J Janick, JN Moore, eds, Advances in Fruit Breeding. Purdue University Press, Lafayette, IN, pp 387–419 [Google Scholar]
- Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E. (2007) Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J Exp Bot 58: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Martínez-Gómez P, Sánchez-Pérez R, Dicenta F. (2008) Fruit and seed development in almond in relation to flowering and ripening dates for fresh market supply. J Am Pomol Soc 62: 82–86 [Google Scholar]
- McCarty CD, Leslie JW, Frost HB. (1952) Bitterness of kernels of almond × peach hybrids and their parents. Proc Am Soc Hortic Sci 59: 254–258 [Google Scholar]
- McCaskill A, Turgeon R. (2007) Phloem loading in Verbascum phoeniceum L. depends on the synthesis of raffinose-family oligosaccharides. Proc Natl Acad Sci USA 104: 19619–19624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzer C, Favre-Bonvin J. (1961) Sur la biogenèse du glucoside cyanogénétique des feuilles de laurier-cerise (Prunus lauro-cerasus). C R Acad Sci (Paris) 253: 1072–1074 [Google Scholar]
- Michiels A, Tucker M, Van den Ende W, Laere AV. (2003) Chromosomal walking of flanking regions from short known sequences in GC-rich plant genomic DNA. Plant Mol Biol Rep 21: 295–302 [Google Scholar]
- Møller BL, Seigler DS. (1991) Biosynthesis of cyanogenic glycosides, cyanolipids and related compounds. In BK Singh, ed, Plant Amino Acids, Biochemistry and Biotechnology. Marcel Dekker, New York, pp 563–609 [Google Scholar]
- Nielsen KA, Tattersall DB, Jones PR, Møller BL. (2008) Metabolon formation in dhurrin biosynthesis. Phytochemistry 69: 88–98 [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schönfelder S, Weiler EW. (2001) The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode beta-cyano-L-alanine hydratase/nitrilase. J Biol Chem 276: 2616–2621 [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Volmer JJ. (2006) Cyanide metabolism in higher plants: cyanoalanine hydratase is a NIT4 homolog. Plant Mol Biol 61: 111–122 [DOI] [PubMed] [Google Scholar]
- Poulton JE. (1990) Cyanogenesis in plants. Plant Physiol 94: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JE, Li CP. (1994) Tissue level compartmentation of (R)-amygdalin and amygdalin hydrolase prevents large-scale cyanogenesis in undamaged Prunus seeds. Plant Physiol 104: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Howad W, Dicenta F, Arús P, Martínez-Gómez P. (2007) Mapping major genes and quantitative trait loci controlling agronomic traits in almond. Plant Breed 126: 310–318 [Google Scholar]
- Sánchez-Pérez R, Howad W, Garcia-Mas J, Arús P, Martínez-Gómez P, Dicenta F. (2010) Molecular markers for bitterness in almond. Tree Genet Genomes 6: 237–245 [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Motawia MS, Dicenta F, Møller BL. (2009) Tissue and cellular localization of individual β-glycosidases using a substrate-specific sugar reducing assay. Plant J 60: 894–906 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Olsen CE, Dicenta F, Møller BL. (2008) Bitterness in almonds. Plant Physiol 146: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmar D. (1994) Translocation of cyanogenic glucosides in cassava. Acta Hortic 375: 61–67 [Google Scholar]
- Shabalina SA, Spiridonov AN, Spiridonov NA, Koonin EV. (2010) Connections between alternative transcription and alternative splicing in mammals. Genome Biol Evol 2: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Suelves M, Puigdomènech P. (1998) Molecular cloning of the cDNA coding for the (R)-(+)-mandelonitrile lyase of Prunus amygdalus: temporal and spatial expression patterns in flowers and mature seeds. Planta 206: 388–393 [DOI] [PubMed] [Google Scholar]
- Swain E, Li CP, Poulton JE. (1992) Tissue and subcellular localization of enzymes catabolizing (R)-amygdalin in mature Prunus serotina seeds. Plant Physiol 100: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Poulton JE. (1994a) Immunocytochemical localization of prunasin hydrolase and mandelonitrile lyase in stems and leaves of Prunus serotina. Plant Physiol 106: 1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain E, Poulton JE. (1994b) Utilization of amygdalin during seedling development of Prunus serotina. Plant Physiol 106: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuenca MC. (1972) Chilling requirements in almond. in Spanish. Anal Estacion Exp Aula Dei 11: 325–329 [Google Scholar]
- Wirthensohn MG, Chin WL, Franks TK, Baldock G, Ford CM, Sedgley M. (2008) Characterizing the flavor phenotypes of almond (Prunus dulcis Mill.) kernels. J Hortic Sci Biotechnol 83: 462–468 [Google Scholar]
- Zhou J, Hartmann S, Shepherd BK, Poulton JE. (2002) Investigation of the microheterogeneity and aglycone specificity-conferring residues of black cherry prunasin hydrolases. Plant Physiol 129: 1252–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.