Abstract
We previously identified an enzyme, phosphatidylcholine diacylglycerol cholinephosphotransferase (PDCT), that plays an important role in directing fatty acyl fluxes during triacylglycerol (TAG) biosynthesis. The PDCT mediates a symmetrical interconversion between phosphatidylcholine (PC) and diacylglycerol (DAG), thus enriching PC-modified fatty acids in the DAG pool prior to forming TAG. We show here that PDCT is required for the efficient metabolism of engineered hydroxy fatty acids in Arabidopsis (Arabidopsis thaliana) seeds. When a fatty acid hydroxylase (FAH12) from castor (Ricinus communis) was expressed in Arabidopsis seeds, the PDCT-deficient mutant accumulated only about half the amount of hydroxy fatty acids compared with that in the wild-type seeds. We also isolated a PDCT from castor encoded by the RcROD1 (Reduced Oleate Desaturation1) gene. Seed-specific coexpression of this enzyme significantly increased hydroxy fatty acid accumulation in wild type-FAH12 and in a previously produced transgenic Arabidopsis line coexpressing a castor diacylglycerol acyltransferase 2. Analyzing the TAG molecular species and regiochemistry, along with analysis of fatty acid composition in TAG and PC during seed development, indicate that PDCT acts in planta to enhance the fluxes of fatty acids through PC and enrich the hydroxy fatty acids in DAG, and thus in TAG. In addition, PDCT partially restores the oil content that is decreased in FAH12-expressing seeds. Our results add a new gene in the genetic toolbox for efficiently engineering unusual fatty acids in transgenic oilseeds.
Vegetable oils consist principally of triacylglycerols (TAGs) and have wide applications in human consumption and more recently in renewable biofuels and industrial materials (Gunstone, 1998; Durrett et al., 2008; Napier and Graham, 2010; Lu et al., 2011). The fatty acid (FA) composition in TAG determines the quality and thus the uses of plant oils. While edible oils should contain as little saturated FAs as possible and a significant proportion of polyunsaturated fatty acids (PUFAs; Riediger et al., 2009), the industrial oils require low PUFAs for desirable oxidative stability or high homogeneity of certain FAs (Dyer and Mullen, 2008; Dyer et al., 2008; Pinzi et al., 2009). For effective improvements of plant oils, we need to understand the fundamental aspects of how plant FAs are synthesized and accumulated in seed oils.
In plants, FA synthesis occurs exclusively in plastids and produces mostly oleic acid (18:1; number of carbons:number of double bonds) and a small amount of palmitic acid (16:0) and stearic acid (18:0) that are esterified to the acyl carrier protein (Ohlrogge and Browse, 1995). In oilseeds, these FAs are almost entirely (more than 95%) exported outside the plastid and are converted into acyl-CoA to be used for glycerolipid synthesis in the endoplasmic reticulum (ER; Browse and Somerville, 1991; Ohlrogge and Browse, 1995). In Arabidopsis (Arabidopsis thaliana) seeds, 18:1-CoA may be elongated into 20:1-CoA to 22:1-CoA by a fatty acid elongase (FAE1) on the ER membrane (Kunst et al., 1992; Kunst and Samuels, 2003) or may be incorporated into the membrane lipid phosphatidylcholine (PC) to be desaturated to linoleic acid (18:2) and subsequently to linolenic acid (18:3) by the oleate desaturase FAD2 and the linoleate desaturase FAD3, also located on the ER (Browse et al., 1993; Sperling et al., 1993; Okuley et al., 1994). These reactions are the major causes of the wide variations of FA composition in TAG of many oilseeds.
The de novo PC synthesis pathway incorporates FAs through diacylglycerol (DAG) by the sequential sn-1 and sn-2 acylations of glycerol-3-phosphate followed by the dephosphorylation of phosphatidic acid (Kennedy, 1961). DAG is converted into PC by a reversible CDP-choline:diacylglycerol cholinephosphotransferase (Slack et al., 1983; Goode and Dewey, 1999). Recent metabolic analyses indicate that the majority of newly synthesized FAs may enter PC through a process called acyl editing, which involves a very rapid cyclic deacylation and reacylation of PC (Williams et al., 2000; Bates et al., 2007, 2009). The genes involved in this process have not been identified, but the enzymes may include the acyl-CoA:lysophosphatidylcholine acyltransferases and phospholipase A2s. The PC-acyl editing cycle incorporates nascent 18:1 into PC and releases modified (e.g. polyunsaturated) 18:2 and 18:3 from PC. This process generates an acyl-CoA pool that is enriched in PUFAs to be utilized for glycerolipid synthesis in the ER.
The TAG synthesis is completed by the acylation of the sn-3 hydroxyl of DAG. This step may be catalyzed by an acyl-coenzyme A:diacylglycerol acyltransferase (DGAT) using the acyl-CoA pool (Hobbs et al., 1999) or by a phospholipid:diacylglycerol acyltransferase (PDAT) that directly transfers the sn-2 acyl group from PC onto DAG to form TAG (Dahlqvist et al., 2000). The DAG precursor may be from de novo synthesis through a series of acylations of glycerol-3-phosphate or may be derived from PC. Recent results demonstrate that the latter provides the predominant source (Bates and Browse, 2011). The high-flux exchange between the PC and DAG pools appears to be mainly catalyzed by the phosphatidylcholine diacylglycerol cholinephosphotransferase (PDCT), although the reverse reaction of CDP-choline:diacylglycerol cholinephosphotransferase may also have some role. The PDCT enzyme, discovered in Arabidopsis encoded by the Reduced Oleate Desaturation1 (ROD1) locus, catalyzes the interconversion between DAG and PC by phosphocholine head group exchange (Lu et al., 2009). Through reactions of PDCT, the acyl groups on DAG enter PC and then return to DAG after they are desaturated or otherwise modified on PC.
This latest model of TAG synthesis described above highlights the central roles of PC as the source of greatly diversified FAs in membrane and storage lipids. This has also been reinforced by results in transgenic plants producing some modified fatty acids (mFAs), such as those containing hydroxy, epoxy, conjugated diene, or cyclopropane groups. These mFAs are synthesized by the FAD2-related enzymes acting on the sn-2 acyl groups on PC (Drexler et al., 2003; Jaworski and Cahoon, 2003; Lu et al., 2006; Napier and Graham, 2010). Despite high levels of mFAs occurring in many native species, attempts to produce high yields of mFAs in a crop species, or in Arabidopsis as a model, have largely been unsuccessful (Jaworski and Cahoon, 2003; Lu et al., 2006; Lu and Kang, 2008). It was shown that inefficient removal of mFAs from PC and incorporation into TAG was responsible for very low contents of conjugated FAs in TAG in transgenic Arabidopsis seeds (Cahoon et al., 2006). This notion was further supported by an experiment showing a castor (Ricinus communis) PDAT that facilitated the transfer of hydroxy fatty acids (HFAs) from PC into TAG in transgenic Arabidopsis (van Erp et al., 2011). The HFA ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid; 18:1OH) constitutes approximately 90% of total FAs in castor seeds. The 18:1OH is produced by the hydroxylation of 18:1 that is esterified to the sn-2 position of PC (Bafor et al., 1991) by the oleate Δ12-hydroxylase (FAH12; van de Loo et al., 1995). Seed-specific expression of FAH12 in Arabidopsis or the oilseed crop camelina (Camelina sativa) only resulted in up to approximately 17% total HFAs including 18:1OH and its derivatives (Broun and Somerville, 1997; Smith et al., 2003; Lu et al., 2006; Lu and Kang, 2008). Coexpressing a PDAT (RcPDAT1A) from castor with FAH12 significantly increased the HFA levels up to approximately 27% (van Erp et al., 2011). Analyzing the FA composition in different lipids during seed development indicated that increased HFA content in TAG was accompanied by a significant decrease of HFAs on PC (van Erp et al., 2011). This result is in agreement with the role of PDAT transferring the HFAs from sn-2 PC onto DAG to form TAG and supports the proposal that PDAT plays an important role in castor for accumulating a high level of HFAs (Bafor et al., 1991; Dahlqvist et al., 2000). In addition, coexpressing FAH12 with RcDGAT2 greatly increased HFA accumulation and also resulted in lowered HFA on PC during seed development, supporting a possible relationship between the efficient removal of mFAs from PC and their incorporation into TAG (Burgal et al., 2008; van Erp et al., 2011).
We now recognize that the flux of acyl groups through PC poses a bottleneck for the accumulation of mFAs in transgenic seeds (Bates and Browse, 2011; van Erp et al., 2011). The PDCT catalyzes the shuffling of acyl groups between PC and DAG, thus providing a potential mechanism for the removal of PUFAs or other mFAs on PC for them to be incorporated into TAG. We show here that PDCT is required for efficient HFA accumulation in transgenic Arabidopsis. Very low levels of HFAs were detected when FAH12 was expressed in seeds of the Arabidopsis rod1 mutant, which contains a lesion in PDCT (Lu et al., 2009). A castor PDCT enzyme encoded by the RcROD1 gene increased HFA levels when coexpressed with FAH12. Our results demonstrate that PDCT plays an important role in seed lipid metabolism and provides a useful tool for engineering HFA and possibly other mFAs as well in transgenic plants.
RESULTS
PDCT Is Required for Efficient HFA Accumulation
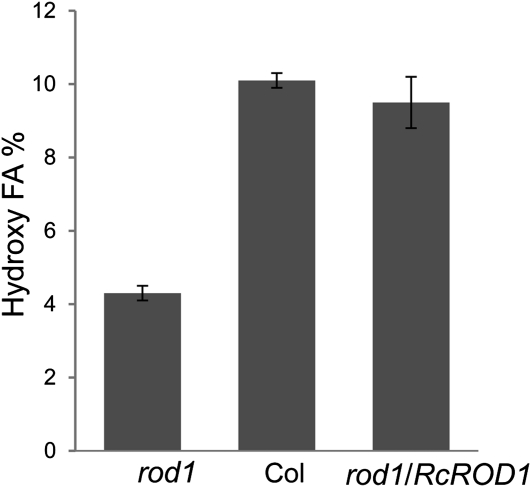
The castor FAH12 acts on PC to convert 18:1 into 18:1OH, which will then be channeled into TAG through several possible pathways (van Erp et al., 2011). The PDCT may play an important role in this process by turning over PC into HFA-containing DAG. To test this possibility, we expressed FAH12 in the Arabidopsis rod1 mutant and the wild type (ecotype Columbia [Col-0]) under the control of the seed-specific phaseolin promoter as described previously (Lu and Kang, 2008). The plasmid used for transformation contained a DsRed marker, and transgenic T1 seeds were selected by screening for red fluorescence. T1 plants were grown, and 10 to 15 transgenic lines from each background were selected based on a ratio of fluorescent to nonfluorescent seeds of 3:1 in the T2 seeds, which suggested single insertions for the transgene FAH12. The FA composition of transgenic red seeds was determined by analyzing fatty acid methyl esters (FAMEs) using gas chromatography (GC). Four novel FAs were detected compared with wild-type seeds, which corresponded to the HFAs previously identified as 18:1OH, densipolic acid (18:2OH), lesquerolic acid (20:1OH), and a small amount of auricolic acid (20:2OH; Broun and Somerville, 1997; Smith et al., 2003). The total HFA contents in each of the transgenic lines ranged from 4% to 5% in the rod1 mutant, compared with approximately 10% in the wild-type background (Fig. 1). Three independent transgenic lines each from the rod1 and wild-type backgrounds were raised to reach homozygosity for the transgene. Analysis of FA composition in the T4 seeds confirmed the results in the T2 seeds (Table I). These results indicated that only about half of the HFAs were incorporated into TAG in the rod1 mutant compared with the wild type and that this inefficiency was presumably caused by the loss of the PDCT function in the rod1 mutant.
Figure 1.
HFA levels in Arabidopsis seeds transformed with the castor FAH12. The lines are rod1 mutant, Col (wild type), and rod1 transformed with RcROD1. The data represent averages ± se from 10 to 15 independent T2 lines.
Table I. FA composition of transgenic Arabidopsis seeds of T4 lines expressing castor RcFAH12 and RcROD1.
The transgenic lines of these genes were obtained from independent transformation events. Data represent measurements of three homozygous lines (means ± se).
| Line | FA Composition |
Total HFAa | ||||||||
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:1 | 18:1OH | 18:2OH | 20:1OH | ||
| wt % of total | ||||||||||
| Col-0 | 8.0 ± 0.1 | 2.7 ± 0.1 | 15.8 ± 0.2 | 30.6 ± 0.2 | 18.7 ± 0.1 | 21.3 ± 0.1 | ||||
| rod1 | 7.9 ± 0.1 | 2.5 ± 0.1 | 33.5 ± 0.2 | 15.5 ± 0.1 | 13.1 ± 0.1 | 25.3 ± 0.1 | ||||
| rod1-RcROD1 | 7.2 ± 0.5 | 3.0 ± 0.5 | 16.2 ± 1.7 | 29.7 ± 1.2 | 17.6 ± 0.4 | 20.7 ± 0.9 | ||||
| rod1-FAH12 | 12.4 ± 0.6 | 5.2 ± 0.2 | 38.3 ± 0.8 | 14.5 ± 0.3 | 5.9 ± 0.1 | 15.4 ± 0.5 | 2.8 ± 0.1 | 1.2 ± 0.1 | 1.5 ± 0.1 | 5.5 ± 0.1 |
| rod1-FAH12/RcROD1 | 10.1 ± 0.1 | 4.8 ± 0.2 | 25.5 ± 0.3 | 24.3 ± 0.2 | 11.6 ± 0.5 | 11.1 ± 1.5 | 6.2 ± 0.2 | 3.0 ± 0.4 | 0.6 ± 0.1 | 9.7 ± 0.6 |
| Col-FAH12 | 10.1 ± 0.4 | 4.7 ± 0.3 | 19.6 ± 0.4 | 22.2 ± 0.2 | 10.7 ± 0.9 | 17.9 ± 0.2 | 6.3 ± 0.1 | 2.1 ± 0.1 | 1.6 ± 0.1 | 9.9 ± 0.3 |
| Col-FAH12/RcROD1 | 10.0 ± 0.5 | 5.1 ± 0.3 | 24.2 ± 0.7 | 17.5 ± 1.2 | 5.2 ± 0.1 | 14.9 ± 0.6 | 12.7 ± 0.2 | 4.6 ± 0.5 | 2.6 ± 0.1 | 19.9 ± 0.5 |
Excluding small amounts of 20:2OH.
A Castor ROD1 with PDCT Activity
The results presented above indicated that PDCT is required for an efficient metabolism of HFAs in transgenic Arabidopsis seeds. This raised the possibility that overexpressing PDCT may increase HFA accumulation in TAG by facilitating the conversion of HFA-containing PC into DAG. Considering that a PDCT enzyme from castor may have been coevolved with FAH12 to contribute to the high content of ricinoleic acid in castor oil (Lu et al., 2006; van Erp et al., 2011), we decided to use the castor PDCT in our transgenic experiments. The Arabidopsis PDCT is encoded by the ROD1 (At3g15820; AtROD1) gene (Lu et al., 2009). We identified a sequence from an EST collection of castor seeds (E. Cahoon, personal communication) that showed homology to AtROD1. The full-length cDNA sequence of this putative castor ROD1, designated RcROD1, was compiled by comparing the castor cDNA (Lu et al., 2007) and genomic (http://castorbean.jcvi.org/index.php) sequences. The deduced RcROD1 (EMBL accession no. EQ973818.1; UniprotKB accession no. B9RV74) contains 285 amino acids, as shown in Supplemental Figure S1. Despite great variations between the two sequences of RcROD1 and AtROD1, especially at the N and C termini, the central regions have over 70% identical residues. The putative transmembrane domains and the catalytic triad (His, His, and Asp) are also conserved in both ROD1 sequences (Supplemental Fig. S1).
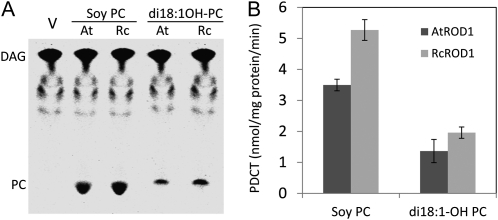
To determine whether RcROD1 possesses PDCT activity, the RcROD1 cDNA sequences were cloned into the yeast expression vector p424 GPD and transinfected into Saccharomyces cerevisiae strain HJ091 (cpt1::LEU2 ept1-) to extract microsomal proteins for enzyme activity assay, as described previously (Lu et al., 2009). When incubated with dioleoyl-[14C]glycerol and nonradioactive soybean (Glycine max) PC, microsomal preparations from HJ091 cells expressing RcROD1 and AtROD1 both were able to synthesize radiolabeled PC. No activity was detected in the control microsomes that were transformed with the empty vector (Fig. 2A). These assays indicate that RcROD1, like AtROD1, has PDCT activity that synthesizes [14C]PC by transferring the phosphocholine head group from PC to [14C]DAG. In our assay conditions, RcROD1 had even higher PDCT activity, at approximately 5.3 nmol mg−1 protein min−1, compared with AtROD1, at approximately 3.5 nmol mg−1 protein min−1 (Fig. 2B). To test whether the castor PDCT prefers HFA-containing substrates, we incubated yeast microsomal preparations of RcROD1 and AtROD1 with dioleoyl-[14C]glycerol and di18:1OH-PC prepared from castor bean (a gift from S. Stymne). As shown in Figure 2, PDCT activity was detected for both enzymes using these PC substrates, but lower activity was found in reactions incubated with di18:1OH-PC. The PDCT activity of AtROD1 and RcROD1 was detected at 1.4 and 2 nmol mg−1 protein min−1, respectively. These results were not indicative of 18:1OH-PC as the preferred substrate for either PDCTs from castor or Arabidopsis in our assay conditions.
Figure 2.
RcROD1 encodes a PDCT enzyme. A, Radio-TLC image of PDCT assays. Microsomes from HJ091 S. cerevisiae cells transfected with p424-AtROD1 (At) or p424-RcROD1 (Rc) were incubated with [14C]glycerol-di18:1-DAG and Soy PC or 18:1OH-PC (1 mm). V, Control with the empty p424 vector. B, PDCT activity measured as the amount of radioactive PC formed. Data represent means of three independent reactions ± se.
The function of RcROD1 as a PDCT was further confirmed by its ability to restore the seed FA composition in the Arabidopsis rod1 mutant to the wild-type level when the RcROD1 cDNA was introduced into the mutant (Table I).
RcROD1 Enhances HFA Accumulation in TAG in Arabidopsis
To determine whether the overexpression of PDCT may increase HFA accumulation, the castor genes RcROD1 and FAH12 were coexpressed in the rod1 mutant and wild-type Col-0 plants under the control of seed-specific promoters (Supplemental Fig. S2). As described above, transgenic seeds were selected based on DsRed expression, and the red T2 seeds were analyzed for FA composition. The levels of HFA content in the rod1 transformants containing both RcROD1 and FAH12 were similar to those in the wild-type background expressing FAH12 alone (Fig. 1; Table I). This is consistent with the results that RcROD1 is able to restore FA composition of the rod1 mutant to the wild-type level. Interestingly, coexpression of RcROD1 with FAH12 in wild-type Arabidopsis increased the HFA content from approximately 10% to approximately 20%, at the expense of mostly 18:2 and 18:3 PUFAs as well as 20:1. The data presented in Table I are from the T4 seeds of three homozygous lines as determined by the segregation ratios of the DsRed marker. These results demonstrated that RcROD1 was able to significantly increase HFA accumulation when coexpressed with the castor FAH12 in Arabidopsis seeds, and this increase was stably inherited over multiple generations.
RcROD1 Increases HFA TAGs and Changes Their Molecular Species
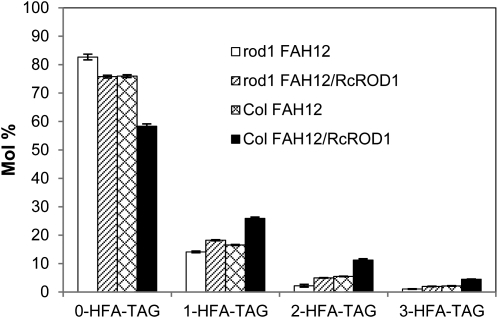
To gain insight in the biochemical mechanisms of the PDCT effects on HFA metabolism in transgenic seeds, we analyzed the HFA-containing TAG and its regiochemical composition in our transgenic lines. There are four molecular species of TAG in FAH12 transgenic Arabidopsis seeds, namely 0-, 1-, 2-, and 3-HFA TAG, according to the number of HFAs esterified onto the glycerol backbone. These TAGs are present in all FAH12-expressing lines in the rod1 mutant or wild-type backgrounds. All classes of HFA TAGs are significantly reduced in the rod1-FAH12 seeds compared with FAH12 expression in wild-type or RcROD1-complemented rod1 seeds. The RcROD1 overexpression in the Col-FAH12 seeds significantly lowers the 0-HFA TAG proportion and increases the amount of all HFA TAG species, especially the 2- and 3-HFA TAGs (11.3% and 4.5%, respectively), which are more than doubled compared with Col-FAH12 (5.5% and 2.1%, respectively; Fig. 3). These results suggest that RcROD1 effectively metabolizes HFAs into TAG in FAH12-expressing Arabidopsis seeds.
Figure 3.
Molecular species composition of HFA-containing TAGs of rod1-FAH12, rod1-FHA12/RcROD1, Col-FAH12, and Col-FAH12/RcROD1 seeds. 0-, 1-, 2-, and 3-HFA represent TAG molecular species with zero, one, two, and three HFAs, respectively (no stereochemistry implied). The HFAs represent the sum of ricinoleate (18:1-OH) and densipolate (18:2-OH) and a small amount of lesquerolic acid (20:1-OH). The data represent averages of three replicates ± se.
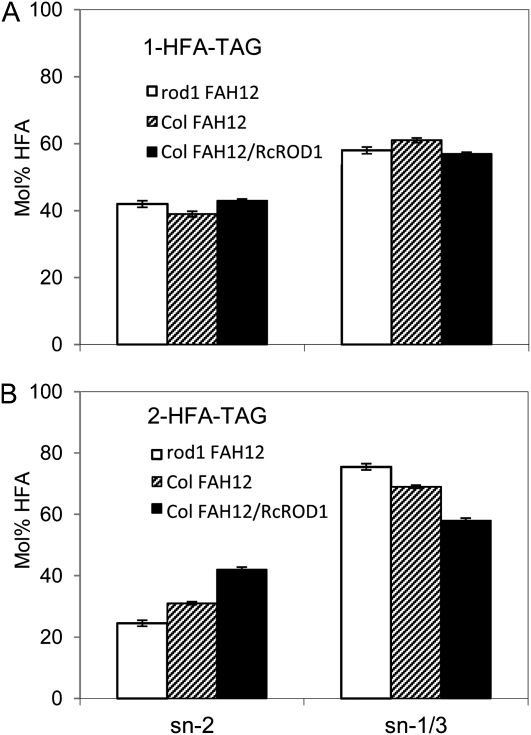
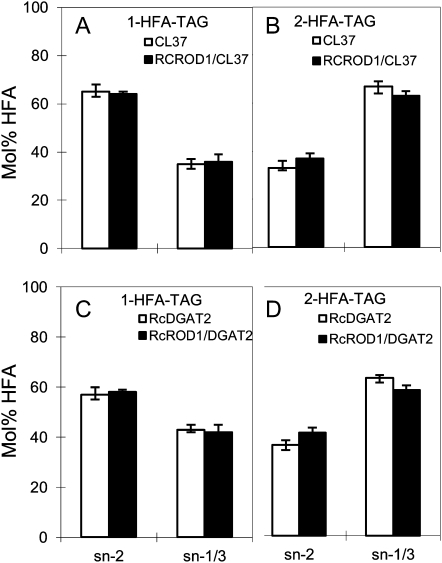
We further analyzed the regiochemical composition of HFA-containing TAGs from the above transgenic lines. Figure 4 shows the HFAs as a percentage of FAs at the sn-2 versus the sn-1/3 position for individual 1- and 2-HFA TAG molecular species. In 1-HFA TAG, PDCT did not significantly affect the percentage of HFAs in the sn-2 and sn-1/3 positions. However, PDCT caused dramatic changes in the 2-HFA TAGs. Compared with Col-FAH12 lines, the percentage of HFAs in the rod1-FAH12 lines decreased in the sn-2 position (from 31% to 24.5%) and increased in the sn-1/3 position (from 69% to 75.5%). Overexpression of RcROD1 in the Col-FAH12 line increased HFA percentage in the sn-2 position from 31% to 42% and decreased it in the sn-1/3 position from 69% to 58% (Fig. 4B). Together with the data in Figure 3, these results indicate that PDCT activity of RcROD1 effectively mobilizes HFAs from PC into DAG and increases HFA contents in TAG of the transgenic Arabidopsis seeds. These results are expected because the hydroxylase works on sn-2 PC, and thus we would expect PDCT to produce sn-2 HFA DAG and thus more sn-2 HFA TAG.
Figure 4.
Regiochemical analysis of 1- and 2-HFA TAG species in rod1-FAH12, Col-FAH12, and Col-FAH12/RcROD1 seeds: mol % HFA at the sn-2 position compared with the sn-1/3 positions in 1- and 2-HFA TAG. A, 1-HFA TAG. B, 2-HFA TAG. The data represent averages of three replicates ± se.
Changes of HFA in PC and TAG during Seed Development
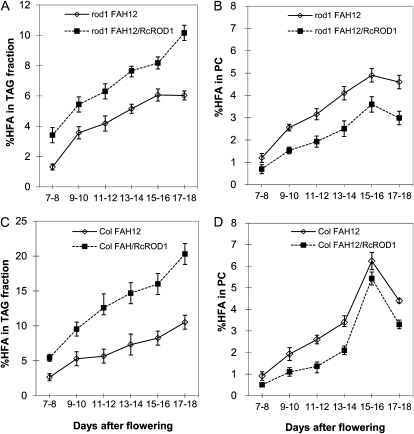
The FAH12 hydroxylase acts on 18:1 at the sn-2 position of the membrane lipid PC (Bafor et al., 1991). Castor seeds can efficiently transfer HFAs into TAG while keeping a very low level of HFAs in membrane lipids. Since PDCT catalyzes the interconversion of PC and DAG, we reasoned that the increased HFA accumulation in TAG by expressing RcROD1 could have resulted from efficient HFA migration from PC to DAG, which was then turned into TAG. Therefore, we measured the HFA contents in PC and TAG in the above transgenic lines during TAG synthesis. Total lipids were extracted from seed samples at 7 to 18 d after flowering (DAF) and separated by thin-layer chromatography (TLC). The PC and neutral lipid (TAG) fractions were recovered from the TLC plates and analyzed by GC. HFA accumulation in TAG increased during seed development and reached the maximum levels of 5% and 10% in 15 to 18 DAF in the rod1 and wild-type backgrounds, respectively (Fig. 5, A and C). As was observed previously in transgenic Arabidopsis expressing FAH12 (Thomaeus et al., 2001), we also found that HFAs transiently accumulated in PC during seed development and reached a maximum of 5% to 6% in the mid to late stage of seed development (approximately 15 DAF; Fig. 5, B and D). There was no significant difference between the rod1 mutant and Col backgrounds.
Figure 5.
Changes in the percentage of HFAs in PC and TAG in transgenic Arabidopsis lines during seed development. A and B, rod1 mutant transformed with FAH12 alone or together with RcROD1. C and D, Wild-type Col transformed with FAH12 or together with RcROD1. The data represent averages of three replicates ± se.
We then analyzed the RcROD1-expressing lines for FA composition in PC and TAG during seed development. As shown in Figure 5, HFA contents in PC decreased significantly in both rod1 and wild-type backgrounds and were apparently associated with increased levels of HFA in TAG. At 15 to 16 DAF, PC in rod1-FAH12 seeds contained 4.9% HFA compared with 3.6% in the seeds of rod1-FAH12-RcROD1 lines (Fig. 5B). At the same time, HFA contents in TAG increased from 6.1% to 8.1% (Fig. 5A). Similarly, PC in Col-FAH12 seeds contained 6.2% HFAs compared with 5.4% in Col-FAH12-RcROD1 seeds (Fig. 5D). The HFAs in TAG increased from 8.3% to 16% during the same period of seed development (Fig. 5C).
Expression of RcROD1 in the CL37 and CL7-RcDGAT2 Backgrounds
It appears that overexpressing RcROD1 may significantly increase the HFA accumulation in transgenic Arabidopsis seeds. This could have resulted from the enhanced conversion of PC into HFA-containing DAG. It has been shown previously that the castor enzyme RcDGAT2 prefers such DAG molecules and dramatically increases HFA accumulation in TAG when coexpressed with FAH12 in Arabidopsis seeds (Burgal et al., 2008). To explore the possibility of synergistic effects of RcROD1 and RcDGAT2 on HFA levels, the above RcROD1 construct was transformed into the CL7-RcDGAT2 line, which was produced by cotransformation of RcDGAT2 and FAH12 on the Arabidopsis fae1 background (Burgal et al., 2008). As a control, RcROD1 was also transformed into the FAH12-containing CL37 line, which was produced on the same background (Lu et al., 2006). Because of the mutation at the FAE1 locus in the fae1 mutant (Kunst et al., 1992), the CL37 and CL7-RcDGAT2 lines do not accumulate very long chain (more than 18) FAs, including 20:1 and 20:1OH, and the seed oils contain 17% and 25% HFAs, respectively, in our analysis (Table II).
Table II. FA composition of transgenic Arabidopsis seeds expressing FAH12 and RcROD1.
CL7 and CL37 are the FHA12-transformed lines in the Arabidopsis fae1 mutant background. The transgenic lines of these genes were obtained from independent transformation events. Data represent measurements of three homozygous lines (means ± se).
| Line | FA Composition |
Total HFA | ||||||
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 18:1OH | 18:2OH | ||
| wt % of total | ||||||||
| CL37 | 12.2 ± 0.3 | 5.6 ± 0.5 | 38.3 ± 1.2 | 19.8 ± 0.9 | 5.3 ± 0.4 | 13.1 ± 0.4 | 4.1 ± 0.2 | 17.2 ± 0.2 |
| CL37-RcROD1 | 11.6 ± 1.1 | 5.7 ± 0.5 | 33.7 ± 1.3 | 19.6 ± 0.9 | 6.1 ± 0.3 | 18.3 ± 0.6 | 4.9 ± 0.3 | 23.3 ± 0.6 |
| CL7-RcDGAT2 | 10.6 ± 0.3 | 5.4 ± 0.4 | 30.1 ± 1.5 | 20.7 ± 1.0 | 6.2 ± 0.4 | 20.1 ± 0.6 | 4.6 ± 0.3 | 24.7 ± 0.6 |
| CL7-RcDGAT2/RcROD1 | 13.1 ± 0.3 | 4.8 ± 0.2 | 23.1 ± 0.5 | 21.4 ± 0.5 | 7.6 ± 0.3 | 23.1 ± 0.4 | 5.4 ± 0.3 | 28.5 ± 1.0 |
The RcROD1 transgenic plants in these backgrounds were grown, and 11 and seven homozygous lines in CL37 and CL7-RcDGAT2 were obtained for analysis by GC to determine their FA composition. The results are shown in Table II, which compares the HFA levels in the CL7-RcDGAT2-RcROD1 triple transgenic line and the CL37-RcROD1 double transgenic line with the CL37 control. Expression of RcROD1 increased HFA levels in the CL37 background from 17% to 22% and further increased in CL7-RcDGAT2 from 25% to 28.5%. These results clearly indicated that RcROD1 may significantly increase HFA accumulation in transgenic Arabidopsis seeds.
We also conducted regiochemical analysis of HFA-containing TAGs in CL37-RcROD1 and CL7-RcDGAT2-RcROD1 and compared CL37 and CL7-RcDGAT2. Similar to the results shown in Figure 4, we found that the percentage of HFAs in the sn-2 position of the 2-HFA TAGs increased in the RcROD1 lines in both CL37 and CL7-RcDGAT2 backgrounds (Fig. 6). These results strongly support the role of the PDCT in transferring HFAs from PC to DAG by phosphocholine head group exchange.
Figure 6.
Regiochemical analysis of 1- and 2-HFA TAG species in CL37, CL37-RcROD1, CL7-RcDGAT2, and CL7-RcDGAT2/RcROD1 seeds: mol % HFA at the sn-2 position compared with the sn-1/3 positions in 1- and 2-HFA TAG. A and C, 1-HFA TAG. B and D, 2-HFA TAG. The data represent averages of three replicates ± se.
The castor PDCT is encoded by the RcROD1 gene, which shows close homology to the Arabidopsis AtROD1. Surprisingly, however, overexpressing AtROD1 using the same vector in the CL7-RcDGAT2 lines did not result in significant increases of HFA contents (data not shown).
RcROD1 Partially Restores Oil Accumulation in FAH12 Seeds
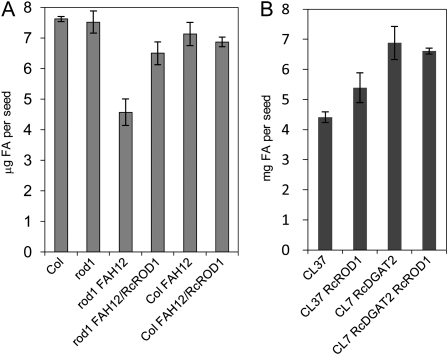
It has been shown that expressing FA hydroxylases in transgenic Arabidopsis caused a significant reduction of oil accumulation in seeds (Dauk et al., 2007; Burgal et al., 2008; Bates and Browse, 2011). It has been demonstrated that the flux of DAG through PC represents a major bottleneck for the accumulation of unusual FAs in TAG of transgenic Arabidopsis seeds (Bates and Browse, 2011). Given its important role in converting PC into DAG, we reasoned that PDCT may help relieve this metabolic bottleneck and increase oil content in HFA-accumulating seeds. We measured seed oil content in RcROD1 transgenic lines and compared that with the FAH12 lines. The CL37 contained a significantly reduced amount of oil, and we measured it at 4.4 μg seed−1. The oil content in CL37-RcROD1 seeds was increased to 5.4 μg seed−1 (Fig. 7B). Although the level is not as high as in the CL7-RcDGAT2 seeds (Burgal et al., 2008; van Erp et al., 2011), this substantial increase indicates that RcROD1 can partially relieve the bottleneck of HFA TAG accumulation. This is further supported by the measurement of oil content in rod1-FAH12 and rod1-FAH12/RcROD1 lines (Fig. 7A). The rod1-FAH12 seeds contain 4.6 μg seed−1, and complementing with RcROD1 in this line increases oil content to 6.5 mg seed−1, nearly as high as in the Col-FAH12 line at 7.4 μg seed−1. These results also support our conclusion that PDCT is required for the efficient metabolism of HFAs in transgenic Arabidopsis seeds.
Figure 7.
Seed oil content in Col, rod1, and FAH12-transformed lines. FAs (total μg seed−1) were determined for each line listed in Tables I and II. The data (averages ± se) represent three replicates for each line.
DISCUSSION
TAGs stored in seeds display an immense diversity of FA composition among different plant species. Efficient utilization of such a rich resource in food and industrial applications has been hindered by a lack of understanding of the mechanisms of FA modification during TAG synthesis. Traditional biochemical and genetic studies continue to yield fruitful insights in this area. One of the recent breakthroughs is the elucidation of the importance of the dynamic metabolism of PC in developing oilseeds. In a metabolic labeling study using developing Arabidopsis seeds, Bates and Browse (2011) demonstrated that the majority of de novo DAG was used for PC synthesis instead of being converted into TAG, suggesting that the major flux of TAG synthesis in Arabidopsis utilizes a de novo DAG-to-PC pathway followed by a PC-derived DAG-to-TAG pathway. Therefore, the PC molecules are not only a major component of cellular membranes but also provide a predominant source of DAG for synthesizing TAG. Since PC is the enzymatic substrate of FA desaturation and other types of modification, such as hydroxylation, the incorporation of FAs into PC and the subsequent removal of modified FAs from PC will greatly affect the final composition of TAG. In support of this, it has been demonstrated that in the castor hydroxylase (FAH12)-expressing Arabidopsis, HFAs containing de novo DAG were not efficiently converted to PC, thus representing a major bottleneck for the accumulation of HFAs in TAG of transgenic Arabidopsis seeds (Bates and Browse, 2011). In another study, it was shown that coexpressing a castor PDAT (RcPDAT1A) with FAH12 significantly increased the HFA accumulation to 27%, compared with 17% when expressing FAH12 alone in transgenic Arabidopsis seeds (van Erp et al., 2011). The PDAT directly transfers the HFAs from the sn-2 position of PC to the sn-3 position of DAG, producing TAG. These studies suggest that efficient mechanisms are required for the acyl fluxes through PC in the genetic engineering of FA modifications in oilseeds.
In our previous study using forward genetics (Lu et al., 2009), we discovered a PDCT in Arabidopsis that controls roughly 40% of 18:1-CoA entering PC for desaturation. The interconversion of PC and DAG mediated by PDCT also transfers the desaturated 18:2 and 18:3 from PC into DAG, which will then form TAG. Our results and those of others indicate that the PDCT reaction is responsible for most of the interconversion of DAG to PC in developing seeds (Bates et al., 2009; Lu et al., 2009). As a result, the PDCT-deficient rod1 mutant in Arabidopsis accumulated increased 18:1 and concomitantly decreased polyunsaturated 18:2 and 18:3 in seed TAG. Here, we show that PDCT is also required for the efficient metabolism of HFAs in transgenic Arabidopsis seeds. Expressing FAH12 in the rod1 mutant only accumulated 5.5% HFAs in TAG, about half of the amount in the wild-type background, which is approximately 10%. Coexpressing FAH12 with the castor PDCT encoded by RcROD1 in wild-type Arabidopsis seeds significantly increased the accumulation of HFAs, at a doubled 20%, compared with expressing FAH12 alone (Table I).
These results are in agreement with the function of PDCT that catalyzes the interconversion between PC and DAG. Overexpressing the castor PDCT (RcROD1) enhances the acyl groups entering PC, where they are desaturated or hydroxylated when expressing FAH12, and then returning to DAG. This symmetrical reaction (one DAG molecule is generated for each DAG consumed) produces 18:1OH-DAG and 18:2OH-DAG, along with the regular 18:2- and 18:3-DAG, resulting in enriched HFA DAGs in the DAG pool and subsequently increasing the accumulation of these modified FAs in TAG. This hypothesis is supported by several lines of evidence obtained in this study.
First, we show that the increased HFA content in TAG of the RcROD1-expressing lines is associated with lowered HFA amounts in PC (Fig. 5), a similar effect to that seen in the RcPDAT1A-expressing lines (van Erp et al., 2011). This indicates that PDCT is required for the efficient removal of HFAs from PC after they are produced.
Second, RcROD1 significantly increases the amount of HFA-containing TAGs, especially the 2- and 3-HFA TAGs, in the Col-FAH12 lines shown by TAG molecular species analysis (Fig. 3). Regiochemical analyses indicate that RcROD1 increases HFAs in the sn-2 position but decreases them in the sn-1/3 positions in the 2-HFA TAGs (Fig. 4). Similar changes are also detected in the CL37 and CL7-RcDGAT2 lines (Fig. 6). This suggests that the stereochemistry of the 1-HFA DAG pool contains more sn-2 HFA in the RcROD1 lines. The HFAs are synthesized from FAH12 acting on acyl groups esterified at the sn-2 position of PC. Overexpression of RcROD1 may enhance the conversion of the sn-2 HFA PC into DAG and therefore may result in more sn-2 HFA DAGs in the DAG pool. The acyltransferase DGAT or PDAT then incorporates a second HFA onto the sn-3 in DAG and forms 2-HFA TAGs. An alternative scenario is that RcROD1 enhances the flux of sn-1 HFA DAG into PC (by passing the bottleneck) and allowing the production of sn-1,2 HFA PC, which is then acylated with a normal FA by DGAT or PDAT.
Third, we noticed that the increased HFAs in the Col-FAH-RcROD1 lines are at the expense of C18 PUFAs (18:2 + 18:3) and 20:1, the products of the desaturation and elongation of 18:1, respectively (Table I). The decrease of PUFAs is expected, because of the competition of FAH12 with FAD2 for the 18:1 substrate. It is interesting that the 20:1 level is also decreased. The very-long-chain FA (20:1) is synthesized from 18:1-CoA outside PC, probably most of which are products directly exported from plastids. Therefore, the elongation and the incorporation into PC are two competing pathways for the 18:1 substrates. We have shown that PDCT is responsible for about 40% of the 18:1 that enters PC (Lu et al., 2009). Overexpressing the castor PDCT may direct more 18:1 to enter PC for desaturation and hydroxylation and may reduce the flow toward the elongation pathway. The elongation pathway is blocked in the CL37 and CL7-RcDGAT2 lines that were generated in the fae1 background. The CL37 seeds already contain 17% HFAs, which is substantially higher than the levels in the Col-FAH12 lines in this study and the lines reported previously (Smith et al., 2003). However, RcROD1 still significantly increases HFA levels in the CL37 and CL7-RcDGAT2 lines (Table II).
Our results with RcROD1 in different transgenic lines, along with previous reports (Smith et al., 2003; Lu et al., 2006; Burgal et al., 2008; van Erp et al., 2011), point out the complexity of the interactions among different mechanisms that channel HFAs into TAG. PDCT enhances the acyl fluxes through PC and thus reduces the pathway toward elongation outside of PC. The rapid PC-DAG turnaround may also have an impact on acyl editing by reducing the HFA-CoA substrates available for DGAT2. In this regard, it is noteworthy that overexpression of RcROD1 results in reduced HFAs in the sn-1/3 positions and increased content in the sn-2 position (Fig. 6). Another interesting factor is the different stereochemistry observed in the 1-HFA TAG for the HFA-expressing Col (and rod1) and the CL37 (and CL7-RcDGAT2) lines (Figs. 4 and 6). The distribution of HFAs in the sn-2 and sn-1/3 positions is opposite between these two groups of lines. It is possible that fae1 in the CL37 and CL7-RcDGAT2 lines at least partly contributes to a shift of more HFAs into the sn-2 position in the 1-HFA TAG. More studies, such as metabolic labeling experiments, are needed to understand the interactions between these competing pathways.
Finally, to support our hypothesis, we show that the RcROD1 partially compensates for the decreased oil accumulation in FAH12-expressing seeds. It has been shown that the FAH12 transgenic seeds contain reduced oil content (Dauk et al., 2007; Burgal et al., 2008; Bates and Browse, 2011). This may have resulted from the buildup of inefficiently used HFAs that cause feedback inhibition of FA synthesis or activate FA β-oxidation (Moire et al., 2004; Bates and Browse, 2011). The rod1-FAH12 lines accumulate higher amounts of HFAs in PC during seed development (Fig. 5), and the seeds contain much reduced HFA and total FAs (Fig. 7). The FA content in CL37 is also significantly reduced (van Erp et al., 2011). Expressing RcROD1 in rod1-FAH12 and CL37 significantly increases seed oil content (Fig. 7), although not to the control levels in wild-type or RcDGAT2-expressing lines. These results suggest that the castor PDCT efficiently converts PC, especially HFA-PC, into DAG and subsequently into TAG.
The castor and Arabidopsis ROD1 homologs both possess PDCT activity. In this study, we have used a castor PDCT in our experiments to coexpress with FAH12, which resulted in increased HFA accumulation in all lines tested, including the wild type (Col), CL37, and CL7-RcDGAT2 (Tables I and II). However, the Arabidopsis PDCT failed to produce such effects. This result was surprising, since we expected that overexpression of either PDCT would enhance the acyl fluxes through PC. We have previously proposed that for the efficient metabolism of HFAs in transgenic seeds, one needs to coexpress other enzymes from the same native species (e.g. castor) that may have coevolved with FAH12 (Lu et al., 2006). This hypothesis has been supported by coexpressing RcDGAT2 and RcPDAT1A with FAH12 in transgenic Arabidopsis seeds (Burgal et al., 2008; van Erp et al., 2011). These results are in agreement with the observations that suggest a preference for HFA-containing substrates of the castor enzymes (Dahlqvist et al., 2000; Burgal et al., 2008). Our results with RcROD1 seemingly also support the coevolution theory, but we cannot demonstrate the substrate preference for HFA-containing PC of the castor PDCT (Fig. 3) due to the limited availability of substrates. However, the symmetrical reaction of DAG-PC conversion may not necessarily require substrate selectivity of the PDCT enzyme (i.e. 18:2/3-PC, 18:1OH-PC over 18:1-PC; 18:1-DAG over 18:2/3-DAG, 18:1OH-DAG), since the desaturases and the hydroxylase will enrich the PC pool with the PUFAs and hydroxylated FAs. On the other hand, although the RcROD1 protein has been detected in castor endosperm (Brown et al., 2012), previous results suggested that the castor PDCT unlikely preferred HFA-containing substrates, since microsomal preparations from castor endosperm rapidly turned the glycerol-labeled di-ricinoleoyl-DAG to TAG, whereas dioleoyl-DAG was preferentially used in PC synthesis (Bafor et al., 1991). Moreover, Dahlqvist et al. (2000) showed that castor bean microsomes preferentially metabolized ricinoleoyl-PC to TAG (probably via PDAT), whereas oleoyl-PC was preferentially metabolized to DAG (probably via PDCT).
Nevertheless, we show in this report that PDCT is required for the efficient metabolism of HFAs in the FAH12 transgenic Arabidopsis seeds. Coexpressing a castor PDCT with FAH12 significantly increases the HFA accumulation. The strong PDCT activity conferred by the RcROD1 gene may enhance the fluxes of FAs through PC, thus partly overcoming the bottleneck of modified FA accumulation in transgenic seeds proposed by previous research (Bates and Browse, 2011).
MATERIALS AND METHODS
Construction of Plasmid Vectors
For coexpressing FAH12 and RcROD1 in Arabidopsis (Arabidopsis thaliana) seeds, the castor (Ricinus communis) open reading frame sequences RcROD1 and RcFAH12 were cloned into the binary vector pBinGlyRed2, kindly provided by Edgar Cahoon (University of Nebraska-Lincoln), which contains a DsRed selection marker (Clontech) behind the constitutive cassava vein mosaic virus promoter (Verdaguer et al., 1996). RcROD1 was placed behind the seed-specific glycinin promoter, and RcFAH12 was driven by a seed-specific napin promoter (Supplemental Fig. S2). To express RcROD1 in the CL7-RcDGAT2 line, the RcROD1 sequences were cloned into the pGATE-PHAS-DsRed vector under the control of the seed-specific phaseolin promoter, as described previously (Lu et al., 2006).
Plant Material and Transformation
Arabidopsis Col-0, mutant rod1 (Lu et al., 2009), transgenic line CL37 expressing FAH12 in the fae1 mutant background (Lu et al., 2006), and CL7-RcDGAT2 coexpressing FAH12 and RcDGAT2 (Burgal et al., 2008) were used in this study. All Arabidopsis plants were grown in controlled-environment chambers at 22°C under a 16-h photoperiod of 150 μmol quanta m−2 s−1 photosynthetically active radiation. Arabidopsis plants were transformed by the floral dipping procedure (Clough and Bent, 1998) following electroporation of individual cDNA clones into Agrobacterium tumefaciens strain GV3101 (pMP90). Transgenic seeds were identified by examination under green illumination using a red filter (Stuitje et al., 2003; Lu et al., 2006).
Lipid Extraction
Total lipids were extracted from mature seeds or developing seeds on ice as described before (Bligh and Dyer, 1959) by suspending in 3 mL of a chloroform:methanol (1:2, v/v) mixture, and then seeds were ground in a mortar. Ground seeds and solvent were transferred to a 10-mL glass tube, and the mixture was made up to 5.8 mL by the addition of 1 mL of chloroform and 1.8 mL of 0.9% NaCl, so the final relative volume of chloroform:methanol:0.9% NaCl is 1:1:0.9. The chloroform phase was transferred to a clean glass tube and evaporated to dryness under a stream of N2. Lipids were resuspended in a small volume of chloroform and stored at −20°C.
PC Analysis
PC was resolved by TLC of the total lipids with a solvent system consisting of chloroform:methanol:water:30% ammonium hydroxide (65:35:3:2.5, v/v/v/v; Cahoon et al., 2006). Lipid bands were visualized under UV light after staining with 0.005% primulin in 80% acetone, and PC was collected for GC analysis as below.
GC Analysis
The FAs of lipid fractions and whole Arabidopsis seeds were derivatized to FAMEs in 1 mL of 2.5% sulfuric acid in methanol at 80°C for 90 min (Browse et al., 1986). FAMEs were injected into a Shimadzu 2010 GC device fitted with a narrow-bore column (HP-Innowax; 30 m × 0.25 mm i.d. × 0.25 μm; Agilent Technologies). The oven temperature was programmed at 190°C initially, followed by an increase of 20°C min−1 to 250°C, and maintained for 9 min.
Characterization of TAG Species, and Regiochemical Analysis of 1- and 2-HFA TAG
Neutral lipids were separated by TLC (silica gel G60 plates; EM Separations Technology) with hexane:diethyl ether:acetic acid (140:60:2, v/v/v; Smith et al., 2003) as the developing solvent system. Castor oil was used to produce standards of TAG species with HFAs as described previously (Smith et al., 2003; van Erp et al., 2011). Lipid bands were visualized under UV light after staining with 0.005% primulin in 80% acetone. The fractions of TAG with zero, one, two, and three HFA residues, namely 0-HFA, 1-HFA, 2-HFA, and 3-HFA TAG, respectively, were scraped from the TLC plate into glass tubes, and FAMEs were prepared for GC analysis as above. Calculation of the percentages of HFAs at the sn-2 position and at the sn-1/3 position was done as described (van Erp et al., 2011).
Seed Oil Content Measurements
Seed oil content was determined by GC analysis of FAMEs following the method of Li et al. (2006) using 30 seeds and 68 μg of triheptadecanoin as a TAG standard.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_002517597.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of deduced amino acid sequences of AtROD1 and RcROD1.
Supplemental Figure S2. The open reading frame nucleotide sequence of RcROD1 and the plasmid construct for coexpressing RcROD1 and FAH12.
Supplementary Material
Acknowledgments
We thank Dr. John Browse (Washington State University) for providing the CL7-RcDGAT2 line and for helpful discussions during this project. We also thank Dr. Ed Cahoon (University of Nebraska-Lincoln) for providing the pBinGlyRed2 binary vector and Dr. Philip Bates (Washington State University) for critical reading of the manuscript.
References
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S. (1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Browse J. (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB, Pollard M. (2007) Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem 282: 31206–31216 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C. (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AP, Kroon JT, Swarbreck D, Febrer M, Larson TR, Graham IA, Caccamo M, Slabas AR. (2012) Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS ONE 7: e30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, McConn M, James D, Jr, Miquel M. (1993) Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate: biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J Biol Chem 268: 16345–16351 [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Browse J, Somerville C. (1991) Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol 42: 467–506 [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J. (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ. (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67: 1166–1176 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97: 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauk M, Lam P, Kunst L, Smith M. (2007) A FAD2 homologue from Lesquerella lindheimeri has predominantly fatty acid hydroxylase activity. Plant Sci 173: 43–49 [Google Scholar]
- Drexler H, Spiekermann P, Meyer A, Domergue F, Zank T, Sperling P, Abbadi A, Heinz E. (2003) Metabolic engineering of fatty acids for breeding of new oilseed crops: strategies, problems and first results. J Plant Physiol 160: 779–802 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Benning C, Ohlrogge J. (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54: 593–607 [DOI] [PubMed] [Google Scholar]
- Dyer JM, Mullen RT. (2008) Engineering plant oils as high-value industrial feedstocks for biorefining: the need for underpinning cell biology research. Physiol Plant 132: 11–22 [DOI] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS. (2008) High-value oils from plants. Plant J 54: 640–655 [DOI] [PubMed] [Google Scholar]
- Goode JH, Dewey RE. (1999) Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol Biochem 37: 445–457 [Google Scholar]
- Gunstone FD. (1998) Movements towards tailor-made fats. Prog Lipid Res 37: 277–305 [DOI] [PubMed] [Google Scholar]
- Hobbs DH, Lu C, Hills MJ. (1999) Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett 452: 145–149 [DOI] [PubMed] [Google Scholar]
- Jaworski J, Cahoon EB. (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6: 178–184 [DOI] [PubMed] [Google Scholar]
- Kennedy EP. (1961) Biosynthesis of complex lipids. Fed Proc 20: 934–940 [PubMed] [Google Scholar]
- Kunst L, Samuels AL. (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kunst L, Taylor DC, Underhill EW. (1992) Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem 30: 425–434 [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG, Browse J. (2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45: 847–856 [DOI] [PubMed] [Google Scholar]
- Lu C, Kang J. (2008) Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep 27: 273–278 [DOI] [PubMed] [Google Scholar]
- Lu C, Napier JA, Clemente TE, Cahoon EB. (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22: 252–259 [DOI] [PubMed] [Google Scholar]
- Lu C, Wallis JG, Browse J. (2007) An analysis of expressed sequence tags of developing castor endosperm using a full-length cDNA library. BMC Plant Biol 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J. (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moire L, Rezzonico E, Goepfert S, Poirier Y. (2004) Impact of unusual fatty acid synthesis on futile cycling through beta-oxidation and on gene expression in transgenic plants. Plant Physiol 134: 432–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier JA, Graham IA. (2010) Tailoring plant lipid composition: designer oilseeds come of age. Curr Opin Plant Biol 13: 330–337 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzi S, Garcia IL, Lopez-Gimenez FJ, de Castro MDL, Dorado G, Dorado MP. (2009) The ideal vegetable oil-based biodiesel composition: a review of social, economical and technical implications. Energy Fuels 23: 2325–2341 [Google Scholar]
- Riediger ND, Othman RA, Suh M, Moghadasian MH. (2009) A systemic review of the roles of n-3 fatty acids in health and disease. J Am Diet Assoc 109: 668–679 [DOI] [PubMed] [Google Scholar]
- Slack CR, Campbell LC, Browse JA, Roughan PG. (1983) Some evidence for the reversibility of cholinephosphotransferase-catalyzed reaction in developing linseed cotyledons in vivo. Biochim Biophys Acta 754: 10–20 [Google Scholar]
- Smith MA, Moon H, Chowrira G, Kunst L. (2003) Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217: 507–516 [DOI] [PubMed] [Google Scholar]
- Sperling P, Linscheid M, Stöcker S, Mühlbach HP, Heinz E. (1993) In vivo desaturation of cis-delta 9-monounsaturated to cis-delta 9,12-diunsaturated alkenylether glycerolipids. J Biol Chem 268: 26935–26940 [PubMed] [Google Scholar]
- Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap JP, Kneppers TJA. (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1: 301–309 [DOI] [PubMed] [Google Scholar]
- Thomaeus S, Carlsson AS, Stymne S. (2001) Distribution of fatty acids in polar and neutral lipids during seed development in Arabidopsis thaliana genetically engineered to produce acetylenic, epoxy and hydroxy fatty acids. Plant Sci 161: 997–1003 [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C. (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92: 6743–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Bates PD, Burgal J, Shockey J, Browse J. (2011) Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol 155: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Beachy RN, Fauquet C. (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Williams JP, Imperial V, Khan MU, Hodson JN. (2000) The role of phosphatidylcholine in fatty acid exchange and desaturation in Brassica napus L. leaves. Biochem J 349: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.